Abstract
Coprophilous fungi inhabit herbivore feces, secreting enzymes to degrade the most recalcitrant parts of plant biomass that have resisted the digestive process. Consequently, the secretomes of coprophilous fungi have high potential to contain novel and efficient plant cell wall degrading enzymes of biotechnological interest. We have used one-dimensional and two-dimensional gel electrophoresis, matrix-assisted laser desorption ionization–time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS/MS), and quadrupole time-of-flight liquid chromatography–tandem mass spectrometry (Q-TOF LC-MS/MS) to identify proteins from the secretome of the coprophilous fungus Doratomyces stemonitis C8 (EU551185) isolated from koala feces. As the genome of D. stemonitis has not been sequenced, cross-species identification, de novo sequencing, and zymography formed an integral part of the analysis. A broad range of enzymes involved in the degradation of cellulose, hemicellulose, pectin, lignin, and protein were revealed, dominated by cellobiohydrolase of the glycosyl hydrolase family 7 and endo-1,4-β-xylanase of the glycosyl hydrolase family 10. A high degree of specialization for pectin degradation in the D. stemonitis C8 secretome distinguishes it from the secretomes of some other saprophytic fungi, such as the industrially exploited T. reesei. In the first proteomic analysis of the secretome of a coprophilous fungus reported to date, the identified enzymes provide valuable insight into how coprophilous fungi subsist on herbivore feces, and these findings hold potential for increasing the efficiency of plant biomass degradation in industrial processes such as biofuel production in the future.
INTRODUCTION
Filamentous fungi exist in a broad range of habitats, fulfilling significant roles in a diversity of ecosystems. Integral to their survival is the ability to secrete enzymes to break down complex materials in the environment into small molecules that can be absorbed into the hyphae and used for nutrition (53). In the past decade, advances in protein identification techniques and genome sequencing have enabled detailed investigation of the secretomes of saprophytic (31, 42, 47, 50, 51), pathogenic (28, 35, 43), and symbiotic fungal species (29), revealing rich and diverse enzyme arrays. The fungal secretomes have been explored to find enzymes and enzyme combinations for various industrial applications, such as paper, textile, and food manufacture (7, 33) and economically and industrially sustainable hydrolysis of plant biomass to fermentable sugars for biofuel production (1, 13, 46, 51).
Coprophilous fungi are a subgroup of saprophytic fungi that can inhabit feces, most commonly herbivore feces (53). As the waste product of the digestive process, herbivore feces are predominantly composed of the most recalcitrant and indigestible parts of the plant: the cell wall polymers cellulose, hemicellulose, and lignin (21). Therefore, the potential for the secretomes of coprophilous fungi to contain novel enzymes for efficient plant cell wall degradation is high. Only one coprophilous fungus, Podospora anserina, has had its genome sequenced to date (10). An impressive array of genes encoding secreted glycosyl hydrolases was revealed, some of which have recently been used to supplement the secretome of the industrially exploited fungus Trichoderma reesei to improve the enzymatic hydrolysis of lignocellulosic biomass (5). To our knowledge, proteomic analysis of the secretome of a coprophilous fungus has not been reported and invites investigation.
Doratomyces stemonitis C8 is a coprophilous fungus isolated from koala feces (38). A koala's diet consists almost entirely of eucalyptus leaves, which are extremely tough and fibrous as a result of the harsh Australian climate and nutrient-poor soils (48). Eucalyptus leaves contain approximately 25% cellulose, 12% lignin, and 15% noncellulose carbohydrates, including hemicellulose and pectin, components that are poorly digested and concentrated in the koala's feces (32, 49) from which D. stemonitis C8 must obtain nutrition. The secretion of enzymes with endoglucanase, xylanase, mannanase, and protease activity by D. stemonitis C8 has been reported previously (40). However, the identity of the full array of enzymes within the D. stemonitis C8 secretome remained to be elucidated.
In the work presented here we have used gel electrophoresis, zymography, and mass spectrometry to identify enzymes within the secretome of D. stemonitis C8. The genome of D. stemonitis has not been sequenced, and consequently protein identification has been challenging, requiring cross-species identification and de novo sequencing (11, 23). The enzymes identified provide an insight into how the coprophilous fungus can subsist on koala feces and also could have potential for development for industrial applications in the future.
MATERIALS AND METHODS
Chemicals and components.
All materials were supplied by Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Fungal strain.
Doratomyces stemonitis C8 (EU551185) was isolated from koala feces in a previous study (38). The strain was maintained on potato dextrose agar (PDA) (Merck, Darmstadt, Germany) slants and stored at 4°C. Prior to liquid culturing, D. stemonitis C8 was inoculated to PDA plates and grown for 7 days at 28°C.
Liquid cultivation.
Four mycelial plugs, 0.5 cm in diameter, were cut from the PDA plates of D. stemonitis C8 and placed into a 250-ml flask with 50 ml of a hydrolase-inducing medium, pH 6.5, containing 2% (wt/vol) Avicel cellulose, 1.5% (wt/vol) soybean flour, 1% (wt/vol) lactose, and minimal salts (37). The culture was incubated on a shaker at 250 rpm for 7 days at 28°C (39, 40) and centrifuged at 4,000 × g for 15 min, and the supernatant was used for secretome analysis. Two additional cultures were grown subsequently in order to visually assess the reproducibility of protein patterns from the supernatants following electrophoresis.
Two-dimensional (2D) gel electrophoresis.
Proteins were precipitated by incubating the supernatant with 20% (wt/vol) trichloroacetic acid (TCA) for 14 h at 4°C. Following centrifugation at 6,000 × g for 15 min at 4°C, the pellet was resuspended in 20 ml cold acetone and incubated on ice for 30 min. Centrifugation was then repeated at 6,000 × g for 15 min at 4°C; the supernatant was removed, and the pellet was air dried prior to resuspension in a solubilization buffer consisting of 8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 1% (wt/vol) dithiothreitol (DTT). The sample was kept on ice and vortexed intermittently over a 2-h period. Desalting of the sample was then carried out in Microcon YM-3 (3,000-nominal-molecular-weight-limit [NMWL]) centrifugal filter devices (Millipore, Bedford, MA) using the same solubilization buffer until the conductivity was reduced to 0.2 mS/cm.
Immobilized pH gradient (IPG) strips (11 cm, pH 4 to 7; GE Healthcare, Sydney, Australia) were rehydrated with approximately 300 μg protein over 6 h and then subjected to isoelectric focusing on an IsoelectriQ focusing machine (Proteome Systems, Sydney, Australia) at 14°C using the following protocol: 300 V for 4 h, 300 to 10,000 V for 8 h, and 10,000 V for 8 h, with a total of approximately 80,000 voltage-hours (80 kVh). The IPG strips were placed in an equilibration solution containing 50 mM Tris HCl, pH 8.8, 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) sodium dodecyl sulfate (SDS), 1% (wt/vol) DTT, and 0.05% (wt/vol) bromophenol blue. Strips were cut in half (each 5.5 cm in length) and placed upon two hand-cast SDS-PAGE gels (12.5% [wt/vol] polyacrylamide) in a running buffer containing 1.4% (wt/vol) glycine, 0.1% (wt/vol) SDS, and 24 mM Tris. Electrophoresis was performed at 5 mA/gel for 20 min and then 12 mA/gel for 3 to 4 h. The gels were fixed in 10% (vol/vol) methanol, 7% (vol/vol) acetic acid for 30 min, stained in Coomassie brilliant blue G-250 (Bio-Rad, Sydney, Australia) for 4 h, and destained overnight in 1% (vol/vol) acetic acid.
