Abstract
Contamination of lakes and ponds plays an essential role as a reservoir of avian influenza A virus (AIV) in the environment. A method to concentrate waterborne AIV is a prerequisite for the detection of virus present at low levels in water. The aim of this study was to develop and validate a method for the concentration and detection of infectious AIV from large volumes of surface water samples. Two filtration systems, glass wool and electropositive NanoCeram filter, were studied. The individual effects of filtration-elution and polyethylene glycol (PEG) concentration parameters on the recovery efficiency of the H1N1 strain from 10-liter surface water samples were assessed. An ultimate 1% recovery rate of infectious viruses was achieved with the optimal protocol, corresponding to filtration through glass wool, followed by a viral elution step and then a PEG concentration. This method was validated for the detection of highly pathogenic H5N1 strains from artificially contaminated larger water volumes, from 10 to up to 50 liters, from different sources. The viral recovery efficiencies ranged from 0.01% to 7.89% and from 3.63% to 13.79% with lake water and rainwater, respectively. A theoretical detection threshold of 2.25 × 102 TCID50 (50% tissue culture infectious dose) in the filtered volume was obtained for seeded lake waters by M gene reverse transcriptase PCR (RT-PCR). Moreover, the method was used successfully in field studies for the detection of naturally occurring influenza A viruses in lake water in France.
INTRODUCTION
Influenza A viruses belong to the Orthomyxoviridae family of negative single-stranded, enveloped RNA viruses. All subtypes of influenza A viruses (H1 to H16 and N1 to N9) have been isolated from wild waterfowls (24, 37). But wild ducks are the main natural reservoir and are generally asymptomatic virus carriers (38). Avian influenza A viruses (AIVs) replicate not only in the respiratory tract but also in the gastrointestinal tract in ducks and are thus shed in high concentrations in the feces (39). Viral transmission occurs most of the time by direct contact between infected birds and essentially the respiratory tract of susceptible hosts. But the role of an indirect waterborne transmission linked to feces-contaminated water has also been confirmed and would be involved in the maintenance of AIVs in ducks (15, 16, 22). AIVs have been isolated from water bodies where waterfowl gather (15, 36, 44) and, moreover, can persist for a long time in water (13, 16). Experimentally, infectious viruses can persist for up to 8 days in bird feces at 22°C (39) and for a few months in cold water (4, 33, 34). All together, these data revealed a mechanism of year-by-year perpetuation of the viruses in the environment where birds breed, especially in cold-climate countries. Contaminated lakes and ponds play essential roles as environmental virus reservoirs. Although most human cases had a history of very close contact with infected poultry, and inhalation of infectious droplets was probably the most common route of infection (3), the oral ingestion or aspiration of contaminated water could be a possible mode of human contamination. To date, there is no clearly defined method to extract and detect influenza viruses in water, although several methods had been employed to determine the viral concentration in water. Based on principles used for enteroviruses, the use of adsorption/elution on electropositive filters was reported and seemed adapted to detect influenza viruses in large volumes of experimentally spiked tap water (27) or naturally contaminated surface water (29). Alternatively, concentration with chicken erythrocytes (16, 18, 27, 29), or with polyethylene glycol (PEG) (22), has been used to concentrate influenza viruses from smaller volumes of tap, lake, or pond water, sometimes in combination with filter adsorption (29). But no recent protocol for the recovery of influenza viruses from surface waters with measured and reported recovery percentages has been published (27).
The objective of the present study was to develop a method for the detection of influenza viruses from large-volume surface water samples, based on the adsorption of the viruses on filters, followed by their elution in the presence of a protein solution and their concentration with polyethylene glycol (PEG). Two filtration systems, glass wool, as used for the detection of enterovirus (2), and NanoCeram electropositive cartridge filter (Argonide), were evaluated. First, a fractional experimental design was conducted to assess individual effects of the filtration system in combination with five filtration-elution parameters (filtration flow rate, elution buffer compositions in beef extract and glycine, elution flow rate, and contact time with elution buffer) and two PEG concentration parameters (precipitation and centrifugation times). Recovery efficiencies were determined for a representative virus of the family of influenza A viruses, namely, the H1N1 A/PR/8/34 strain. The concentration method was validated for the detection of highly pathogenic H5N1 strains in artificially contaminated waters from different sources. Moreover, the method was used on natural surface water samples suspected to be contaminated with influenza A viruses in H5N1 outbreak-related places from Cambodia and France.
MATERIALS AND METHODS
Influenza A virus propagation.
Influenza A virus subtype H1N1 (A/PR/8/34) was purchased from the American Type Culture Collection (LGC Promochem, Strasbourg, France). Two highly pathogenic avian influenza virus (HPAIV) H5N1 strains, namely, A/HK/156/97 (clade 0) and A/DK/CAM/67F8/2008 (clade 1), were used for experiments performed in France and in Cambodia, respectively. Methods for the propagation of the A/PR/8/34 and A/HK/156/97 influenza viruses on MDCK cells were used in France to prepare inoculums, as previously described (21, 43). In Cambodia, A/DK/CAM/67F8/2008 influenza virus stock was obtained after propagation in specific-pathogen-free (SPF) 9- to 11-day-old embryonated hen eggs, as previously described (12). The viruses were stored at −80°C until further use.
Infectivity assays.
