Abstract
Nesterenkonia strain AN1 was isolated from a screening program for nitrile- and amide-hydrolyzing microorganisms in Antarctic desert soil samples. Strain AN1 showed significant 16S rRNA sequence identity to known members of the genus. Like known Nesterenkonia species, strain AN1 was obligately alkaliphilic (optimum environmental pH, 9 to 10) and halotolerant (optimum environmental Na+ content, 0 to 15% [wt/vol]) but was also shown to be an obligate psychrophile with optimum growth at approximately 21°C. The partially sequenced genome of AN1 revealed an open reading frame (ORF) encoding a putative protein member of the nitrilase superfamily, referred to as NitN (264 amino acids). The protein crystallized readily as a dimer and the atomic structure of all but 10 amino acids of the protein was determined, confirming that the enzyme had an active site and a fold characteristic of the nitrilase superfamily. The protein was screened for activity against a variety of nitrile, carbamoyl, and amide substrates and was found to have only amidase activity. It had highest affinity for propionamide but demonstrated a low catalytic rate. NitN had maximal activity at 30°C and between pH 6.5 and 7.5, conditions which are outside the optimum growth range for the organism.
INTRODUCTION
Antarctic desert soils, which are restricted to localized regions of Antarctica's coastal margins, have been shown to harbor substantial populations of prokaryotes and lower eukaryotes (12). These communities exhibit much higher species diversity than suspected previously (8, 32). A high proportion of the phylotypic signals represent uncultured species, and these extremophilic habitats are thus validly considered to be a source of novel cold-active microorganisms and gene products (43).
Amidases (EC 3.5.1.4) hydrolyze amides with the production of the corresponding carboxylic acids and ammonia and are the most widely used amide-hydrolyzing enzymes in industry. The predominant industrial application of amidases is in the synthesis of natural and nonnatural amino acids from amino acid amides. Technology developed by both the Nitto Chemical Industry Company (Japan) and DSM (Netherlands) exploits the enantioselective amidase activities of various Pseudomonas strains (21, 35).
Cold-active amidases have yet to find commercial application, but there remains substantial interest in the applications of cold-active enzymes. The focus of such interest is in industrial sectors where low-temperature processing is beneficial, most commonly in the food and beverage industry and in some chemical biotransformations (9).
These enzymes belong either to the amidase signature (AS; CATH no. 3.90.1300.10) or nitrilase (CATH no. 3.60.110.10) superfamily (10, 16, 29). Although amidases vary in substrate specificities and biological functions, their catalysis is typically mediated by an S–cis-S–K catalytic triad (28, 36) for the AS superfamily and Glu, Lys, Glu, and Cys catalytic residues for the nitrilase superfamily (7, 27).
Aliphatic amidases belong with the branch 2 enzymes of the nitrilase superfamily according to the classification of Pace and Brenner (29) and typically hydrolyze short-chain aliphatic amides. Members of the superfamily have subunits of 30 to 44 kDa and exist as active dimers, tetramers, hexamers, octamers, and longer oligomeric spirals (19, 39).
Enzymes of the nitrilase superfamily are classified into 13 branches, although the substrate specificity is known for only 9 branches (7). Only branch 1 has nitrilase or cyanide hydratase activity, and eight of the remaining branches have amidase or amide condensation activities (7).
Eleven crystal structures of various members of the nitrilase superfamily, including three amidases, are available and all show high structural homology. Despite low sequence conservation, they all display the characteristic αββα fold (39). Monomers associate along a conserved A interface (39), forming a dimeric αββα-αββα sandwich architecture.
In this study, we characterize a member of the nitrilase superfamily with amidase activity derived from a novel psychrophilic strain of the genus Nesterenkonia, isolated from Antarctica Dry Valley soils. Its structure is shown to differ from the other three amidase structures in ways that may play a role in cold adaptation.
MATERIALS AND METHODS
Sample source and microbial isolation.
Samples of mineral soil were collected from the Miers Valley, McMurdo region, eastern Antarctica (GPS position 78°05.930′S, 163°48.174′E) during the 2005-2006 austral summer. All soil samples were stored below 0°C during transport and at −80°C in the laboratory. Nitrile- and amide-degrading microorganisms were isolated by plating soil suspensions (in distilled water [dH2O]) onto various nitrogen-free media supplemented with 10 mM acetonitrile and incubating the samples at 18°C for 7 days. Colonies were subcultured to clonal purity on media enriched with tryptone.
Bacterial strains, media, and plasmids.
Nesterenkonia strain AN1 was grown at 25°C in modified Castenholz medium enriched with tryptone (5). Escherichia coli GeneHog (Invitrogen) was the host strain for cloning, and strain BL21(DE3)pLysS (Stratagene) was used for expression of the recombinant protein. The expression vector pET28a(+) (Novagen) was used for overexpression of the N-terminally hexahistidine-tagged fusion protein.
16S rRNA identification and genome sequencing.
