Abstract
Curli are adhesive fimbriae of Enterobacteriaceae and are involved in surface attachment, cell aggregation, and biofilm formation. Here, we report that both inter- and intrastrain variations in curli production are widespread in enterohemorrhagic Escherichia coli O157:H7. The relative proportions of curli-producing variants (C+) and curli-deficient variants (C−) in an E. coli O157:H7 cell population varied depending on the growth conditions. In variants derived from the 2006 U.S. spinach outbreak strains, the shift between the C+ and C− subpopulations occurred mostly in response to starvation and was unidirectional from C− to C+; in variants derived from the 1993 hamburger outbreak strains, the shift occurred primarily in response to oxygen depletion and was bidirectional. Furthermore, curli variants derived from the same strain displayed marked differences in survival fitness: C+ variants grew to higher concentrations in nutrient-limited conditions than C− variants, whereas C− variants were significantly more acid resistant than C+ variants. This difference in acid resistance does not appear to be linked to the curli fimbriae per se, since a csgA deletion mutant in either a C+ or a C− variant exhibited an acid resistance similar to that of its parental strain. Our data suggest that natural curli variants of E. coli O157:H7 carry several distinct physiological properties that are important for their environmental survival. Maintenance of curli variants in an E. coli O157:H7 population may provide a survival strategy in which C+ variants are selected in a nutrient-limited environment, whereas C− variants are selected in an acidic environment, such as the stomach of an animal host, including that of a human.
INTRODUCTION
Bacterial pathogens have evolved various mechanisms to maximize their survival fitness in a challenging environment, including in a human host. Genetic variants produced by adaptive mutations, genome rearrangements, or gene deletions and acquisitions can confer upon a subpopulation of cells certain phenotypes that would provide them survival advantages in a particular niche (5). Additionally, it is known that phenotypic variations or cell individualities are present among cells in a genetically homogeneous population, which provides bacteria another source of intrapopulation diversity. Such variations include the expression of bacterial cell surface antigens, resistance to antimicrobial treatment, bacterial swimming motility, and the induction of prophages (10). Although molecular mechanisms underlying bacterial cell individuality are not fully understood, epigenetic regulation and stochastic gene expression are proven important forces in generating population heterogeneity (2). The entire bacterial population could benefit by creation of variant subpopulations that have the potential to be better equipped to survive in stressful conditions or to adapt rapidly to new niches.
Enterohemorrhagic Escherichia coli (EHEC) strains include a diverse group of Shiga toxin-producing E. coli (STEC) strains that cause severe disease, including hemolytic uremic syndrome (HUS). E. coli O157:H7 is the most common EHEC serotype and contributes significantly to human infections and outbreaks in North America. EHEC strains are present commonly in livestock intestines and can be transmitted to humans via food, including raw meat and fresh vegetables. In a recent study of tracking E. coli O157:H7 in a major produce production region in California, E. coli O157:H7 was isolated from various environmental sources, including water, soil, sediment, and wild animals (8). Persistence of this pathogen in an agricultural environment poses potential risks for preharvest contamination and human health (25). E. coli O157:H7 strains vary greatly in their genomic and virulence repertoires. Strains linked to the 2006 U.S. spinach outbreak resulted in higher rates of hospitalization and HUS than the average rates for 350 previous outbreaks that occurred from 1982 to 2002 (26, 31). Furthermore, there is evidence of increased infection frequency caused by strains closely related to the 2006 spinach outbreak strains in the past few years, suggesting that strains with increased survival fitness and virulence are selected in natural habitats and/or in infected humans (26).
Curli are the thin aggregative fimbriae produced in many Enterobacteriaceae (46) and have been recognized as an important colonization factor due to their adhesive properties (28). Curli production is also an important virulence factor due to its roles in biofilm formation, attachment to eukaryotic cells, and induction of host inflammatory response (4, 14, 20, 23, 38, 41). Recently, curli have been shown also to provide cells protection from heavy metals (18). Typically, curli are expressed in response to low temperature, low oxygen, low osmolarity, and nutrient limitation (3). Biogenesis of curli requires the function of two operons, csgBA and csgDEFG, which are transcribed divergently (15). CsgA is the curli subunit protein, whereas CsgD is a transcriptional activator of the csgBA operon. CsgB is a nucleator protein, which, together with an outer membrane lipoprotein (CsgG) and two curli assembly factors (CsgE and CsgF), is required for production of mature curli (7, 16, 17, 32, 43). Variation in curli expression has been reported for natural E. coli isolates originating from diverse environments (27, 36). Increased curli production by an E. coli O157:H7 strain that caused the first outbreak linked to meat was associated with enhanced virulence in mice and eukaryotic cell invasion (39–41). Variation in curli production in this strain has been linked to mutations in the csgD promoter (40). Here, we hypothesize that the 2006 U.S. spinach outbreak strains possess distinct physiological properties that enhance their environmental survival compared with that of other E. coli O157:H7 outbreak strains. We assembled a set of E. coli O157:H7 strains associated with several U.S. outbreaks, from both clinical and environmental sources, with our primary focus on the 2006 spinach outbreak. We then isolated curli-producing variants (C+) and curli-deficient variants (C−) from each strain and examined the physiological properties of curli variants that are important for E. coli O157:H7 survival in both host and nonhost environments. We found that C+ variants grew better under nutrient-limited conditions than the corresponding C− variants, whereas C− variants were more acid resistant than C+ variants. This physiological divergence between E. coli O157:H7 curli variants may provide the entire E. coli O157:H7 population with a survival strategy that, by shifting the dynamics of population structure, allows the pathogen to rapidly adapt to diverse habitats.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The E. coli O157:H7 isolates used in this study and their sources are listed in Table 1. Clinical E. coli O157:H7 isolates are RM6607 (associated with the 1993 U.S. hamburger outbreak), RM6011, RM6049, RM6069, and RM6654 (associated with the 2006 U.S. spinach outbreak), and RM6535 (associated with a 2006 lettuce outbreak). Outbreak strains isolated from bagged spinach (RM6067, RM9994, RM9998, and RM10000) were kindly provided by E. Hÿttia-Trees (CDC, Atlanta, GA). The outbreak strain isolated from bagged iceberg lettuce (RM6535) was kindly provided by B. Juni (Minnesota Department of Public Health). Details of the isolation and characterization of the E. coli O157:H7 environmental isolates from San Benito and Monterey counties in California have been described previously (8). The serotype of each strain was confirmed by immunoblot analysis using an anti-O157 monoclonal antibody (kindly provided by J. Keen) (44). Strains were grown routinely in Luria-Bertani (LB) broth or modified LB broth containing various concentrations of salt as detailed in each section. LB-full salt contains 10 g of NaCl per liter (LBFS), whereas LB-no salt contains no NaCl (LBNS).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Supplier/reference/accession no. |
|---|---|---|
| Strains | ||
| E. coli O157:H7 | ||
| RM6011 | Human isolate (WI) associated with the 2006 U.S. spinach outbreak | T. Monson |
| RM6049 | Human isolate (CA) associated with the 2006 U.S. spinach outbreak | M. Janda |
| RM6069 | Human isolate (PA) associated with the 2006 U.S. spinach outbreak | W. Chmielecki |
| RM6654 | Human isolate (NM) associated with the 2006 U.S. spinach outbreak | E. Hÿttia-Trees |
| RM6067 | Bagged baby spinach isolate (PA) associated with the 2006 U.S. spinach outbreak | E. Hÿttia-Trees |
| RM9994 | Bagged baby spinach isolate (UT) associated with the 2006 U.S. spinach outbreak | E. Hÿttia-Trees |
| RM9998 | Bagged baby spinach isolate (NV) associated with the 2006 U.S. spinach outbreak | E. Hÿttia-Trees |
| RM10000 | Bagged baby spinach isolate (AZ) associated with the 2006 U.S. spinach outbreak | E. Hÿttia-Trees |
| RM6535 | Human isolate (MN) associated with a 2006 U.S. iceberg lettuce outbreak | B. Juni |
| RM6416 | Water isolate (CA) associated with a 2006 U.S. iceberg lettuce outbreak | R. Mandrell |
| RM6607 | Human isolate associated with the 1993 hamburger outbreak | 44 |
| RM6608 | Ground beef isolate associated with the 1993 hamburger outbreak | 44 |
| MQC108 | In-frame deletion of csgA in strain RM6067 C+ variant (Kmr) | This study |
| MQC111 | In-frame deletion of csgA in strain RM6067 C− variant (Kmr) | This study |
| MQC114 | In-frame deletion of csgA in strain RM6535 C+ variant (Kmr) | This study |
| MQC116 | In-frame deletion of csgA in strain RM6535 C− variant (Kmr) | This study |
| MQC118 | In-frame deletion of csgA in strain RM6607 C+ variant (Kmr) | This study |
| MQC120 | In-frame deletion of csgA in strain RM6607 C− variant (Kmr) | This study |
| E. coli non-O157 | ||
| MC4100 | Parental strain of LSR1, a derivative of E. coli K-12 | 6 |
| LSR1 | MC4100csgG::Tn105 (Cmr) | 32 |
| Plasmids | ||
| pKD46 | Red recombinase plasmid (Ampr) | AY048746 (9) |
| pKD4 | Template plasmid (Kmr) | AY048743 (9) |
Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Ampr, ampicillin resistance. Two-letter U.S. state designations shown in parentheses indicate sources of spinach and lettuce outbreak strains.
