Abstract
Myc oncoproteins promote cell cycle progression in part through the transcriptional up-regulation of the cyclin D2 gene. We now show that Myc is bound to the cyclin D2 promoter in vivo. Binding of Myc induces cyclin D2 expression and histone acetylation at a single nucleosome in a MycBoxII/TRRAP-dependent manner. Down-regulation of cyclin D2 mRNA expression in differentiating HL60 cells is preceded by a switch of promoter occupancy from Myc/Max to Mad/Max complexes, loss of TRRAP binding, increased HDAC1 binding, and histone deacetylation. Thus, recruitment of TRRAP and regulation of histone acetylation are critical for transcriptional activation by Myc.
Keywords: Gene transcription, chromatin, oncogene, differentiation, HAT, HDAC
The c-myc proto-oncogene encodes a transcription factor, Myc, that is an evolutionary conserved regulator of cell proliferation and cell growth (Grandori et al. 2000). Myc activates a diverse group of genes as part of a heterodimeric complex with a partner protein, Max (e.g., Coller et al. 2000; Guo et al. 2000; O'Hagan et al. 2000a). The bHLHLZ DNA-binding/dimerization domain of Myc is localized at the C terminus of the protein whereas the N-terminal 150 amino acids have the capacity of transactivate transcription and are required for activation of target genes (Grandori et al. 2000). Within this region, two highly conserved elements (so-called Myc boxes) have been identified. Mutations in MycBoxI (MBI) have differential effects in biological assays, suggesting that this domain possesses regulatory activity. Deletion of the N terminus including MBI affects activation of some reporter genes and cripples Myc function in primary but not in established cells, pointing to the possibility that this domain also contributes to the regulation of some target genes (Xiao et al. 1998; Hirst and Grandori 2000).
In contrast, mutations in MBII abolish all biological functions of Myc, suggesting that MBII is an essential effector domain of Myc (for discussion, see McMahon et al. 1998). Indeed, deletion of MBII generates a dominant-negative allele of Myc that blocks transformation by Bcr-Abl (Sawyers et al. 1992). A simple hypothesis therefore would predict that MBII is important for modulation of expression of critical target genes by Myc. Curiously, however, deletion of MBII has varying and assay-dependent effects on the ability of Myc to modulate target gene expression, and no consistent view as to how it contributes to gene regulation has emerged. Recently, MBII has been shown to bind to TRRAP, a component of the TIP60 and PCAF/GCN5 histone acetyl transferase (HAT) complexes, suggesting that recruitment of complexes with HAT activity and modification of histone acetylation may contribute to the regulation of gene expression by Myc in vivo (Grant et al. 1998; McMahon et al. 1998, 2000; Ikura et al. 2000). The findings raise the possibility that results obtained using reporter assays may not reflect accurately the role of MBII and that the contribution of MBII to gene regulation may be unraveled only by the analysis of activation of endogenous genes that are embedded in chromatin.
Although Myc controls diverse biological functions, one effector pathway has been studied extensively over the last years. Activation of Myc induces cyclin E/Cdk2 kinase by antagonizing the function of the cell cycle inhibitor p27 (Amati et al. 2001). Four direct target genes of Myc have been implicated in this pathway: Myc stimulates expression of the cyclin D2 and Cdk4 genes, leading to sequestration of p27 in cyclin D2/Cdk4 complexes (Bouchard et al. 1999; Coller et al. 2000; Dey et al. 2000; Hermeking et al. 2000). Subsequently, p27 is degraded, and the transcriptional activation of the Cul-1 and Cks genes by Myc has been implicated in this process (Coller et al. 2000; O'Hagan et al. 2000b). Cyclin D2−/− fibroblasts have an impaired proliferative response to Myc, showing that cyclin D2 is an essential downstream effector of Myc (Bouchard et al. 1999). Conversely, depletion of p27 partially compensates for the defect in cellular proliferation caused by deletion of the c-myc gene (O'Hagan et al. 2000b). Taken together, the data define one pathway by which Myc regulates cellular proliferation.
