Abstract
Thraustochytrids, marine protists known to accumulate polyunsaturated fatty acids (PUFAs) in lipid droplets, are considered an alternative to fish oils as a source of PUFAs. The major fatty acids produced in thraustochytrids are palmitic acid (C16:0), n − 6 docosapentaenoic acid (DPA) (C22:5n − 6), and docosahexaenoic acid (DHA) (C22:6n − 3), with eicosapentaenoic acid (EPA) (C20:5n − 3) and arachidonic acid (AA) (C20:4n − 6) as minor constituents. We attempted here to alter the fatty acid composition of thraustochytrids through the expression of a fatty acid Δ5 desaturase gene driven by the thraustochytrid ubiquitin promoter. The gene was functionally expressed in Aurantiochytrium limacinum mh0186, increasing the amount of EPA converted from eicosatetraenoic acid (ETA) (C20:4n − 3) by the Δ5 desaturase. The levels of EPA and AA were also increased by 4.6- and 13.2-fold in the transgenic thraustochytrids compared to levels in the mock transfectants when ETA and dihomo-γ-linolenic acid (DGLA) (C20:3n − 6) were added to the culture at 0.1 mM. Interestingly, the amount of EPA in the transgenic thraustochytrids increased in proportion to the amount of ETA added to the culture up to 0.4 mM. The rates of conversion and accumulation of EPA were much higher in the thraustochytrids than in baker's yeasts when the desaturase gene was expressed with the respective promoters. This report describes for the first time the finding that an increase of EPA could be accomplished by introducing the Δ5 desaturase gene into thraustochytrids and indicates that molecular breeding of thraustochytrids is a promising strategy for generating beneficial PUFAs.
INTRODUCTION
n − 3 polyunsaturated fatty acids (PUFAs), such as eicosapentaenoic acid (EPA) (C20:5n − 3) and docosahexaenoic acid (DHA) (C22:6n − 3), have attracted increasing attention in the development of medicines and nutritional supplements based on their serological and cardiovascular benefits (2, 26). DHA is thought to be integral to the development of neural tissues and the retina (8), and EPA is believed to have anticancer effects (22). n − 3 PUFAs are generally obtained from fish oils, but fish stocks have been gradually decreasing due to overfishing and environmental pollution (13). The need for fish oil substitutes has stimulated efforts by plant biotechnologists to accumulate beneficial PUFAs in seed oils of transgenic plants (5). An alternative approach to producing n − 3 PUFAs involves thraustochytrids, eukaryotic marine protists, which accumulate large amounts of PUFAs in their droplets (3, 4, 9, 27). However, basic information and tools for genetic manipulation are still lacking for thraustochytrids.
In animals and plants, PUFAs are generated in a standard (desaturase/elongase) pathway, whereas in thraustochytrids and some marine bacteria, they are mainly generated in a polyketide-like fatty acid synthesis pathway (PUFA synthase) (16, 18). Interestingly, fatty acid desaturases and elongases which could be involved in the standard pathway are also found in some thraustochytrids (19, 28).
The major fatty acids produced in thraustochytrids are palmitic acid (C16:0), n − 6 docosapentaenoic acid (DPA) (C22:5n − 6), and DHA, while EPA and arachidonic acid (AA) (C20:4n − 6) are minor constituents (27). Thraustochytrids are therefore considered suitable for the production of DHA and DPA but not EPA or AA. EPA and AA are generated from eicosatetraenoic acid (ETA) (C20:4n − 3) and dihomo-γ-linolenic acid (DGLA) (C20:3n − 6), respectively, by fatty acid Δ5 desaturase, which inserts a double bond at position 5 between the preexisting double bond and the carboxyl end of the fatty acid (7, 10, 12, 28), although it is still unclear whether the enzyme functions in thraustochytrids to produce the PUFAs.
In this study, a fatty acid Δ5 desaturase isolated from Thraustochytrium aureum ATCC 34304 was expressed in Aurantiochytrium limacinum mh0186 (6, 23) using an expression system composed of the ubiquitin promoter and terminator, both isolated from T. aureum ATCC 34304. The gene was transcribed into the desaturase mRNA, and the product functioned as a fatty acid Δ5 desaturase, resulting in an increase of EPA in the thraustochytrid. It is worth noting that the rates of conversion and accumulation of EPA were much higher in thraustochytrids than in yeasts driven by the respective promoters. These results indicate that thraustochytrids are suitable for molecular breeding to produce PUFAs using the gene expression system described in this study.
MATERIALS AND METHODS
Materials.
T. aureum ATCC 34304 was purchased from the American Type Culture Collection. A. limacinum mh0186 was identified based on the sequence of the 18S ribosomal DNA (rDNA) (DDBJ accession number AB362211). The restriction enzymes and T4 DNA ligase were purchased from Nippon Gene (Tokyo, Japan). Synthetic oligonucleotides were obtained from Hokkaido System Science (Hokkaido, Japan) and Genenet (Fukuoka, Japan). The antibiotic neomycin (G418) was purchased from Nacalai Tesque (Kyoto, Japan). Eicosatetraenoic acid (ETA) (C20:4n − 3), dihomo-γ-linoleic acid (DGLA) (C20:3n − 6), docosapentaenoic acid (DPA) (C22:5n − 3) and docosatetraenoic acid (DTA) (C22:4n − 6) were purchased from Cayman Chemical Co. (Michigan). Eicosadienoic acid (EDA) (C20:2n − 6), linoleic acid (LA) (C18:2n − 6), and α-linolenic acid (ALA) (C18:3n − 3) were obtained from Sigma. Eicosatrienoic acid (ESA) (C20:3n − 3) was purchased from Biomol. Sealife was obtained from Marinetech (Tokyo, Japan). All other reagents were of the highest purity available.
