Abstract
A library of engineered promoters of various strengths is a useful genetic tool that enables the fine-tuning and precise control of gene expression across a continuum of broad expression levels. The methylotrophic yeast Pichia pastoris is a well-established expression host with a large academic and industrial user base. To facilitate manipulation of gene expression spanning a wide dynamic range in P. pastoris, we created a functional promoter library through mutagenesis of the constitutive GAP promoter. Using yeast-enhanced green fluorescent protein (yEGFP) as the reporter, 33 mutants were chosen to form the functional promoter library. The 33 mutants spanned an activity range between ∼0.6% and 19.6-fold of the wild-type promoter activity with an almost linear fluorescence intensity distribution. After an extensive characterization of the library, the broader applicability of the results obtained with the yEGFP reporter was confirmed using two additional reporters (β-galactosidase and methionine adenosyltransferase [MAT]) at the transcription and enzyme activity levels. Furthermore, the utility of the promoter library was tested by investigating the influence of heterologous MAT gene expression levels on cell growth and S-adenosylmethionine (SAM) production. The extensive characterization of the promoter strength enabled identification of the optimal MAT activity (around 1.05 U/mg of protein) to obtain maximal volumetric SAM production. The promoter library permits precise control of gene expression and quantitative assessment that correlates gene expression level with physiologic parameters. Thus, it is a useful toolbox for both basic and applied research in P. pastoris.
INTRODUCTION
The creation of a library of engineered promoters of various strengths has enabled the precise control of gene expression in a broad range of activities required for detailed genotype-phenotype characterization and the identification of an optimal gene expression level for the desired product yield (1, 4, 12, 20, 22, 29, 35). For example, a library of promoters has been used to assess the influence of phosphoenolpyruvate carboxylase levels on growth yield and deoxyxylulose-P synthase levels on lycopene production and to demonstrate that the optimal expression levels of deoxyxylulose-P synthase are different in a strain preengineered to produce lycopene and in the parental strain (1). In another example, random mutagenesis of the Pm promoter has been applied to improve production of the medically important granulocyte-macrophage colony-stimulating factor in Escherichia coli at an industrial level (4). Furthermore, promoter libraries coupled with in silico modeling have been used to guide predictable gene network construction in Saccharomyces cerevisiae (13). Therefore, the promoter library constitutes a valuable addition to the genetic toolbox for the study of metabolic engineering, functional genomics, and synthetic biology, as well as for various biotechnology applications. However, this powerful tool is implemented mainly in bacteria (2, 4, 12, 20, 35, 36). There are only limited examples in which a promoter library has been created in yeast (1, 29, 30), and only one promoter library has been created for fine-tuning of gene expression in the methylotrophic yeast Pichia pastoris (15).
P. pastoris combines the advantages of eukaryotes, such as efficient protein secretion and posttranslational modifications, with fast growth on economical salt-based media (11). P. pastoris is by far the most commonly used yeast species in the production of recombinant proteins for both basic research and commercial purposes. Hartner et al. (15) constructed a library consisting of 46 mutant promoters through deletion and duplication of putative transcription factor binding sites within the inducible AOX1 promoter (PAOX). The constructed library spanned from ∼6% to >160% of the native promoter activity when induced by methanol. Hartner et al. also designed short synthetic promoters by combining PAOX-derived cis-acting elements with different basal promoters, because short promoters usually enable a more efficient PCR-based generation of expression cassettes or mutant libraries than do long promoters. However, all five synthetic promoters exhibited only about 10% of the activity of the wild-type PAOX.
Inducible promoters can allow for a continuous control of gene expression. Practical applications of these systems, however, are limited by transcriptional heterogeneity at the single-cell level (21, 34), inducer toxicity, and inducer-mediated pleiotropic effects. These problems highlight the need for creating a promoter library based on a constitutive promoter, which would permit steady-state gene expression control and ensure transcriptional homogeneity without an inducer. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key enzyme in glycolysis. Its promoter (PGAP) provides strong constitutive expression on glucose at a level comparable to that seen with PAOX (41) and has been widely used for constitutive expression of heterologous proteins in P. pastoris (45). The additional advantage of using PGAP, particularly for industrial fermentation, is that methanol is not required for induction. This feature makes strain growth more straightforward without carbon source shifts and eliminates the hazards and costs related to the storage and delivery of a large volume of methanol. Meanwhile, because its size (477 bp) is less than half of that of PAOX (960 bp), PGAP is more suitable for the efficient generation of expression cassettes or mutant libraries.