Activation of cyclin E/Cdk2 kinase by Myc critically depends on the integrity of MBII in addition to domains involved in binding to Max and to DNA. Indeed, transient transfection experiments have suggested that MBII is required for up-regulation of cyclin D2 promoter activity by Myc. In addition, inhibition of histone deacetylase activates expression of cyclin D2 in quiescent cells (Bouchard et al. 1999). We therefore decided to explore in detail the role of MBII, histone acetylation, and TRRAP in transcriptional activation of the cyclin D2 gene by Myc. Our data strongly support a model in which Myc recruits acetylase complexes to the cyclin D2 promoter in vivo and thereby contributes to activation of the gene locus.
Results
Activation of MycER proteins by addition of 4-hydroxytamoxifen (4-OHT) rapidly induces transcription of the cyclin D2 gene; the induction is resistant to inhibition of protein synthesis, showing that Myc acts directly on the cyclin D2 promoter (Bouchard et al. 1999). In transient transfection experiments, regulation by Myc is mediated by an evolutionary conserved E-box element (E3); in contrast, a second E-box (E4) is dispensable for regulation (see Fig. 1A; Bouchard et al. 1999).
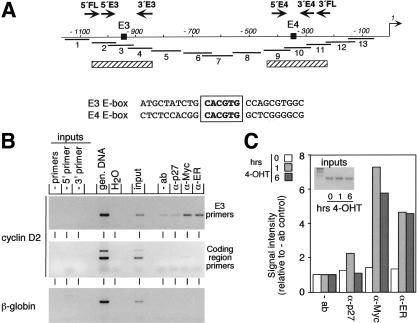
Figure 1.
In vivo binding of MycER proteins at the mouse cyclin D2 promoter. (A) Schematic drawing of the mouse cyclin D2 promoter from −1100 bp to the transcriptional start site (+1). (Black boxes) Two E-boxes (E3 and E4); (arrows) primer pairs (FL, E3, and E4); (thick lines) primer sets used for mapping; (hatched bars) position of nucleosomes. (B) ChIP assays using α-Myc antibodies or antibodies directed against the hormone-binding domain of the estrogen receptor (ER). As control, either no antibody or irrelevant control antibody (α-p27) were used. Immunoprecipitated DNA was analyzed by PCR using the indicated primer pairs. Inputs and genomic DNA (gen. DNA) are shown as control. (C) Quantitation of MycER binding to the cyclin D2 promoter after addition of hormone. MycER cells were serum-starved for 72 h and either left untreated or treated for 1 or 6 h with 250 nM 4-OHT, respectively. Subsequently, ChIP assays were conducted as described in (B). The signals obtained in the PCR were quantified and plotted relative to the signal obtained from −ab precipitation. (4-OHT) 4-hydroxytamoxifen.
To show that Myc is bound to the cyclin D2 promoter in vivo, we conducted chromatin immunoprecipitations (ChIP). Chromatin was prepared using a formaldehyde-cross-linking protocol (Strutt and Paro 1999; Eberhardy et al. 2000) from murine fibroblasts stably expressing a MycER protein 6 h after the addition of 250 nM 4-OHT. Under these conditions, antibodies directed against either human Myc or the hormone-binding domain of the murine estrogen receptor precipitated DNA encompassing the E3 E-box of the cyclin D2 promoter (Fig. 1A,B). In contrast, DNA fragments located downstream in the cyclin D2 gene or in the β-globin gene were not precipitated (Fig. 1B). Controls using either no antibody or antibodies directed against p27 revealed that the precipitation was specific.
The hormone-binding domain of the estrogen receptor interferes with binding of MycER to Max in absence of ligand (M. Eilers, unpubl.). Because binding to Max is required for Myc to bind to DNA, the findings suggested that DNA binding might be regulated for MycER proteins. Indeed, time course experiments (Fig. 1C; see also Fig. 3E) revealed that MycER was not bound to the cyclin D2 promoter in the absence of 4-OHT. Binding was detected as early as 1 h after addition of 4-OHT, and longer incubation did not lead to a further increase in binding. In parallel experiments, we were unable to detect binding of Myc to the E4 E-box of the cyclin D2 promoter (data not shown). We concluded that MycER proteins interact in vivo with the E3 E-box of the cyclin D2 promoter in a hormone-regulated manner.