Isolation of E. coli O157:H7 curli variants.
The frozen glycerol stock of each E. coli O157:H7 strain was scraped using an inoculating loop and resuspended in 1 ml LBNS broth. Ten-fold serial dilutions were prepared from these cell suspensions, and 50 μl of each dilution was plated onto Congo red indicator (CRI) plates (LBNS plates supplemented with 40 μg/ml of Congo red dye and 10 μg/ml of Coomassie brilliant blue). The plates were incubated at 28°C for 2 to 3 days. Colonies with various colors, e.g., red, brown, or white, were picked and patched onto CRI plates for further characterization. To confirm that the variants on CRI plates were different in curli production, we determined the curli production of E. coli O157:H7 variants by immunoblot analysis by using an antibody specific to the curli subunit protein CsgA (kindly provided by M. Chapman). Briefly, E. coli O157:H7 variant cells grown on CRI plates were resuspended in saline solution (0.85% NaCl) and the cell density was adjusted to an optical density at 600 nm (OD600) of 5/ml. After boiling for 5 min, 5 μl of each cell lysate was deposited on a nitrocellulose membrane (Invitrogen). The membrane was blocked first with 1% bovine serum albumin (BSA) at 4°C overnight and then incubated with anti-CsgA antibody at a dilution of 1:3,000 in 1% BSA at room temperature for 1 h followed by incubation with the alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (Thermo Scientific) at a dilution of 1:1,000 in 1% BSA at room temperature for 1 h. The binding of the cell lysate to the anti-CsgA antibody was detected using the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) substrate kit (Sigma, St. Louis, MO). E. coli strain MC4100 and its mutant strain LSR1, which is defective in curli assembly due to the inactivation of CsgG protein (32), were used as positive and negative controls, respectively.
Genotyping of E. coli O157:H7 curli variants by MLVA.
Multilocus variable-repeat analysis (MLVA) was performed using capillary electrophoresis methods described previously (8). The 11 loci used in this study were described previously (21, 24). Briefly, 11 loci (designated Vhec1 to Vhec7 and O157-17, -19, -25, and -37) were amplified by PCR directly from whole-cell lysates using fluorescent primers in three multiplex PCRs. The PCR products were pooled and diluted 1:50 using sterile water. One microliter of diluted PCR products was mixed with 12 μl of HiDi formamide containing 0.08 μl of DNA molecular weight marker (MapMarker 1000 [X-rhodamine labeled, 50 to 1,000 bp]; Bioventures Inc., Murfreesboro, TN). The sample mixture was denatured at 95°C for 5 min prior to electrophoresis (ABI 3130xl genetic analyzer). The sizes of the DNA fragments were determined using GeneMapper software (ABI) and converted to a number of tandem repeats (TR) by subtraction of the number of amplified, nonrepeat sequences and then division by the repeat size.
Proportions of E. coli O157:H7 curli variants under various growth conditions.
A single colony of an E. coli O157:H7 curli variant from the CRI plate was inoculated into 1.0 ml of LBNS or LBFS broth and grown at 28°C or 37°C with rotation (150 rpm) or statically in an anaerobic jar (Oxoid, United Kingdom) flushed with Bioblend gas (Praxair Inc., Richmond, CA) (80% N2, 10% H2, 10% CO2) for 1 to 2 days. Ten-fold serial dilutions were made from each culture, and 100 μl of each dilution was plated onto CRI plates and incubated at 28°C for 2 to 3 days. The proportions of C+ and C− variants in each culture at the end of incubation were expressed as the percentage of C+ variants in the total number of E. coli O157:H7 cells. Each data set was the average of results from at least three biological replicates.
Biofilm formation.
E. coli O157:H7 curli variants were grown in LBNS broth at 28°C overnight. Each culture was diluted in fresh LBNS broth at a concentration of 103 cells/ml. One milliliter of diluted culture was dispensed into a borosilicate glass tube and incubated statically either at 28°C aerobically or at 37°C anaerobically in an anaerobic jar (Oxoid) for 2 days as described above. After incubation, the planktonic cells were removed gently, and the tubes were rinsed twice with 1 ml sterile distilled water and then stained with 1 ml of 0.1% crystal violet at room temperature for 30 min based on a method by O'Toole et al. (29). The dye was removed gently, and the glass tubes were washed twice with sterile distilled water. The crystal violet bound to the glass tube was solubilized in 0.5 ml of 95% ethanol, and the absorbance was determined at 570 nm using a microplate reader (SpectraMax 340; Molecular Devices, Sunnyvale, CA). Each data set was the average of results from at least three biological replicates.
Survival in diluted peptone water.
E. coli O157:H7 curli variants were grown in LB broth at 37°C overnight on a shaker (150 rpm). The cells were collected by centrifugation at 10,000 × g for 3 min. After being washed once in 0.01% peptone water, the cells were resuspended in 0.01% peptone water at a concentration of 105 cells/ml, and the cultures were incubated at 15°C for 5 to 7 days. The viable cells were recovered by plating onto LB agar plates using an automated spiral plater (Autoplate4000; Spiral Biotech, Norwood, MA), and the concentration of viable cells was compared with that of the initial inoculum to estimate the growth of curli variants in diluted peptone water. In parallel, cells were plated onto CRI plates and incubated at 28°C for 2 to 3 days. The proportions of C+ and C− variants in the culture under this growth condition were expressed as the percentage of C+ variants over the total number of E. coli O157:H7 cells recovered on CRI plates.
Acid challenge assay.
E. coli O157:H7 curli variants were grown in LBNS broth at 28°C overnight on a shaker (150 rpm). The overnight cultures were diluted to a concentration of 106 cells/ml in LBNS broth acidified to pH 2.5 by addition of HCl, and cultures were incubated at 37°C for 2 to 6 h. The viable cells were recovered by plating onto LB agar plates by using an automated spiral plater (Autoplate4000; Spiral Biotech), and the concentration of viable cells was compared with that of the initial inoculum to estimate the acid resistance of E. coli O157:H7 curli variants. The proportions of C+ and C− variants were assessed as described above.
Gene disruption.