In the present study, a functional library of constitutive promoters covering a wide range of activities was created through the mutagenesis of PGAP using error-prone PCR (EP-PCR). After an extensive characterization of the promoter library employing the yeast-enhanced green fluorescent protein (yEGFP) gene, promising sequences were tested using two additional reporters, β-galactosidase (Gal) and methionine adenosyltransferase (MAT), at both transcript and protein levels. Finally, the library was used to assess the effect of MAT levels on S-adenosylmethionine (SAM) production and cell growth and to identify an optimal MAT level for product formation in recombinant P. pastoris.
MATERIALS AND METHODS
Unless otherwise stated, all reagents were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Unless specifically mentioned, all culture media and ingredients were prepared according to the protocol from the Pichia expression kit (Invitrogen).
Bacterial strains, plasmids, and growth conditions.
The plasmids used for this study and their sources are described in Table 1. E. coli DH5α competent cells (Takara Biotechnology, Dalian, China) were propagated in Luria-Bertani broth at 37°C, and zeocin was used for positive selection at 25 μg/ml. P. pastoris GS115 (his4) was routinely grown in yeast extract-peptone-dextrose (YPD) medium at 30°C. P. pastoris recombinant strains were selected on minimal dextrose (MD) plates with 0.4 μg/ml biotin. The cultivation of recombinant P. pastoris strains in 250-ml shake flasks containing 25 ml YPD medium was similar to that described previously (14): sterile glycerol was added to a final concentration of 1.0% (wt/vol) every 12 h. The cultivation process for SAM production in recombinant P. pastoris strains was performed according to a previously described procedure (17), with some modifications in the SAM production phase: buffered glycerol-complex medium was used during the production phase, glycerol concentration was measured and supplemented to a final concentration of 1.0% (wt/vol) every 12 h, and sterile l-methionine (Bafeng, China) was added every 24 h to a final concentration of 0.1% (wt/vol) to initiate SAM synthesis.
Table 1.
Plasmids used in this study
| Plasmid | Description | Source and/or reference |
|---|---|---|
| pPICZAgfp | PCR product yEGFP cut with NotI/XhoI was cloned into the NotI/SalI sites of pPICZ A; Zeor; 4.0 kb | This study |
| pGAPgfp | Derivative of pPICZAgfp in which the BglII/NotI PAOX region was replaced by the PCR product PGAP; Zeor; 3.5 kb | This study |
| pGHg | Derivative of pGAPgfp in which the BamHI/BglII fragment from pAO815 was inserted into the BglII site; Zeor; 7.6 kb | This study |
| pGH | Derivative of pGHg in which the egfp gene was removed by HindIII digestion; Zeor; 6.8 kb | This study |
| pSAOH5 | Expression vector containing the E. coli lacZ gene coding for β-galactosidase; Ampr; 10 kb | 39 |
| pDS56 | Vector containing the methionine adenosyltransferase (MAT) gene (ds56) obtained through DNA shuffling; Ampr; 3.5 kb | 17 |
DNA manipulation and sequencing.
Plasmid DNA isolation was performed with an AxyPrep Plasmid Miniprep kit (Axygen Biosciences, Union, CA). DNA was extracted from agarose gel slabs using an AxyPrep DNA gel extraction kit (Axygen Biosciences, Union, CA). After linearization of the plasmid with BspEI, P. pastoris GS115 cells were transformed using the standard electroporation protocol according to the instruction manual of the Pichia expression kit (Invitrogen), leading to the insertion of the plasmid into the chromosomal his4 locus. Correct genomic integration of the plasmids in positive clones was confirmed by PCR. The DNA sequence of the mutated PGAP region was determined at Takara Biotechnology (Dalian, China).