Cloning of cDNA encoding TauΔ5des.
To obtain a DNA fragment encoding the fatty acid Δ5 desaturase (TauΔ5des) from T. aureum ATCC 34304, a set of primers was designed based on the sequence of the fatty acid Δ5 desaturase from Thraustochytrium sp. ATCC 26185 (DDBJ accession number FJ821482): 3F (5′-TAC TGG AAG AAC CAG CAC AGC AAG CAC CAC-3′)/1RNES (5′-CGC CGT GGG GAA GAG GTG GTG CTC GAT CTG-3′). PCR was then performed with the T. aureum ATCC 34304 cDNA library as a template using Advantage 2 polymerase mix (Clontech, California). The cycling parameters for PCR were 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and the number of cycles was 30. A 550-bp PCR product was subcloned into the pGEM-T Easy vector (Promega Corporation, Madison, WI) and sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, California) and a DNA sequencer (ABI model 3130; Applied Biosystems). The sequence of an insert, showing high identity with the Δ5 desaturase gene of Thraustochytrium sp. ATCC 26185, was used to design the primers for 5′ and 3′ rapid amplification of cDNA ends (RACE). A 3′-RACE product was obtained using RACEd5F (5′-TGT CCT GCT TCC TGG TTG GTC TC-3′) and RACEd5FNES (5′-TCT GGA CCC TGT TTC TGC ACC CGC-3′). The cycling parameters for PCR were as described above. A 5′-RACE product, however, was not amplified by this method, and thus, the LA PCR in vitro cloning kit (Takara Bio, Japan) was used along with GSP1 (5′-ACCGCAAAGTTGGTGAAGATG-3′) and GSP2 (5′-CAAAGCCAAAGGTGGCCATGTAGAGAC-3′). PCR was then carried out with the T. aureum ATCC 34304 genome (BglII digest) as a template using the Advantage 2 polymerase mix. The cycling parameters for PCR were 94°C for 30 s, 60°C for 30 s, and 72°C for 3 min, and the number of cycles was 30. As a result, we obtained a 2.5-kbp PCR product corresponding to the 5′ region of TauΔ5des. Finally, the open reading frame (ORF) of the TauΔ5des gene was determined by comparing the ORF of the fatty acid Δ5 desaturase gene of Thraustochytrium sp. ATCC 26185.
Phylogenetic analysis.
Amino acid sequences of various fatty acid desaturases, including TauΔ5des, were aligned using the ClustalW 1.81 software program (24), and a phylogenetic tree was constructed with the neighbor-joining methods using the MEGA4 program (www.megasoftware.net). The percentage of replicate trees was calculated by the bootstrap test (1,000 replicates).
Generation of S. cerevisiae harboring the TauΔ5des gene.
The ORF of TauΔ5des was amplified by PCR using d5fulllengthF, containing an EcoRI site (5′-CGA ATT CAT GGG ACG CGG CGG CGA AGG TCA G-3′), and d5fulllengthR, containing a XhoI site (5′-GCT CGA GTT GGG TCG GGA TAA AAT AAA TGG C-3′). PrimeSTAR HS DNA polymerase (Takara Bio) was used for amplification. The cycling parameters for PCR were the same as described above. The PCR products were digested with EcoRI and XhoI and cloned into the same sites of pYES2/CT (Invitrogen, California). The TauΔ5des expression vector, designated pYpΔ5Des, was introduced into Saccharomyces cerevisiae INVSc1 (Invitrogen) using the lithium acetate method (25). The transformants were selected on agar plates lacking uracil (0.67% Difco yeast nitrogen base without amino acid, 2% glucose, 0.059% CSM [complete supplement mixture minus adenine, histidine, leucine, tryptophan, and uracil], 0.002% adenine, 0.002% l-histidine, 0.01% l-leucine, 0.002% l-tryptophan). The transformants harboring the TauΔ5des gene were designated scΔ5ura, and the mock transfectants were designated scura.
Generation of A. limacinum mh 0186 harboring the TauΔ5des gene.
To express the TauΔ5des gene in thraustochytrids, a linear DNA cassette containing a TauΔ5neor gene driven by the ubiquitin promoter/terminator and a neomycin resistance (Neor) gene driven by the EF1α promoter/terminator was constructed. For mock transformants, the TauΔ5des gene with the ubiquitin promoter/terminator was omitted from the cassette. The EF1-α promoter/terminator and ubiquitin promoter/terminator were cloned from T. aureum ATCC 34304. The DNA linear cassette was introduced into A. limacinum mh0186 cells by electroporation. Cells (5 × 106) and 5 μg DNA in 80 μl of Nucleofector solution L (Amaxa Biosystems, Maryland) were transferred to a 0.1-cm-gap cuvette and electroporated (pulsed conditions: 50 μF, 50 Ω, 7.5 kV/cm, 2 times) by using a Gene Pulser system (Bio-Rad, CA). The cells were then immediately resuspended in 1 ml of GY medium (3% glucose, 1% yeast extract, and 1.75% Sealife), incubated at 25°C for 1 day, and spread on PD agar plates (0.48% potato dextrose broth, 1.75% Sealife, and 1.5% agar) containing neomycin at 0.5 mg/ml. After incubation at 25°C for 2 to 5 days, colonies appearing on the plates were regarded as transformants. The transformants harboring the TauΔ5des gene were designated mhΔ5neor, and the mock transfectants were designated mhneor.
RT-PCR analysis.