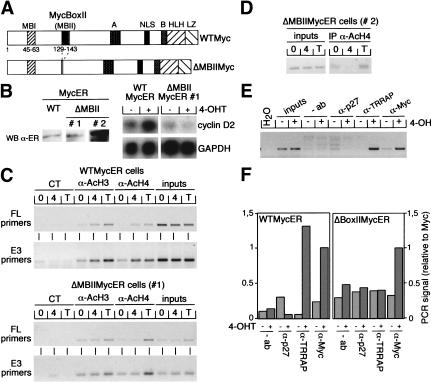
Figure 3.
Essential role of MycBoxII in activation of mouse cyclin D2 expression. (A) Schematic representation of wild-type Myc (WTMyc) and the MycBoxII (MBII) deletion mutant (ΔMBIIMyc) used in this study. (NLS) Nuclear localization signal; (HLH) helix–loop–helix; (LZ) leucine zipper. (B) (left) Western blot with α-ER antibodies documenting expression levels of MycER proteins in WTMycER and ΔMBIIMycER clones (designated #1 and #2). (right) Northern blots documenting expression of cyclin D2 and GAPDH mRNAs in WTMycER and ΔMBIIMycER#1 clones before (−) and 6 h after (+) addition of 250 nM 4-OHT. (WT) wild type; (WB) Western blot. (C) ChIP assays using either α-AcH3, α-AcH4, or control antibodies from serum-starved WTMycER and ΔMBIIMycER#1 clones either left untreated (0) or incubated for 6 h with 250 nM 4-OHT (4) or 100 ng/mL trichostatin A (T). Precipitated DNA was analyzed by PCR with primer pairs spanning either both E-boxes (FL) or the E3 E-box of the cyclin D2 promoter. (CT) control. (D) ChIP assays (performed as described in C) from a clone expressing high levels of ΔMBIIMycER (clone #2, see B). (E) ChIP assays with the indicated antibodies from serum-starved MycER cells either left untreated or treated for 6 h with 250 nM 4-OHT. PCRs were performed with primer pairs specific for the E3 E-box. (F) Quantitation of PCR assays performed with the E3 primers and chromatin immunoprecipitated from WTMycER and ΔMBIIMycER#1 clones before (−) and after (+) addition of 250 nM 4-OHT.
Inhibition of histone deacetylation using trichostatin A induces expression of cyclin D2 in quiescent cells to the same level as activation of Myc, raising the possibility that Myc may control expression of cyclin D2 by affecting histone acetylation (Bouchard et al. 1999). Therefore, we used antibodies directed against acetylated histones H3 and H4 for ChIP experiments. Activation of MycER resulted in an increase in acetylation of both histones H3 and H4 at the cyclin D2 promoter (Fig. 2A). Similar increases were observed after treatment of cells with trichostatin A (Fig. 2A), whereas total levels of acetylated histones H3 and H4 were unchanged (data not shown). When primers that span individual E-boxes were used, the changes observed after activation of Myc were most pronounced for acetylation of histone H4 at the E3 E-box; in comparison, changes in acetylation of both histones H3 and H4 at the E4 E-box and of histone H3 at the E3 E-box were less pronounced (Fig. 2B,C). Taken together, the data show that binding of Myc is sufficient to trigger histone acetylation at the cyclin D2 promoter.