One-dimensional (1D) SDS-PAGE.
Supernatant from a 7-day culture of D. stemonitis C8 (as described above) was subjected to 1D SDS-PAGE on a 1D NuPAGE Novex 4 to 12% Bis-Tris gel (Invitrogen, Thornton, Australia). To ensure enough protein for identification, three samples were prepared, each containing 20 μl of supernatant (approximately 8 μg of protein) and 5 μl of loading buffer (0.5 M Tris HCl, pH 6.8, 50% [vol/vol] glycerol, 10% [wt/vol] SDS, 5% [vol/vol] β-mercaptoethanol, and 0.05% [wt/vol] bromophenol blue). The samples were boiled for 3 min, centrifuged for 30 s, and cooled before being loaded onto three lanes of the gel. Electrophoresis was carried out at 100 V for 60 min using a 3-(N-morpholino)propanesulfonic acid (MOPS) running buffer containing 50 mM MOPS (Bio-Rad, Sydney, Australia), 50 mM Tris, 0.1% SDS, and 0.03% (wt/vol) EDTA. The gel was fixed and stained as described above.
1D zymograms.
In an attempt to directly equate protein identification with displayed enzyme activity in the D. stemonitis C8 supernatant, 1D endoglucanase, xylanase, mannanase, β-glucosidase, and protease zymograms were prepared as described by Peterson et al. (40), and protein bands exhibiting enzyme activity were excised and subjected to mass spectrometry using a technique similar to that described by Peterson et al. (39; see below).
2D mannanase zymogram.
To seek identification of the D. stemonitis C8 proteins with mannanase activity, a 2D mannanase zymogram was used. Protein precipitation, solubilization, desalting, isoelectric focusing, and 2D electrophoresis were carried out as described for the 2D gel above, except that the SDS-PAGE gels contained 0.05% (wt/vol) locust bean gum (galactomannan). Following electrophoresis, the gels were soaked in 2.5% (vol/vol) Triton X for 4 h, washed thoroughly in Milli-Q water, and incubated overnight at 30°C in 50 mM citrate buffer, pH 5.5. The gels were washed in Milli-Q water, stained in 1% (wt/vol) Congo red for 10 min, and destained in 1 M NaCl for 20 min to reveal protein spots with mannanase activity.
Identification of proteins from the 2D gel.
A total of 120 protein spots were cut from the 2D gel, destained with 50% acetonitrile, 50 mM ammonium bicarbonate, and incubated overnight at 37°C with 15 ng/μl trypsin. The samples were desalted and concentrated using C18 zip-tips (Eppendorf, Hamburg, Germany) and eluted onto an AnchorChip sample plate (Bruker Daltonics, Billerica, MA) with 1 μl of matrix (1 mg/ml α-cyano-4-hydroxy cinnamic acid in 90% [vol/vol] acetonitrile, 0.1% [vol/vol] trifluoroacetic acid). Matrix-assisted laser desorption ionization (MALDI)–mass spectrometry (MS) was performed using an Applied Biosystems (Foster City, CA) 4800 proteomics analyzer. An Nd:YAD laser (355 nm) was used to irradiate the sample, and spectra were acquired in reflectron mode in the mass range of 700 to 3,500 Da. The eight strongest peptides from each MS scan were isolated and fragmented by collision-induced dissociation (CID) and subjected to tandem time-of-flight mass analysis (TOF/TOF MS/MS). Peak lists were generated by DataExplorer (Applied Biosystems, Foster City, CA) and searched against fungal proteins in the NCBI nonredundant database (NCBInr 20100813) using the Mascot search engine (Matrix Sciences, London, United Kingdom). Protein identifications were assigned where the Mowse scores were significant (P < 0.05). Searches were also made against all proteins in NCBInr, and peptides matching known contaminants (e.g., trypsin, keratin, and collagen) were eliminated from the analysis. Candidate de novo sequences for all good-quality unassigned spectra were generated using Mascot Distiller (Matrix Sciences, London, United Kingdom) and manually BLAST searched (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against proteins in NCBInr and NCBInr (Fungi). Identifications were made where two or more de novo peptides matched to a fungal protein in the database (Expect < 0.001). Ten of the largest spots from a replicate 2D gel of D. stemonitis C8 supernatant were reanalyzed by Q-TOF LC-MS/MS using the procedure outlined below for one-dimensional (1D) gel bands.
Identification of proteins from the 1D SDS-PAGE gel.
Eight horizontal slices were cut across the three lanes of the 1D SDS-PAGE gel of D. stemonitis C8 supernatant. Each slice was chopped finely, destained, and digested with trypsin as described above. The peptides were extracted by sonication for 2 × 15 min with 0.1% (vol/vol) trifluoroacetic acid (TFA), followed by incubation with 50% (vol/vol) acetonitrile–0.1% (vol/vol) TFA for 5 min at room temperature (RT) and incubation with 100% acetonitrile for a further 5 min at RT. Each sample was then resuspended in 80 μl 0.1% (vol/vol) TFA. The resulting peptides were separated by nano-liquid chromatography (LC) using the Tempo nano-LC system (Applied Biosystems, Foster City, CA). Forty microliters of each sample was injected onto a Michrom peptide Captrap (Michrom Bioresources, Auburn, CA) for preconcentration and desalting with 0.1% (vol/vol) formic acid, 2% (vol/vol) acetonitrile at 8 μl/min. The peptide trap was then switched on line with the analytical column containing C18 RP silica (SGE ProteCol C18, 300 A, 3 μm, 150 μm × 10 cm; SGE, Ringwood, Australia). Peptides were eluted from the column using a linear solvent gradient, with steps, from H2O:CH3CN (90:10; plus 0.1% formic acid) to H2O:CH3CN (0:100; plus 0.1% formic acid) at 500 nl/min over a 50-min period. The LC eluent was subjected to positive ion nanoflow electrospray analysis on an Applied Biosystems (Foster City, CA) QSTAR Elite mass spectrometer. From each TOF-MS survey scan (m/z 400 to 1,600, 0.5 s), the three largest multiply charged ions (counts > 25) were sequentially subjected to MS/MS analysis. Peak lists were generated using Mascot Distiller (Matrix Sciences, London, United Kingdom). Mascot searches against the NCBInr database and de novo sequencing of all good-quality unassigned spectra were conducted as outlined above.
Identification of proteins from the 2D gel.
Bands and spots were excised from the zymogram gels, destained, reduced and alkylated, and digested with trypsin, as described by Peterson et al. (39), and then subjected to Q-TOF LC-MS/MS and spectral analysis for protein identification as described for the 1D SDS-PAGE gel (above).
RESULTS
2D gel electrophoresis of secreted proteins.
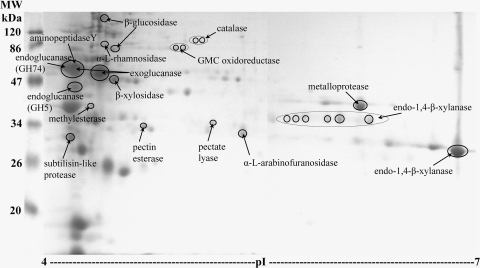
Proteins in the D. stemonitis C8 culture supernatant were separated by 2D gel electrophoresis (Fig. 1). Preliminary studies using pH 3 to 10 IPG strips revealed that all proteins that were visible on 2D gels when stained with Coomassie blue had pIs between pH 4 and pH 7, so pH 4 to 7 IPG strips were used to produce gels for the MS analysis described here. Five visually similar 2D gel maps were produced from the three separate cultures of D. stemonitis C8. Over 150 distinct spots were visible on each gel following staining with Coomassie blue, and most of the secreted protein had a pI of 4 to 5 (Fig. 1).