Total RNA was prepared from mhΔ5neor and mhneor with Sepazol RNA I Super (Nacalai Tesque), the RNeasy Mini kit (Qiagen, Tokyo, Japan), and DNase I (TaKaRa Bio) and reverse transcribed to cDNA using PrimeScript reverse transcriptase (RT) (TaKaRa Bio). Then, PCR amplification was performed using, for the TauΔ5des gene, D5EVF (5′-TTG ATA TCA TGG GAC GCG GCG GCG AAG GTC AGG T-3′) and D5EVR (5′-TTG ATA TCC TAA GCG GCC TTG GCC GCC GCC TG-3′), and for the Neor gene, proGF (5′-GCG ACC TAA GCA ACA CTA GCC AAC ATG ATT GAA CAG GAC GGC CTT CAC-3′) and GterR (5′-AGT ATA GCA CAT ACT ACA GAT AGC TCA AAA GAA CTC GTC CAG GAG GC-3′). The cycling parameters for PCR were 98°C for 10 s, 60°C for 30 s, and 72°C for 1.5 min, and the number of cycles was 30. For semiquantitative RT-PCR, the template concentration and cycle number of PCR were adjusted by using respective glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes as a standard. PCR amplifications of GAPDH genes of thraustochytrids and yeasts were performed using mhg3pdhF2 (5′-CAC CGG CTC TGA CTA CGT TGT GG-3′) plus mhg3pdhR2 (5′-CTT CAT GGC GGC GCA GAT CTC CTC-3′) and scg3pdhF1 (5′-TAT GCT GCT TAC ATG GTC AAG TAC G-3′) plus scg3pdhR1(5′-ACA ACG GCA TCT TCG GTG TAA CCC-3′), respectively. The cycling parameters for PCR were 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and the number of cycles was 30 for the GAPDH gene and 25 for the TauΔ5des gene.
Southern blotting.
After digestion of the genomic DNAs of mhΔ5neor and mhneor with the restriction enzyme BglII, the digestion products were subjected to electrophoresis using a 0.7% agarose gel and then transferred to the Biodyne membrane (PALL Gelman Laboratory). Part (600 bp) of the TauΔ5des gene was amplified by PCR and used as a probe after labeling with digoxigenin (DIG). The DIG probe was prepared by using a PCR DIG probe synthesis kit (Roche, Germany). The PCR was carried out using D5ORFcomF (5′-GAC GCG GCG GCG AAG GTC AGG-3′) and D5ORFcomR (5′-CTT GCT GTG CTG AAC GCG CCA C-3′) as primers. The cycling parameters for PCR were 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and the number of cycles was 30.
Fatty acid analysis.
Fatty acid methyl esters (FAMEs) were prepared as described previously (20). FAMEs were analyzed by gas-liquid chromatography (GC) and GC-mass spectrometry (GC-MS) using a Shimadzu GC-MS QP-5000 instrument (Shimadzu Co., Kyoto, Japan) as reported previously (1). The rate of conversion of the substrate into the product was calculated as follows: conversion rate (percent) = [GC area for the product/(GC area for the product plus GC area for the substrate)] × 100.
Conversion of fatty acids in transgenic S. cerevisiae and A. limacinum mh 0186.
The scΔ5ura and scura cells were cultured for 2 days at 25°C in 5 ml of Ura− medium containing 2% glucose, harvested (1,500 × g for 3 min), and washed with distilled water. To induce the expression of the TauΔ5des gene, the cells were suspended in 10 ml of Ura− medium containing 2% galactose. The cells were harvested and then recultured at 25°C for 1 day in Ura− medium containing various fatty acids at the concentrations indicated. The mhΔ5neor and mhneor cells were cultured for 2 days at 25°C in 5 ml of GY medium, harvested (3,500 × g for 5 min), and washed with 1.75% Sealife. The cells were cultured at 25°C for 1 day in PD medium containing various fatty acids at the concentrations indicated. Total fatty acids were extracted, and FAMEs were analyzed by GC and GC-MS.
RESULTS
cDNA cloning of the fatty acid Δ5 desaturase (TauΔ5des) from T. aureum ATCC 34304.
To isolate the cDNA encoding TauΔ5des, PCR primers were designed based on the consensus sequence of the Δ5 desaturase gene of Thraustochytrium sp. ATCC 26185. The TauΔ5des gene was found to possess an open reading frame consisting of 1,320 bp encoding 439 deduced amino acid residues, which contained a cytochrome b5 domain (HPGGSI) and three histidine boxes (HECGH, HSKHH, and QIEHH). To analyze the sequence identity/similarity with other fatty acid Δ5 desaturases, we constructed a phylogenetic tree (see Fig. S1 in the supplemental material). TauΔ5des was most closely related to the Thraustochytrium sp. enzyme, and the identity/similarity of two enzymes were 58.0%/69.4%. However, TauΔ5des seemed to be evolutionally different from another thraustochytrid (Oblongichytrium sp.) enzyme.
Functional analysis of the TauΔ5des gene in S. cerevisiae.
To reveal the substrate specificity of TauΔ5des, yeast cells containing TauΔ5des cDNA (scΔ5ura) or an empty vector (scura) under the control of the GAL1 promoter were cultured in Ura− medium containing 2% galactose and various fatty acids at 0.1 mM. The conversion rates of various fatty acids added were then examined. As shown in Table 1, 19.9% ± 2.2% of ETA (C20:4n − 3) and 22.9% ± 2.3% of DGLA (C20:3n − 6) were converted to EPA (C20:5n − 3) and AA (C20:4n − 6), respectively, in the scΔ5ura cells, whereas these substrates were not changed in the scura cells (data not shown). Meanwhile, α-linolenic acid (C18:3n − 3), linoleic acid (C18:2n − 6), eicosatrienoic acid (C20:3n − 3), eicosadienoic acid (C20:2n − 6), docosapentaenoic acid (C22:5n − 3), and docosatetraenoic acid (C22:4n − 6) were not effective substrates (Table 1). Comparison of the GC-MS spectra indicated that spectra of EPA and AA from scΔ5ura were identical to those of the corresponding authentic standards (data not shown). These results clearly indicated that TauΔ5des cloned in this study was a Δ5 desaturase which inserts a double bond at position 5 of ETA and DGLA, generating EPA and AA, respectively.