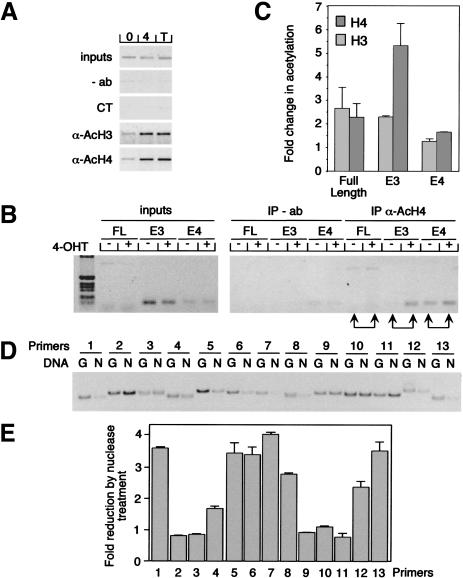
Figure 2.
Myc-regulated histone acetylation at the cyclin D2 promoter. (A) Immunoprecipitation of chromatin from serum-starved MycER cells either left untreated (0) or treated for 6 h with 250 nM 4-OHT (4) or 100 ng/mL trichostatin A (T) using anti-acetylated histones H3 (AcH3) or H4 (AcH4) antibodies. As control, either no antibody (−ab) or irrelevant control antibody (α-p27, labeled CT) was used. Precipitated chromatin was PCR-amplified using FL primers. (B) Anti-AcH4 immunoprecipitation (IP) of chromatin isolated from serum-starved MycER cells either left untreated (−) or treated for 6 h (+) with 250 nM 4-OHT and analyzed by PCR using three different primer pairs FL, E3, and E4 (depicted in Fig. 1A). (C) Quantitation of the average changes of histone acetylation observed in several independent experiments. (D) Mapping of nucleosomal positions in the cyclin D2 promoter. Chromatin was incubated with micrococcal nuclease to digest internucleosomal DNA. Nucleosomal DNA then was purified. Parallel PCRs were conducted with the primer pairs depicted in Fig. 1A from either undigested (G) or nucleosomal DNA (N) and analyzed on a polyacrylamide gel. (E) Quantitative representation of C documenting the fold reduction in signal intensity by nuclease treatment for each primer pair.
To obtain information about the nucleosomal organization of the cyclin D2 promoter, chromatin was isolated and treated with micrococcal nuclease to digest internucleosomal DNA. Subsequently, nucleosomal DNA was isolated and amplified using overlapping and closely spaced primer pairs (Fig. 1A). Comparison of the signals obtained from either undigested or digested DNA revealed that the E3 and the E4 E-boxes, but not the region between them, are protected from digestion and thus localized within nucleosomes (Fig. 2D,E). The findings define two nucleosomes on the cyclin D2 promoter, with Myc inducing the preferential hyperacetylation of histone H4 at a single nucleosome.
The Myc-associated protein TRRAP is a component of the PCAF/GCN5 and TIP60 HAT complexes (see Introduction), suggesting that histone acetylation might be mediated by recruitment of TRRAP to the cyclin D2 promoter. Binding of TRRAP to Myc requires the integrity of MBII in the N terminus of the protein (McMahon et al. 1998). To determine whether this domain has a role in activation of the cyclin D2 gene, we infected mouse 3T3 fibroblasts with retroviruses encoding either WTMycER or ΔMBIIMycER proteins (Fig. 3A); multiple individual clones were isolated after each infection. Up-regulation of cyclin D2 mRNA in response to addition of 4-OHT was observed in multiple WTMycER clones but in none of the ΔMBIIMycER clones (Fig. 3B and data not shown). For the subsequent analysis, a pair of clones was selected that express equivalent amounts of either protein (Fig. 3B, left; ΔMBII clone #1). In parallel experiments, histone acetylation at the cyclin D2 promoter was observed after activation of the wild-type MycER protein, but not on activation of the ΔMBIIMycER mutant (Fig. 3C). In contrast, addition of trichostatin A induced hyperacetylation of histones in both cell lines to a similar extent. Thus, Myc-dependent induction of cyclin D2 expression and histone acetylation both require the integrity of MBII.