Construction of the PGAP mutant libraries.
To construct the yEGFP expression vector pGHg, the plasmid pKT127 (Euroscarf) was used as a template to amplify the yEGFP fragment through the primer pair GFP-1f/GFP-1r (see Table S1 in the supplemental material). NotI/HindIII sites were added at the 5′ end, and HindIII/XhoI sites were added at the 3′ end. Next, the yEGFP fragment was cloned into the NotI/SalI sites of pPICZ A, generating plasmid pPICZAgfp. For expression of the yEGFP reporter gene under PGAP control, the primers GAP-1f and GAP-1r (Table S1) were used for PCR amplification of the PGAP fragment from plasmid pGAPZ A. The PGAP fragment was cut with BglII/NotI and used to replace the PAOX region of pPICZAgfp, yielding the plasmid pGAPgfp. Afterward, the BamHI/BglII DNA fragment harboring the HIS4 gene from pAO815 was inserted into the BglII site of pGAPgfp, generating the plasmid pGHg (Fig. 1A). After confirmation of the correct orientation, the vector pGHg was digested by HindIII and then self-ligated to obtain the control vector pGH. To construct the reporter strain G/GHg, plasmid pGHg was digested by BspEI and transformed into P. pastoris GS115. As a blank control, G/GH was constructed by transforming GS115 with pGH.
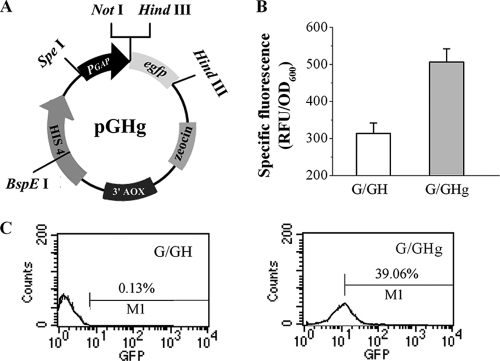
Fig. 1.
Expression of egfp driven by PGAP in P. pastoris. (A) Reporter plasmid pGHg. (B) The yEGFP expression level was measured by using a microplate reader. Reporter strain G/GHg (gray) and control strain G/GH (white) transformed with the empty vector pGH were grown in BMD medium in shake flasks for 60 h. The values represent the averages of three independent cultivations. (C) yEGFP expression level illustrated by flow cytometry histograms.
EP-PCR (7) was carried out to obtain the PGAP mutants using plasmid pGHg as the template, along with the primers GAP-EPf and GAP-Epr (see Table S1 in the supplemental material). The conditions for EP-PCR random mutagenesis were as follows: a 100-μl reaction mixture containing 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 0.5 mM MnCl2, 1 mM dCTP, 1 mM dTTP, 0.2 mM dATP, 0.2 mM dGTP, 2 μM (each) primer, 20 ng template DNA, and 2.5 units Taq DNA polymerase. PCR was performed in a PCR machine (MyCycler Thermal Cycler; Bio-Rad) for 25 cycles. One cycle consisted of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C. To increase mutational diversity and reach an appropriate error rate, three consecutive EP-PCRs were conducted. Finally, a mixture of three consecutive EP-PCR products was digested with SpeI/NotI and inserted into pGHg cleaved with the same enzymes. The plasmid mixture was transformed into P. pastoris GS115 after linearization with BspEI. The transformants were plated onto selective MD plates for subsequent screening.
Screening for PGAP mutants and determination of yEGFP fluorescence.
PGAP mutants were screened based on the fluorescence signal in recombinant P. pastoris grown in 48-deep-well plates under the following standard conditions: 28°C, agitation at 340 rpm, and 80% humidity. The cultivation consisting of three phases was similar to that previously described (15), with some modifications. A master plate, which contained strains bearing the PGAP variant-yEGFP expression cassette, was generated by inoculating 900 μl YPD with single colonies of strains from the selective MD plates. After a 24-h incubation period, 150 μl of 60% glycerol was added. The glycerol stock master plates were stored at −80°C after further incubation for 30 min and used for all subsequent experiments. A preculture in buffered minimal dextrose (BMD) medium (with 0.2% glucose) was inoculated with a 30-μl culture from the master plate and grown for 36 h to obtain uniform cell densities in all individual wells. To determine promoter activity, the main culture plate containing 900 μl of BMD (with 1% glucose) medium per well was inoculated with 30 μl of the preculture and incubated for 36 h.