Fig. 1.
2D gel produced from the electrophoresis of protein (approximately 300 μg) from the supernatant of D. stemonitis C8 following growth in a hydrolase-inducing liquid medium for 7 days. Proteins were visualized on a 12.5% SDS-PAGE gel stained with Coomassie blue and identified by MALDI-TOF/TOF MS/MS or Q-TOF LC-MS/MS as indicated in Table 1. MW, molecular mass in kilodaltons. The subtilisin-like protease and α-l-arabinofuranosidase were identified by de novo sequencing, and peptide matches to the α-l-rhamnosidase and pectin esterase had Mascot scores below significance level (P < 0.05); however, all four identifications were confirmed subsequently by Q-TOF LC-MS/MS of protein bands from the 1D SDS-PAGE gel.
Protein identifications from the 2D gel.
Identifications assigned to proteins in the D. stemonitis C8 secretome from MALDI-TOF/TOF MS/MS and Q-TOF LC-MS/MS analysis of spots on the 2D gel are listed in Table 1, together with identifications of proteins from the 1D SDS-PAGE gel and 2D zymogram (see below). The proteins are listed according to their function, as predicted in the NCBI database (www.ncbi.nlm.nih.gov/), the family group to which the protein belongs, based on amino acid sequence similarity determined by the CAZy (www.cazy.org/) or MEROPS (www.merops.ac.uk/) database, and the substrates upon which they act. Peptide sequences are provided in Tables S1 to S4 in the supplemental material.
Table 1.
Assignment of proteins in the secretome of Doratomyces stemonitis C8 to fungal proteins in the NCBI databasea
| Enzyme category, protein name/functionb | Target substrate | Species | Accession no. | Predicted SigPc | No. of peptide matches (no. of unique peptides)d |
Total no. of peptide matches (no. of unique matches)e | |||
|---|---|---|---|---|---|---|---|---|---|
| 2D gel |
1D gel LC | 2D zym LC | |||||||
| MALDI | LC | ||||||||
| Glycosyl hydrolases (GH) | |||||||||
| Cellobiohydrolase I (GH7) | Cellulose | Magnaporthe oryzae | gi|39971383 | Y | 9 (1) | 9 (1) | 18 (1) | ||
| Cellobiohydrolase I (GH7) | Cellulose | Humicola grisea | gi|4204214 | Y | 4 (1) | 5 (1) | 8 (1) | ||
| Cellobiohydrolase I (GH7) | Cellulose | Podospora anserina | gi|171676762 | Y | 2 (1) | 2 (1) | |||
| Endoglucanase I (GH5) | Cellulose | Robillarda sp. | gi|6855474 | Y | 7 (2) | 2 (1) | 9 (2) | ||
| β-Glucosidase (GH3) | Cellulose | Coprinopsis cinerea okayama | gi|299755823 | Y | 3 (2) | 3 (2) | |||
| β-Glucosidase (GH3) | Cellulose | Botryotinia fuckeliana | gi|154302511 | N | 4 (1) | 4 (1) | |||
| β-Glucosidase (GH3) | Cellulose | Ajellomyces capsulatus | gi|671684 | Y | 2 (1) | 2 (1) | |||
| β-Glucosidase (GH3) | Cellulose | Magnaporthe oryzae | gi|39966399 | Y | 1 (1) | 1 (1) | |||
| Endoglucanase (GH74) | Xyloglucan | Hypocrea jecorina | gi|31747160 | Y | 1 (1) | 2 (2) | 3 (2) | ||
| Endo-1,4-β-xylanase (GH10) | Xylan | Chaetomium globosum | gi|116179352 | Y | 6 (2) | 1 (1) | 53 (4) | 60 (4) | |
| Endo-1,4-β-xylanase (GH10) | Xylan | Aspergillus oryzae | gi|74582795 | Y | 1 (1) | 1 (1) | |||
| β-Xylosidase (GH43) | Xylan | Aspergillus fumigatus | gi|70982855 | Y | 1 (1) | 1 (1) | 2 (1) | ||
| α-l-Arabinofuranosidase (GH62) | Xylan | Hypocrea jecorina | gi|31747156 | Y | 2 (2)f | 2 (1) | 4 (3) | ||
| α-l-Arabinofuranosidase (GH43) | Xylan | Magnaporthe oryzae | gi|39940636 | Y | 1 (1) | 1 (1) | |||
| Endo-1,4-β-mannanase (GH5) | Mannan | Magnaporthe oryzae | gi|145606609 | Y | 1 (1) | 29 (1) | 30 (2) | ||
| Endo-1,4-β-mannanase (GH5) | Mannan | Podospora anserina | gi|171677227 | Y | 11 (1) | 11 (1) | |||
| Endo-1,4-β-mannanase (GH5) | Mannan | Chaetomium globosum | gi|116205649 | Y | 2 (1) | 2 (1) | |||
| α-l-Rhamnosidase (GH78) | Pectin | Podospora anserina | gi|171694950 | Y | 1 (1)g | 3 (2) | 4 (2) | ||
| Polysaccharide lyases (PL), carbohydrate esterases (CE) | |||||||||
| Pectate lyase B (PL1) | Pectin | Verticillium albo-atrum | gi|261353894 | Y | 2 (2) | 2 (2) | |||
| Rhamnogalacturonan lyase (PL4) | Pectin | Pyrenophora tritici-repentis | gi|189193225 | N | 4 (1) | 4 (1) | |||
| Pectin esterase (CE8) | Pectin | Podospora anserina | gi|171682268 | Y | 1 (1)g | 2 (1) | 3 (1) | ||
| 4-O-Methyl-glucuronoyl-methylesterase (CE15) | Xylan/lignin | Schizophyllum commune | gi|300099970 | Y | 1 (1) | 1 (1) | |||
| Oxidoreductases | |||||||||
| GMC oxidoreductase | Lignin/cellulose | Gibberella zeae | gi|46115568 | Y | 3 (2) | 3 (2) | |||
| GMC oxidoreductase | Lignin/cellulose | Podospora anserina | gi|171683764 | Y | 2 (1) | 3 (1) | 5 (1) | ||
| GMC oxidoreductase | Lignin/cellulose | Podospora anserina | gi|171691498 | Y | 2 (1) | 2 (1) | |||
| GMC oxidoreductase | Lignin/cellulose | Moniliophthora perniciosa | gi|238607896 | Y | 2 (1) | 2 (1) | |||
| Catalase | Hydrogen peroxide | Aspergillus clavatus | gi|121712100 | Y | 2 (2) | 2 (2) | |||
| Catalase B | Hydrogen peroxide | Paracoccidioides brasiliensis | gi|225683111 | Y | 1 (1) | 1 (1) | |||
| Isoamyl alcohol oxidase | Unknown | Gibberella zeae | gi|82779927 | Y | 1 (1) | 1 (1) | |||
| FAD/FMN-containing dehydrogenase | Unknown | Penicillium chrysogenum | gi|211589700 | Y | 1 (1) | 1 (1) | |||
| Proteases | |||||||||
| Metalloprotease (M36) | Protein | Microsporum canis | gi|238845207 | Y | 6 (3) | 4 (2) | 10 (3) | ||
| Subtilisin protease; proteinase K-like (S8) | Protein | Neotyphodium lolii | gi|170674493 | Y | 2 (2)f | 1 (1) | 3 (3) | ||
| Aminopeptidase Y; PA_ScAPY-like | Protein | Phaeosphaeria nodorum | gi|169611336 | Y | 1 (1) | 3 (1) | 4 (1) | ||
| Aminopeptidase Y; PA_ScAPY-like | Protein | Podospora anserina | gi|171677143 | Y | 1 (1) | 1 (1) | |||
| Miscellaneous | |||||||||
| Glyceraldehyde-3-phosphate dehydrogenase | Paracoccidioides brasiliensis | gi|30580398 | N | 3 (2) | 3 (2) | ||||
| Major facilitator superfamily | Gibberella zeae | gi|46121949 | N | 1 (1) | 1 (1) | ||||
| Histidine phosphatase | Magnaporthe oryzae | gi|145608592 | N | 1 (1) | 1 (1) | ||||
| Acetate kinase | Pyrenophora tritici-repentis | gi|189206552 | N | 1 (1) | 1 (1) | ||||
| Nuclear pore complex subunit | Candida glabrata | gi|50288161 | N | 1 (1) | 1 (1) | ||||
| Hypothetical protein | Neurospora crassa | gi|164426947 | Y | 1 (1) | 1 (1) | ||||
| Hypothetical protein | Coccidioides immitis | gi|119187541 | N | 1 (1) | 1 (1) | ||||
| Hypothetical protein | Laccaria bicolor | gi|170112093 | N | 1 (1) | 1 (1) | ||||
| Hypothetical protein | Gibberella zeae | gi|46132978 | N | 1 (1) | 1 (1) | ||||
| Hypothetical protein | Yarrowia lipolytica | gi|50556336 | N | 1 (1) | 1 (1) | ||||
Assignment of proteins to those in the NCBI database (www.ncbi.nlm.nih.gov/) was determined by MALDI-TOF/TOF MS/MS and/or Q-TOF LC-MS/MS of protein spots or bands of the 2D gel, 1D gel, or 2D mannanase zymogram. Proteins are grouped according to their predicted enzymatic function as defined by NCBI (www.ncbi.nlm.nih.gov/), the protein families to which they belong based on amino acid sequence similarities defined by the CAZy database (www.cazy.org/) and Merops database (http://merops.sanger.ac.uk), and the substrates upon which they act.