To exclude that we had accidentally picked a clone in which the amounts of the ΔMBIIMycER protein were limiting, we repeated the analysis with a clone that expressed ∼10-fold higher levels of ΔMBIIMycER protein (Fig. 3B; clone #2). In this clone, addition of 4-OHT suppressed histone acetylation at the E3 E-box (Fig. 3D); thus, even at very high expression levels, ΔMBIIMycER proteins did not induce histone acetylation and instead acted as a dominant negative inhibitor of acetylation.
To show directly that TRRAP is recruited to the cyclin D2 promoter, we raised a polyclonal antiserum directed against TRRAP and performed ChIP assays before and after activation of MycER using primers that span the E3 E-box (Fig. 3E). The corresponding fragment was amplified from both α-Myc and α-TRRAP immunoprecipitations if chromatin was prepared from cells in which MycER had been activated by addition of 4-OHT, but not if chromatin was prepared from nonstimulated cells. Furthermore, experiments using WTMycER and ΔMBIIMycER chimeras in parallel revealed that both proteins interacted with the cyclin D2 promoter, but TRRAP was recruited to the promoter only if MBII was intact (Fig. 3F). No binding of TRRAP was detected at the E4 E-box either before or after activation of Myc (data not shown). The results show that TRRAP is recruited to the cyclin D2 promoter on activation of Myc and that the integrity of MBII is required for recruitment of TRRAP.
To investigate how histone acetylation and TRRAP binding mediated by endogenous Myc protein may contribute to regulation of cyclin D2 under physiological circumstances, we chose the human promyelocytic leukemia cell line HL60. These cells differentiate toward macrophages in response to the phorbolester TPA. During differentiation, the DNA-binding activity of Myc/Max complexes decreases. In contrast, Mad1, a Max binding partner and Myc antagonist, is induced (Sommer et al. 1998). Northern blot analysis revealed that cyclin D2 mRNA expression is strongly down-regulated during TPA-induced differentiation of HL60 cells (Fig. 4A).
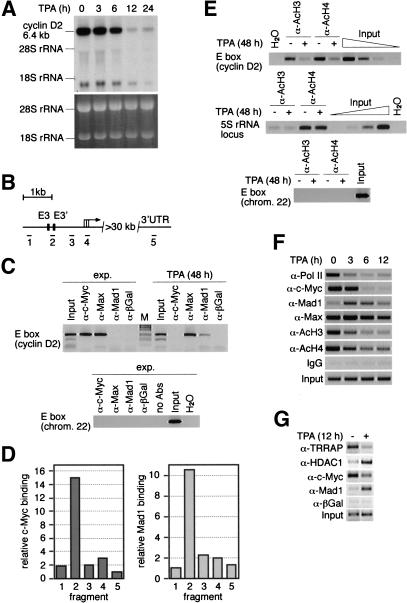
Figure 4.
Interaction of endogenous Myc/Max/Mad with the cyclin D2 promoter and regulation of histone acetylation in HL60 cells. (A) Northern blot documenting cyclin D2 expression on treatment of HL60 with TPA for the indicated times. The ethidium bromide-stained gel is displayed as control. (B) Schematic drawing of the human cyclin D2 locus. (Black boxes) The two E-boxes located within the first 2.5 kb of the cyclin D2 promoter. (Black bars) Location of the amplified fragments. (UTR) untranslated region. (C) ChIP assays documenting interaction of endogenous Myc, Max, and Mad1 proteins with the cyclin D2 promoter. HL60 cells were treated as in A, and ChIP assays were conducted with the indicated antibodies; precipitates were analyzed using primers specific for fragment 2 (top). Primers specific for a chromosome 22 E box were used as control (bottom). (exp.) Exponentially growing cells. (D) Quantitation of Myc binding in exponentially growing HL60 cells and of Mad1 binding in TPA differentiated HL60 cells to the five fragments of the cyclin D2 promoter as depicted in (B). Relative DNA binding defines the signal intensities obtained from the ChIP assays normalized to the input signals. (E) ChIP assays using either α-AcH3 or α-AcH4 antibodies of either untreated or TPA-treated (48 h) HL60 cells. Primers specific for fragment 2 of human cyclin D2 (top), a 5S rRNA locus (middle), and chromosome 22 E-box (bottom) were used. The input lanes show a dilution series in fourfold steps documenting the linearity of the assays. (F) Time course experiment documenting changes in promoter occupancy and histone acetylation on treatment of HL60 cells with TPA for the indicated time periods (fragment 2). Pol II occupancy was determined with fragment 4. (G) Regulated binding of TRRAP and HDAC1 to the cyclin D2 promoter. The experimental setup was as above.