Genomic DNA was extracted from AN1 cells by a method described previously (43). The strain was identified using universal bacterial 16S rRNA primers E9F (5′-GAG TTT GAT CCT GGC TCA-3′) (15) and U1510R (5′-GGT TAC CTT GTT ACG T-3′) (33) in a standard PCR. The 16S rRNA sequence was compared with GenBank database entries (www.ncbi.nlm.nih.gov/GenBank/index.html) using the basic local alignment search tool (BLAST) algorithm (2). The genome of AN1 was sequenced using Illumina DNA sequencing technology. The partially sequenced genome was used as the database for local BLAST analysis with known protein sequences of the nitrilase superfamily as protein queries.
Effects of temperature, pH, and salinity on the growth of AN1.
To determine the effect of temperature on the growth of strain AN1, a confluent culture was used to inoculate flasks containing 1/5 volumes of media to a final optical density at 600 nm (OD600) of 0.1. Incubation was carried out at 16, 21, 25, and 30°C with shaking at 150 rpm. To determine the effect of pH on growth, the ratios of NaHCO3 and Na2CO3 (500 mM final concentrations) in the media were adjusted to give pH values of 7, 8.5, 9, 9.5, 10, and 10.8. The media were supplemented with NaCl to maintain the total Na+ content at 500 mM. Incubation was carried out at 21°C with shaking. The effect of salinity on growth was determined at 0.75, 1.3, 1.7, 2.2, and 2.6 M NaCl. All growth experiments were performed in triplicate, and the OD600 was monitored spectrophotometrically.
Cloning and expression of NitN.
The primers used for the PCR amplification of the NitN open reading frame (ORF) were NitF (5′-CAT ATG CGA ATC GCG CTG ATG CAG CAC ACC G-3′) and NitR (5′-CTC GAG CTA CCA GGA GCG GCT CTT CGG GCA CTT CG-3′). NdeI (underlined) and XhoI (double-underlined) recognition sequences were used for the cloning of the NitN ORF into the same sites on pET28a(+). E. coli cell cultures harboring the resulting plasmid were incubated at room temperature for 4 h after induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside).
Ni chelation chromatography.
The His · Bind resin and buffer kit (Novagen) was used for the Ni chelation chromatography purification of His-tagged fusion protein. Cell lysates were prepared using BugBuster protein extraction reagent and benzonuclease (Novagen), and protein purification was performed according to the manufacturer's instructions. The protein was eluted in a solution of 500 mM imidazole, 500 mM NaCl, and 20 mM Tris-HCl (pH 7.9). Final concentrations of 10% glycerol and 2 mM dithiothreitol (DTT) were added to the eluate, and the mixture was dialyzed overnight in a ratio of 1:400 against a solution of 50 mM KH2PO4-K2HPO4 (pH 7.6), 150 mM NaCl, 10% glycerol, and 2 mM DTT. The purified protein was stored at 4°C. Fractions from each step of Ni chelation chromatography were analyzed by SDS-PAGE (24).
Protein analytical methods.
The molecular mass of the purified recombinant protein was determined by size exclusion chromatography on a TSK-gel G4000 PWXL column (Tosoh BioScience). The column was equilibrated with 50 mM KH2PO4-K2HPO4 (pH 7.6) containing 10% glycerol and 2 mM DTT. Protein standards used were thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) (Sigma-Aldrich). Fractions with maximum protein absorbance were analyzed using SDS-PAGE.
Amide-hydrolyzing activity assay.
Amidase activity was determined by the release of ammonia using the phenol-hypochlorite ammonia detection method (41). Amide (1 M) stocks were prepared in distilled water, except for l-alaninamide hydrochloride (prepared in 29% [wt/vol] NaOH) and hexanoamide (prepared in 50% [vol/vol] ethanol). A standard reaction mixture (100 μl) contained 50 mM KH2PO4-K2HPO4 (pH 7.6), 150 mM NaCl, 2 to 500 mM substrate, and 2.5 μg of enzyme. The reaction was terminated by the addition of 3.5 volumes of reagent A (0.59 M phenol and 1 mM sodium nitroprusside). Color was developed by the addition of a volume of reagent B (2.0 M sodium hydroxide and 0.11 M sodium hypochlorite) equal to the volume of the reaction mixture. Absorbance at 600 nm was measured after 5 min of incubation at 55°C. Control reactions were carried out without enzyme. Standards were prepared using NH4Cl. One unit of enzyme activity was defined as the amount of enzyme that catalyzed the release of 1 μmol of NH3 per min under standard assay conditions. All assays were performed in triplicate.
Determination of kinetic constants.
Apparent Km values for acetamide and propionamide were determined using the standard reaction assay mixture (100 μl) containing various substrate concentrations (2.5 to 500 mM), 50 mM KH2PO4-K2HPO4, 150 mM NaCl, and 2.5 μg of enzyme. Reactions were carried out at 25°C for 30 min. Direct linear plots (13) were used to calculate Km and Vmax values. kcat values were calculated using a molecular mass of 27.95 kDa, assuming one active site per monomer.
Determination of the thermolability, optimal temperature, and optimal pH.
To determine the optimal temperature, reactions were carried out at 4, 10, 20, 30, 40, 50, and 60°C. Aliquots of the standard reaction mixture (100 μl) containing 100 mM acetamide, 50 mM KH2PO4-K2HPO4 (pH 7.6), and 150 mM NaCl were incubated at the specified temperature for 5 min before the addition of the enzyme. Reactions were carried out for 30 min.