Names or predicted functions of conserved domains in proteins in the NCBInr database (www.ncbi.nlm.nih.gov/) to which significant peptide matches (P < 0.05) were made using the Mascot search engine.
Signal peptide prediction by SignalP (www.cbs.dtu.dk/services/SignalP/): Y, yes; N, no.
Mass spectrometry method: MALDI-TOF/TOF MS/MS (MALDI) for proteins from 2D gel, Q-TOF LC-MS/MS (LC) for proteins from 2D gel, 1D gel, and 2D zymogram (2D zym).
Total number of peptide matches to the identified protein, derived using the methods and gels indicated in adjacent columns.
Identification made from MALDI-TOF/TOF MS/MS spectra by de novo sequencing.
Mascot score was below significance level (P > 0.05) for the match between the peptide derived from the MALDI-TOF/TOF MS/MS data and the protein in the NCBI database.
From the 120 spots excised from the 2D gel (Fig. 1), seven proteins were identified using MALDI-TOF/TOF MS/MS and Mascot searches against the NCBI database: glycosyl hydrolase family 10 (GH10) endo-1,4-β-xylanase, GH3 β-glucosidase, GH43 β-xylosidase, polysaccharide lyase family 1 (PL1) pectate lyase, carbohydrate esterase family 15 (CE15) 4-O-methyl-glucuronoyl-methylesterase, catalase, and glucose/methanol/choline (GMC) oxidoreductase (Fig. 1; Table 1, 2D gel MALDI). A further two protein identifications were made by de novo sequencing: a GH62 α-l-arabinofuranosidase and an S8 subtilisin-like protease (Fig. 1; Table 1). An additional two D. stemonitis C8 proteins yielded single peptide matches to proteins in the NCBI database with Mascot scores below significance level, but significant matches to the same proteins were later obtained using Q-TOF LC-MS/MS of the 1D gel (α-l-rhamnosidase and pectin esterase; Fig. 1; Table 1). Single de novo peptides that matched GH7 cellobiohydrolases, GH74 endoglucanases, pectin esterases, and GH5 endo-1,4-β-mannanases in the NCBI database (Expect < 0.001) were generated from other protein spots (see Table S1 in the supplemental material).
Additional protein identifications could be made when 10 of the large spots from a replicate 2D gel were subjected to Q-TOF LC-MS/MS (Table 1, 2D gel LC). GH7 cellobiohydrolase and GH10 endo-1,4-β-xylanase were identified from the largest protein spots (Fig. 1, Table 1). Other proteins identified included GH5 endoglucanase, GH74 endoglucanase, M36 metalloprotease, and aminopeptidase Y (Fig. 1; Table 1) De novo sequencing of Q-TOF LC-MS/MS data from protein spots at approximately 38 to 40 kDa, pI 4.2 to 4.3; 27 kDa, pI 4.3; 25 kDa, pI 4.5; and 24 kDa, pI 4.6, on the 2D gel (see Fig. S1 and Table S2 in the supplemental material; spots 25, 8, 4, and 5) revealed numerous peptides bearing similarity to GH6 cellobiohydrolase (cellobiohydrolase II), Cip1, GH45 endoglucanase, and GH11 endo-1,4-beta-xylanase, respectively, but no peptide matches could be made above significance level. Most of the assigned identifications were to enzymes in the NCBI database whose molecular masses and pIs were similar to those of the protein spots from which they were identified on the 2D gel (see Tables S1 to S4 in the supplemental material).
Protein identifications from the 1D SDS-PAGE gel.
More proteins in the D. stemonitis C8 supernatant were identified by Q-TOF LC-MS/MS analysis of protein bands from the 1D SDS-PAGE gel than had been identified from protein spots on the 2D gel. The collective total of significant peptide matches made from protein in the eight bands of the 1D SDS-PAGE gel to proteins in the NCBI database is shown in Table 1 (1D gel LC). The peptide sequences and the bands from which they were derived are provided in Table S3 in the supplemental material.
Most of the proteins that had been identified in the D. stemonitis C8 secretome from spots on the 2D gel (Fig. 1; Table 1, 2D gel MALDI) were also identified by Q-TOF LC-MS/MS of bands of a similar molecular mass on the 1D gel (Table 1, 1D gel LC; see Table S3 in the supplemental material). Peptides assigned to the GH10 endo-1,4-β-xylanase (gi|116179352) dominated the spectra, with a total of 53 matches made to four unique peptides (Table 1, 1D gel LC). Next in abundance were peptides assigned to GH7 cellobiohydrolases (16 hits to three unique peptides from three different proteins).
Protein identifications achieved from the 1D SDS-PAGE gel by Q-TOF LC-MS/MS that had not been identified from the 2D gel included GH43 α-l-arabinofuranosidase, GH5 endo-1,4-β-mannanase, PL4 rhamnogalacturonan lyase, isoamyl alcohol oxidase, and FAD/FMN-containing dehydrogenase (Table 1, 1D gel LC). The molecular mass of the SDS-PAGE gel band from which the D. stemonitis C8 proteins were derived correlated well with the molecular masses of the proteins to which they were assigned in the NCBI database (see Table S3 in the supplemental material). De novo sequencing of unassigned spectra resulted in an additional 35 single peptide matches (Expect < 0.001) to proteins in the NCBI database that had been identified by the Mascot search or had the same predicted function. A further 74 de novo sequences could be tentatively assigned to glycosyl hydrolases, polysaccharide lyases, and proteases in the NCBI database with sequence similarities below significance level (Expect > 0.001) (see Table S3 in the supplemental material).