Inactivation of the gene encoding the curli subunit protein CsgA in E. coli O157:H7 curli variants was achieved by replacing the open reading frame of the csgA gene with a kanamycin cassette by using the standard lambda red-mediated gene replacement method (9). Briefly, the kanamycin cassette was amplified directly from the template plasmid pKD4 (Table 1) using the composite primers csgA_UFP1 (ACGTTAATTTCCATTCGACTTTTAAATCAATCCGATGGGGGTTTTACATGGTGTAGGCTGGAGCTGCTTC) and csgA_DRP2 (GAAAAAAAACAGGGCTTGCGCCCTGTTTCTTTAATACAGATGATGTATTAATGGGAATTAGCCATGGTCC). The PCR products (about 1.6 kb) were gel purified and transformed in curli variants carrying the helper plasmid pKD46 (Table 1) by electroporation. The Kmr transformants were selected on LB agar plates containing 50 μg/ml of kanamycin. The csgA deletion mutants were verified by colony PCR using primers flanking the csgA gene (csg_F4 and csg_R1) (see Table S1 in the supplemental material). Mutants were then colony purified at 37°C and tested for ampicillin sensitivity for loss of helper plasmid pKD46.
Statistical analysis.
Statistical tests were performed with SigmaPlot version 11.0 (Systat Software, Inc.).
RESULTS
Isolation of E. coli O157:H7 curli variants.
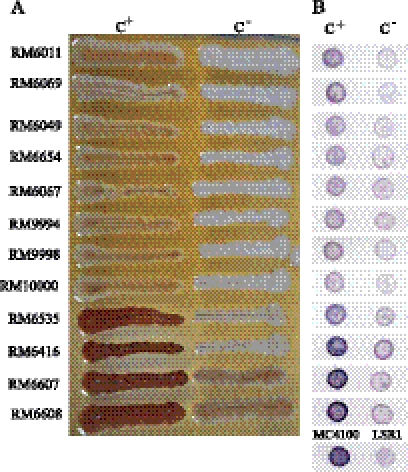
Curli, also known as bacterial amyloid, can bind to diazo dye Congo red (CR); therefore, CR is often used to detect and isolate curliated bacteria. When E. coli O157:H7 strains were grown on CRI plates, strains displayed different colors, such as red, brown, or white, indicating that there was interstrain variation in curli production. Moreover, when a single strain was grown on CRI plates, colonies with various colors often arose. Depending on the strain, some were mixtures of red and white colonies, and others were dominated by red, brown, or white colonies with a few variants, suggesting that there was intrastrain variation in curli production. To investigate the full spectrum of curli variation in E. coli O157:H7, we assembled a group of 12 E. coli O157:H7 strains, of which eight are associated with the 2006 U.S. spinach outbreak, two with a 2006 U.S. iceberg lettuce outbreak, and two with the 1993 U.S. hamburger outbreak (Table 1). To avoid bias introduced by the source of isolation, we included strains from both humans (clinical) and nonhost environments, including bagged spinach, lettuce, hamburger, and field water (Table 1). We isolated both C+ and C− variants from each strain (Fig. 1A). The C+ variants displayed stronger pigmentation than their corresponding C− variants. Furthermore, C+ variants derived from strains associated with a 2006 lettuce outbreak and the 1993 hamburger outbreak displayed stronger pigmentation than C+ variants derived from strains associated with the 2006 spinach outbreak (Fig. 1A). In addition to curli fimbriae, there are several outer membrane proteins reported to bind to CR (22, 34). Therefore, we used immunoblot analysis with an antibody specific to the curli subunit protein, CsgA, to confirm that the variants isolated from CRI plates differed in curli production. Consistent with the pigmentation of colonies on CRI plates, C+ variants displayed stronger binding of anti-CsgA antibody than their corresponding C− variants; thus, these E. coli O157:H7 variants were termed curli variants (Fig. 1B). For curli variants derived from the same strain, C+ designated variants that produced more curli than the C−-designated variants, which produced no or minimal curli. C+ variants derived from strains associated with a 2006 lettuce outbreak (RM6535 and RM6416) and the 1993 hamburger outbreak (RM6607 and RM6608) showed stronger binding of the anti-CsgA antibody than all eight C+ variants derived from strains associated with the 2006 spinach outbreak (RM6011, RM6069, RM6049, RM6654, RM6067, RM9994, RM9998, and RM10000) (Fig. 1B), suggesting that considerable interstrain variations in curli production also existed in these E. coli O157:H7 populations.
Fig. 1.
Isolation of E. coli O157:H7 curli variants. Curli variants of E. coli O157:H7 grown on CRI plates (A) and immunoblot of E. coli O157:H7 variants probed with anti-CsgA antibody (B). The correlation between colony pigmentation on CRI plates and binding of anti-CsgA-specific antibody indicates that color variants on CRI plates differ in curli production. E. coli strain LSR1 was not subjected to heat treatment because it is defective in curli assembly but not in the production of CsgA.
Genotypes of E. coli O157:H7 curli variants determined by MLVA.
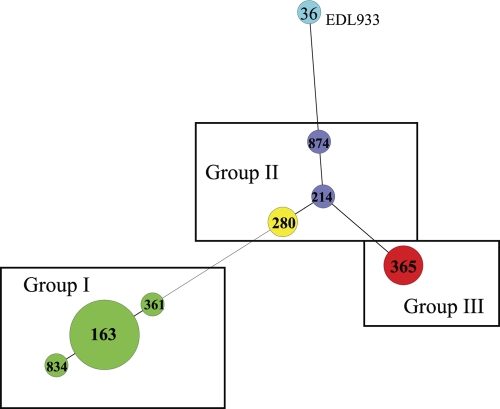
To confirm the intrastrain relationship of the curli variants derived from the same strain, we performed strain typing by MLVA for 24 curli variants. Different tandem repeat (TR) numbers were detected for curli variants derived from three strains at either Vhec4 or Vhec3 (see Table S2 in the supplemental material). RM6011W, a C− variant derived from a clinical strain RM6011 from the 2006 spinach outbreak (Fig. 2, MLVA 834), had 18 TR at Vhec4 compared with 17 in the corresponding C+ variant RM6011R (Fig. 2, MLVA 163). Similarly, RM6069W, a C− variant derived from another RM6069 clinical isolate from the 2006 spinach outbreak (Fig. 2, MLVA 361), had 16 TR at Vhec4 compared with 17 in the corresponding C+ variant, RM6069R. A Vhec4 TR is composed of 6 nucleotides (AAATAG). In E. coli O157:H7 strain EDL933, Vhec4 contains 10 TR and is located upstream of a gene encoding an IS30 transposase (Z3936). For Vhec3, the curli variants derived from strain RM6416 had 9 and 12 TR in the C+ and C− variants, respectively (Fig. 2, MLVA 874 and 214). This strain was isolated from a water sample collected from a farm in the southern central valley of California during a farm investigation following a December 2006 outbreak associated with bagged iceberg lettuce. The MLVA type of RM6416 is related closely to a clinical strain associated with this outbreak (RM6535) (Fig. 2, MLVA 280). Vhec3 TR also contains a 6-nucleotide repeat (AAGGTG), and in strain EDL933 (Fig. 2, MLVA 36), this locus contains 9 TR, the same number we detected for C+ variant RM6416R. Regardless of these minor changes in the number of TR observed at Vhec4 or Vhec3, curli variants derived from the same strain showed either identical (9 pairs) or highly related (3 pairs) MLVA types (Fig. 2). Curli variants derived from these 12 E. coli O157:H7 strains grouped into three classes: group I included the variants derived from the 2006 U.S. spinach outbreak strains, group II included the variants derived from a human and a water isolate associated with a 2006 iceberg lettuce outbreak, and group III included the variants derived from the 1993 hamburger outbreak strains (Fig. 2).
Fig. 2.