To evaluate thermostability, aliquots of the enzyme were assayed at 25°C for 30 min after incubation at 4, 10, 20, 30, 40, 50, and 60°C for 10, 20, 30, and 60 min. Plots of log relative activity versus time were used to determine the half-life of enzyme activity.
For optimal pH, the following buffers were used: CH3COOHNa (pH 4.5), MES (morpholineethanesulfonic acid; pH 5.5 to 6.5), KH2PO4-K2HPO4 (pH 6.5 to 8), Na2B4O7-H3BO3 (pH 8 to 9), NaOH glycine (pH 9 to 9.5), and Na2CO3-NaHCO3 (pH 9.5 to 11). Standard reaction mixtures (100 μl) contained a 100 mM concentration of the respective buffer (90 mM in the case of borate buffer), 150 mM NaCl, and 100 mM acetamide. Reactions were initiated by the addition of 2.5 μg of enzyme, and mixtures were incubated at 25°C for 30 min.
Crystallization and structure determination.
Purified protein was crystallized without removal of the His tag in a mixture of 0.1 M sodium acetate trihydrate and 2 M ammonium sulfate, pH 4.6, by vapor diffusion using the hanging-drop method at a temperature of 293 K. Diffraction data were collected on beamline BM14 at the European Synchrotron Radiation Facility (ESRF) using a single wavelengh of 0.9778 Å at a temperature of 123 K. The data were integrated using d*Trek (31), and the structure was resolved using Phenix (38) for both molecular replacement and initial map fitting. The template used for molecular replacement was the A chain of the hypothetical protein PH0642 from Pyrococcus horikoshii (Protein Data Bank [PDB] code 1j31) (34). This protein has 24.4% identity to NitN. Manual building was carried out using Coot (14), and refinement was done with both Phenix (38) and REFMAC5 (40). The publication-quality molecular graphics were prepared with UCSF Chimera (30).
Accession numbers.
The atomic coordinates and structure factors of NitN were deposited in the PDB under code 3hkx. The 16S rRNA sequence of strain AN1 (1,496 bp) was deposited in GenBank under accession no. GQ153973. The sequence of the ORF encoding NitN was deposited in GenBank under accession no. ACS35546.
RESULTS AND DISCUSSION
Characterization of strain AN1.
Strain AN1 was isolated on nitrogen-free Castenholz medium supplemented with acetonitrile. The organism formed red-orange colonies on tryptone-enriched Castenholz medium and appeared as coccoid cells when viewed by light microscopy. The 16S rRNA sequence (1,496 bp) of AN1 shared 99% identity with the corresponding sequences of three known strains of the Nesterenkonia genus: N. halotolerans (GenBank accession no. [GB] AY588277) (26), N. sandarakina (GB AY588277) (25), and N. jeotgali (GB AY928901) (42).
Nesterenkonia strain AN1 showed optimal growth below 25°C (Fig. 1A), with the shortest doubling time at approximately 21°C. Above 25°C, the growth rate decreased significantly. This growth temperature profile supports the classification of this isolate as psychrotolerant, making it the first reported psychrotolerant member of the genus Nesterenkonia. This isolate was also shown to be moderately alkaliphilic and halotolerant, with optimal growth at pH 9.6 (Fig. 1B) and rapid growth in media with up to 15% (wt/vol) Na+ (Fig. 1C). The very narrow pH range for growth (pH 9 and 10) suggested that the organism was an obligate alkaliphile.
Fig. 1.
Effects of temperature (A), pH (B), and salt concentration (C) on the growth of strain AN1.
Identification of a member of the nitrilase superfamily from the genomic sequence.
BLAST (2) analysis of the partially sequenced AN1 genome for nitrilase sequence homologs detected an ORF with a product designated NitN (264 amino acids, corresponding to GB ACS35546). NitN showed significant similarity to the conserved protein family pfam00795 (6), identified as nonpeptide carbon-nitrogen hydrolases of the nitrilase superfamily. BLAST analysis of NitN against the NCBI protein database showed that it shared significant sequence conservation (∼50% identity) with two putative protein sequences (GB ABK04917 and ABK02500) of Arthrobacter sp. FB24 (GB CP000454) which were both ambiguously labeled as nitrilases, cyanide hydratases, and/or apolipoprotein N-acyltransferases. No signature amidase genes were found in the region of the genome that was sequenced.
A search of the Protein Data Bank using GenTHREADER (20) identified all members of the nitrilase superfamily with known structures as homologs having between 17 and 32.9% sequence identity. A structural alignment of the NitN protein sequence with members having known structures confirmed the presence of the conserved catalytic residues E41, K111, E119, and C145 (39) (Fig. 2). The greatest sequence identities were to the formamidase AmiF from Helicobacter pylori, 32.9% (PDB code 2dyu) (18); the amidases from Pseudomonas aeruginosa, 30.6% (PDB code 2uxy) (4), and Geobacillus pallidus RAPc8, 29.9% (PDB code 2plq) (22); and the hypothetical protein PH0642 from P. horikoshii, 24.4% (PDB code 1j31) (34). In addition, the predicted fold of NitN superimposed on the αββα monomer fold typical of the nitrilase superfamily enzymes (Fig. 2).