Infectivity of influenza H1N1 and H5N1 viruses was determined for experiments conducted in France by using a microtiter endpoint titration, as previously described (21). Infectivity was calculated by the Spearman and Karber method (14) and expressed as the 50% tissue culture infectious dose (TCID50) per milliliter, as described in the European standard NF EN 14476 (1). For experiments conducted in Cambodia, an infectivity assay for influenza H5N1 virus was performed on 9- to 11-day-old embryonated chickens eggs, followed by a hemagglutination assay using amnioallantoic fluid as described previously (12, 42). The Reed-Muench method (26) was used to calculate 50% egg infectious dose (EID50). Comparisons were made between the number of EID50 and TCID50 units for the Cambodian H5N1 virus in order to be able to express results in both units (data not shown).
Water samples and sampling sites.
Experiments for development and validation were conducted with water matrix representatives of water bodies where waterfowl gather. A total of 10 to 50 liters of surface waters were collected at approximately 2 m from the waterside. First, to optimize the conditions used with the concentration method, the surface water was sampled from different places in a pond in northern France in autumn. Second, natural water samples were used to validate the method for the detection of experimentally added highly pathogenic H5N1 viruses in water from different sources and to assess the sensitivity of the method. Lake waters were sampled from an ornithological park in northern France and from lakes located in the Kampong Cham and Prey Veng provinces (Cambodia). Rainwater samples were also collected in Cambodia. Finally, natural surface water samples suspected to be contaminated with influenza A viruses from Cambodia and France were used to assess the method for the detection of influenza A viruses from large water volumes. Water samples were previously sampled from the surrounding vicinities of H5N1-infected patients' households in Cambodia in April 2007 (Ponhea Kraek, Kampong Cham province) and December 2008 (Kandal Steung, Kandal province) and stored at −80°C. Water specimens were also collected from Boeung Thom Lake (Kampong Cham) between April and July 2009 and from a wet zone near Prey Trakhob village (Prey Veng) where H5N1 outbreaks occurred in poultry in 2006 (25, 40). Other water samples were collected from three different ponds in France (Dombes region) during the mass migration of birds in autumn 2009, because many birds tested positive for highly pathogenic H5N1 in early 2006.
Filter media and preparation.
Glass wool filters were prepared as previously described for the concentration of enteroviruses from water (2, 35). Stainless steel pressure holders (47 mm diameter, 200 ml) were used (Sartorius, France). Fifty grams of oiled sodocalcic glass wool (Saint-Gobain, France) was packed into the holders. NanoCeram electropositive cartridge filters, manufactured by Argonide Corp., Sanford, FL, are ready to use. Pleated filter cartridges are 63 mm wide by 127 mm long. The filter media correspond to multilayer nano alumina fibers, which give them electropositive charges that are dispersed throughout a cellulose and polyester fiber matrix, with a 2 μm average pore size.
Development of the concentration method.
The proposed protocol, using a filtration-elution step followed by polyethylene glycol (PEG) concentration, was adapted from existing methods for detection of enteric and influenza A viruses. To obtain an optimal protocol, a fractional experimental design was conducted to evaluate the influence of eight factors on the viral recovery efficiency (Table 1). Four different water samples (A, B, C, and D), corresponding to different sampling dates, were used. Two trials were implemented for each parameter combination. A/PR/8/34 (H1N1) virus stock was added to a final concentration of approximately 1 × 107 TCID50 in 10 liters of surface water. After being mixed, a small volume was immediately sampled for the subsequent virus infectivity assay. The seeded water was pumped by a peristaltic pump from a large plastic jerry can through the tested filter at an average flow rate of 10 or 30 liters/h, as previously used (8, 9, 28). Two elution procedures were evaluated for elution of viruses from the glass wool and the NanoCeram cartridge filter with 300 ml and 500 ml of an eluting solution, respectively. The latter consisted of 1.5 or 3% (wt/vol) beef extract (Becton, Dickinson and Company, Le Pont-de-Claix, France) solution (pH 9.5), containing 0 or 0.05 M glycine (Sigma-Aldrich, St. Louis, MO), as previously described (2, 9, 10, 17, 19, 28). Each elution was performed at an average flow rate of 10 or 30 liters/h, as previously used (8, 9, 28). The elution buffer was kept in contact with the filter for 5 or 10 min (corresponding to three successive contact times of 1.5 or 3.5 min with 100 ml elution buffer in the case of the use of the glass wool filter or to a contact time of 5 or 10 min with the entire 500-ml portion of the solution in the case of the use of the NanoCeram filter). The total elution volume was evacuated with air and collected. Filter eluents (corresponding to approximately 400 ml and 500 ml for glass wool and NanoCeram filters, respectively) were neutralized to pH 7 to 7.5 with 1 N HCl. A sample of 5 ml was collected. Viruses present in the filter eluent were concentrated using modifications of the viral concentration method based on PEG precipitation, as previously described (6, 7). A 50% (wt/vol) PEG 6000 (Promega, Madison, WI)-1.5 M NaCl solution was added to obtain a final concentration of 10% (vol/vol), homogenized by shaking, and then incubated at 4°C for 2 h or overnight. The mixture was centrifuged at 10,000 × g for 1 or 2 h at 4°C. The supernatant was discarded, and the pellet was resuspended in 3 ml of phosphate-buffered saline (PBS) solution. Each filter eluent and virus concentrate were analyzed separately by plaque assay to determine the infectious virus recovery.
Table 1.