Genomic relationship of E. coli O157:H7 curli variants determined by MLVA. Tandem repeats (TR) at 11 loci (detailed in Table S2 in the supplemental material) of E. coli O157:H7 curli variants were imported into Bionumerics, and a minimal spanning tree was created with an MST algorithm using the number of TR at each of 11 loci as categorical coefficients. All coefficients were equally weighed. E. coli O157:H7 strain EDL933 (MLVA 36) was included as a reference. Numbers within circles designate MVLA types. The size of a circle represents the number of isolates for each MVLA type.
E. coli O157:H7 population structure determined by the proportion of curli variants under various growth conditions.
The expression of curli is regulated by many environmental factors, including temperature, salt, oxygen, and available nutrients. Therefore, we examined whether growth conditions impacted the population structure of E. coli O157 cultures initiated with either C+ or C− variants. The growth conditions examined were temperature (28°C versus 37°C), salt (LBFS versus LBNS), oxygen (atmospheric oxygen versus oxygen less than 0.0001%), and nutrients (early stationary growth phase versus late stationary growth phase). No difference in growth rates was observed between curli variants derived from the same strain grown in LBFS or LBNS broth at either 28°C or 37°C (data not shown). For curli variants belonging to group I and II based on MLVA data (see above and Fig. 2), conversion from C+ to C− variants was observed infrequently under all four culture conditions (Table 2), and the population structure remained stable relative to the initial start culture (100% C+ variants). For group III C+ variants (RM6607R and RM6608R), anaerobic growth resulted in a mixed population of C+ and C− variants (Table 2 [37°C, anaerobiosis, LBFS]), and the percentage of C− variants increased from 6.0 and 3.8% at 24 h of incubation to 24.0 and 23.3% at 48 h of incubation for RM6607R and RM6608R, respectively (Table 2). In contrast, C− variants, especially those from group I (derived from the 2006 spinach outbreak strains), appeared unstable, yielding a mixed population of C+ and C− variants after 24 or 48 h of incubation (Table 2). This change did not appear to be regulated by a low salt concentration, since the percentage of C+ variants was significantly higher when C− variants were grown in LBFS than in LBNS broth (Table 2 [28°C, aerobiosis, LBNS versus 28°C, aerobiosis, LBFS]) nor by low temperature or low oxygen tension, because the percentage of C+ variants was the highest when C− variants were grown in LBFS at 37°C aerobically (Table 2 [37°C, aerobiosis, LBFS]). Rather, the change correlated with age of the culture at sampling time (Table 2 [24 h and 48 h]). Therefore, nutrient limitation in the late stationary phase is likely an environmental factor triggering the shift from C− to C+ variants. It is noteworthy that the C− variants of group III undergo a pronounced shift to C+ during culture under anaerobiosis in LBFS at 37°C, suggesting that there is interstrain variation in controlling the switch between C+ and C− in E. coli O157:H7.
Table 2.
Percentage of C+ variants in the total E. coli O157:H7 population under various culture conditionsa
| Variantb | Sourcec | MLVA type (group) | % of C+ variants in E. coli O157:H7 population |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 28°C, aerobiosis, LBNS |
28°C, aerobiosis, LBFS |
37°C, aerobiosis, LBFS |
37°C, anaerobiosis, LBFS |
|||||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |||
| RM6011R | HS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6011W | HS | 834 (I) | 0.1 | 16.2 | 0.3 | 41.5 | 0.0 | 67.8 | 10.1 | 24.8 |
| RM6069R | HS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6069W | HS | 361 (I) | 0.0 | 26.0 | 1.1 | 36.8 | 0.0 | 71.2 | 19.3 | 19.7 |
| RM6049R | HS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6049W | HS | 163 (I) | 0.1 | 5.3 | 0.1 | 26.7 | 0.0 | 67.5 | 11.7 | 20.9 |
| RM6654R | HS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6654W | HS | 163 (I) | 0.1 | 9.2 | 0.0 | 33.2 | 0.0 | 67.0 | 11.5 | 26.3 |
| RM6067R | SS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6067W | SS | 163 (I) | 0.1 | 9.7 | 0.1 | 25.4 | 0.1 | 48.1 | 0.6 | 28.0 |
| RM9994R | SS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM9994W | SS | 163 (I) | 0.0 | 12.3 | 0.1 | 32.1 | 0.2 | 54.6 | 4.8 | 25.4 |
| RM9998R | SS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM9998W | SS | 163 (I) | 0.1 | 13.7 | 0.4 | 13.2 | 0.0 | 51.3 | 10.6 | 27.0 |
| RM10000R | SS | 163 (I) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM10000W | SS | 163 (I) | 0.1 | 26.1 | 0.1 | 46.7 | 1.9 | 62.0 | 2.2 | 26.6 |
| RM6535R | HL | 280 (II) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6535W | HL | 280 (II) | 0.1 | 0.0 | 0.2 | 0.0 | 0.0 | 0.4 | 0.0 | 0.4 |
| RM6416R | WL | 874 (II) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| RM6416W | WL | 214 (II) | 0.2 | 0.6 | 0.9 | 0.0 | 0.0 | 0.1 | 0.1 | 0.3 |
| RM6607R | HH | 365 (III) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 94.0 | 76.0 |
| RM6607W | HH | 365 (III) | 0.2 | 0.4 | 0.9 | 1.0 | 0.0 | 1.4 | 1.3 | 81.1 |
| RM6608R | MH | 365 (III) | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 96.2 | 76.7 |
| RM6608W | MH | 365 (III) | 0.1 | 0.4 | 0.5 | 0.6 | 0.0 | 2.4 | 3.4 | 31.0 |
Percentages of C+ variants in the total population recovered on CRI plates.
The suffixes “R” and “W” in the variant names refer to the C+ and C− variants, respectively.
HS, human isolates associated with the 2006 spinach outbreak; SS, bagged spinach isolates associated with the 2006 spinach outbreak; HL, human isolates associated with a 2006 lettuce outbreak; WL, water isolates associated with a 2006 lettuce outbreak; HH, human isolates associated with the 1993 hamburger outbreak; MH, meat isolates associated with the 1993 hamburger outbreak.
Biofilm formation of E. coli O157:H7 curli variants.
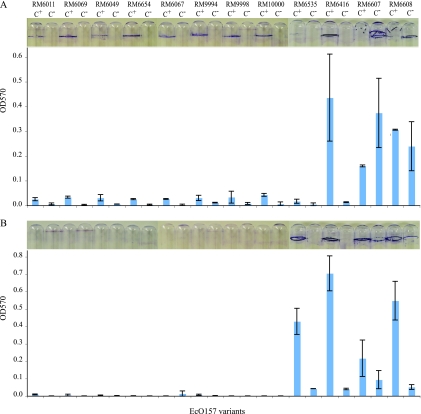
Biofilm formation is presumed to be beneficial to E. coli O157:H7 survival in a natural environment and also in a human host. Hence, we investigated whether C+ variants were better biofilm producers than their corresponding C− variants by measuring their attachment to borosilicate glass in two distinct growth conditions: aerobiosis at 28°C and anaerobiosis at 37°C. All eight group I C+ variants produced visible rings on borosilicate culture tubes following 2 days of incubation at 28°C (Fig. 3A), but no visible biofilm formation was observed when they were grown at 37°C anaerobically (Fig. 3B). No or minimal visible biofilm formation was observed for the eight group I C− variants at both growth conditions. For group II curli variants, a C+ variant derived from clinical isolate RM6535 displayed greater attachment than the corresponding C− variant only under anaerobiosis at 37°C, whereas C+ derived from water isolate RM6416 displayed greater attachment than the corresponding C− variant at both growth conditions (Fig. 3). For group III variants (MLVA type 365), C+ variants appeared to attach to borosilicate glass better than their corresponding C− variants when they were grown at 37°C anaerobically (Fig. 3) but not when they were grown at 28°C aerobically. The latter conditions resulted in no significant difference in attachment between the curli variants derived from the same strain (Fig. 3). These results confirm that biofilm formation of E. coli O157:H7 also is highly strain specific. Among all 12 strains examined, the eight 2006 spinach outbreak strains (RM6011, RM6069, RM6049, RM6654, RM6067, RM9994, RM9998, and RM10000) were the weakest biofilm producers regardless of their source, i.e., spinach or human. These data suggest also that curli promotes biofilm formation in E. coli O157:H7 since C+ variants generally are better in attachment than their corresponding C− variants. However, this characteristic depends largely on the physiological state of the cells.