From each well of the main culture plates, 30 μl culture broth was taken and added to 220 μl of phosphate-buffered saline (PBS) in a 96-well plate. The yEGFP expression in recombinant P. pastoris was analyzed by fluorescence measurements at an excitation wavelength of 395 nm and an emission wavelength of 509 nm in a Synergy 2 Multi-Detection microplate reader (BioTek, Winooski, VT). Optical density at 600 nm (OD600) was monitored with a Spectramax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA). The yEGFP fluorescence was calculated by subtracting the blank value (G/GH grown and measured under the same conditions) from the sample value.
For flow cytometry, cells at the exponential growth phase were harvested and diluted with PBS to a final OD600 of around 0.2. Flow cytometry was performed on a FACSCalibur flow cytometer (Becton Dickinson) using CellQuest software.
Enzyme assays and analysis of SAM accumulation in P. pastoris.
β-Galactosidase assays were performed according to the protocol supplied with the Pichia expression kit (Invitrogen). One unit of enzyme was defined as the amount of enzyme required to hydrolyze 1 nmol o-nitrophenyl-β-d-galactopyranoside (ONPG) per minute at 28°C. SAM concentration was determined as previously described by He et al. (16), and MAT activity was assayed according to the method reported by Hu et al. (17). One unit of enzyme was defined as the quantity of enzyme needed to catalyze the conversion of 1 μmol l-Met to SAM per hour at 37°C.
Determination of gene copy number and transcript level by real-time PCR.
The copy number of the integrated target genes (egfp, lacZ, and ds56) was determined by quantitative real-time PCR using the P. pastoris GAPDH gene as the reference gene. Reaction conditions were established as recommended by the SYBR Premix Ex Taq manual (Takara Biotechnology, Dalian, China) with 1 ng of genomic DNA as the template. For amplification of the GAPDH gene, the primer pair GAPDH-RTf/GAPDH-RTr (see Table S1 in the supplemental material) was used. Primers GFP-RTf and GFP-RTr were used for egfp, primer pair lacZ-RTf/lacZ-RTr was used for lacZ, and primers DS56-RTf and DS56-RTr (Table S1) were used for ds56. All real-time quantitative PCRs were run in triplicate wells in a StepOnePlus real-time PCR system (ABI) using the following program: 95°C for 3 min, 45 cycles of 95°C for 15 s, and 60°C for 30 s.
Total RNA was extracted from cells harvested at the exponential growth phase in shake flasks (20 ml YPD medium in 250-ml shake flasks). Quantification of the egfp, lacZ, and ds56 transcripts was performed using the PrimeScript RT reagent kit with SYBR Premix Ex Taq (Takara Biotechnology, Dalian, China) according to the manufacturer's instructions in a StepOnePlus instrument (ABI), using real-time quantitative PCR and the threshold cycle (2−ΔΔCT) method (24, 31). Quantitative real-time PCR was performed using cDNA reverse transcribed from 100 ng RNA as template and the same primer pairs for copy number determination at 0.2 μM, with the following program: 10 min at 96°C, followed by 40 cycles of 30 s at 95°C, 60 s at 55°C, and 30 s at 72°C. Amplification for each sample was carried out in triplicate wells.
Nucleotide sequence accession numbers.
The mutant PGAP sequences were deposited in GenBank under accession numbers JF414578 to JF414584.
RESULTS
Construction of a PGAP mutant library for efficient screening.