Recovery of infectious H1N1 from large volumes of surface water using different filtration, elution, and PEG concentration conditions
| Water sample | Level from factorial design | Parameter used for: |
TCID50 recovered (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Filtration |
Elution |
PEG concn |
|||||||||
| Filtera | Flow rate (liters/h) | Buffer composition |
Contact time (min) | Flow rate (liters/h) | Precipitation time (h) | Centrifugation time (h) | Eluate | PEG concentrate | |||
| Beef extract (%) | Glycine (M) | ||||||||||
| A | 1 | Glass W | 10 | 1.5 | /b | 10 | 30 | O/Nc | 2 | 2.3 | 1.2 |
| 2 | NanoC | 10 | 3 | 0.05 | 10 | 30 | O/N | 1 | <0.1 | <0.1 | |
| 3 | NanoC | 10 | 1.5 | / | 10 | 10 | 2 | 1 | <0.1 | <0.1 | |
| 4 | Glass W | 10 | 3 | 0.05 | 10 | 10 | 2 | 2 | 1.0 | 0.3 | |
| 5 | NanoC | 30 | 1.5 | / | 5 | 30 | O/N | 1 | 0.9 | <0.1 | |
| 6 | Glass W | 30 | 3 | 0.05 | 5 | 30 | O/N | 2 | 0.6 | 0.3 | |
| 7 | Glass W | 30 | 1.5 | / | 5 | 10 | 2 | 2 | 1.0 | 0.7 | |
| 8 | NanoC | 30 | 3 | 0.05 | 5 | 10 | 2 | 1 | 0.2 | <0.1 | |
| B | 9 | Glass W | 10 | 1.5 | 0.05 | 5 | 10 | O/N | 1 | <0.1 | 0.1 |
| 10 | NanoC | 10 | 3 | / | 5 | 10 | O/N | 2 | 0.8 | <0.1 | |
| 11 | NanoC | 10 | 1.5 | 0.05 | 5 | 30 | 2 | 2 | <0.1 | <0.1 | |
| 12 | Glass W | 10 | 3 | / | 5 | 30 | 2 | 1 | 1.8 | <0.1 | |
| 13 | NanoC | 30 | 1.5 | 0.05 | 10 | 10 | O/N | 2 | 0.1 | <0.1 | |
| 14 | Glass W | 30 | 3 | / | 10 | 10 | O/N | 1 | 5.4 | 0.5 | |
| 15 | Glass W | 30 | 1.5 | 0.05 | 10 | 30 | 2 | 1 | 6.4 | 0.8 | |
| 16 | NanoC | 30 | 3 | / | 10 | 30 | 2 | 2 | 0.1 | <0.1 | |
| C | 1 | Glass W | 10 | 1.5 | / | 10 | 30 | O/N | 2 | <0.1 | <0.1 |
| 2 | NanoC | 10 | 3 | 0.05 | 10 | 30 | O/N | 1 | 0.1 | <0.1 | |
| 3 | NanoC | 10 | 1.5 | / | 10 | 10 | 2 | 1 | <0.1 | <0.1 | |
| 4 | Glass W | 10 | 3 | 0.05 | 10 | 10 | 2 | 2 | 4.1 | 1.0 | |
| 5 | NanoC | 30 | 1.5 | / | 5 | 30 | O/N | 1 | 0.2 | <0.1 | |
| 6 | Glass W | 30 | 3 | 0.05 | 5 | 30 | O/N | 2 | 1.3 | 0.4 | |
| 7 | Glass W | 30 | 1.5 | / | 5 | 10 | 2 | 2 | 3.9 | 1.7 | |
| 8 | NanoC | 30 | 3 | 0.05 | 5 | 10 | 2 | 1 | <0.1 | 0.1 | |
| D | 9 | Glass W | 10 | 1.5 | 0.05 | 5 | 10 | O/N | 1 | 1.2 | <0.1 |
| 10 | NanoC | 10 | 3 | / | 5 | 10 | O/N | 2 | <0.1 | <0.1 | |
| 11 | NanoC | 10 | 1.5 | 0.05 | 5 | 30 | 2 | 2 | 0.1 | <0.1 | |
| 12 | Glass W | 10 | 3 | / | 5 | 30 | 2 | 1 | 2.4 | 0.4 | |
| 13 | NanoC | 30 | 1.5 | 0.05 | 10 | 10 | O/N | 2 | <0.1 | <0.1 | |
| 14 | Glass W | 30 | 3 | / | 10 | 10 | O/N | 1 | 10.5 | 0.3 | |
| 15 | Glass W | 30 | 1.5 | 0.05 | 10 | 30 | 2 | 1 | 14.9 | 3.9 | |
| 16 | NanoC | 30 | 3 | / | 10 | 30 | 2 | 2 | <0.1 | <0.1 | |
Glass W, glass wool filter; NanoC, NanoCeram filter.
/, without glycine.
O/N, overnight.
Evaluation of H5N1 virus recovery and detection threshold of the method.
The optimal method, corresponding to the factor combination for which the highest level of recovery was previously predicted, was evaluated using two H5N1 strains seeded in natural waters from different sources. H5N1 virus (A/HK/156/97) was seeded into a 10- or 50-liter lake water sample to be tested at high and low final loads of approximately 1 × 107 TCID50 and 1 × 103 TCID50 per sample, respectively. Three or four trials were conducted for each condition used. The limit of detection of the viral concentration method was assessed. Quantification cycle (Cq) values obtained by reverse transcriptase PCR (RT-PCR) were plotted against the inoculated viral concentration in water. A calibration curve was then built using linear regression. The total number of PCR cycles was 50; it was then checked that a positive result was obtained when a full amplification curve could be observed, which corresponded to detection occurring in less than 42 PCR cycles (Cq ≤ 42) (data not shown). The detection threshold was thus the viral concentration for which the probability of obtaining a positive result was 0.95. In parallel, eight experiments were performed by seeding 1 × 105 TCID50 H5N1 virus A/DK/CAM/67F8/2008 in 10-liter volumes of lake waters sampled in Cambodia. Moreover, to compare recovery values obtained for waters with different physicochemical characteristics, the A/DK/CAM/67F8/2008 virus was seeded into 10 liters of rainwater. Two or three experiments were conducted, with final virus loads ranging from 1 × 103 to 1 × 106 TCID50. Virus stocks were used undiluted or diluted in PBS on the day of the experiment and then mixed into the entire volume of water to be filtered. The viral solution was immediately tested with a virus infectivity assay and RT-PCR quantification to determine the seeding viral concentration. A sample of 5 ml was collected just after the elution step to determine the efficiency of virus elution. Elution solutions and PEG concentrates were stored at −80°C until further use. Samples were analyzed by virus titration and by real-time RT-PCR to determine the percentages of virus recovery.