Table 1.
Substrate specificity of TauΔ5des in yeastsa
| Substrate added | Product generated | Conversion rate (%) |
|---|---|---|
| C18:3n−3 | C18:4n−3 | <0.5 |
| C18:2n−6 | C18:3n−6 | <0.5 |
| C20:4n−3 | C20:5n−3 | 19.9 ± 2.2 |
| C20:3n−6 | C20:4n−6 | 22.9 ± 2.3 |
| C20:3n−3 | C20:4n−3 | <0.5 |
| C20:2n−6 | C20:3n−6 | <0.5 |
| C22:5n−3 | C22:6n−3 | <0.5 |
| C22:4n−6 | C22:5n−6 | <0.5 |
To examine the specificity of TauΔ5des, various fatty acids were added to the culture of scΔ5ura. After 1 day of culture at 25°C in Ura− medium containing 2% galactose, fatty acids were extracted from scΔ5ura cells and FAMEs were analyzed by GC. The conversion rate (%) was calculated as follows: [GC area for the product/(GC area for the substrate + GC area for the product)] × 100. The values are the means of triplicate determinations with SD.
Construction of DNA cassette for expression of TauΔ5des in thraustochytrids.
We prepared an expression construct using the promoter and terminator regions of housekeeping genes of thraustochytrids. Among the several candidates tested, we found that the promoter and terminator regions of two housekeeping genes were suitable for the expression of target genes, i.e., an 812-bp promoter region and a 589-bp terminator region of ubiquitin and a 633-bp promoter region and a 1,229-bp terminator region of EF-1α, all of which were cloned from the T. aureum ATCC 34304 genome library. The TauΔ5des gene fused with the ubiquitin promoter/terminator was connected to a Neor gene fused with the EF-1α promoter/terminator. The construct containing the two genes was inserted into the pUC18 vector to generate a circular DNA construct (pUBneorΔ5), as shown in Fig. S2a in the supplemental material. In this study, however, a linear DNA cassette containing the TauΔ5des and Neor genes (TauΔ5neor; see Fig. S2b) and that containing the Neor gene (neor in Fig. S2c), amplified using pUBneorΔ5 as a template, were used for the transformation of thraustochytrids. The mh0186 strain transformed with TauΔ5neor or the Neor gene was designated mhΔ5neor or mhneor, respectively.
RT-PCR and Southern blotting analysis of transgenic A. limacinum mh0186.
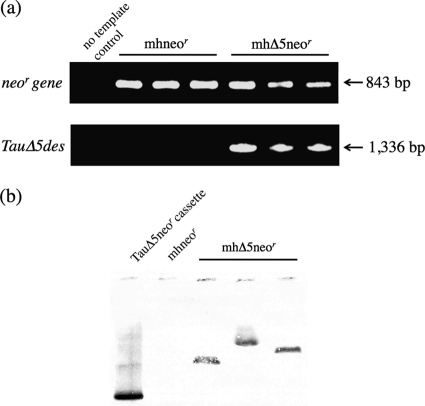
The expression of TauΔ5des and Neor mRNA in the transgenic thraustochytrids was analyzed by reverse transcriptase (RT)-PCR. A Neor gene transcript was detected in both mhΔ5neor and mhneor, while a TauΔ5des gene transcript was detected in mhΔ5neor (Fig. 1). Thus, both the TauΔ5des and Neor genes were transcribed in the thraustochytrids as expected. To examine the copy number of the TauΔ5des gene inserted into the genome of mhΔ5neor, Southern blotting was performed using a DIG-labeled TauΔ5des gene fragment as a probe. As a result, each hybridized band with a different size was detected in the genome DNA of three different mhΔ5neor transformants after digestion with BglII (Fig. 1b), indicating one copy of the linear DNA cassette harboring the TauΔ5des gene was inserted into the mhΔ5neor genome at random positions.
Fig. 1.
RT-PCR of the Neor and TauΔ5des genes (a) and Southern blotting of the TauΔ5des gene (b) expressed in A. limacinum mh0186. (a) Total RNA, extracted from mhneor and mhΔ5neor, was transcribed to cDNA and subjected to RT-PCR. The transcripts were detected using specific primers for the Neor and TauΔ5des genes (n = 3). The left lane shows the no-template control. (b) Genomic DNAs of three different mhΔ5neor transformants and mhneor, digested with the restriction enzyme BglII, were subjected to Southern blotting using the DIG-labeled TauΔ5des gene as a probe. Details are described in Materials and Methods.
Conversion of fatty acids in mhΔ5neor.