Fig. 2.
Structural alignment of the sequence of NitN (PDB code 3hkx) with the aliphatic amidase from G. pallidus (PDB code 2plq) and the hypothetical protein PH0642 from P. horikoshii (PDB code 1j31). The secondary structural elements identified for NitN are indicated in the bottom line of each row. The conserved catalytic residues (E41, K111, E119, and C145) are highlighted in bold text. Parts of the structure not visible in the electron density map are underlined. The asterisks in the top row indicate conserved (nonglycine) residues, and # indicates the locations of conserved glycine residues. Sequences corresponding to parts of the PDB code 3hkx structure that have relatively high temperature factors, indicating high mobility, are italicized. The sequence numbering shown for PDB code 3hkx differs from that of the deposited coordinates by 20 to account for the 20-amino-acid His tag which was included in the crystallized protein.
Expression, purification, and characterization of NitN.
The N-terminally hexahistidine-tagged NitN fusion protein could be expressed in soluble form in E. coli cells. NitN was purified to near homogeneity (estimated >95%) by Ni chelation chromatography. The estimated molecular mass of the major band on SDS-PAGE was consistent with the molecular mass estimated from the deduced protein sequence (30.1 kDa). The purified NitN protein eluted as a single symmetric peak with an apparent molecular mass of 45.4 kDa. However, SDS-PAGE analysis of active fractions confirmed the presence of 30-kDa bands. Given that the predicted molecular mass for a dimer would be ∼60 kDa, we conclude that NitN does not show classical Stokes elution behavior, possibly due to interaction with the gel.
Enzyme activity.
NitN did not show any detectable activity on nitrile (acetonitrile, acrylonitrile, benzonitrile, and 3-cyanopyridine) or carbamoyl (N-carbamoyl-dl-alanine and N-carbamoyl-dl-α-amino-n-butyric acid) substrates. However, the purified enzyme was active on short-chain aliphatic amide substrates, with a strong preference for propionamide (Table 1).
Table 1.
Substrate specificity for NitN
| Substrate | Relative activitya (%) |
|---|---|
| Propionamide | 100 |
| Fluoroacetamide | 19 |
| Butyramide | 16 |
| Acetamide | 10 |
| Urea | 3 |
| Acrylamide | 1 |
| Hexanamide | 1 |
| Lactamide | 1 |
| Nicotinamide | 1 |
| Isobutyramide | 0 |
| l-Alaninamide | 0 |
| l-Asparagine | 0 |
| l-Glutamine | 0 |
| Diacetamide | 0 |
| Benzamide | 0 |
| Formamide | 0 |
Activities are expressed as percentages relative to the maximum activity of acetamide (100%).
Kinetic parameters for NitN (Table 2) were determined for acetamide and propionamide by using direct linear plots of initial velocity (Vi) versus substrate concentration. While propionamide (C3) is clearly the favored substrate on the basis of rate, substrate affinity, and catalytic-constant data, these values all fall far below equivalent values for most known aliphatic amidases.
Table 2.
Kinetic parameters for NitN on acetamide and propionamide
| Substrate | Kma (mM) | Sp acta (U mg−1) | kcat (s−1) | kcat/Km ratio (mM−1 s−1) |
|---|---|---|---|---|
| Acetamide | 137.7 ± 1.9 | 3.67 ± 0.34 | 1.71 | 0.012 |
| Propionamide | 38.6 ± 1.24 | 8.53 ± 0.27 | 3.97 | 0.103 |
Values are means ± SD.
Aliphatic amidases of the nitrilase superfamily are known to rapidly hydrolyze short-chain aliphatic amides (16). However, comparison of NitN catalytic rates with those of other amidases should take into account that amidases are usually assayed using the acyl transfer assay (17) but that no acyl transfer activity was found for this enzyme. Acyl transfer rates vary widely among different amidases and for different donor substrates. Specific activities for the bacterial signature amidase were approximately 2 μmol · mg−1 · min−1 (23), compared to over 4,000 μmol · mg−1 · min−1 for the G. pallidus amidase (27).
Known aliphatic amidases typically show higher affinity constants for acetamide (with 2 carbon atoms) than for propionamide (with 3 carbon atoms). Km (millimolar) values of known aliphatic amidases for acetamide and propionamide are 3.9 and 20 for H. pylori (37) and 1 and 21 for P. aeruginosa (4). These amidases have little to no activity on linear amides containing fewer than 2 (e.g., formamide) and more than 4 (e.g., butyramide and hexanamide) carbon atoms.
Activity as a function of temperature.