Detection of influenza viruses from environmental samples.
Pond and lake waters suspected to be contaminated with AIVs were tested. Ten-liter samples were filtered for validation experiments as described above. Viruses were quantified by RT-PCR.
Viral RNA isolation and quantification.
The following protocol was used for the method development, validation of experiments with the A/HK/156/97 H5N1 strain, and detection of virus in environmental samples from France. RNA was extracted from 140 μl of the concentrate using QIAamp viral RNA minikit (Qiagen, Valencia, CA), according to the manufacturer's instructions. The extracted RNA was recovered in 60 μl elution buffer. Quantitative RT-PCRs were performed on a LightCycler 2.0 (Roche Diagnostics, Mannheim, Germany). Two RT-PCR systems designed for the detection of matrix (M) and hemagglutinin (HA) genes were used separately to detect all subtypes of influenza A viruses and avian influenza viruses H5, respectively. An RT-PCR for each amplification system was performed in a 15-μl reaction mixture, containing 6 mM MgCl2 with 5 μl of extracted RNA, using the Invitrogen SuperScript III Platinum one-step quantitative RT-PCR system and the standard cycling program and TaqMan probes reaction mix protocols recommended by the manufacturer. The sources of the primers and TaqMan probes and their final concentrations used in the present study were as follows: generic M gene (32) and avian H5 gene (30), 500 nM primers and 200 nM probes. A second protocol was used in Cambodia for validation of the experiments with the A/DK/CAM/67F8/2008 H5N1 strain and detection in environmental waters. Viral RNA was extracted from 200 μl of viral concentrate and eluted in 60 μl using the MagNA Pure nucleic acid isolation kit (Roche Diagnostics) on a MagNA Pure Light Cycler instrument (Roche Diagnostics), according to the manufacturer's instructions. One-step RT-PCR using the TaqMan probe was performed on the iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA) to measure the quantity of the HA gene of the H5N1 avian influenza virus using quantified synthetic RNA. Sequences of the nucleotides and probes used, reaction mixtures using the QuantiTect probe RT-PCR kit (Qiagen), and RT-PCR temperature cycling conditions were previously described (41).
The absence of RT-PCR inhibitors was controlled in water samples by addition of an M or H5 gene RNA control (virus-specific internal standard) to each analyzed sample just before RNA amplification.
A standard curve was obtained for each real-time RT-PCR by analyzing 10-fold serial dilutions of viral RNA extracted from the seeding viral concentration for which the infectious titer was determined by infectivity assay. The quantities of viruses in the dilutions were expressed in the TCID50 by reference to the logarithmic value of the viral concentration used for viral RNA extraction. The obtained standard curves were used to estimate the quantities of infectious viruses, expressed as equivalent TCID50 values, detected in samples. The slope (s) of the standard curve was used to calculate the amplification efficiency (E) of the RT-PCR in conformity with the formula, E = 10 (−1/s) − 1 (5). Amplification efficiencies from 85% to 115% were considered acceptable.
Virus recovery efficiency.
The percent virus recovery was calculated as follows:
Statistical analysis.
Statistical computations and tests were done using S-PLUS statistical software (MathSoft, Seattle, WA). Analysis of variance was performed to evaluate individual effects of the studied factors on virus recovery. Effects were considered significant when P values were <0.05. Linear regression analysis was performed to evaluate the limit of detection.
RESULTS
Optimal concentration method.
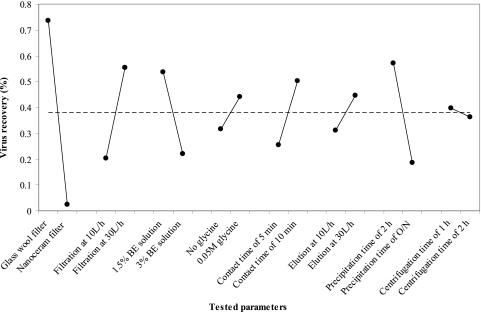
Recovery rates after adsorption-elution alone and after the subsequent PEG concentration were separately determined for H1N1 virus by titration of the infectious particles on MDCK cells (Table 1). The experimental design enabled the reduction of the experiments to 16 combinations of parameters. Parameter combination 15 (described in Table 1) was identified to be optimal to preserve virus infectivity. Average recoveries of 10.6% and 2.3% of infectious particles were obtained from 10 liters of experimentally contaminated water after elution and after PEG concentration, respectively. The main effects of tested parameters on viral recovery were analyzed and represented in Fig. 1. There was a highly significant difference (P < 0.05) in virus recovery by use of glass wool for filtration rather than NanoCeram filters (data not shown). Accordingly, the optimal protocol corresponded to a continuous filtration at 30 liters/h through glass wool, followed by a contact time of 10 min with 300 ml of 1.5% beef extract buffer (pH 9.5) containing 0.05 M glycine and a viral elution step at 30 liters/h, and then a PEG concentration with a precipitation step for 2 h and a centrifugation time of 1 h at 4°C.