With the exception of one β-glucosidase and the rhamnogalacturonan lyase (Table 1), all of the above-described identification assignments were to proteins possessing signal peptides and predicted to be secreted by SignalP (www.cbs.dtu.dk/services/SignalP/). Five hypothetical proteins of unknown function were also identified, one of which was predicted to be secreted, and a further five identifications were made to proteins that are intracellular in other fungal species, suggesting that a small amount of cell lysis may have occurred in culturing or centrifugation.
1D zymograms.
Many of the protein bands exhibiting enzyme activity on the xylanase, endoglucanase, β-glucosidase, and protease zymograms (Fig. 2) were of a molecular mass similar to those of protein spots from the 2D gel (Fig. 1) from which enzymes of appropriate activity had been identified. The approximately 30-kDa band on the xylanase zymogram (Fig. 2, lane Xy) aligned with the GH10 endo-1,4-β-xylanase identified at approximately 30 kDa, pI 7, on the 2D gel (Fig. 1). The faint bands at approximately 40 kDa and 80 kDa on the endoglucanase zymogram (Fig. 2, lane En) aligned roughly to protein identified on the 2D gel (Fig. 1) as a GH5 endoglucanase (approximately 42 kDa, pI 4.2) and a GH74 endoglucanase (approximately 50 to 75 kDa, pI 4.2). Similarly, the high-molecular-mass band (>120 kDa) on the β-glucosidase zymogram (Fig. 2, lane βg) corresponded to a β-glucosidase identified at >120 kDa on the 2D gel (Fig. 1). No endoglucanases were identified from protein of <38 kDa using the Mascot search engine to account for the large band at approximately 20 kDa on the endoglucanase zymogram. However, de novo sequencing of Q-TOF LC-MS/MS data from a protein spot at approximately 25 kDa, pI 5, on the 2D gel (see Fig. S1 in the supplemental material; spot 4), revealed numerous peptides bearing similarity to GH45 endoglucanases in the NCBI database (see Table S2 in the supplemental material; spot 4), although matches were below significance level. No β-glucosidases were identified from protein of 30 to 50 kDa to account for the band on the β-glucosidase zymogram at approximately 40 kDa (Fig. 2, lane βg). The approximately 26-kDa band on the protease zymogram (Fig. 2, lane Pr) roughly aligned to the serine protease at approximately 30 kDa, pI 4.2, on the 2D gel, and the large 34- to 45-kDa band on the protease zymogram to the metalloprotease at approximately 42 kDa, pI 6, on the 2D gel (Fig. 1). Each of the above proteins that had been identified from the 2D gel was also identified from bands of approximately the same molecular mass from the 1D SDS-PAGE gel (see Table S3 in the supplemental material). The 33- to 40-kDa bands on the mannanase zymogram (Fig. 2, lane Mn) corresponded to the GH5 endo-1,4-β-mannanases (Table 1) that had been identified from bands of similar molecular mass on the 1D SDS-PAGE gel (see Table S3 in the supplemental material), but no mannanases were identified from the 2D gel.
Fig. 2.
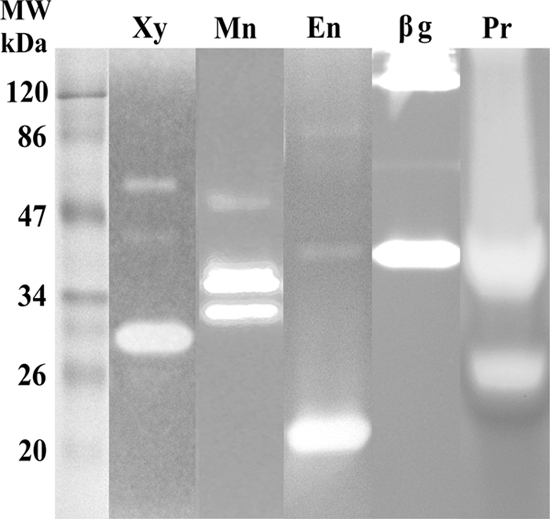
1D zymograms displaying xylanase (lane Xy), mannanase (lane Mn), endoglucanase (lane En), β-glucosidase (lane βg), and protease (lane Pr) activity of proteins in the supernatant of D. stemonitis C8. A 30-μl sample of supernatant (approximately 12 μg of protein) was loaded per lane. MW, molecular mass in kilodaltons.
Protein identifications from the 1D zymograms.
Protein identifications could be made from bands excised from the 1D xylanase, mannanase, and endoglucanase zymograms (Fig. 2) using Q-TOF MS/MS. The substrates and stains used in these zymogram gels did not appear to affect the MS analysis. However, in most instances, protein identifications from the zymogram bands did not match with enzymes with the expected activity. Instead, identifications were the same as those of the most abundant proteins of similar molecular masses, specifically the GH10 xylanase (gi|116179352) and GH7 cellobiohydrolase (gi|39971383, gi|4204214), which had already been identified from the 1D SDS-PAGE and 2D gel (Table 1; Fig. 1). No fungal proteins could be identified from the protease zymogram because the substrate casein dominated the MS/MS spectra. Identification of fungal proteins from the β-glucosidase zymogram bands was also not possible, although trypsin could be identified from the samples, indicating that the MS/MS process was still able to function correctly in the presence of the β-glucosidase substrate 4-MU-β-d-glucopyranoside that was contained in the gel (40).
2D mannanase zymogram.
The 2D mannanase zymogram produced from the supernatant of D. stemonitis C8 is shown in Fig. 3. Mannanase activity was apparent from protein at approximately 35 to 38 kDa, pI 4.3 to 4.5 (Fig. 3, spot 2), and from a slightly smaller spot at approximately 34 kDa, pI 4.5 (Fig. 3, spot 3). A chain of smaller spots indicating mannanase activity was visible at approximately 47 kDa and pI 4.5 to 5 (Fig. 3, spot 1). The molecular mass of the proteins displaying enzyme activity on the 2D mannanase zymogram correlated well with bands on the 1D mannanase zymogram (Fig. 2, lane Mn).
Fig. 3.

Mannanase zymogram (pI 4 to 5.5) produced after 2D gel electrophoresis of protein (approximately 300 μg) from the supernatant of D. stemonitis C8. Spots on the zymogram reveal proteins with mannanase activity. No mannanase activity was detected above pI 5.5. Protein in spots 1 and 2 could be assigned to endo-1,4-β-mannanases in the NCBInr database by Q-TOF LC-MS/MS analysis (Table 1). Spot 3 could not be identified. MW, molecular mass in kilodaltons.
Protein identifications from the 2D mannanase zymogram.
Separating proteins in the D. stemonitis C8 supernatant by isoelectric point as well as by molecular mass was effective in isolating the proteins with mannanase activity from other abundant proteins occurring at the same molecular mass. Consequently, confident identification of two proteins with mannanase activity could be made using Q-TOF LC-MS/MS: 29 peptides from protein forming the chain of small spots (Fig. 3, spot 1) matched to an endo-1,4-β-mannanase from Magnaporthe oryzae (gi|145606609), and 11 peptides from the large protein spot (Fig. 3, spot 2) matched to an endo-1,4-β-mannanase from the coprophilous fungus Podospora anserina (gi|171677227) (Table 1, 2D zym LC; see Table S4 in the supplemental material). The Magnaporthe oryzae endo-1,4-β-mannanase had been identified by a single peptide match from the 1D gel (Table 1, 1D gel LC; see Table S3 in the supplemental material) with a confidence score (Mowse score) of 48, but identification from the 2D zymogram could be made with a confidence score of 394 (see Table S4 in the supplemental material). Identification of the Podospora anserina endo-1,4-β-mannanase was achieved only from the 2D mannanase zymogram (Table 1, 2D zym LC; see Table S4 in the supplemental material). The protein in the other spot exhibiting mannanase activity on the 2D zymogram (Fig. 3, spot 3) could not be identified.