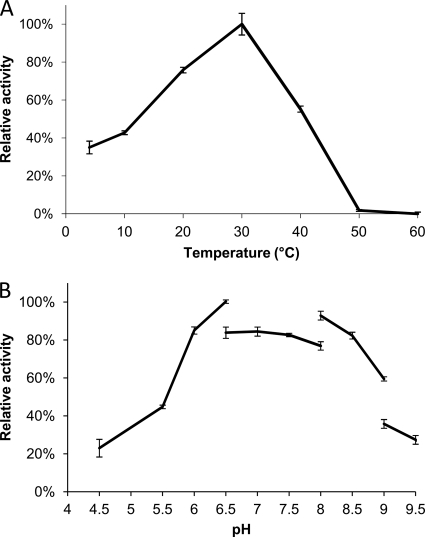
NitN activity was determined for a temperature range of 4 to 60°C. The highest activity was at 30°C (Fig. 3A). This value is approximately 10°C above the optimal growth temperature of the psychrophilic Nesterenkonia strain AN1 (∼21°C). NitN retained 35% activity at 4°C. The relatively high level of activity at low temperatures, the low-temperature activity optimum, and the evidence of inactivation at temperatures above 30°C are consistent with the psychrophilic origins of the enzyme and its parent organism.
Fig. 3.
(A) Effect of temperature on NitN activity. Activities are expressed as percentages relative to the maximum activity at 30°C (100%). (B) Effect of pH on the activity of NitN. Activities are expressed as percentages relative to the maximum activity at pH 7 (100%).
Enzyme thermostability.
Thermostability profiles for NitN were broadly consistent with those of other cold-active enzymes (Fig. 3A). Under standard incubation conditions, NitN was stable at 30°C but rapidly inactivated at 40°C and above. Linear semilog plots of activity versus time showed half-lives of 3 h at 40°C and 3.7 min at 60°C. Inactivated NitN enzyme incubated at 0°C did not regain activity. These results should be contrasted with those for the nitrilase superfamily amidase from the moderate thermophile G. pallidus, which has a half-life in excess of 3 h at 60°C (27).
Determination of optimal pH.
The effect of pH on NitN activity was determined over a range of pH values from 4.5 to 11 (Fig. 3B). Tris-HCl and CAPS (N-cyclohexyl-3-aminopropanesulfonic acid) buffers, used for the pH range of 8 to 11, were found to inhibit NitN activity.
NitN showed optimal activity between pH 6.5 and 7.5 and retained ≥50% activity between pH 6 and 9 (Fig. 3B). The optimal pH of the enzyme is lower than the optimal pH for growth of Nesterenkonia (pH 9.6), which suggests that the intracellular pH is maintained near neutrality. Other aliphatic amidases typically have pH optima around 7 and less than 50% activity at pH 9 (11, 17, 27, 37). The lower point of inflection is at pH 5.8, indicating that a residue which is mechanistically important has a pKa at approximately this value, possibly E41 or E119. The reason for the falloff in activity at high pH demands further analysis.
Crystallization and structure determination.
Crystals of NitN formed within 2 h in “sitting-drop” crystallization experiments with unbuffered 2 M ammonium sulfate but had a multilamellar appearance and were unsuitable for X-ray diffraction. The addition of 0.1 M sodium acetate trihydrate (pH 4.6) slowed the crystal growth time to 3 weeks and resulted in good-quality, oblate hexagonal crystals. The statistics of data collected from one of these crystals are shown in Table 3. The structure was resolved by molecular replacement and refined to a resolution of 1.66 Å.
Table 3.
Crystallographic data for NitN
| Parameter | Value(s) for N-terminally His (20-amino-acid)-tagged NitN |
|---|---|
| Data collection statistics | |
| Wavelength (Å) | 0.979 |
| Space group | C2221 |
| Unit cell dimensions | |
| a, b, c (Å) | 76.00, 115.64, 65.32 |
| α = β = γ (°) | 90.00 |
| Unit cell vol (Å3) | 574,074 |
| Mosaicity | 1.45 |
| Resolution range (Å) (outer shell) | 38.00–1.66 (1.72–1.66) |
| Total no. of observations | 240,821 |
| Total no. of unique observations | 34,232 |
| Completeness (%) (outer shell) | 99.6 (98.9) |
| Redundancy (outer shell) | 7.03 (6.92) |
| Signal-to-noise ratio [I/σ(I)]a (outer shell) | 15.6 (4.2) |
| Rmerge (outer shell) | 0.046 (0.366) |
| Reduced χ2 (outer shell) | 0.94 (1.33) |
| Wilson plot avg B-factor (Å2) | 24.53 |
| Matthew's coefficient | 2.21 |
| Solvent content (%) | 45.0 |
| Refinement statistics | |
| Amino acid no. | 8–0 (His tag), 1–238, 249–263 |
| Protein molecular massb (Da) | 30,085.86 |
| No. of nonhydrogen atoms | 1,980 |
| No. of water atoms | 196 |
| Reflection data | |
| No. of reflections in working set | 32,495 |
| % of reflections in test set | 5.05 |
| Rfactor (%) (overall) | 18.64 |
| Rfree (%) (overall) | 21.88 |
| Overall figure of merit | 0.8589 |
| RMSc deviations from ideality | |
| Bond length (Å) | 0.0220 |
| Bond angle (°) | 1.802 |
| Avg B value (Å2) | 26.37 |
I, intensity of a reflection; (I), average intensity.
The value is for the average isotopic composition of the entire expressed peptide.
RMS, root mean square.