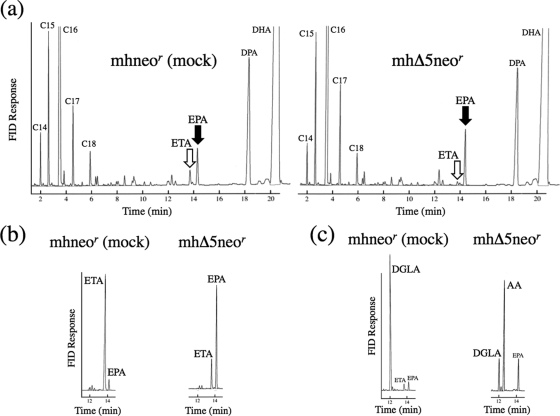
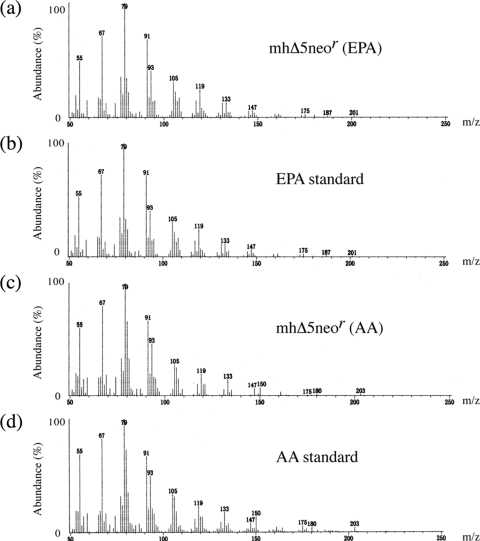
To examine whether the Δ5 desaturase in thraustochytrids was functional, the fatty acid composition of mhΔ5neor was compared with that of mhneor. Total fatty acids were extracted from transgenic thraustochytrids, and their FAME derivatives were analyzed by GC. In mhneor, ETA was present but DGLA was hardly detected (Table 2). Both acted as the substrates for TauΔ5des when expressed in the yeasts (Table 1). In mhΔ5neor, ETA almost disappeared, and simultaneously, EPA increased by 1.4-fold compared to that in mhneor, indicating that endogenous ETA was converted to EPA by TauΔ5des expressed in mhΔ5neor (Fig. 2a and Table 2). However, the increase in AA was not observed in the mhΔ5neor cells, possibly because endogenous DGLA was hardly detected (Table 2). Interestingly, EPA and AA levels were increased by 4.6- and 13.2-fold in mhΔ5neor compared to mhneor when ETA and DGLA were added at 0.1 mM (Fig. 2b and c and Table 2). In mhΔ5neor, the rates of conversion of ETA to EPA and of DGLA to AA were calculated to be 75.2% and 66.4%, respectively, while almost no conversion of these substrates was observed in mhneor under the conditions used (Table 2). The GC-MS spectra of EPA and AA from mhΔ5neor were identical to those of corresponding authentic standards (Fig. 3). These results indicate that TauΔ5des was functionally expressed in A. limacinum mh0186 and converted ETA and DGLA to ETA and AA, respectively.
Table 2.
Fatty acid compositions in mhneor and mhΔ5neor,a
| Fatty acid | Proportion (%) in culture of: |
|||||
|---|---|---|---|---|---|---|
| mhneor | mhΔ5neor | mhneor + ETA | mhΔ5neor + ETA | mhneor + DGLA | mhΔ5neor + DGLA | |
| C14:0 | 2.23 ± 0.05 | 2.32 ± 0.03 | 2.26 ± 0.10 | 2.43 ± 0.07 | 2.22 ± 0.06 | 2.28 ± 0.16 |
| C15:0 | 2.43 ± 0.62 | 2.97 ± 0.96 | 2.48 ± 0.64 | 3.04 ± 0.91 | 2.53 ± 0.63 | 2.96 ± 0.79 |
| C16:0 | 55.2 ± 1.83 | 52.1 ± 3.15 | 54.6 ± 1.56 | 51.8 ± 3.56 | 53.5 ± 2.36 | 52.0 ± 3.41 |
| C17:0 | 0.97 ± 0.22 | 1.19 ± 0.42 | 0.96 ± 0.23 | 1.17 ± 0.41 | 0.99 ± 0.21 | 1.19 ± 0.41 |
| C18:0 | 1.54 ± 0.03 | 1.39 ± 0.13 | 1.56 ± 0.02 | 1.4 ± 0.13 | 1.56 ± 0.03 | 1.42 ± 0.13 |
| DGLA | — | — | — | — | 3.92 ± 0.21 | 1.09 ± 0.7 |
| AA | 0.18 ± 0.04 | 0.21 ± 0.02 | 0.15 ± 0.02 | 0.22 ± 0.02 | 0.14 ± 0.01 | 1.85 ± 0.24 |
| ETA | 0.32 ± 0.02 | 0.04 ± 0.04 | 3.27 ± 0.44 | 0.94 ± 0.5 | 0.39 ± 0.04 | 0.08 ± 0.05 |
| EPA | 0.65 ± 0.04 | 0.94 ± 0.13 | 0.62 ± 0.03 | 2.85 ± 0.35 | 0.60 ± 0.04 | 1.15 ± 0.29 |
| DPA | 5.17 ± 0.05 | 5.61 ± 1.00 | 4.92 ± 0.06 | 5.35 ± 0.97 | 4.92 ± 0.11 | 5.44 ± 0.89 |
| DHA | 31.3 ± 0.93 | 33.2 ± 2.44 | 29.2 ± 0.53 | 30.8 ± 2.52 | 29.3 ± 1.32 | 30.5 ± 1.94 |
mhneor and mhΔ5neor cells were cultured at 25°C for 1 day in PD medium supplemented with 0.1 mM ETA, 0.1 mM DGLA, or neither. Total fatty acids were extracted, and FAMEs were analyzed by GC. The values, means of triplicate determinations with SD, represent percentages. —, <0.03%.
Fig. 2.