Fig. 1.
Individual effects of parameters on infectious H1N1 recovery. The mean recovery by factor level (dot) and global mean recovery (dashed line) are represented. BE, beef extract; O/N, overnight.
Comparison of H5N1 virus recovery from lake water and rainwater and assessment of the sensitivity of the method.
The optimal concentration method (described above) was evaluated for H5N1 virus recovery from 10 to 50 liters of seeded lake water and rainwater, and virus concentration efficiencies were compared (Table 2). The amounts of infectious particles added to samples were determined by an infectivity assay. Infectious virus was also recovered from concentrated samples when they were seeded with 1 × 107 TCID50 but not when experiments were performed using seeds at 1 × 103 TCID50. Consequently, to increase sensitivity for the lower seed levels, virus titers in eluate and concentrate samples were quantified by real-time RT-PCR using a standard curve. For each analyzed sample, comparable Cq values obtained for the RNA control diluted in elution and PEG samples or in water allowed us to check for the absence of RT-PCR inhibitors. In cases of the presence of RT-PCR inhibitors, the results of RT-PCR amplification for 10-fold diluted samples were considered after verification that no inhibition remained after dilution. The overall recovery efficiencies of influenza virus measured in this way ranged from less than 0.01% to 7.89% for 10 to 50 liters of lake waters and from 3.63% to 13.79% for 10 liters of rainwaters. Especially considering the experiments performed with lake waters in France, average recoveries of about 1.9% and 1.0% were obtained by M and H5 gene-specific detection, respectively. Slightly higher values of virus recovery were obtained from 10-liter samples of lake waters than from 50-liter samples. Indeed, no virus was detectable from the 50 liter volume in two out of three trials when 1 × 103 TCID50 was seeded and the H5 gene targeted. Moreover, in most experiments in which a low virus concentration was seeded, no virus was recovered in elution samples before the concentration step, whether the H5 or M gene was targeted. The data also suggest that interfering substances in the larger samples affected recovery at the PEG step. In the 10-liter samples for which data are available for both the elution and PEG-concentrated samples, there is a mean 7.7-fold reduction in the percent recovered between the two. For the 50-liter samples, this difference increases to 20.5-fold. Comparatively, an average overall recovery of 0.37% was obtained from 10 liters of lake waters sampled in Cambodia and experimentally seeded with 1 × 105 TCID50. Higher values of virus recovery were obtained from rainwaters than from lake waters. The average recovery was about 7.63%.
Table 2.
Validation of H5N1 influenza A virus recovery from large volumes of lake water and rainwater
| Type of water/country | Seeded virus strain | Date of samplinge | Expt no. | Vol of sample (liters) | Gene-specific RT-PCR | TCID50 of virus added to samplec | TCID50 recovered (%)d |
|
|---|---|---|---|---|---|---|---|---|
| Eluate | PEG concentrate | |||||||
| Lake/France | A/HK/156/97 | Oct 2008 (1) | 1 | 10 | M | 1.71 × 107 | 34.91 | 7.89 |
| H5 | 6.12 × 106 | 12.45 | 3.51 | |||||
| 2 | 10 | M | 1.71 × 107 | 17.60 | 3.26 | |||
| H5 | 6.12 × 106 | 6.27 | 1.21 | |||||
| Oct 2008 (2) | 3 | 10 | M | 9.46 × 107 | 4.37 | 0.36 | ||
| H5 | 3.38 × 107 | 4.85 | 0.22 | |||||
| 4 | 10 | M | 9.46 × 107 | 5.12 | 1.08 | |||
| H5 | 3.38 × 107 | 5.27 | 1.22 | |||||
| Nov 2008 | 5 | 10 | M | 2.66 × 103 | NDa | 1.88 | ||
| H5 | 1.70 × 103 | ND | 0.47 | |||||
| 6 | 10 | M | 2.66 × 103 | ND | 1.59 | |||
| H5 | 1.70 × 103 | ND | 1.36 | |||||
| Dec 2008 | 7 | 10 | M | 4.94 × 103 | ND | 2.25 | ||
| H5 | 8.44 × 103 | ND | 2.59 | |||||
| 8 | 10 | M | 4.94 × 103 | ND | 0.45 | |||
| H5 | 8.44 × 103 | ND | ND | |||||
| Jan 2009 | 9 | 50 | M | 2.93 × 107 | 0.20 | 0.004 | ||
| H5 | 2.52 × 106 | ND | 0.34 | |||||
| 12 | 50 | M | 4.87 × 103 | 35.52 | 2.07 | |||
| H5 | 2.21 × 103 | ND | 0.18 | |||||
| Feb 2009 | 10 | 50 | M | 5.09 × 106 | 6.95 | 0.25 | ||
| H5 | 7.76 × 106 | 1.80 | 0.35 | |||||
| 13 | 50 | M | 4.30 × 103 | 34.42 | 1.90 | |||
| H5 | 2.27 × 103 | ND | ND | |||||
| March 2009 | 11 | 50 | M | 5.88 × 106 | 8.61 | 0.80 | ||
| H5 | 4.70 × 107 | 0.29 | 0.02 | |||||
| 14 | 50 | M | 3.05 × 103 | ND | 2.50 | |||
| H5 | NDa | ND | ND | |||||
| Lake/Cambodia (Kampong Cham) | A/DK/CAM/67F8/2008 | April 2009 | 15 | 10 | H5 | 1.74 × 105 | NTb | 0.17 |