DISCUSSION
The coprophilous fungus Doratomyces stemonitis C8, isolated from koala feces (38), was grown in a hydrolase-inducing liquid medium, and secreted proteins were identified using gel electrophoresis and mass spectrometry. The genome sequence of D. stemonitis was not available, so cross-species identification, de novo sequencing, and zymography assisted the analysis. Most identified proteins within the D. stemonitis C8 secretome (Table 1) were enzymes involved in the degradation of the plant cell wall polymers cellulose, hemicellulose, pectin, and lignin. A diverse set of proteases were also identified, as well as some proteins of unknown function.
Cellulose is the largest single component of eucalyptus leaves and is partially broken down by microbial degradation in a koala's hindgut (48). However, the bulk of the cellulose fibers are released in the koala's feces (16). Consequently, it is not surprising that the D. stemonitis C8 secretome contains the three main classes of enzymes involved in cellulose degradation that have been commonly identified in the secretomes of other saprophytic fungi (13, 31, 42, 47, 50, 51): cellobiohydrolases that attack the ends of glucose chains forming the cellulose fibers to release glucose dimers (cellobiose), endoglucanases that cleave within the chains to expose more sites of attack for the cellobiohydrolases, and β-glucosidases that break down cellobiose to release glucose (26).
The most abundant of the cellulose-degrading enzymes in the D. stemonitis C8 secretome was cellobiohydrolase from the glycosyl hydrolase (GH) family 7 (www.cazy.org/), identified from two of the largest spots on the 2D gel (Fig. 1; Table 1). Also known as cellobiohydrolase I, GH7 cellobiohydrolases cleave cellobiose molecules progressively from the reducing ends of cellulose chains (54). In similarity to the secretome of D. stemonitis C8, cellobiohydrolase I forms a large proportion of the secretome of the industrially exploited fungus Trichoderma reesei, in which cellobiohydrolase I accounts for up to 60% of the secreted protein (30).
Two endoglucanases were identified in the D. stemonitis C8 secretome, one from GH family 5 and the other from GH family 74 (Table 1). Whereas GH5 endoglucanases typically cleave within cellulose chains, GH74 endoglucanases tend to be specific for xyloglucan (12). Some endoglucanase activity was shown by faint bands on the 1D endoglucanase zymogram (approximately 40 kDa and 80 kDa; Fig. 2, lane En) at the approximate molecular masses of the identified endoglucanases on the 2D gel (Fig. 1). However, the enzyme responsible for the prominent band at approximately 20 kDa on the endoglucanase zymogram (Fig. 2, lane En) could only be tentatively identified as a GH45 endoglucanase based on de novo sequencing of Q-TOF LC-MS/MS data from a protein spot at approximately 25 kDa, pI 5, on the 2D gel (Fig. 1; see Fig. S1 and Table S2, spot 4, in the supplemental material). As GH45 endoglucanases are typically of a low molecular mass, bearing an unusually small catalytic domain of approximately 20 kDa (17), the identification seems quite plausible and warrants further investigation. GH45 endoglucanases are highly sought after in the textile, detergent (44), and biofuel (14) industries due to their dispersing effect on solid celluloses, increasing the overall activity in a cellulolytic enzyme array.
To complete the breakdown of cellulose to glucose, the D. stemonitis C8 secretome contained protein bearing homology to four different GH3 β-glucosidases in the NCBI database. Multiple assignments to enzymes of the same function could be an indication that single D. stemonitis C8 enzymes bear similarity to different proteins in the NCBI database at different parts of their amino acid sequences. Alternatively, there could be redundancy in enzyme type within the secretome, a common phenomenon in filamentous fungi that may increase metabolic flexibility and chances of survival under different environmental conditions (2). Evidence for the activity of the high-molecular-mass β-glucosidase identified from the 2D gel (Fig. 1) was provided by a band on the β-glucosidase zymogram above 120 kDa (Fig. 2, lane βg).
Xylan is a poorly digested hemicellulose that is particularly abundant in the leaves of dicotyledons such as eucalyptus (20) and requires the action of numerous enzymes for its complete degradation: endo-1,4-β-xylanases to cleave within the xylose sugar backbone, β-xylosidases to degrade xylose oligosaccharides, and several accessory enzymes to remove sugar, acid, or ether containing side chains (41). A GH10 endo-1,4-β-xylanase was the most abundant protein identified in the secretome of D. stemonitis C8, with the greatest number of all peptide assignments from protein in the 1D and 2D gels (Table 1), and its activity was displayed on the xylanase zymogram (Fig. 2, lane Xy). Xylanases from GH family 10 are known to have broad substrate specificity and can also degrade short-chain cellulose oligosaccharides (3). The train of spots identified as endo-1,4-β-xylanase from pI 5.5 to 6.5 on the 2D gel suggested that isoforms of the enzyme were present in the secretome, possibly due to posttranslational modifications (19). The D. stemonitis C8 secretome also contained a β-xylosidase and two α-l-arabinofuranosidases from GH families 62 (Abf2) and 43 (Table 1), which can release arabinofuranosyl side chains from monosubstituted and disubstituted xylose residues, respectively (15). The high degree of specialization in the D. stemonitis C8 secretome to the degradation of xylan is also a feature of the predicted secretome of the coprophilous fungus Podospora anserina (10).
The hemicellulose mannan is another indigestible component of the cell wall matrix of dicotyledonous leaves (9), and the D. stemonitis C8 secretome contained protein bearing homology to three different GH5 endo-1,4-β-mannanases in the NCBI database, which cleave within the mannose backbone of the mannan polymer. The endo-1,4-β-mannanases could not be identified from the 2D gel, and only three peptide matches could be made to endo-1,4-β-mannanases from the 1D gel, suggesting that the quantity of this enzyme was low in the supernatant. However, mannanase activity was clearly displayed on the 1D and 2D zymogram gels (Fig. 2, lane Mn, and Fig. 3) and 2D zymography proved to be a particularly effective technique to enable separation and confident identification of the mannanase by mass spectrometry (Table 1, 2D zym LC), without interference from other highly abundant proteins in the secretome.
Cellulose and hemicellulose are covalently linked with lignin within the plant cell wall, increasing their resistance to enzymatic attack. D. stemonitis C8 secreted numerous GMC oxidoreductases (Table 1), which produce hydroxyl radicals to modify the phenolic units of lignin and increase the access of hydrolytic enzymes to cellulose and hemicellulose polymers (45). Similarly, the predicted secretome of the coprophilous fungus P. anserina contains numerous GMC oxidoreductases (10). In addition, a 4-O-methyl-glucuronoyl-methylesterase was identified in the D. stemonitis C8 secretome that can hydrolyze the covalent ester linkages between 4-O-methyl-d-glucuronic acid residues in xylan side chains and the alcohols in the lignin polymer (22). A homologous enzyme, Cip2, from Trichoderma reesei has been of interest recently for its ability to increase the efficiency of plant biomass degradation (1, 55).