Fig. 3.
Biofilm formation by E. coli O157:H7 curli variants. Variants were grown at 28°C statically in 1 ml of LBNS broth with atmospheric oxygen (A) or at 37°C in an anaerobic jar (B). To assess the attachment of cells on the glass wall, the cells were stained with 0.1% crystal violet, which was then solubilized in 95% ethanol to determine the absorbance at 570 nm. Bars represent the average OD570 from three independent biological replicates, whereas the images of glass tubes stained with crystal violet are of one representative replicate per variant.
C+ variants grow better under nutrient limitation than C− variants.
Since nutrient limitation is one of the most common stresses enteric bacteria encounter in a nonhost environment, we assessed the ability of E. coli O157:H7 curli variants in coping with this stress by comparing the growth of C+ and C− variants in 0.01% peptone water at 15°C. Group I C+ variants grew better than C− variants under this nutrient-limited condition regardless of their source (clinical or environmental). On average, the total viable cells of these C+ variants increased 8.4- and 9.2-fold after 5 and 7 days of incubation in 0.01% peptone water, respectively, whereas all eight C− variants did not display any change in total viable cells (Table 3). Similarly, group III C+ variants (MLVA type 365) grew better than the corresponding C− variants under this nutrient-limited condition. In contrast to group I C− variants, which didn't grow in 0.01% peptone water, group III C− variants exhibited slow growth (Table 3).
Table 3.
Growth and stability of E. coli O157:H7 curli variants in 0.01% peptone watera
| Variantb | Sourcec | MLVA type (group) | Fold increase in viable CFU/mld |
Stability of curli variants (% C+)e |
||
|---|---|---|---|---|---|---|
| 5 days | 7 days | 5 days | 7 days | |||
| RM6011R | HS | 163 (I) | 5.2 ± 2.3 | 6.2 ± 2.6 | 100.0 | 100.0 |
| RM6011W | HS | 834 (I) | 1.0 ± 0.0 | 0.9 ± 0.1 | 1.7 | 6.0 |
| RM6069R | HS | 163 (I) | 8.0 ± 1.2 | 9.7 ± 2.5 | 100.0 | 100.0 |
| RM6069W | HS | 361 (I) | 1.0 ± 0.1 | 1.1 ± 0.2 | 2.3 | 19.8 |
| RM6049R | HS | 163 (I) | 6.0 ± 1.0 | 6.6 ± 1.2 | 100.0 | 100.0 |
| RM6049W | HS | 163 (I) | 1.0 ± 0.1 | 1.1 ± 0.3 | 5.4 | 20.2 |
| RM6654R | HS | 163 (I) | 7.9 ± 2.1 | 10.5 ± 3.9 | 100.0 | 100.0 |
| RM6654W | HS | 163 (I) | 1.0 ± 0.1 | 1.0 ± 0.3 | 0.4 | 12.0 |
| RM6067R | SS | 163 (I) | 7.3 ± 3.0 | 7.1 ± 3.5 | 100.0 | 100.0 |
| RM6067W | SS | 163 (I) | 1.1 ± 0.1 | 1.1 ± 0.0 | 0.0 | 0.4 |
| RM9994R | SS | 163 (I) | 9.5 ± 1.2 | 10.1 ± 1.3 | 100.0 | 100.0 |
| RM9994W | SS | 163 (I) | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.2 | 4.8 |
| RM9998R | SS | 163 (I) | 11.4 ± 2.0 | 11.9 ± 2.4 | 100.0 | 100.0 |
| RM9998W | SS | 163 (I) | 1.1 ± 0.2 | 1.1 ± 0.1 | 0.2 | 6.2 |
| RM10000R | SS | 163 (I) | 11.9 ± 0.9 | 11.2 ± 1.7 | 100.0 | 100.0 |
| RM10000W | SS | 163 (I) | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.2 | 2.1 |
| RM6535R | HL | 280 (II) | 9.3 ± 3.5 | 7.4 ± 1.4 | 100.0 | 100.0 |
| RM6535W | HL | 280 (II) | 10.3 ± 2.0 | 11.0 ± 1.0 | 0.0 | 0.0 |
| RM6416R | WL | 874 (II) | 6.8 ± 1.7 | 7.4 ± 1.4 | 100.0 | 100.0 |
| RM6416W | WL | 214 (II) | 12.4 ± 3.2 | 10.7 ± 1.7 | 0.0 | 0.0 |
| RM6607R | HH | 365 (III) | 6.9 ± 0.6 | 7.3 ± 1.4 | 100.0 | 100.0 |
| RM6607W | HH | 365 (III) | 1.7 ± 0.8 | 2.5 ± 1.7 | 0.1 | 0.2 |
| RM6608R | MH | 365 (III) | 8.2 ± 2.6 | 8.5 ± 0.4 | 100.0 | 99.9 |
| RM6608W | MH | 365 (III) | 2.1 ± 0.2 | 6.1 ± 4.2 | 0.0 | 1.0 |
E. coli O157:H7 curli variants were inoculated in 0.01% peptone water and incubated at 15°C for up to a week.
The suffixes “R” and “W” in the variant names refer to the C+ and C− variants, respectively.
HS, human isolates associated with the 2006 spinach outbreak; SS, bagged spinach isolates associated with the 2006 spinach outbreak; HL, human isolates associated with a 2006 lettuce outbreak; WL, water isolates associated with a 2006 lettuce outbreak; HH, human isolates associated with the 1993 hamburger outbreak; MH: meat isolates associated with the 1993 hamburger outbreak.
CFU counts were determined on LB plates.
Curli variants were recovered on CRI plates.
Consistent with our previous observation, C+ variants were very stable under this growth condition and no conversion to C− variants was observed. In contrast, C− variants were highly unstable, yielding a mixed population of C+ and C− variants at both time points examined. The percentage of C+ variants arising from these C− variants increased significantly from 1.3 to 8.9% on average (t test, P < 0.005) when incubation time increased from 5 to 7 days (Table 3). Moreover, the frequency of the switch from C− to C+ in the group I variants appeared to be associated with the source of the strain. For example, there was a significantly greater percentage of C+ variants arising from C− variants derived from the clinical isolates (RM6011W, RM6069W, RM6049W, and RM6654W) than those derived from environmental isolates (RM6067W, RM9994W, RM9998W, and RM10000W) (t test, P = 0.02 [5 days] and P = 0.03 [7 days]). The conversion of group III C− to C+ was observed at 7 days; however, the percentage of C+ variants was much lower than that in group I C− variants (Table 3). Unlike either group I or group III C− variants, group II C− variants appeared very stable under this nutrient-limited condition, resulting in no observable conversion to C+ variants (Table 3).
C− variants are more acid resistant than C+ variants.