| May 2009 | 16 | 10 | H5 | 1.74 × 105 | NT | 0.49 | ||
| June 2009 | 17 | 10 | H5 | 1.74 × 105 | NT | 0.09 | ||
| July 2009 | 18 | 10 | H5 | 1.74 × 105 | NT | 0.08 | ||
| Lake/Cambodia (Prey Veng) | A/DK/CAM/67F8/2008 | April 2009 | 19 | 10 | H5 | 1.74 × 105 | NT | 1.66 |
| May 2009 | 20 | 10 | H5 | 1.74 × 105 | NT | 0.04 | ||
| June 2009 | 21 | 10 | H5 | 1.74 × 105 | NT | 0.06 | ||
| July 2009 | 22 | 10 | H5 | 1.74 × 105 | NT | ND | ||
| Rain/Cambodia | A/DK/CAM/67F8/2008 | March 2009 | 23 | 10 | H5 | 1.74 × 106 | NT | 7.57 |
| 24 | 10 | H5 | 1.74 × 106 | NT | 8.68 | |||
| 25 | 10 | H5 | 1.74 × 105 | NT | 9.31 | |||
| 25 | 10 | H5 | 1.74 × 105 | NT | 8.79 | |||
| 26 | 10 | H5 | 1.74 × 104 | NT | 3.33 | |||
| 27 | 10 | H5 | 1.74 × 104 | NT | 13.79 | |||
| 28 | 10 | H5 | 1.74 × 104 | NT | 5.98 | |||
| 29 | 10 | H5 | 1.74 × 103 | NT | ND | |||
| 30 | 10 | H5 | 1.74 × 103 | NT | ND | |||
| 31 | 10 | H5 | 1.74 × 103 | NT | 3.63 | |||
ND, not detected.
NT, not tested.
Results obtained by using an infectivity assay.
Results based upon viral quantifications by real-time RT-PCR.
Oct, October; Nov, November; Dec, December; Jan, January; Feb, February.
The detection threshold, corresponding to the amount of infectious particles below which no detection by RT-PCR can be obtained, was evaluated in lake waters by linear regression. Figure 2 shows the M and H5 gene-specific RT-PCR detection results obtained during the experimental scheme to assess the concentration method. A theoretical detection threshold of 2.25 × 102 TCID50 in the filtered volume was obtained for the detection of H5N1 virus in lake water by M gene-specific RT-PCR. In addition, a detection threshold of 1.74 × 104 TCID50 was obtained by RT-PCR detection of the H5 gene.
Fig. 2.
Assessment of theoretical detection thresholds of the concentration method by M (A) and H5 (B) gene-specific RT-PCR detections from seeded lake water. The linear regression model (solid line) was built with experimental Cq values obtained by RT-PCR and plotted against inoculated viral loads in water (circles). For a given viral load, the Cq value has a 0.95 probability to fall below the diagonal dashed line. A positive detection occurs for Cq values lower than or equal to 42 (horizontal dotted line). The detection threshold, corresponding to the viral load for which a positive detection is obtained with a probability of 0.95, is thus found at the intersection of these two lines.
Detection of influenza A viruses in environmental waters.
No influenza virus subtype H5 was detected in waters collected from places involved in previous outbreaks in Cambodia. However, the concentration method was efficient for the detection of influenza A virus in Dombes ponds (France) during fall migration in 2009. AIVs, ranging from 3 × 102 to 9 × 104 TCID50 in 10 liters, were detected by M gene RT-PCR in 4 out of 9 water samples, corresponding to samples collected from 1 pond in October and from 3 different ponds in November 2009.
DISCUSSION
During influenza A outbreaks, avian influenza viruses can be isolated from unconcentrated lake water (15, 36, 44). However, since water contamination by AIV possibly occurs at low levels or decreases over time, sensitive methods were needed to detect the presence of these pathogens in natural waters. In contrast to previous studies (16, 18, 22, 27, 29), the present study was conducted using large-volume samples with water matrix representatives of water bodies where waterfowl gather. This type of water sample was used to take into account the impact of water composition (in terms of suspended solids and soluble organic compounds) on virus adsorption to the filter and on flow rate due to filter clogging.
It was important for the concentration method to be appropriate for the detection of infectious viral particles. Therefore, the optimization of the concentration method was performed by evaluating the percent virus recovery from eluent and concentrate samples by using infectious titers obtained by endpoint titration. Moreover, the described procedure was then confirmed as adapted to concentrate infectious H5N1 cultures from large volumes of surface waters. However, the major drawback to the cell culture assay is that it is time-consuming and requires days of incubation, whereas molecular techniques are rapid, highly sensitive, and specific. Therefore, RT-PCR detection methods were preferentially used as the quantification method of determining the equivalent infectious viral load in environmental waters.