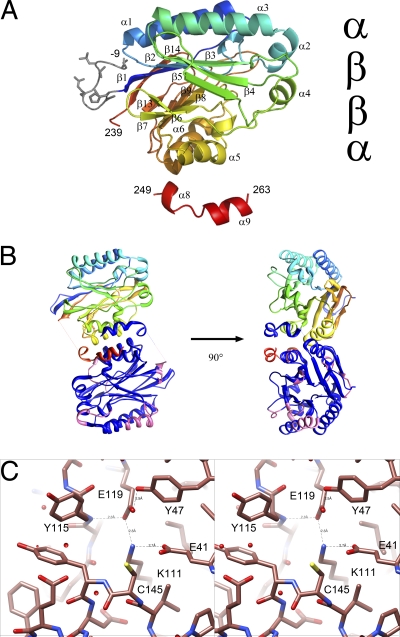
The overall features of the monomer in the resulting atomic model clearly identify NitN as a typical member of the nitrilase superfamily, having the typical αββα fold (Fig. 4A). The density of most of the map was, in general, easily interpretable. However, no interpretable density for residues 239 to 248 (underlined in Fig. 2) was visible. This region, corresponding to helix α7 in other structures, is usually well defined. The dimer interface which is conserved in all known members of the nitrilase superfamily is located with its 2-fold axis coincident with a crystal 2-fold axis. Six salt bridges stabilize this interface, including those formed between E152 and R155 on helix α5, R186 and E189 on helix α6, and R254 and the terminal carboxyl (263). Further electrostatic interactions across the dimer interface are formed by D253, which interacts with both Y146 (the residue following the active-site cysteine) and K120 in the opposing subunit. These interface interactions are particularly interesting as they probably play a role in constraining the conformation of the active site. No other extensive interfaces were apparent in the crystal, strongly suggesting that the biological unit is a dimer (Fig. 4B).
Fig. 4.
Fold of NitN. (A) The fold of the monomer is shown. The visible His tag residues are shown in gray, and the sequence is colored from blue to red. The characteristic αββα fold is clearly seen. (Note that the numbering is displaced by 20 from that given in the PDB code 3hkx deposit to account for the His tag.) (B) Structure of the catalytically active dimer. One monomer is drawn as in panel A, whereas the other highlights the regions of high-temperature factor in pink. The average temperature factor of the Nesterenkonia sp. enzyme is 24, compared to 14 in the case of the PDB code 1j31 enzyme and 9 in the case of the PDB code 2plq enzyme. (C) Stereoview of the catalytic site of NitN. Hydrogen bonding between E41, K111, E119, and Y47 will alter the pKas of the catalytic residues.
The active-site cysteine lies in a solvent-accessible pocket. Parts of residues E41, Y47, K111, Y115, G116, P117, E119, C145, Y146, D147, E149, A170, L171, A172, G173, E175, V178, and F208 contribute to the pocket (Fig. 4C). A side of the pocket is dominated by the aromatic ring of tyrosine 115. Unlike the amidase from G. pallidus (PDB code 2plq), which has the active-site entrance obstructed by W138 (in the same relative location as Y115) and M193, NitN seems to have little to constrain the size of the substrate that could be accepted.
The reasons for the substrate activity profile are not immediately obvious. We suggest that none of the compounds tested are likely to closely represent the in vivo substrate.
The feature which may account for the cold adaptation of NitN is apparent from a comparison with the hexameric structures of the mesophilic amidases and the hypothetical nitrilase from the thermophile P. horikoshii. The relatively high (55°C) thermostability of the mesophilic P. aeruginosa aliphatic amidase was attributed to the extended C-terminal regions that interlock the monomers across the dimer interface, which is preserved in NitN (4). In addition, monomers in the hexameric form of the P. aeruginosa aliphatic amidase interact across a further interface, not present in NitN, and the cumulative effect may contribute to the thermostability of this amidase relative to NitN (4). The structural differences that result in NitN being psychrophilic are probably also cumulative. It is possible to identify changes relative to the hypothetical protein PH0642 from P. horikoshii (PDB code 1j31) which could contribute to the increased thermostability of this enzyme. One example of such a change is the substitution of a leucine (L5) in NitN for a tyrosine (Y60) at the corresponding position in the PDB code 1j31 protein. The hydroxyl of Y6 is hydrogen bonded to the hydroxyl of S221 (corresponding to G223 in NitN). This hydrogen bond stabilizes the core region of PH0642 by linking β1 to β13. Furthermore, it is clear that several regions of NitN have relatively high temperature factors in the normally flexible regions compared to other members of the superfamily, including: C53 to Q55, S66 to R75, D100 to V105, W135 to R138, L171 to S177, E224 to G230, and P237 to D238. Presumably, this increased flexibility allows the enzyme to function at low temperatures. In seeming contradiction, the structure is less flexible than the PDB code 1j31 structure in the sequence following the active-site glutamate (E41), where the longer helix α2 (V48 to Q55) contributes to the rigidity of this region.