The immediate challenge faced by gastrointestinal pathogens, like E. coli O157:H7, in an infected human host is survival in the extreme acidic environment present in the stomach. Therefore, we compared the survival of E. coli O157:H7 curli variants following exposure to acidified LB broth (pH 2.5). The viable cells were recovered on LB agar plates after incubation in acidified broth at 37°C for 2 and 6 h, and the number of CFU was compared with that of the initial inoculum (CFU/ml at time zero). It is noteworthy that all C+ variants were significantly more sensitive to acidic pH than their corresponding C− variants, regardless of their genotypes (Table 4). For group I, II, and III, the average decreases in viable cells of C+ variants were 2.2, 5.7, and 5.6 log, respectively, and those of C− variants were 0.1, 0.6, and 0.0 log following a 2-h acid challenge (Table 4). When the acid challenge was increased to 6 h, the numbers of viable cells of most C+ variants decreased by over 5.0 log, with the exception of that of RM6011R, which decreased by 1.1 log (Table 4).
Table 4.
Survival and stability of E. coli O157:H7 curli variants following acid challenge
| Varianta | Sourceb | MLVA type (group) | Fold decrease in log CFU/mlc |
Stability of curli variants (% C+)d |
||
|---|---|---|---|---|---|---|
| 2 h | 6 h | 2 h | 6 h | |||
| RM6011R | HS | 163 (I) | 0.4 ± 0.3 | 1.1 ± 0.2 | 100.0 | NA |
| RM6011W | HS | 834 (I) | 0.1 ± 0.1 | 0.3 ± 0.1 | 0.0 | 0.0 |
| RM6069R | HS | 163 (I) | 1.6 ± 0.1 | 5.4 ± 0.8 | 100.0 | 96.3 |
| RM6069W | HS | 361 (I) | 0.0 ± 0.0 | 0.1 ± 0.0 | 0.0 | 0.0 |
| RM6049R | HS | 163 (I) | 3.1 ± 0.2 | 5.9 ± 1.1 | 99.9 | 37.3 |
| RM6049W | HS | 163 (I) | 0.0 ± 0.0 | 0.3 ± 0.0 | 0.0 | 0.0 |
| RM6654R | HS | 163 (I) | 2.2 ± 0.4 | 5.8 ± 1.2 | 100.0 | 95.0 |
| RM6654W | HS | 163 (I) | 0.2 ± 0.1 | 0.3 ± 0.0 | 0.0 | 0.0 |
| RM6067R | SS | 163 (I) | 2.0 ± 0.2 | 6.6 ± 0.1 | 100.0 | NA |
| RM6067W | SS | 163 (I) | 0.0 ± 0.0 | 0.5 ± 0.5 | 0.0 | 0.0 |
| RM9994R | SS | 163 (I) | 2.6 ± 0.7 | 6.5 ± 0.2 | 100.0 | 100.0 |
| RM9994W | SS | 163 (I) | 0.1 ± 0.1 | 0.4 ± 0.2 | 0.0 | 0.0 |
| RM9998R | SS | 163 (I) | 2.8 ± 0.8 | 6.5 ± 0.0 | 100.0 | NA |
| RM9998W | SS | 163 (I) | 0.3 ± 0.1 | 1.0 ± 0.5 | 0.0 | 0.0 |
| RM10000R | SS | 163 (I) | 2.7 ± 0.5 | 6.6 ± 0.1 | 100.0 | NA |
| RM10000W | SS | 163 (I) | 0.2 ± 0.0 | 0.4 ± 0.1 | 0.0 | 0.0 |
| RM6535R | HL | 280 (II) | 4.8 ± 0.1 | 5.0 ± 0.1 | 0.0 | 0.0 |
| RM6535W | HL | 280 (II) | 0.6 ± 0.0 | 3.7 ± 0.5 | 0.0 | 0.0 |
| RM6416R | WL | 874 (II) | 6.6 ± 0.0 | 6.6 ± 0.1 | NA | NA |
| RM6416W | WL | 214 (II) | 0.5 ± 0.1 | 3.3 ± 0.2 | 0.1 | 0.0 |
| RM6607R | HH | 365 (III) | 5.8 ± 1.0 | 5.9 ± 1.0 | NA | NA |
| RM6607W | HH | 365 (III) | 0.0 ± 0.0 | 0.6 ± 0.2 | 0.1 | 0.1 |
| RM6608R | MH | 365 (III) | 5.4 ± 1.6 | 5.8 ± 1.0 | NA | NA |
| RM6608W | MH | 365 (III) | 0.0 ± 0.1 | 0.5 ± 0.3 | 0.2 | 0.1 |
The suffixes “R” and “W” in the variant names refer to the C+ and C− variants, respectively.
HS, human isolates associated with the 2006 spinach outbreak; SS, bagged spinach isolates associated with the 2006 spinach outbreak; HL, human isolates associated with a 2006 lettuce outbreak; WL, water isolates associated with a 2006 lettuce outbreak; HH, human isolates associated with the 1993 hamburger outbreak; MH, meat isolates associated with the 1993 hamburger outbreak.
CFU counts were determined on LB plates following incubation in acidified broth and then compared with the CFU counts prior to acid challenge in order to assess survival.
Stability of curli variants was examined by measuring the percentage of C+ variants in the total E. coli O157:H7 population recovered on CRI plates. NA, no cells were recovered on CRI plates.
Interstrain variation in acid tolerance among C− variants was observed also. For example, C− variants in group II (RM6535W and RM6416W) were much more sensitive to acidic broth than group I C− variants (derived from the 2006 spinach outbreak strains) and group III C− variants (derived from the 1993 hamburger outbreak strains). The average decreases in viable cells for group II C− variants were 0.6 and 3.5 log, compared with 0.1 and 0.4 log for group I C− variants, and 0.0 and 0.6 log for group III C− variants, following 2 and 6 h of acid challenge, respectively (Table 4).
As expected, prolonged incubation in acidified broth enriched for C− variants in E. coli O157:H7 populations. For example, following 6 h under acidic conditions, cells recovered on CRI plates were a mixture of both C+ and C− for three of the group I C+ variants (RM6069R, 4.7% C−; RM6049R, 62.7% C−; RM6654R, 95.0% C−), although no C− variants were detected in the initial culture (Table 4). For the majority of C+ variants, we were unable to evaluate the final population structure due to high acid sensitivity of C+ variants and the poor recovery of viable cells on CRI plates (Table 4). In contrast to growth in 0.01% peptone water, C− variants in acidified broth were very stable. No detectable conversion from C− to C+ was observed for either group I or group II C− variants, whereas a low frequency of conversion (0.1%) was observed for the group III C− variants (Table 4).
Survival of E. coli O157:H7 csgA deletion mutants following acid challenge.
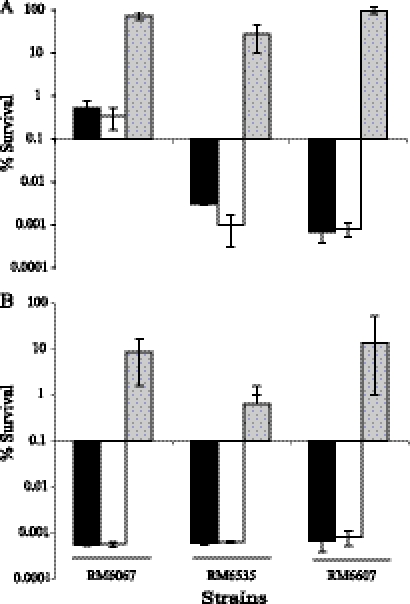
The significant difference in acid resistance between the curli variants of E. coli O157:H7 led us to investigate whether the presence of curli fimbriae per se was the cause of the acid sensitivity in E. coli O157:H7 C+ variants. We generated a csgA deletion mutant in three pairs of E. coli O157:H7 curli variants, RM6067R and RM6067W, RM6535R and RM6535W, and RM6607R and RM6607W, which represented the three groups of curli variants examined in this study (Fig. 2). We examined the survival of CsgA-deficient mutants derived from C+ variants following acid challenge and compared the survival of mutants with the parental strains (C+ variants) and the corresponding C− variants. All three csgA deletion mutants displayed a survival rate similar to their parental C+ variants following either 2 h (Fig. 4A) or 6 h (Fig. 4B) of acid challenge and were much more acid sensitive than their corresponding C− variants. Similarly, deletion of csgA in C− variants did not have any impact on acid resistance in any of the three C− variants examined (data not shown). These data suggest that production of curli fimbriae in E. coli O157:H7 C+ variants is not the cause of their acid sensitivity.