To create a PGAP mutant library, the reporter plasmid pGHg (Fig. 1A) was first constructed and used to test the expression of the yEGFP gene (egfp) under the control of the constitutive promoter PGAP in the recombinant strain G/GHg. With the microplate reader, yEGFP expression in G/GHg could be easily distinguished from the background fluorescence signal in the control strain (Fig. 1B). Meanwhile, the reporter expression level and monovariate distribution of fluorescence were also tested by flow cytometry (Fig. 1C). Therefore, the reporter plasmid pGHg could be used to construct a PGAP mutant library. Alterations in specific fluorescence detected by the microplate reader reflected changes in the strength of PGAP mutants. Homogeneity of the expression level was also further examined with flow cytometry. The control strain had a fluorescence signal (about 300 relative fluorescence units [RFU]/OD600), and the fluorescence intensities emitted by the fresh medium and the cell-free broth after 60 h of growth were 106 RFU and 215 RFU, respectively. Thus, background fluorescence can be attributed to interference from culture media, cellular components, and metabolites such as riboflavin (37). Consequently, yEGFP fluorescence intensity was calculated by subtracting the blank value (G/GH grown and measured under the same conditions) from the sample value. The specific fluorescence referred to in the present study is the ratio of fluorescence intensity to the optical density at 600 nm of the same culture broth.
With the reporter plasmid pGHg, a PGAP mutant library was generated by replacing the original PGAP with mutated PGAP variants obtained through EP-PCR. To increase mutational diversity and reach an appropriate error rate, three consecutive rounds of EP-PCR were carried out under the same conditions. After random sequencing of 10 PCR products from each round, an overall range of mutation rates from 1.1% to 4.0% was observed (Table 2). Sequencing results also revealed that the mutations were randomly distributed throughout the amplified sequence without obvious nearest-neighbor effects (data not shown). The mixture of PGAP mutant plasmids was then transformed into P. pastoris GS115, and the recombinant cells were screened based on the fluorescence intensity of the culture broth, as measured with the microplate reader.
Table 2.
Mutation rates of the PGAP fragments of three consecutive EP-PCRsa
| EP-PCR | No. of point mutations | Mutation rate (%) |
|---|---|---|
| First round | 5.0 ± 2.8 | 1.1 ± 0.6 |
| Second round | 10.7 ± 2.5 | 2.2 ± 0.2 |
| Third round | 19.0 ± 3.6 | 4.0 ± 0.2 |
Reaction conditions are described in Materials and Methods. Mutation rate refers to the mean number of mutations in 10 PCR products from each round of EP-PCR.
Characterization of the PGAP mutant collection.
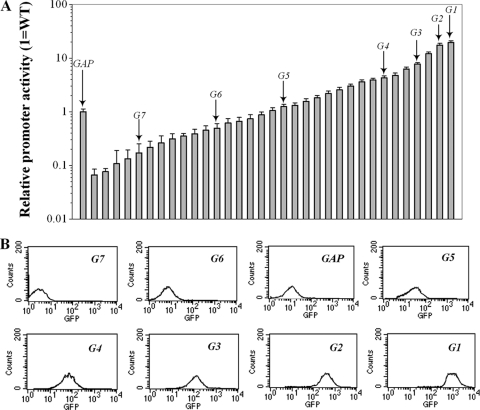
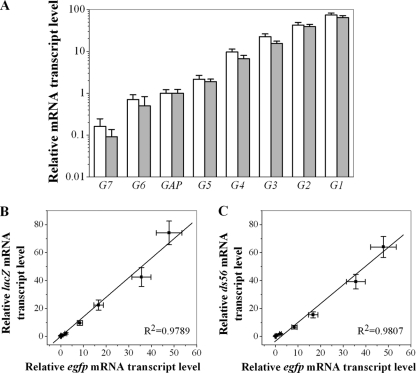
Nearly 30,000 promoter mutants, spanning 3 orders of magnitude of yEGFP fluorescence, were screened using the high-throughput screening approach in 48-deep-well plates. Among them, 33 mutants, spanning a 296-fold range of fluorescence intensity, were finally chosen to form a functional promoter library (Fig. 2A).
Fig. 2.