The feature previously identified by Pace and Brenner (29) as a tool for classifying members of the nitrilase superfamily is sequence similarity in the immediate vicinity of the three then-identified active-site residues. NitN, which displays only amidase activity, does not conform to their suggested pattern. The highest similarity was to NitFhit (29), which has no known activity. In particular, the sequence at the active-site cysteine is CYD and not CDD, which is more typical of the aliphatic amidases. In both NitFhit and NitN, the tyrosine hydroxyl group forms a hydrogen bond across the dimer interface. In the aliphatic amidase represented by the structure with PDB code 2plq, the corresponding aspartate, D167, is also involved in an interaction across the dimer interface, in this case the formation of a salt bridge with a lysine, K278 (22). This indicates a general principle in which the formation of the dimer interface and the active site are connected rather than the requirement of a specific property for the residue following the active-site cysteine.
Conclusion.
A novel psychrophilic isolate of the haloalkaliphilic bacterial genus Nesterenkonia encodes a member of the nitrilase superfamily. The recombinantly expressed protein, NitN, is an aliphatic amidase with activity on short-chain amides, which would place it in branch 2 of the classification scheme of Pace and Brenner (29), but the sequence at the active-site cysteine does not conform to the standard pattern.
The protein, which is active as a dimer, also differs from previously described hexameric aliphatic amidases from H. pylori (18), P. aeruginosa (4), and G. pallidus (1). In addition, NitN lacked the 35-amino-acid C-terminal sequence found in the aliphatic amidases of G. pallidus (22) and P. aeruginosa (3).
We suggest that the small aliphatic amides used as experimental substrates are almost certainly not the natural substrates for NitN, due to the high Km values and low catalytic rates. No detailed comparison of the catalytic rate with those of known aliphatic amidases was possible, since kinetic parameters for these enzymes were obtained using the acyl transfer assay in the presence of hydroxylamine (16, 17). Amidases typically have high catalytic rates in this assay, since hydroxylamine is a better acyl acceptor than water (17). No detectable activity was obtained for NitN under the assay conditions reported for other amidases.
Despite the low catalytic rates of NitN, the functional properties, in particular the apparent temperature optimum and thermostability characteristics, were generally consistent with the psychrotrophic origins of the enzyme.
The simple structural form of NitN and the ease of expression and purification are attractive features for further studies of this member of the nitrilase superfamily. For example, the potential roles of the additional catalytic glutamate residue E142 in the G. pallidus aliphatic amidase (22, 39) and the conserved catalytic residues (EKEC), which may dictate either amide- or nitrile-hydrolyzing activity, are all targets for mutagenic and coordinated crystallographic analyses.
ACKNOWLEDGMENTS
We thank Brandon Weber and Robert N. Thuku (Electron Microscope Unit, University of Cape Town) for the assistance in size exclusion chromatography and protein sequence alignments. We also thank J. Rees and J.-M. Celton (Genome Sequencing Faculty, University of the Western Cape) for providing the nucleotide sequences of the AN1 genome for in silico gene mining.
The National Research Foundation (NRF), South Africa, supported this work in the form of a grant.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Agarkar V. B., Kimani S. W., Cowan D. A., Sayed M. F., Sewell B. T. 2006. The quaternary structure of the amidase from Geobacillus pallidus RAPc8 is revealed by its crystal packing. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ambler R. P., Auffret A. D., Clarke P. H. 1987. The amino acid sequence of the aliphatic amidase from Pseudomonas aeruginosa. FEBS Lett. 215:285–290 [DOI] [PubMed] [Google Scholar]
- 4. Andrade J., Karmali A., Carrondo M. A., Frazão C. 2007. Structure of amidase from Pseudomonas aeruginosa showing a trapped acyl transfer reaction intermediate state. J. Biol. Chem. 282:19598–19605 [DOI] [PubMed] [Google Scholar]
- 5. Atlas R. M. 2005. Handbook of media for environmental microbiology. Chapman & Hall, New York, NY [Google Scholar]
- 6. Bork P., Koonin E. V. 1994. A new family of carbon-nitrogen hydrolases. Protein Sci. 3:1344–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brenner C. 2002. Catalysis in the nitrilase superfamily. Curr. Opin. Struct. Biol. 12:775–782 [DOI] [PubMed] [Google Scholar]
- 8. Cary S., McDonald I., Barrett J., Cowan D. 2010. On the rocks: microbial ecology of Antarctic cold desert soils. Nat. Rev. Microbiol. 8:129–138 [DOI] [PubMed] [Google Scholar]
- 9. Cavicchioli R., Siddiqui K. S., Andrews D., Sowers K. R. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 3:253–261 [DOI] [PubMed] [Google Scholar]
- 10. Chebrou H., Bigey F., Arnaud A., Galzy P. 1996. Study of the amidase signature group. Biochim. Biophys. Acta 1298:285–293 [DOI] [PubMed] [Google Scholar]
- 11. Cheong T. K., Oriel P. J. 2000. Cloning of a wide-spectrum amidase from Bacillus stearothermophilus BR388 in Escherichia coli and marked enhancement of amidase expression using directed evolution. Enzyme 26:152–158 [DOI] [PubMed] [Google Scholar]