The adsorption of viral particles to a membrane is due to electrostatic interactions but depend on both the environmental characteristics and surface properties of the virus (20). Studies showed that charges of influenza viruses above their isoelectric point, which was approximately 5 (11, 23), were negative. Therefore, the natural pH of the treated water, ranging from 7.95 to 8.2 (data not shown), favored virus adsorption on electropositive filters but also persistence of virus in environmental samples (33). When glass wool and NanoCeram filters were compared in this study, glass wool gave significantly higher recoveries of infectious H1N1 virus (Fig. 1; Table 1). Higher recoveries may have been possible with NanoCeram filters if the protocol published after our study was complete had been used (17), but this protocol, which uses two elution steps, has not been tested with influenza. In selection of a two-step concentration procedure to increase sensitivity, it was important to consider the virus concentration efficiency, preservation of infectivity, and compatibility with the filtration and RT-PCR detection methods. Erythrocyte adsorption procedures were often reported as the reference method used for AIVs, when followed by the isolation of influenza viruses in embryonated chicken eggs or tissue culture (16, 27, 29). PCR amplification of the M gene of the influenza virus could be used after this concentration method, but sensitivity was lower than the detection of viral concentrations by embryonated chicken egg isolation (18), probably due to PCR inhibitors produced by red blood cell lysis. In this study, the PEG conditions used were chosen and optimized according to previously described methods for the concentration of infectious enteric viruses from water and vegetables (6, 7). Moreover, the PEG method enabled us to reduce the final volume to 3 ml, which can be assayed by RT-PCR. The results showed the usefulness and concentration efficiency of this method, especially since it was possible to detect virus after PEG concentration, while no virus had been recovered in elution samples from 1 × 103 TCID50-seeded waters. However, some weaknesses of the system became evident when virus concentration from larger volumes was attempted or when large amounts of organic matter were present in the water. Humic acid and other organic compounds were also concentrated from water onto filters. These compounds were eluted from the filters along with the virus and formed a precipitate when eluting solution was concentrated by the PEG method. They probably affected the viral recovery at the PEG step in the larger samples, reducing the recovery percentage in the PEG-concentrated samples and probably interfering with molecular detection of H5 gene when seeding low concentrations and processing 50 liters of lake water. The efficiency of the concentration method was dependent on water characteristics, with more effectiveness for virus concentration using cleaner waters. H5N1 virus recoveries were 5- to 50-fold higher in rainwater than in surface water samples (Table 2). Water characteristics and high levels of soluble organic compounds could significantly affect enteric virus adsorption to electropositive filters (19, 31). Moreover, the method described here is sensitive enough to detect an H5N1 presence in quantities as low as 2.25 × 102 TCID50 in 50-liter water volumes by M gene-specific RT-PCR, while erythrocyte methodology enabled the detection of 3.0 × 102 EID50 of influenza H1N1 virus in 1 liter of river water (18).
The concentration procedure outlined in this study will facilitate rapid detection of influenza viruses and, moreover, can be used as a quantification method of determining the infectious viral load in environmental waters. Indeed, this system was used successfully in field studies for the detection of naturally occurring influenza A viruses in lake water.
ACKNOWLEDGMENTS
This work was supported by Sixth Framework Program grant FP6-2005-SSP-5-B-INFLUENZA (Resistance of Influenza Viruses in Environmental Reservoirs and Systems RIVERS) from the European Union.
We thank Jean-Claude Manuguerra (Institut Pasteur, Paris, France) and Philippe Sabatier (Ecole Veterinaire, Lyon, France) for providing the virus H5N1 A/HK/156/97 and the natural water samples collected in Dombes ponds (Ain, France), respectively.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1. AFNOR 2005. NF EN 14476—antiseptiques et désinfectants chimiques. Essai virucide quantitatif de suspension pour les antiseptiques et désinfectants chimiques utilisés en médecine humaine. AFNOR, Paris, France [Google Scholar]
- 2. AFNOR 1996. XP T 90-451—essais des eaux. Recherche des Entérovirus. Méthode par concentration sur laine de verre et détection par culture cellulaire. AFNOR, Paris, France [Google Scholar]
- 3. Brankston G., Gitterman L., Hirji Z., Lemieux C., Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect. Dis. 7:257–265 [DOI] [PubMed] [Google Scholar]
- 4. Brown J., Swayne D., Cooper R., Burns R., Stallknecht D. 2007. Persistence of H5 and H7 avian influenza viruses in water. Avian Dis. 51:285–289 [DOI] [PubMed] [Google Scholar]
- 5. Bustin S., et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 6. Dubois E., et al. 2002. Modified concentration method for the detection of enteric viruses on fruits and vegetables by reverse transcriptase-polymerase chain reaction or cell culture. J. Food Prot. 65:1962–1969 [DOI] [PubMed] [Google Scholar]
- 7. Dubois E., et al. 2007. Detection and quantification by real-time RT-PCR of hepatitis A virus from inoculated tap-waters, salad vegetables, and soft fruits: characterization of the method performances. Int. J. Food Microbiol. 117:141–149 [DOI] [PubMed] [Google Scholar]
- 8. Gantzer C., Maul A., Audic J. M., Schwartzbrod L. 1998. Detection of infectious enteroviruses, enterovirus genomes, somatic coliphages, and bacteroides fragilis phages in treated wastewater. Appl. Environ. Microbiol. 64:4307–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gantzer C., Senouci S., Maul A., Levi Y., Schwartzbrod L. 1997. Enterovirus genomes in wastewater: concentration on glass wool and glass powder and detection by RT-PCR. J. Virol. Methods 65:265–271 [DOI] [PubMed] [Google Scholar]