Promoter activity as represented by the fluorescence intensity of yEGFP reporter. (A) Reporter clones were cultured in 48-well deep-well plates. The relative promoter activities were normalized to the activity obtained with the wild-type (WT) GAP promoter. The data represent averages ± standard deviations of single measurements of three independent cultivations in separate deep-well plates. (B) The selected clones harboring the promoter variants as indicated were grown in shake flasks and showed a uniform yEGFP expression level as illustrated by representative flow cytometry histograms.
To ensure that the results obtained by bulk population-averaged measurements reflected single-cell behavior, transformants containing seven mutant promoters (G1, G2, G3, G4, G5, G6, and G7) and the original strain G/GHg (with the wild-type PGAP) were selected to test the level of yEGFP expression and fluorescence distributions by flow cytometry in shake flask culture. Only clones with clean monovariate distributions of fluorescence (Fig. 2B) were retained for further analysis.
All seven selected PGAP mutants (G1, G2, G3, G4, G5, G6, and G7) were sequenced. The sequencing results demonstrated that all seven clones represented a unique promoter sequence with high variance in both locations and densities of the mutated residues. The mutation rate ranged from 1.0% (G4) to 3.4% (G5), and mutations seemed to be randomly distributed throughout the promoter sequence (see Fig. S1 in the supplemental material).
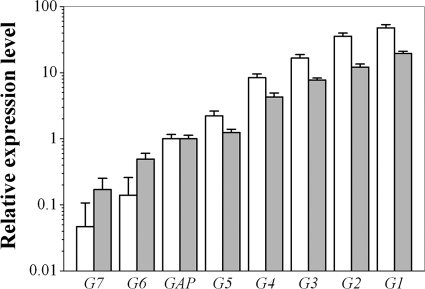
Gene expression under the control of seven selected PGAP variants was further investigated at the transcriptional level. The relative mRNA levels of egfp in the selected transformants cultured in shake flasks were measured and normalized to the value obtained with the unmutated PGAP (Fig. 3). The relative level of the egfp transcript was found to span almost 3 orders of magnitude (955-fold range) (Table 3) and to correlate with the corresponding yEGFP fluorescence level. These results support the hypothesis that the various levels of reporter gene expression in all clones were due to differences in basal transcriptional activity, i.e., caused by various strengths of the different PGAP mutants. Thus, the obtained series of promoters can fine-tune the expression of the reporter gene egfp.
Fig. 3.
Characterization of promoter strengths as determined from transcript quantification and fluorescence intensity of the yEGFP gene. The transformants containing selected GAP promoter mutants were grown in shake flasks (20 ml YPD medium in 250-ml shake flasks) for 60 h. The relative fluorescence intensities (gray) shown are mean values of at least two independent experiments and were normalized to the intensity obtained with the wild-type GAP promoter. Total RNA was extracted from cells harvested from the logarithmic phase during growth in shake flasks. Promoter performance was measured directly at egfp mRNA transcript levels by quantitative real-time PCR. Relative mRNA transcript levels (white) were normalized to the value obtained with the wild-type PGAP.
Table 3.