Increase of EPA in mhΔ5neor. (a) GC analysis of FAMEs from mhneor and mhΔ5neor. Transgenic mh0186 cells were cultured in GY medium at 25°C for 2 days without fatty acids. Total fatty acids were extracted, and FAMEs were analyzed by GC-MS. White and black arrows indicate the methyl ester derivatives of ETA and EPA, respectively. (b) Conversion of EPA from ETA in mhneor (upper) or mhΔ5neor (lower). (c) Conversion of AA from DGLA in mhneor (upper) or mhΔ5neor (lower). Transgenic mh0186 cells were precultured in GY medium at 25°C for 2 days, harvested, and washed with 1.75% Sealife. The cells were further cultured at 25°C for 1 day in a PD medium supplemented with ETA (b) or DGLA (c) at a concentration of 0.1 mM. FID, flame ionization detector.
Fig. 3.
GC-MS of EPA and AA generated in mhΔ5neor. mhΔ5neor was precultured in a GY medium at 25°C for 2 days, harvested, and washed with 1.75% Sealife. The cells were further cultured at 25°C for 1 day in PD medium supplemented with 0.1 mM ETA or DGLA. Total fatty acids were extracted, and FAMEs were analyzed by GC-MS. (a) FAMEs from mhΔ5neor corresponding to the EPA methyl ester on GC. (b) EPA methyl ester standard. (c) FAMEs from mhΔ5neor corresponding to the AA methyl ester on GC. (d) AA methyl ester standard.
Conversion and accumulation of EPA in transgenic thraustochytrids and yeasts.
To compare the rates of conversion of EPA from ETA and accumulation of EPA in the transgenic thraustochytrids with those in the transgenic yeasts, increasing amounts of ETA were added to the cultures of mhΔ5neor and scΔ5ura. The conversion rate was much higher in the transgenic thraustochytrids than in the transgenic yeasts at various concentrations of ETA, i.e., it reached 60 to 90% in mhΔ5neor but remained below 25% in the scΔ5ura (Fig. 4a). Almost no conversion of ETA to EPA was observed in mock transformants of thraustochytrids and yeasts. Interestingly, the amount of EPA accumulated in mhΔ5neor increased in proportion with the amount of ETA added to the culture up to 0.4 mM, while that of EPA in scΔ5ura reached a plateau at 0.1 mM ETA (Fig. 4b). These results indicate the thraustochytrids to be superior to the yeasts as a transgenic host to accumulate EPA. The conversion and accumulation of EPA were still found to occur using transformants stored at −80°C for 1 year. As a result of semiquantitative RT-PCR, the transcript level of the TauΔ5des gene in yeasts was found to be higher than that in thraustochytrids (Fig. 4c).
Fig. 4.
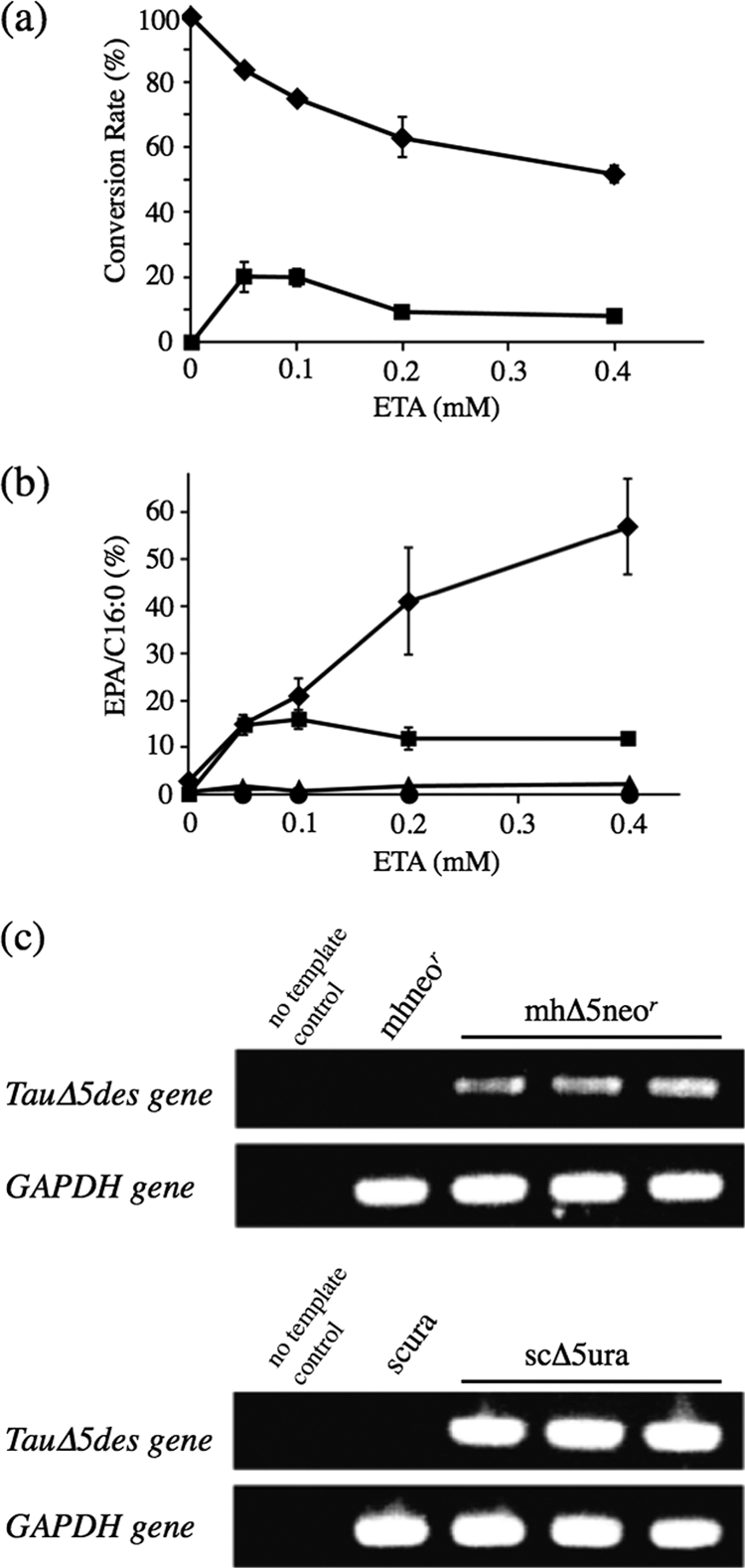
Comparison of the conversion and accumulation rates of EPA in mhΔ5neor and scΔ5ura. The mhΔ5neor and scΔ5ura cells were cultured at 25°C for 2 days in GY medium and Ura− medium containing 2% glucose, respectively. Cells were harvested and washed with 1.75% Sealife and distilled water, respectively. The transgenic thraustochytrid and yeast cells were further cultured at 25°C for 1 day in PD medium and Ura− medium containing 2% galactose, respectively. Both media were supplemented with ETA at a concentration of 0.05 to 0.4 mM, as indicated. Fatty acids were extracted, and their FAMEs were analyzed by GC. (a) Rates of conversion of EPA from ETA at the different concentrations of ETA. The values were calculated as follows: conversion rate (%) = [GC area for the product/(GC area for the product + GC area for the substrate)] × 100. ♦, mhΔ5neor; ■, scΔ5ura. (b) Rates of accumulation of EPA. The values