Pectin is a largely indigestible polysaccharide in leaves and consists of a fairly homogeneous region of galacturonic acid that can be acetylated or methylated and a more heterogeneous region of xylogalacturonan and rhamnogalacturonan substituted with l-arabinose, feruloyl, and acetyl residues (52). Numerous enzymes were identified in the D. stemonitis C8 secretome (Table 1) that are involved in pectin degradation: a pectate lyase that cleaves within galacturonic acid chains (24), a rhamnogalacturonan lyase that cleaves between rhamnose and galacturonic acid residues in rhamnogalacturonan (8), an α-l-rhamnosidase that releases terminal α-l-rhamnosidase residues (6), and a pectin esterase that releases methyl residues (36). None of these enzymes are found in the secretome of the industrially exploited T. reesei (13, 25, 29, 51), which has a very low pectin-degrading ability in comparison to other industrial fungi such as Aspergillus species (34, 47). The presence of the pectin-degrading enzymes in the secretome of D. stemonitis C8 could be reflective of the high levels of pectin contained within eucalyptus leaves, compared to the lower pectin levels found in decaying wood and dead plant material, which forms the natural habitat of many Trichoderma species (18). Consequently, the enzymes may be of interest as supplements to the Trichoderma reesei secretome, particularly for degradation of freshly harvested plant materials, such as corn stover, that form a cheap and abundant resource for biofuel production (46).
Three kinds of proteases were identified in the secretome of D. stemonitis C8 (Table 1) that could allow the fungus to utilize protein in the koala feces. The zinc-dependent metalloprotease from the M36 family (Table 1; Fig. 1; Fig. 2, lane Pr, prominent band at 34 to 45 kDa) and the serine protease of the subtilase superfamily S8 (Table 1; Fig. 1; Fig. 2, lane Pr, approximately 26-kDa band) are endoproteases that cleave within amino acid chains. The aminopeptidases, which bear homology to the vacuolar protease aminopeptidase Y of Saccharomyces cerevisiae (aminopeptidase Y; PA_ScAPY-like; Table 1), are exoproteases that cleave at the amino termini of proteins or peptides (27). Proteases have been found in the secretomes of most filamentous fungi explored to date, including pathogenic (27, 28, 35, 43) and saprophytic (13, 31, 42, 47) species. The proteases in D. stemonitis could be predominantly involved in the degradation of microbial bodies or sloughed epithelial cells in the koala feces (32) but may also play a role in increasing the efficiency of the degradation of the plant cell wall matrix (4).
As the genome of D. stemonitis has not been sequenced, all protein assignments in the D. stemonitis C8 secretome were made by cross-species identification, based on sequence similarities to proteins from other fungal species in the NCBI database (Table 1). The greatest number of assignments (seven) were to proteins from Podospora anserina, the only coprophilous fungus that has had its genome sequenced to date (10). Although D. stemonitis and P. anserina belong to the same fungal class (Sordariomycetes), the two species belong to different subclasses (Hypocreomycetes and Sordariomycetidae, respectively) and orders (Microascales and Sordariales, respectively), and are therefore quite distantly related phylogenetically (www.ncbi.nlm.nih.gov/Taxonomy). Many of the other protein assignments were also from fungi from the class Sordariomycetes, but the only species that are in the subclass Hypocreomycetes, along with D. stemonitis, are Hypocrea jecorina (anamorph, Trichoderma reesei; two identifications; Table 1) and Gibberella zeae (four identifications; Table 1). The high number of similarities between the D. stemonitis and P. anserina enzymes could be a result of convergent evolution, each species adapting to the coprophilous environment in a similar way. This can be investigated further as enzymes are identified from the secretomes of more coprophilous fungi in the future.
Over 20 different types of enzymes have been identified in the secretome of D. stemonitis C8 as a result of our work (Table 1). The GH10 endo-1,4-β-xylanase and GH7 cellobiohydrolase dominated the secretome (Fig. 1; Table 1). In addition, numerous other enzymes that break down cellulose, hemicellulose, lignin, pectin, and protein were identified (Table 1). However, it is important to recognize that there were many small protein spots on the 2D gel (Fig. 1) and good-quality MS/MS spectra that could not be confidently assigned to any known protein in the NCBI database. Proteins that remained unidentified in the D. stemonitis C8 secretome might be enzymes that have complementary activity to identified enzymes in the array and may increase the access of the identified enzymes to their target substrates.
In summary, the techniques of gel electrophoresis, zymography, mass spectrometry, cross-species identification, and de novo sequencing have allowed us to identify proteins in the secretome of the coprophilous fungus D. stemonitis C8, for which no genome sequence was available. In the first proteomic analysis of the secretome of a coprophilous fungus, the array of enzymes identified provide significant insight into how coprophilous fungi can degrade recalcitrant plant cell wall polymers in herbivore feces. Furthermore, the enzymes could have potential for development to increase the efficiency of industrial processes involving plant biomass degradation in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Macquarie University Research Excellence Scholarship to R.P. in the Department of Chemistry and Biomolecular Sciences, Macquarie University, Sydney, Australia, and access to the Australian Proteome Analysis Facility (APAF), Sydney, Australia, an initiative of the Australian Government as part of the National Collaborative Research and Infrastructure Strategy (NCRIS).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Banerjee G., Scott-Craig J. S., Walton J. D. 2010. Improving enzymes for biomass conversion: a basic research perspective. Bioenerg. Res. 3:82–92 [Google Scholar]
- 2. Bouws H., Wattenberg A., Zorn H. 2008. Fungal secretomes—nature's toolbox for white biotechnology. Appl. Microbiol. Biotechnol. 80:381–388 [DOI] [PubMed] [Google Scholar]
- 3. Collins T., Gerday C., Feller G. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3–23 [DOI] [PubMed] [Google Scholar]
- 4. Cosgrove D. J. 2005. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6:850–861 [DOI] [PubMed] [Google Scholar]
- 5. Couturier M., et al. 2011. Podospora anserina hemicellulases potentiate the Trichoderma reesei secretome for saccharification of lignocellulosic biomass. Appl. Environ. Microbiol. 77:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui Z., Maruyama Y., Mikami B., Hashimoto W., Murata K. 2007. Crystal structure of glycoside hydrolase family 78 α-L-rhamnosidase from Bacillus sp. GL1. J. Mol. Biol. 374:384–398 [DOI] [PubMed] [Google Scholar]
- 7. Demain A. L., Velasco J., Adrio J. L. 2005. Industrial mycology: past, present and future. p. 1-26 In An Z. (ed.), Handbook of industrial mycology. CRC Press, New York, NY [Google Scholar]
- 8. de Vries R. P., McCann M. C., Visser J. 2005. Modification of plant cell wall polysaccharides using enzymes from Aspergillus, p. 613–644 In Yarema K. J. (ed.), Handbook of carbohydrate engineering. Taylor and Francis, Boca Raton, FL [Google Scholar]
- 9. Dey P. M., Harbome J. B. 1997. Plant biochemistry. Academic Press, San Diego, CA [Google Scholar]
- 10. Espagne E., et al. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grinyer J., McKay M., Nevalainen H., Herbert B. 2004. Fungal proteomics: initial mapping of biological control strain Trichoderma harzianum. Curr. Genet. 45:163–169 [DOI] [PubMed] [Google Scholar]