- 10. Gassilloud B., Duval M., Schwartzbrod L., Gantzer C. 2003. Recovery of feline calicivirus infectious particles and genome from water: comparison of two concentration techniques. Water Sci. Technol. 47:97–101 [PubMed] [Google Scholar]
- 11. Gerba C. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133–168 [DOI] [PubMed] [Google Scholar]
- 12. Grimes S. E. 2002. A basic laboratory manual for the small-scale production and testing of I-2 Newcastle disease vaccine, p. 136 In FAO Regional Office for Asia and the Pacific publication, Bangkok, Thailand [Google Scholar]
- 13. Halvorson D., et al. 1983. Epizootiology of avian influenza—simultaneous monitoring of sentinel ducks and turkeys in Minnesota. Avian Dis. 27:77–85 [PubMed] [Google Scholar]
- 14. Hamilton R. R. C., Thurston R. V. 1977. Trimmed Spearman-Karber method for estimating median lethal concentrations in toxicity bioassays. Environ. Sci. Technol. 11:714–719 [Google Scholar]
- 15. Hinshaw V., Webster R., Turner B. 1979. Water-bone transmission of influenza A viruses? Intervirology 11:66–68 [DOI] [PubMed] [Google Scholar]
- 16. Ito T., et al. 1995. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Arch. Virol. 140:1163–1172 [DOI] [PubMed] [Google Scholar]
- 17. Karim M., Rhodes E., Brinkman N., Wymer L., Fout G. 2009. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 75:2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khalenkov A., Laver W., Webster R. 2008. Detection and isolation of H5N1 influenza virus from large volumes of natural water. J. Virol. Methods 149:180–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambertini E., et al. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langlet J., Gaboriaud F., Gantzer C., Duval J. 2008. Impact of chemical and structural anisotropy on the electrophoretic mobility of spherical soft multilayer particles: the case of bacteriophage MS2. Biophys. J. 94:3293–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lénès D., et al. 2010. Assessment of the removal and inactivation of influenza viruses H5N1 and H1N1 by drinking water treatment. Water Res. 44:2473–2486 [DOI] [PubMed] [Google Scholar]
- 22. Markwell D., Shortridge K. 1982. Possible waterborne transmission and maintenance of influenza viruses in domestic ducks. Appl. Environ. Microbiol. 43:110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller G. L., Lauffer M. A., Stanley W. M. 1944. Electrophoretic studies on PR8 influenza virus. J. Exp. Med. 80:549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munster V., et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. OIE 2009, posting date. Summary of outbreaks of highly pathogenic avian influenza in Cambodia. OIE, Paris, France: http://www.oie.int/wahis/public.php?page=disease_outbreak_summary&summary_country=KHM&reportid=3768 [Google Scholar]
- 26. Reed L. J., Muench L. H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 27. Roepke D., Halvorson D., Goyal S., Kelleher C. 1989. An adsorption-elution technique for the recovery of influenza virus from water. Avian Dis. 33:649–653 [PubMed] [Google Scholar]
- 28. Senouci S., Maul A., Schwartzbrod L. 1996. Comparison study on three protocols used to concentrate poliovirus type 1 from drinking water. Zentralbl. Hyg. Umweltmed. 198:307–317 [PubMed] [Google Scholar]
- 29. Sivanandan V., Halvorson D., Laudert E., Senne D., Kumar M. 1991. Isolation of H13N2 influenza A virus from turkeys and surface water. Avian Dis. 35:974–977 [PubMed] [Google Scholar]
- 30. Slomka M. J., et al. 2007. Validated H5 Eurasian real-time reverse transcriptase-polymerase chain reaction and its application in H5N1 outbreaks in 2005–2006. Avian Dis. 51:373–377 [DOI] [PubMed] [Google Scholar]
- 31. Sobsey M., Glass J. 1984. Influence of water quality on enteric virus concentration by microporous filter methods. Appl. Environ. Microbiol. 47:956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spackman E., et al. 2002. Development of a real-time reserve transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stallknecht D., Kearney M., Shane S., Zwank P. 1990. Effects of pH, temperature, and salinity on persistence of avian influenza viruses in water. Avian Dis. 34:412–418 [PubMed] [Google Scholar]
- 34. Stallknecht D., Shane S., Kearney M., Zwank P. 1990. Persistence of avian influenza viruses in water. Avian Dis. 34:406–411 [PubMed] [Google Scholar]
- 35. Vilaginès P., Sarette B., Husson G., Vilagenes R. 1993. Glass wool for virus concentration at ambient water pH level. Water Sci. Technol. 27:299–306 [Google Scholar]
- 36. Vong S., Ly S., Mardy S., Holl D., Buchy P. 2008. Environmental contamination during influenza A virus (H5N1) outbreaks, Cambodia, 2006. Emerg. Infect. Dis. 14:1303–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Webby R., Webster R., Richt J. 2007. Influenza viruses in animal wildlife populations. Curr. Top. Microbiol. Immunol. 315:67–83 [DOI] [PubMed] [Google Scholar]
- 38. Webster R., Bean W., Gorman O., Chambers T., Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webster R., Yakhno M., Hinshaw V., Bean W., Murti K. 1978. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology 84:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. WHO 2010, posting date Confirmed human cases of avian influenza A(H5N1). WHO, Geneva, Switzerland: http://www.who.int/csr/disease/avian_influenza/country/en/ [Google Scholar]
- 41. WHO 2007, posting date Recommendations and laboratory procedures for detection of avian influenza (H5N1) in specimens from suspected human cases. WHO, Geneva, Switzerland: http://www.who.int/csr/disease/avian_influenza/guidelines/RecAIlabtestsAug07.pdf [Google Scholar]
- 42. WHO 2005, posting date Recommended laboratory tests to identify avian influenza A virus in specimens from humans. WHO, Geneva, Switzerland: http://www.who.int/csr/disease/avian_influenza/guidelines/avian_labtests2.pdf [Google Scholar]
- 43. WHO 2002. WHO manual on animal influenza diagnosis and surveillance. WHO/CDS/CSR/NCS/2002.5 Rev. 1 WHO, Geneva, Switzerland [Google Scholar]
- 44. Zhang G., et al. 2006. Evidence of influenza a virus RNA in Siberian lake ice. J. Virol. 81:2538. [DOI] [PMC free article] [PubMed] [Google Scholar]