Impact of promoter strength on the expression of different genesa
| Promoter |
ds56 |
egfp |
lacZ |
||||
|---|---|---|---|---|---|---|---|
| Transcript level | MAT activity (U/mg of protein) | Volumetric production of SAM (g/liter) | Transcript level | yEGFP fluorescence (RFU/OD600) | Transcript level | Gal activity (U/mg of protein) | |
| G7 | 0.09 ± 0.04 | 0.10 ± 0.02 | 0.04 ± 0.01 | 0.05 ± 0.06 | 0.17 ± 0.08 | 0.16 ± 0.08 | 0.26 ± 0.05 |
| G6 | 0.50 ± 0.17 | 0.34 ± 0.03 | 0.24 ± 0.05 | 0.14 ± 0.12 | 0.49 ± 0.11 | 0.71 ± 0.21 | 0.5 ± 0.07 |
| GAP | 1.00 ± 0.24 | 1.00 ± 0.13 | 1.00 ± 0.11 | 1.00 ± 0.16 | 1.00 ± 0.13 | 1.00 ± 0.22 | 1.00 ± 0.14 |
| G5 | 1.89 ± 0.29 | 1.31 ± 0.15 | 1.34 ± 0.38 | 2.22 ± 0.42 | 1.25 ± 0.15 | 2.17 ± 0.53 | 1.35 ± 0.25 |
| G4 | 6.72 ± 1.31 | 2.29 ± 0.28 | 5.33 ± 0.52 | 8.43 ± 1.22 | 4.29 ± 0.64 | 9.72 ± 1.74 | 5.48 ± 0.64 |
| G3 | 15.52 ± 2.12 | 3.99 ± 0.64 | 8.63 ± 0.78 | 16.74 ± 2.08 | 7.75 ± 0.57 | 22.52 ± 3.76 | 18.95 ± 2.74 |
| G2 | 39.18 ± 5.18 | 4.72 ± 0.36 | 11.04 ± 0.40 | 35.64 ± 4.24 | 12.14 ± 1.24 | 42.57 ± 6.76 | 51.38 ± 4.85 |
| G1 | 64.04 ± 7.47 | 5.70 ± 0.34 | 10.56 ± 0.21 | 47.78 ± 5.72 | 19.58 ± 1.43 | 74.14 ± 8.52 | 96.16 ± 7.78 |
Relative yEGFP fluorescence, Gal specific activities, MAT specific activities, volumetric production of SAM, and transcript levels of reporters were normalized to the value obtained with the wild-type PGAP. Wild-type values were set to 1.
PGAP mutants also confer various expression levels of two alternative reporter genes.
To test the broader applicability of the results obtained with the reporter yEGFP, eight selected members (G1, G2, G3, G4, G5, GAP, G6, and G7) of the promoter library were used to express two additional reporters: the enzyme β-galactosidase gene (lacZ) amplified from plasmid pSAOH5 (39) with the primers LacZ-f and LacZ-r and a recombinant MAT gene (ds56) amplified from plasmid pDS56 (17) with the primers DS56-f and DS56-r. These two reporter genes were placed under the control of the selected promoters to construct the expression plasmids, which were then integrated into the P. pastoris GS115 chromosome for further characterization.
First, the copy numbers of the egfp, lacZ, and ds56 expression cassettes were measured in the recombinant strains. As the recombinant strains possessed only one copy of the expression cassette, no gene dosage effect complicated the result.
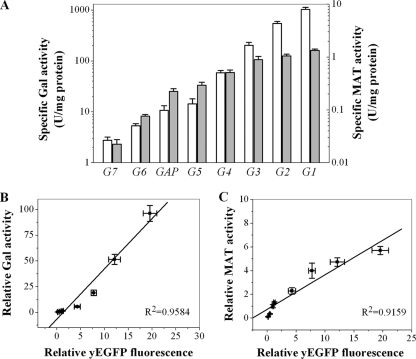
Next, the Gal and MAT activities were measured. For these two enzymes expressed under the control of the eight promoters, the same trend was observed as for the yEGFP expression driven by the same promoters (Fig. 4A). The specific activities of Gal and MAT spanned a 370- and a 57-fold range, respectively (Table 3). When the relative expression level of the reporter genes was normalized to the value obtained with the wild-type PGAP, the enzyme activities correlated well with the yEGFP fluorescence (Fig. 4B and C).
Fig. 4.
Enzyme activity of β-galactosidase (Gal) and methionine adenosyltransferase (MAT) under the control of 8 selected PGAP library members. (A) Specific activity of Gal (white) and MAT (gray) under the control of 8 different promoters as indicated. Enzyme specific activity is given in U/mg protein in the cell lysates. Experiments were carried out in shake flask cultures for 60 h in buffered minimal glycerol (BMGY) medium. Samples for measuring enzyme activity and biomass were taken at the end of the fermentation. (B) Relative Gal activity under the control of 8 different promoters as a function of the yEGFP fluorescence controlled by the same promoter. (C) Relative MAT activity under the control of 8 different promoters as a function of the yEGFP fluorescence controlled by the same promoter. The relative enzyme activities and yEGFP fluorescence were normalized to the value obtained with the wild-type PGAP. Data represent means ± standard deviations of three independent cultivations.