- 12. Cowan D., Russels N., Mamais A., Sheppard D. 2002. Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 6:431–436 [DOI] [PubMed] [Google Scholar]
- 13. Eisenthal R., Cornish-Bowden A. 1974. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem. J. 139:715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Farrelly V., Rainey F. A., Stackebrandt E. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fournand D., Arnaud A. 2001. Aliphatic and enantioselective amidases: from hydrolysis to acyl transfer activity. J. Appl. Microbiol. 91:381–393 [DOI] [PubMed] [Google Scholar]
- 17. Fournand D., Arnaud A., Galzy P. 1998. Study of the acyl transfer activity of a recombinant amidase overproduced in an Escherichia coli strain. Application for short-chain hydroxamic acid and acid hydrazide synthesis. J. Mol. Catal. B Enzym. 4:77–90 [Google Scholar]
- 18. Hung C.-L., et al. 2007. Crystal structure of Helicobacter pylori formamidase AmiF reveals a cysteine-glutamate-lysine catalytic triad. J. Biol. Chem. 282:12220–12229 [DOI] [PubMed] [Google Scholar]
- 19. Jandhyala D., et al. 2003. CynD, the cyanide dihydratase from Bacillus pumilus: gene cloning and structural studies. Appl. Environ. Microbiol. 69:4794–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones D. T. 1999. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 287:797–815 [DOI] [PubMed] [Google Scholar]
- 21. Kamphuis J., et al. 1992. The production and uses of optically pure natural and unnatural amino acids, p. 187–208 In Collins A. N., Sheldrake G. N., Crosby J. (ed.), Chirality in industry. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 22. Kimani S. W., Agarkar V. B., Cowan D. A., Sayed M. F.-R., Sewell B. T. 2007. Structure of an aliphatic amidase from Geobacillus pallidus RAPc8. Acta Crystallogr. D Biol. Crystallogr. 63:1048–1058 [DOI] [PubMed] [Google Scholar]
- 23. Ko H. J., et al. 2010. Molecular characterization of a novel bacterial aryl acylamidase belonging to the amidase signature enzyme family. Mol. Cells 29:485–492 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 15:680–685 [DOI] [PubMed] [Google Scholar]
- 25. Li W. J., et al. 2005. Nesterenkonia sandarakina sp. nov. and Nesterenkonia lutea sp. nov., novel actinobacteria, and emended description of the genus Nesterenkonia. Int. J. Syst. Evol. Microbiol. 55:463–466 [DOI] [PubMed] [Google Scholar]
- 26. Li W. J., et al. 2004. Nesterenkonia halotolerans sp. nov. and Nesterenkonia xinjiangensis sp. nov., actinobacteria from saline soils in the west of China. Int. J. Syst. Evol. Microbiol. 54:837–841 [DOI] [PubMed] [Google Scholar]
- 27. Makhongela H. S., et al. 2007. A novel thermostable nitrilase superfamily amidase from Geobacillus pallidus showing acyl transfer activity. Appl. Microbiol. Biotechnol. 75:801–811 [DOI] [PubMed] [Google Scholar]
- 28. Neumann S., Granzin J., Kula M.-R., Labahn J. 2002. Crystallisation and preliminary X-ray data of the recombinant peptide amidase from Stenotrophomonas maltophilia. Acta Crystallogr. D Biol. Crystallogr. 58:333–335 [DOI] [PubMed] [Google Scholar]
- 29. Pace H. C., Brenner C. 2001. The nitrilase superfamily: classification, structure and function. Genome Biol. 2:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersen E. F., et al. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 31. Pflugrath J. W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55:1718–1725 [DOI] [PubMed] [Google Scholar]
- 32. Pointing S., et al. 2009. Highly specialized microbial diversity in hyper-arid polar desert. Proc. Natl. Acad. Sci. U. S. A. 106:19964–19969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reysenbach A. L., Pace N. R. 1995. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction, p. 101–105 In Robb F. T., Place A. R. (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Sakai N., Tajika Y., Yao M., Watanabe N., Tanaka I. 2004. Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6 Å resolution. Proteins 57:869–873 [DOI] [PubMed] [Google Scholar]
- 35. Sakashita K., Nakamura T., Watanabe I. June 1993. Process for producing l-amino acids. U.S. patent 5,215,897
- 36. Shin S., et al. 2002. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 21:2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skouloubris S., Labigne A., De Reuse H. 2001. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol. Microbiol. 40:596–609 [DOI] [PubMed] [Google Scholar]
- 38. Terwilliger T. C., et al. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thuku R. N., Brady D., Benedik M. J., Sewell B. T. 2009. Microbial nitrilases: versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 106:703–727 [DOI] [PubMed] [Google Scholar]
- 40. Vagin A. A., et al. 2004. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60:2184–2195 [DOI] [PubMed] [Google Scholar]
- 41. Weatherburn M. V. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971–974 [Google Scholar]
- 42. Yoon J. H., Jung S. Y., Kim W., Nam S. W., Oh T. K. 2006. Nesterenkonia jeotgali sp. nov., isolated from jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 56:2587–2592 [DOI] [PubMed] [Google Scholar]
- 43. Zhou J., Bruns M. A., Tiedje J. M. 1996. DNA recovery from soils of diverse compositions. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]