In the human cyclin D2 promoter, the position and sequence surrounding the E3 E-box are conserved compared with the murine promoter whereas E4 is not. A second CACGTG E-box, termed E3′, is present in the human promoter that corresponds to a CACATG in the murine promoter (cf. Figs. 1A and 4B; see Bouchard et al. 1999). Thus, the human promoter contains two closely spaced E-boxes that were analyzed with PCR primers that amplify a fragment (#2) that lies between these two E-boxes (Fig. 4B). ChIP assays showed binding of Myc and Max, but not Mad1, to this fragment in exponentially growing HL60 cells (Fig. 4C, top). Binding was specific, because no PCR signal was obtained from immunoprecipitates using control antibodies and because little binding to other fragments of the D2 promoter was seen (Fig. 4C,D, left). In differentiated cells, Mad1 and Max bound, but Myc did not (Fig. 4C, top). Similar to the findings with Myc, binding of Mad1 was specific for the E-box containing region of the cyclin D2 promoter (Fig. 4D, right). In control experiments, no binding of either Myc, Max, or Mad1 was detected to a Myc consensus E-box localized on chromosome 22; this E-box is located in a chromosomal region without any detectable gene (Fig. 4C, bottom). Thus, endogenous proteins of the Myc/Max/Mad network bind specifically to one or two closely spaced E-boxes of the human cyclin D2 promoter, and during HL60 differentiation there is a switch of cyclin D2 promoter occupancy from Myc/Max to Mad/Max complexes.
Down-regulation of cyclin D2 expression was accompanied by a large decrease in both histone H3 and H4 acetylation at the cyclin D2 promoter (Fig. 4E). This change was specific for the cyclin D2 locus as no difference was observed when a fragment of a ribosomal gene locus was analyzed (Fig. 4E, middle). In addition, the region surrounding the E-box located on chromosome 22 showed background levels of histone H3 and H4 acetylation in both growing and differentiated HL60 cells (Fig. 4E, bottom). The decrease in acetylation on TPA treatment of HL60 cells involved both histones H3 and H4 and occurred throughout the human cyclin D2 promoter. In contrast, acetylation was constitutively low in the coding region of the cyclin D2 gene (O. Dittrich, K. Dohmann, and B. Lüscher, unpubl.).
Time course experiments revealed that changes in histone acetylation were detectable within 3 h and complete by 6 h after TPA addition, preceding changes in mRNA expression by at least 3 h (Fig. 4, cf. A and F). The changes were paralleled by a loss of RNA polymerase II (Pol II) loading and of Myc binding and an increase in Mad1 binding to the cyclin D2 promoter (Fig. 4F). Consistent with the idea that binding of Myc recruits TRRAP to promoters in vivo, binding of TRRAP to the cyclin D2 promoter was easily detectable in untreated HL60 cells, whereas little binding was detected in cells treated for 12 h with TPA. In contrast, HDAC1 was recruited to the cyclin D2 promoter during differentiation, in agreement with the suggestion that HDAC1 mediates repression by Mad1 (Fig. 4G; Alland et al. 1997; Hassig et al. 1997; Laherty et al. 1997; Sommer et al. 1997).
Taken together, the results show that changes in the occupancy of the cyclin D2 promoter by proteins of the Myc/Max/Mad network, alterations in histone acetylation, and alterations in mRNA expression occur as an ordered chain of events. The findings strongly support a model, in which endogenous Myc/Max and Mad1/Max complexes contribute to the regulation of cyclin D2 expression through recruitment of HAT- and HDAC-containing complexes, respectively.