were calculated as follows: accumulation rate = (GC area for EPA/GC area for palmitic acid, C16:0) × 100. Values are the means of triplicate determinations with SD. ♦, mhΔ5neor; ■, scΔ5ura; ▴, mhneor; ●, scura. (c) Determination of the transcription level of the TauΔ5des gene in thraustochytrid and yeast transformants by semiquantitative RT-PCR. The template concentration and the cycle number of PCR were adjusted by using respective thraustochytrid and yeast GAPDH genes as a standard. Details are described in Materials and Methods.
DISCUSSION
Our aim is to develop a new source of PUFAs, especially EPA, as a substitute for fish oils. We attempted here to increase the EPA levels by introducing a Δ5 desaturase gene into thraustochytrids, which possess DHA and n − 6 DPA but not EPA as major PUFAs. First, we isolated a Δ5 desaturase gene from T. aureum ATCC 34304 and confirmed using baker's yeast that the gene product catalyzed the introduction of a double bond at position 5 of both ETA and DGLA, producing EPA and AA, respectively. To our knowledge, there have been very few reports on the method for expression of target genes in thraustochytrids (14, 15). We thus first developed a new system for the expression of the target gene in thraustochytrids. We found that the promoter and terminator regions of housekeeping genes of T. aureum ATCC 34304, such as the ubiquitin and EF-1α genes, could drive expression of the target gene in the thraustochytrids. In the present study, we generated a linear DNA cassette composed of a Neor gene and a Δ5 desaturase gene, driven with the promoter/terminator regions of ubiquitin and EF-1α, respectively. As expected, this DNA cassette was shown to function efficiently in the thraustochytrids, resulting in the conversion to EPA of endogenous as well as exogenous ETA. It is worth noting, however, that no AA increased in the mhΔ5neor cells when DGLA was not added to the culture. This result may stem from the lack of sufficient endogenous substrate in the thraustochytrids, i.e., DGLA was hardly detected by GC in the A. limacinum mh0186 cells cultured under the conditions described.
Although ETA and DGLA were almost completely converted to EPA and AA, respectively, in the mhΔ5neor cells, these substrates remained intact in the mhneor cells (Fig. 2b and c and Table 2). This result may indicate that a Δ5 desaturase was not present in strain mh0186 or does not actually function under the conditions used in this study.
Comparison of the rates of conversion and accumulation of EPA in A. limacinum with those in S. cerevisiae revealed the thraustochytrids to be markedly superior. It is plausible that thraustochytrids have many more lipid droplets, in which PUFAs are accumulated mainly as triacylglycerol, than do yeasts (11, 17, 21). It was found that the transcription level of the TauΔ5des gene in yeasts was higher than that in thraustochytrids (Fig. 4c). However, the possibility that TauΔ5des mRNA was not efficiently transcribed to the protein in yeasts is not ruled out at present, because two rare codons (CGG coding for arginine) for yeasts were found in the TauΔ5des gene.
A. limacinum mh0186 was selected as a host for the expression of fatty acid Δ5 desaturase in this study because of the strain's rapid growth and good transformation efficiency. The growth and biomass of mhΔ5neor were found to be almost equal to those of mhneor under the conditions used in this study.