Fig. 4.
Survival of E. coli O157:H7 csgA deletion mutants following acid challenge. Curli variants and the csgA deletion mutants were incubated in acidified broth (pH 2.5) at 37°C for 2 h (A) and 6 h (B). The viable cells were recovered on LB agar plates and compared in CFU with the viable cells at time zero. Black columns represent C+ variants RM6067R, RM6535R, and RM6607R; white columns represent CsgA-minus mutants (strains MQC108, MQC114, and MQC118) derived from the three C+ variants; and grey columns are corresponding C− variants, RM6067W, RM6535W, and RM6607W.
DISCUSSION
Curli, also known as bacterial amyloid, is an important virulence factor involved in initial surface attachment, biofilm formation, and induction of the inflammatory response. Regulation of curli production involves a complex network of regulatory proteins, including the stationary-phase sigma factor RpoS, small protein Crl, histone-like protein H-NS, integration host factor IHF, and the signaling molecule cyclic di-GMP (c-di-GMP) (30, 35, 37). Also, there are three two-component regulatory systems involved in curli expression. The OmpR/EnvZ system responds to changes in osmolarity, whereas both the CpxA/R and Rcs systems respond to cell envelope stress (11, 12, 19). Our results confirmed considerable inter- and intrastrain variations in curli production among different E. coli O157:H7 strains related to different outbreaks. C+ variants isolated from strains linked to a large 2006 U.S. spinach outbreak produced the least amount of curli compared with C+ variants isolated from strains linked to a 2006 iceberg lettuce outbreak and a 1993 hamburger outbreak (Fig. 1). It has been reported that in the E. coli O157:H7 human hamburger outbreak strain EDL933, mutations in the csgD promoter regions were correlated with higher curli expression, whereas in Salmonella, mutations in the afgD promoter led to constitutive expression of curli fimbriae (33, 40). Both inter- and intrastrain variations in curli production observed in our study could not be explained simply by mutations in the csgD promoter, since all 24 curli variants shared the identical DNA sequence spanning csgA to csgD (data not shown). Considering the complexity of the regulation of curli expression, genetic change and/or stochastic gene expression in any of those transcriptional regulators may lead to variations in curli production among individual E. coli O157:H7 cells in the population. Indeed, it was reported that a single point mutation in the ompR gene in E. coli K-12 resulted in the increased transcription of the csgA gene, and the production of curli became independent of RpoS function in this mutant strain (42). Thus, it is reasonable to hypothesize that mutation in any of the genes encoding either direct or indirect regulators of curli genes will affect the transcription of curli genes and can be potential causes of the inter- or intrastrain variations observed in our study. Substantial genetic analyses of these variants are required to facilitate our understanding of observed physiological changes at the molecular level.
The proportions of the two subpopulations, C+ variants and C− variants, in a final E. coli O157:H7 cell population initiated either from a C+ variant or a C− variant appeared strain specific and were affected by culture conditions. Most C+ variants that we examined in this study were very stable and reverted to C− phenotype infrequently, regardless of their growth conditions. The exceptions were the two C+ variants derived from the 1993 hamburger outbreak strain (RM6607R and RM6608R), in which a considerable proportion of C− variants arose in the culture initiated from a C+ variant during anaerobic growth (Table 2). Furthermore, the group I C− variants (derived from strains associated with the 2006 U.S. spinach outbreak) reverted to C+ variants frequently, compared with either group II or group III C− variants, resulting in a high percentage of C+ variants in the final E. coli O157:H7 population. These observations suggest that the putative mechanism that controls the shift between the two subpopulations, C+ and C− variants, may differ among E. coli O157:H7 strains. The shift can be either bidirectional or unidirectional, and the proportions of the two subpopulations can be strain specific. The highest percentage of C+ variants was observed in aged cultures initiated with C− variants, suggesting that nutrient limitation or starvation is a major factor promoting the conversion from the C− to the C+ phenotype. Decreasing either the culture temperature or the salt concentration in culture broth did not increase the shift from C− to C+, suggesting that environmental signals that stimulate curli gene expression are not identical to those that trigger the conversion between C+ and C− variants in E. coli O157:H7. Overall, our data indicate that conversion between the C+ and C− variants in E. coli O157:H7 is not a random process but is linked tightly to particular environmental signals.
It is well documented that curli play an important role in biofilm formation, especially in the initial stage of biofilm development, in both E. coli and Salmonella (1, 23). Recent research in our group on interactions between Salmonella and the fungus Aspergillus niger suggests that curli are also important in maintaining the structure of a mature biofilm (Maria Brandl, personal communication). Biofilms enhance bacterial persistence in natural environments and in infected human hosts by providing physical protection as well as metabolic benefits for the microbial community embedded in the extracellular matrix. Therefore, the ability to produce biofilms would benefit bacteria in certain niches. Biofilm formation by E. coli O157:H7 depends on multiple factors, including cell appendages, such as pili and flagella, as well as production of extracellular polysaccharides. The E. coli O157:H7 strains examined in this study varied greatly in their ability to form a biofilm on a glass surface. Strains related to the 2006 spinach outbreak were the poorest biofilm producers of all strains examined (Fig. 3). Deletion of csgA in a C+ variant of this group (RM6067R) did not further decrease the surface attachment, suggesting that factors other than curli may be more important for biofilm formation on glass for this group of E. coli O157:H7 strains. In contrast, deletion of the csgA gene in the C+ variant RM6607R (group III variants) led to about 60% reduction in biofilm formation on glass, suggesting that curli is indeed an important colonization factor in this variant (data not shown). Our data suggest also that biofilm formation by E. coli O157:H7 is additionally dependent on the physiological state of the cells. The role of curli in biofilm formation also is strain specific. It is noteworthy that considerable intrastrain variations occurred in biofilm formation in E. coli O157:H7, suggesting that individual cells within a pure culture of E. coli O157:H7 differ in their ability to attach to surfaces and form a biofilm. The role of this phenotypic variation in the survival and virulence of E. coli O157:H7 in its natural habitats remains elusive. The ability to switch from biofilm growth to planktonic cells is also important in providing bacteria a mechanism to escape from an environmental or host niche that is in decline.
Natural environments and animal hosts, including humans, are two major and distinct habitats for enteric pathogens, including E. coli O157:H7. To be able to establish colonization in a host, pathogens must survive the acidic conditions in the stomach before they reach the intestine. Once the pathogens are shed from an animal host, they must overcome multiple adverse environmental stresses, such as limited nutrients, fluctuating temperature, desiccation, and radiation from UV light, to be capable of causing an infection. Curli variants derived from the same E. coli O157:H7 strain carry several distinct physiological properties that are important for survival in both host and nonhost environments. For example, acid resistance is important for survival in host environments, whereas the ability to scavenge nutrients is important for survival in a natural habitat with limited nutrients. We report here that C− variants are much more acid resistant than their corresponding C+ variants, averaging about 105-fold-greater viable counts following a 6-h incubation in acidified broth. On the other hand, C+ variants, especially those from group I, grow better than their corresponding C− variants under nutrient-limited conditions, averaging 8.4-fold-greater viable counts following a 7-day incubation in 0.01% peptone water. These physiological divergences between the natural curli variants isolated from the same strain appeared not to be linked directly to the production of curli fimbriae, since the survival of a csgA deletion mutant derived from either a C+ variant or a C− variant was similar to that of its parental strain (Fig. 4). Our data indicate that divergences between the curli variants may be broader than previously thought. The observed phenotypes, including curli production and acid resistance, could be the consequences of changes in the regulatory cassettes involved in biogenesis of curli and activation of acid-resistant systems or proteins. Indeed, there are several common master transcriptional regulators involved in both curli biogenesis and regulation of a glutamate-dependent acid resistance system (AR2), including RpoS and H-NS (13). The AR2 system is known to provide E. coli cells with the highest level of protection at pH 2.5 compared with those of the RpoS-dependent acid resistance system (AR1) and the arginine-dependent acid resistance system (AR3).