Discussion
The aim of this study was to resolve the role of MBII and TRRAP in gene activation by Myc, using an endogenous target gene of Myc, cyclin D2, as model. We now report that upon binding to the cyclin D2 promoter, Myc recruits TRRAP and induces the preferential acetylation of histone H4 at a single nucleosome. Conversely, loss of endogenous Myc binding correlates with histone deacetylation and loss of TRRAP binding during the TPA-induced differentiation of a human promyelocytic cell line, HL60. The integrity of MBII is required for TRRAP recruitment, histone acetylation, and transcriptional activation at the cyclin D2 locus. Therefore, previous suggestions that MBII has no role in transcriptional activation based on transient reporter assays need to be reevaluated. Deletion of the entire N terminus of Myc up to MBII (s-Myc; Xiao et al. 1998) rendered Myc unable to induce cell cycle progression and expression of either cyclin A or cyclin D2 in 3T3 fibroblasts (Bouchard et al., unpubl.), consistent with recent results that the N terminus of Myc is required for regulation of proliferation and induction of gene expression in a cell-type-dependent manner (Hirst and Grandori 2000). Most likely, this is because stable association with TRRAP requires sequences in the N terminus of Myc in addition to MBII (McMahon et al. 1998).
Mad proteins are thought to antagonize the function of Myc by recruiting a repressor complex that contains histone deacetylase activity (Alland et al. 1997; Hassig et al. 1997; Laherty et al. 1997; Sommer et al. 1997). Several observations suggest that this model applies to the cyclin D2 promoter. First, repression of the cyclin D2 promoter by Mad1 requires the integrity of an N-terminal domain, which mediates recruitment of histone deacetylase complexes through interaction with Sin3A (Ayer et al. 1996; Bouchard et al. 1999). Second, during HL60 differentiation, Mad1 and HDAC1 are corecruited to the cyclin D2 promoter, correlating with histone deacetylation of both histones H3 and H4 at the cyclin D2 promoter. Taken together, our data strongly support a model in which endogenous Myc/Max and Mad/Max complexes contribute to the regulation of transcription of the cyclin D2 gene through their antagonistic effects on histone acetylation. In addition these findings show the functional relevance of the switch between Myc/Max and Mad/Max complexes during differentiation of hematopoietic cells as proposed previously (Ayer and Eisenman 1993; Larsson et al. 1997). Recent work on the gene encoding the catalytic subunit of telomerase, htert, suggests that this model also may apply to this promoter (Xu et al. 2001).
Up-regulation of the CAD gene by Myc does not involve changes in histone acetylation. Instead, high levels of histone acetylation at the promoter were found in both quiescent and proliferating cells (Eberhardy et al. 2000), showing that Myc can control at least one step in addition to histone acetylation to promote active transcription. Additional proteins have been identified that bind to different domains of Myc and that are candidates for such an activity: for example, the C terminus of Myc binds to Ini1, a component of Swi/Snf family of chromatin-remodeling complexes (Cheng et al. 1999). Clearly, a detailed analysis of the role of Myc in activation of individual promoters will be required before the role of each interaction in Myc biology can be resolved fully.
Materials and methods
Tissue culture
The established mouse 3T3 cell line expressing MycER proteins was described elsewhere (Bouchard et al. 1999). For the experiments, 70% confluent cells were serum-starved in 0.1% FCS for 72–96 h, then either untreated or incubated with 250 nM 4-OHT (Sigma) or 100 ng/mL trichostatin A (Calbiochem). Differentiation of HL60 cells has been described previously (Sommer et al. 1998).