- 12. Grishutin S. G., et al. 2004. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 1674:268–281 [DOI] [PubMed] [Google Scholar]
- 13. Herpoël-Gimbert I., et al. 2008. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Himmel M. E., et al. 2010. Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323–341 [Google Scholar]
- 15. Hinz S. W. A., et al. 2009. Hemicellulase production in Chrysosporium lucknowense C1. J. Cereal Sci. 50:318–323 [Google Scholar]
- 16. Hume I. J. 1984. Microbial fermentation in herbivorous marsupials. Bioscience 34:435–440 [Google Scholar]
- 17. Igarashi K., Ishida T., Hori C., Samejima M. 2008. Characterization of an endoglucanase belonging to a new subfamily of glycoside hydrolase family 45 of the basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 74:5628–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jennings D. H. 1995. The physiology of fungal nutrition. Cambridge University Press, Cambridge, England [Google Scholar]
- 19. Kannicht C. (ed.). 2002. Methods in molecular biology, vol. 194. Posttranslational modification of proteins: tools for functional proteomics. Humana Press, Totowa, NJ [Google Scholar]
- 20. Knox J. P. 2008. Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11:308–313 [DOI] [PubMed] [Google Scholar]
- 21. Krug J. C., Benny G. L., Keller H. W. 2004. Coprophilous fungi, p. 467–500 In Mueller G. M., Bills G. F., Foster M. S. (ed.), Biodiversity of fungi: inventory and monitoring methods. Elsevier, San Diego, CA [Google Scholar]
- 22. Li X.-L., Spániková S., de Vries R. P., Biely P. 2007. Identification of genes encoding microbial glucuronoyl esterases. FEBS Lett. 581:4029–4035 [DOI] [PubMed] [Google Scholar]
- 23. Liska A. J., Shevchenko A. 2003. Expanding the organismal scope of proteomics: cross-species protein identification by mass spectrometry and its implications. Proteomics 3:19–28 [DOI] [PubMed] [Google Scholar]
- 24. Marin-Rodriguez M. C., Orchard J., Seymour G. B. 2002. Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53:2115–2119 [DOI] [PubMed] [Google Scholar]
- 25. Martinez D., et al. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553–560 [DOI] [PubMed] [Google Scholar]
- 26. Martins L. F., Kolling D., Camassola M., Dillon A. J. P., Ramos L. P. 2008. Comparison of Penicillium echinulatum and Trichoderma reesei cellulases in relation to their activity against various cellulosic substrates. Bioresour. Technol. 99:1417–1424 [DOI] [PubMed] [Google Scholar]
- 27. Monod M., et al. 2002. Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 292:405–419 [DOI] [PubMed] [Google Scholar]
- 28. Mueller O., et al. 2008. The secretome of the maize pathogen Ustilago maydis. Fungal Genet. Biol. 45:S63–S70 [DOI] [PubMed] [Google Scholar]
- 29. Nagendran S., Hallen-Adams H. E., Paper J. M., Aslam N., Walton J. D. 2009. Reduced genomic potential for secreted plant cell-wall-degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Genet. Biol. 46:427–435 [DOI] [PubMed] [Google Scholar]
- 30. Nummi M., Niku-Paavola M.-L., Lappalainen A., Enari T.-M., Raunio V. 1983. Cellobiohydrolase from Trichoderma reesei. Biochem. J. 215:677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oda K., et al. 2006. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 72:3448–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Osawa R. 1991. An investigation of streptococcal flora in feces of koalas. J. Wildl. Manage. 55:623–627 [Google Scholar]
- 33. Østergaard L. H., Olsen H. S. 2010. Industrial applications of fungal enzymes, p. 269-290 In Hofrichter M. (ed.), The mycota. A comprehensive treatise on fungi as experimental systems for basic and applied research, vol. 10. Industrial applications. Springer-Verlag, Berlin, Germany [Google Scholar]
- 34. Pain A., Hertz-Fowler C. 2008. Genomic adaptation: a fungal perspective. Nat. Rev. Microbiol. 6:572–573 [DOI] [PubMed] [Google Scholar]
- 35. Paper J. M., Scott-Craig J. S., Adhikari N. D., Cuomo C. A., Walton J. D. 2007. Comparative proteomics of extracellular proteins in vitro and in planta from the pathogenic fungus Fusarium graminearum. Proteomics 7:3171–3183 [DOI] [PubMed] [Google Scholar]
- 36. Pelloux J., Rustérucci C., Mellerowicz E. J. 2007. New insights into pectin methylesterase structure and function. Trends Plant Sci. 12:267–277 [DOI] [PubMed] [Google Scholar]
- 37. Penttilä M., Nevalainen H., Rättö M., Salminen E., Knowles J. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155–164 [DOI] [PubMed] [Google Scholar]
- 38. Peterson R. A., Bradner J. R., Roberts T. H., Nevalainen K. M. H. 2009. Fungi from koala (Phascolarctos cinereus) faeces exhibit a broad range of enzyme activities against recalcitrant substrates. Lett. Appl. Microbiol. 48:218–225 [DOI] [PubMed] [Google Scholar]
- 39. Peterson R., Grinyer J., Joss J., Khan A., Nevalainen H. 2009. Fungal proteins with mannanase activity identified directly from a Congo Red stained zymogram by mass spectrometry. J. Microbiol. Methods 79:374–377 [DOI] [PubMed] [Google Scholar]
- 40. Peterson R., Grinyer J., Nevalainen H. 2011. Extracellular hydrolase profiles of fungi isolated from koala faeces invite biotechnological interest. Mycol. Prog. 10:207–218 [Google Scholar]
- 41. Saha B. C. 2000. α-L-arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol. Adv. 18:403–423 [DOI] [PubMed] [Google Scholar]
- 42. Sato S., Liu F., Koc H., Tien M. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shah P., et al. 2009. Comparative proteomic analysis of Botrytis cinerea secretome. J. Proteome Res. 8:1123–1130 [DOI] [PubMed] [Google Scholar]
- 44. Shimonaka A., et al. 2006. Amino acid regions of family 45 endoglucanases involved in cotton defibrillation and in resistance to anionic surfactants and oxidizing agents. Biosci. Biotechnol. Biochem. 70:2460–2466 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka H., Itakura S., Enoki A. 1999. Hydroxyl radical generation by an extracellular low-molecular-weight substance and phenol oxidase activity during wood degradation by the white-rot basidiomycete Trametes versicolor. J. Biotechnol. 75:57–70 [DOI] [PubMed] [Google Scholar]
- 46. Teter S. A., Cherry J. R. 2005. Improving cellulose hydrolysis with new cellulase compositions, p. 12027–12033 In AIChE Annual Meeting Conference Proceedings, Cincinnati, OH [Google Scholar]
- 47. Tsang A., Butler G., Powlowski J., Panisko E. A., Baker S. E. 2009. Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet. Biol. 46:S153–S160 [DOI] [PubMed] [Google Scholar]
- 48. Tyndale-Biscoe H. 2005. Life of marsupials. CSIRO, Collingwood, Victoria, Australia [Google Scholar]
- 49. Ulrey D. E., Robinson P. T., Whetter P. A. 1981. Eucalyptus digestibility and digestible energy requirements of adult male koalas, Phascolarctos cinereus (Marsupialia). Aust. J. Zool. 29:847–852 [Google Scholar]
- 50. Vanden Wymelenberg A., et al. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vinzant T. B., et al. 2001. Fingerprinting Trichoderma reesei hydrolases in a commercial cellulase preparation. Appl. Biochem. Biotechnol. 91–93:99–107 [DOI] [PubMed] [Google Scholar]
- 52. Voragen A., Coenen G.-J., Verhoef R., Schols H. 2009. Pectin, a versatile polysaccharide present in plant cell walls. Struct. Chem. 20:263–275 [Google Scholar]
- 53. Webster J., Weber R. W. S. 2007. Introduction to fungi. Cambridge University Press, Cambridge, England [Google Scholar]
- 54. Wei H., et al. 2009. Natural paradigms of plant cell wall degradation. Curr. Opin. Biotechnol. 20:330–338 [DOI] [PubMed] [Google Scholar]
- 55. Wood S. J., et al. 2008. Crystallization and preliminary X-ray diffraction analysis of the glucuronoyl esterase catalytic domain from Hypocrea jecorina. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:255–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.