The marked differences in the physiology of E. coli O157:H7 curli variants observed in this study provide an example of how bacterial pathogens modulate the dynamics of population behavior by controlling the relative proportions of two subpopulations, C+ and C− variants, to improve their survival fitness. We speculate that C+ variants dominate E. coli O157:H7 populations in natural environments with low nutrient levels, such as field water, soil, or sediment. Our results suggest that the number of E. coli O157:H7 C+ variant cells ingested by an animal host may decline during passage through gastric juice as a result of their high acid sensitivity. It remains unclear whether curli production of E. coli O157:H7 would enhance their colonization in the host intestine. C+ variants examined in this study all are able to produce curli at 37°C, suggesting a putative role of E. coli O157:H7 curli in host infection as demonstrated in other clinical E. coli strains (4). However, curli are known to stimulate host innate immune responses (38), and in a recent study in Salmonella enterica serovar Typhimurium, the curli-deficient strain appeared to be more virulent than the curli-producing strain in mice (45). The results of our study provide an example of how bacterial pathogens maintain a dynamic population of cells to ensure the presence of phenotypes that facilitate survival and rapid adaptation of a subpopulation to a given niche. A niche-specific phenotype would be selected and amplified if the environmental conditions become favorable for growth of the surviving phenotype. Therefore, phenotypic variations and/or cell individualities provide strategies that ensure the optimal survival of the pathogens exposed to diverse niches.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to C. Ball, W. Chmielecki, E. Hÿttia-Trees, M. Janda, B. Juni, T. Monson, and FDA for providing E. coli O157:H7 strains. We thank Matthew Chapman for providing anti-CsgA antibody and E. coli strains MC4100 and LSR1.
This work was supported in part by USDA-ARS CRIS project 5325-42000-044-00D and by National Research Initiative Competitive Grant numbers 2006-55212-16927 and 2007-35212-18239, from the USDA National Institute of Food and Agriculture.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Austin J. W., Sanders G., Kay W. W., Collinson S. K. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295–301 [DOI] [PubMed] [Google Scholar]
- 2. Avery S. V. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4:577–587 [DOI] [PubMed] [Google Scholar]
- 3. Barnhart M. M., Chapman M. R. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bian Z., Brauner A., Li Y., Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602–612 [DOI] [PubMed] [Google Scholar]
- 5. Brzuszkiewicz E., Gottschalk G., Ron E., Hacker J., Dobrindt U. 2009. Adaptation of pathogenic E. coli to various niches: genome flexibility is the key. Genome Dyn. 6:110–125 [DOI] [PubMed] [Google Scholar]
- 6. Casadaban M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 7. Chapman M. R., et al. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cooley M., et al. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One 2:e1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidson C. J., Surette M. G. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253–268 [DOI] [PubMed] [Google Scholar]
- 11. Dorel C., Vidal O., Prigent-Combaret C., Vallet I., Lejeune P. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169–175 [DOI] [PubMed] [Google Scholar]
- 12. Ferrieres L., Clarke D. J. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50:1665–1682 [DOI] [PubMed] [Google Scholar]
- 13. Foster J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898–907 [DOI] [PubMed] [Google Scholar]
- 14. Gophna U., et al. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammar M., Arnqvist A., Bian Z., Olsen A., Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661–670 [DOI] [PubMed] [Google Scholar]
- 16. Hammar M., Bian Z., Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:6562–6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hammer N. D., Schmidt J. C., Chapman M. R. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U. S. A. 104:12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hidalgo G., Chen X., Hay A. G., Lion L. W. 2010. Curli produced by Escherichia coli PHL628 provide protection from Hg(II). Appl. Environ. Microbiol. 76:6939–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jubelin G., et al. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kai-Larsen Y., et al. 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 6:e1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keys C., Kemper S., Keim P. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98:928–940 [DOI] [PubMed] [Google Scholar]
- 22. Khurana R., Uversky V. N., Nielsen L., Fink A. L. 2001. Is Congo red an amyloid-specific dye? J. Biol. Chem. 276:22715–22721 [DOI] [PubMed] [Google Scholar]
- 23. Kikuchi T., Mizunoe Y., Takade A., Naito S., Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49:875–884 [DOI] [PubMed] [Google Scholar]
- 24. Lindstedt B. A., Heir E., Gjernes E., Vardund T., Kapperud G. 2003. DNA fingerprinting of Shiga-toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mandrell R. E. 2009. Enteric human pathogens associated with fresh produce: sources, transport and ecology, p. 5–41 In Fan X., Niemira B. A., Doona C. J., Feeherry F. E., Gravani R. B. (ed.), Microbial safety of fresh produce. IFT Press/Wiley-Blackwell Publishing, Ames, IA [Google Scholar]
- 26. Manning S. D., et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maurer J. J., Brown T. P., Steffens W. L., Thayer S. G. 1998. The occurrence of ambient temperature-regulated adhesins, curli, and the temperature-sensitive hemagglutinin tsh among avian Escherichia coli. Avian Dis. 42:106–118 [PubMed] [Google Scholar]
- 28. Olsen A., Jonsson A., Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655 [DOI] [PubMed] [Google Scholar]
- 29. O'Toole G. A., Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 30. Pratt L. A., Silhavy T. J. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225–1236 [DOI] [PubMed] [Google Scholar]
- 31. Rangel J. M., Sparling P. H., Crowe C., Griffin P. M., Swerdlow D. L. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson L. S., Ashman E. M., Hultgren S. J., Chapman M. R. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59:870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romling U., Sierralta W. D., Eriksson K., Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 34. Smalley J. W., Birss A. J., McKee A. S., Marsh P. D. 1995. Congo red binding by Porphyromonas gingivalis is mediated by a 66 kDa outer-membrane protein. Microbiology 141(Pt. 1):205–211 [DOI] [PubMed] [Google Scholar]
- 35. Sommerfeldt N., et al. 2009. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331 [DOI] [PubMed] [Google Scholar]
- 36. Szabo E., et al. 2005. Curli expression of enterotoxigenic Escherichia coli. Folia Microbiol. (Praha) 50:40–46 [DOI] [PubMed] [Google Scholar]
- 37. Tagliabue L., Maciag A., Antoniani D., Landini P. 2010. The yddV-dos operon controls biofilm formation through the regulation of genes encoding curli fibers' subunits in aerobically growing Escherichia coli. FEMS Immunol. Med. Microbiol. 59:477–484 [DOI] [PubMed] [Google Scholar]
- 38. Tukel C., et al. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58:289–304 [DOI] [PubMed] [Google Scholar]
- 39. Uhlich G. A., Gunther N. W., Bayles D. O., Mosier D. A. 2009. The CsgA and Lpp proteins of an Escherichia coli O157:H7 strain affect HEp-2 cell invasion, motility, and biofilm formation. Infect. Immun. 77:1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uhlich G. A., Keen J. E., Elder R. O. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uhlich G. A., Keen J. E., Elder R. O. 2002. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vidal O., et al. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X., Chapman M. R. 2008. Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 380:570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Westerman R. B., He Y., Keen J. E., Littledike E. T., Kwang J. 1997. Production and characterization of monoclonal antibodies specific for the lipopolysaccharide of Escherichia coli O157. J. Clin. Microbiol. 35:679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. White A. P., et al. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1048–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zogaj X., Bokranz W., Nimtz M., Romling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.