Chromatin immunoprecipitation
ChIP assays were performed as described previously by others (Strutt and Paro 1999; Eberhardy et al. 2000) using the following antibodies: anti-acetylated histones H3 (nos. 06–599) and H4 (nos. 06–866; Upstate Biotechnologies), Myc (N-262), Mad1 (C-19), Max (C-17), HDAC1 (H-51), ER α (sc-542), p27 (sc-528) (Santa Cruz), Pol-II (8WG16; Berkeley Antibody Company), a rabbit antiserum raised against TRRAP or antibodies specific for TRRAP obtained from M. Cole (MacMahon et al. 1998). For each experiment, PCRs were performed with different numbers of cycles or with dilution series of input DNA to determine the linear range of the amplification; all results shown fall within this range. All PCR signals were quantified using National Institutes of Health Image software. Primer sequences are available on request.
Nucleosomal mapping
Digestion of isolated nuclei were performed with 375 units of micrococcal S7 nuclease (no. 107921; Roche) at 37°C for 10 min. PCRs were performed using 5 ng of purified mononucleosomes or undigested genomic DNA as control. PCR products were run on a nondenaturating 16% polyacrylamide gel and analyzed by ethidium bromide staining.
Northern analysis
For Northern blot analysis, total RNA was isolated (RNeasy; QIAGEN) from HL-60 cells at the indicated time points after TPA addition. Total RNA (20 μg/lane) was run on a 0.9% formaldehyde/agarose gel and blotted to a Hybond-N filter. Hybridization was performed in church buffer using a radiolabeled 0.7-kb PstI fragment of the human cyclin D2 cDNA.
Acknowledgments
We thank Renato Paro and Ralph Herrera for providing ChIP protocols and protocols for nucleosomal mapping, respectively. We thank Hartmut Land and Michael Cole for gifts of anti-TRRAP antibodies. This work was supported by an AICR grant to Caroline Bouchard, and by grants from the Deutsche Forschungsgemeinschaft (to B.L. and M.E.), the European Community (to M.E.), and the Fonds der Chemischen Industrie (B.L. and M.E).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The paper by Frank et al. (this issue) shows Myc-dependent changes in histone acetylation at, and TRRAP recruitment to, several different loci. These findings are consistent with our work on the cyclin D2 gene.
Footnotes
E-MAIL luescher@ruth-aachen.de; FAX 49-241-8888427.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.907901.
References
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes & Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Ayer D, Laherty CD, Lawrence QA, Armstrong AP, Eisenman RN. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SW, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Coller H A, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, She H, Kim L, Boruch A, Guris DL, Carlberg K, Sebti SM, Woodley DT, Imamoto A, Li W. Colony-stimulating factor-1 receptor utilizes multiple signaling pathways to induce cyclin D2 expression. Mol Biol Cell. 2000;11:3835–3848. doi: 10.1091/mbc.11.11.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, D'Cunha CA, Farnham PJ. Direct examination of histone acetylation on Myc target genes using chromatin immunoprecipitation. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR, III, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray . Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst SK, Grandori C. Differential activity of conditional MYC and its variant MYC-S in human mortal fibroblasts. Oncogene. 2000;19:5189–5197. doi: 10.1038/sj.onc.1203904. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang W-M, Sun J-M, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Larsson L-G, Bahram F, Burkhardt H, Lüscher B. Analysis of the DNA-binding activities of Myc–Max–Mad network complexes during induced differentiation of U-937 monoblasts and F9 teratocarcinoma cells. Oncogene. 1997;15:737–748. doi: 10.1038/sj.onc.1201390. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c- Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG, Jr, DePinho RA. Myc-enhanced expression of cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes & Dev. 2000a;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan RC, Schreiber-Agus N, Chen K, David G, Engelman JA, Schwab R, Alland L, Thomson C, Ronning DR, Sacchettini JC, et al. Gene-target recognition among members of the myc superfamily and implications for oncogenesis. Nat Genet. 2000b;24:113–119. doi: 10.1038/72761. [DOI] [PubMed] [Google Scholar]
- Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Lüscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- Sommer A, Bousset K, Kremmer E, Austen M, Lüscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol. 1999;119:455–467. doi: 10.1385/1-59259-681-9:455. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann SR. Transactivation-defective c-mycS retains the ability to regulate proliferation and apoptosis. Genes & Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]