In conclusion, we have shown an increase of EPA in thraustochytrids transformed with the Δ5 desaturase gene driven by the ubiquitin promoter. This study could facilitate the molecular breeding of thraustochytrids enriched with specific PUFAs by expressing target genes involved in PUFA synthesis and open the door to an era of tailor-made production of PUFA in thraustochytrids.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Abe E., et al. 2006. A novel phosphatidylcholine which contains pentadecanoic acid at sn-1 and docosahexaenoic acid at sn-2 in Schizochytrium sp. F26-b. J. Biochem. 140:247–253 [DOI] [PubMed] [Google Scholar]
- 2. Biondo P. D., Brindley D. N., Sawyer M. B., Field C. J. 2008. The potential for treatment with dietary long-chain polyunsaturated n-3 fatty acids during chemotherapy. J. Nutr. Biochem. 19:787–796 [DOI] [PubMed] [Google Scholar]
- 3. Bongiorni L., Jain R., Raghukumar S., Aggarwal R. K. 2005. Thraustochytrium gaertnerium sp. nov.: a new thraustochytrid stramenopilan protist from mangroves of Goa, India. Protist 156:303–315 [DOI] [PubMed] [Google Scholar]
- 4. Burja A. M., Radianingtyas H., Windust A., Barrow C. J. 2006. Isolation and characterization of polyunsaturated fatty acid producing Thraustochytrium species: screening of strains and optimization of omega-3 production. Appl. Microbiol. Biotechnol. 72:1161–1169 [DOI] [PubMed] [Google Scholar]
- 5. Graham I. A., Larson T., Napier J. A. 2007. Rational metabolic engineering of transgenic plants for biosynthesis of omega-3 polyunsaturates. Curr. Opin. Biotechnol. 18:142–147 [DOI] [PubMed] [Google Scholar]
- 6. Hayashi M., Yukino T., Watanabe F., Miyamoto E., Nakano Y. 2007. Effect of vitamin B12-enriched thraustochytrids on the population growth of rotifers. Biosci. Biotechnol. Biochem. 71:222–225 [DOI] [PubMed] [Google Scholar]
- 7. Hoffmann M., et al. 2007. A small membrane-peripheral region close to the active center determines regioselectivity of membrane-bound fatty acid desaturases from Aspergillus nidulans. J. Biol. Chem. 282:26666–26674 [DOI] [PubMed] [Google Scholar]
- 8. Innis S. M. 2008. Dietary omega 3 fatty acids and the developing brain. Brain Res. 1237:35–43 [DOI] [PubMed] [Google Scholar]
- 9. Jeh E. J., Kumaran R. S., Hur B. 2008. Lipid body formation by Thraustochytrium aureum (ATCC 34304) in response to cell age. Korean J. Chem. Eng. 25:1103–1109 [Google Scholar]
- 10. Kang D. H., Anbu P., Kim W., Hur B. 2008. Coexpression of elo-like enzyme and Δ5, Δ4 desaturases derived from Thraustochytrium aureum ATCC 34304 and the production of DHA and DPA in Pichia pastoris. Biotechnol. Bioprocess Eng. 13:483–490 [Google Scholar]
- 11. Kohlwein S. D. 2010. Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285:15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumon Y., et al. 2008. Isolation and characterization of a Δ5 desaturase from Oblongichytrium sp. Biosci. Biotechnol. Biochem. 72:2224–2227 [DOI] [PubMed] [Google Scholar]
- 13. Lee J. H., O'Keefe J. H., Lavie C. J., Harris W. S. 2009. Omega-3 fatty acid: cardiovascular benefits, sources and sustainability. Nat. Rev. Cardiol. 6:753–758 [DOI] [PubMed] [Google Scholar]
- 14. Lippmeier J. C., et al. 2009. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids 44:621–630 [DOI] [PubMed] [Google Scholar]
- 15. Metz J. G., et al. 2009. Biochemical characterization of polyunsaturated fatty acid synthesis in Schizochytrium: release of the products as free fatty acids. Plant Physiol. Biochem. 47:472–478 [DOI] [PubMed] [Google Scholar]
- 16. Metz J. G., et al. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293 [DOI] [PubMed] [Google Scholar]
- 17. Morita E., Kumon Y., Nakahara T., Kagiwada S., Noguchi T. 2006. Docosahexaenoic acid production and lipid-body formation in Schizochytrium limacinum SR21. Mar. Biotechnol. 8:319–327 [DOI] [PubMed] [Google Scholar]
- 18. Okuyama H., Orikasa Y., Nishida T., Watanabe K., Morita N. 2007. Bacterial genes responsible for the biosynthesis of eicosapentaenoic and docosahexaenoic acids and their heterologous expression. Appl. Environ. Microbiol. 73:665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qiu X. 2003. Biosynthesis of docosahexaenoic acid (DHA, 22:6-4,7,10, 13,16,19): two distinct pathways. Prostaglandins Leukot. Essent. Fatty Acids 68:181–186 [DOI] [PubMed] [Google Scholar]
- 20. Qiu X., Hong H., MacKenzie S. L. 2001. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 276:31561–31566 [DOI] [PubMed] [Google Scholar]
- 21. Raghukumar S. 2008. Thraustochytrid marine protists: production of PUFAs and other emerging technologies. Mar. Biotechnol. 10:631–640 [DOI] [PubMed] [Google Scholar]
- 22. Spencer L., et al. 2009. The effect of omega-3 fatty acids on tumor angiogenesis and their therapeutic potential. Eur. J. Cancer 45:2077–2086 [DOI] [PubMed] [Google Scholar]
- 23. Taoka Y., et al. 2009. Influences of culture temperature on the growth, lipid content and fatty acid composition of Aurantiochytrium sp. strain mh0186. Mar. Biotechnol. 11:368–374 [DOI] [PubMed] [Google Scholar]
- 24. Thompson J. D., Higgins D. G., Gibson T. J. 1994. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venancio A., Domingues L., Lima N. 1999. Transformation of a flocculating Saccharomyces cerevisiae using lithium acetate and pYAC4. J. Basic Microbiol. 39:37–41 [PubMed] [Google Scholar]
- 26. von Schacky C., Harris W. S. 2007. Cardiovascular benefits of omega-3 fatty acids. Cardiovasc. Res. 73:310–315 [DOI] [PubMed] [Google Scholar]
- 27. Ward O. P., Singh A. 2005. Omega-3/6 fatty acids: alternative sources of production. Process Biochem. 40:3627–3652 [Google Scholar]
- 28. Warude D., Joshi K., Harsulkar A. 2006. Polyunsaturated fatty acids: biotechnology. Crit. Rev. Biotechnol. 26:83–93 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.