Finally, promoter performance was also evaluated at the transcriptional level by quantitative reverse transcription-PCR (qRT-PCR). The relative levels of the lacZ and ds56 transcripts under the control of the eight selected promoters spanned a 463- and a 711-fold range, respectively (Table 3). A similar trend was observed in the relative egfp mRNA levels (Fig. 5). Therefore, the transcription of two alternative reporters was also fine-tuned by the representative members of the promoter library.
Fig. 5.
Promoter performance was measured directly at mRNA transcript levels of the β-galactosidase gene (lacZ) and the methionine adenosyltransferase gene (ds56). (A) Relative mRNA transcript levels of lacZ (white) and ds56 (gray) under the control of 8 different promoters as indicated. Total RNA was extracted from cells harvested from the logarithmic phase during growth in shake flasks (20 ml YPD medium in 250-ml shake flasks) and measured by quantitative real-time PCR. Relative mRNA transcript levels of the reporters were normalized to the value obtained with the wild-type PGAP. (B) Relative lacZ transcript under the control of 8 different promoters as a function of the egfp transcript driven by the same promoter. (C) Relative ds56 transcript under the control of 8 different promoters as a function of the egfp transcript driven by the same promoter. Data represent means ± standard deviations of three independent cultivations.
From the results presented above, it was seen that the promoter activities in the functional library were fairly independent of the reporters tested. These results strongly supported the idea that the various levels of reporter gene expression in all clones originated from differences in basal transcriptional activity, i.e., caused by the various strengths of different PGAP mutants.
Use of PGAP variants to study the relationship between MAT expression and SAM accumulation in P. pastoris.
SAM, an important molecule for normal cell function and survival, is synthesized from ATP and l-methionine (l-Met) catalyzed by MAT. Considerable interest in SAM production has arisen because SAM can effectively treat affective disorders (43), liver disease (8), and neurological disorders (5). Overexpression of the heterologous MAT gene (ds56) driven by PAOX has been conducted to enhance SAM accumulation in P. pastoris. However, the degree of increased MAT activity was much higher than that of SAM production (17). To investigate the influence of MAT activity on SAM production and determine the critical or optimum MAT activity for SAM synthesis, the PGAP library was applied to introduce precise transcriptional control of ds56 in strains containing various-strength promoter-ds56 expression cassettes.
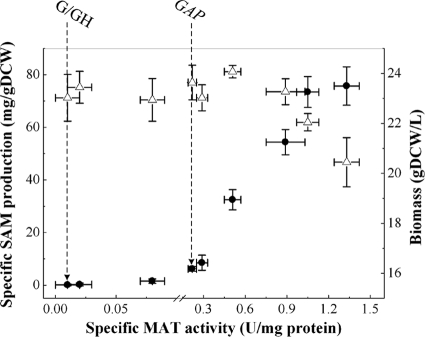
Eight members of the PGAP library (G1, G2, G3, G4, G5, GAP, G6, and G7), including the strongest and extremely weak promoter mutants, as well as the original PGAP, were used to construct ds56 expression plasmids and the corresponding recombinant strains. Both biomass and SAM production were measured in the eight integrated transformants, along with the control strain (G/GH), and plotted as a function of specific MAT activity (Fig. 6). Almost no SAM accumulation was detected in the control strain in which no heterologous MAT gene was introduced. Higher MAT activity and specific SAM production were obtained with increased promoter strength, demonstrating that ds56 expression level has a positive effect on the SAM production. A nearly linear relationship was observed when ds56 was driven by G7, G6, GAP, G5, G4, G3, and G2, with the MAT activity below 1.05 U/mg of protein, suggesting that MAT is a rate-limiting factor in SAM production in these recombinant strains. However, when a higher MAT activity (1.33 U/mg of protein) was obtained with the mutant promoter G1, the increase in SAM production reached a plateau, implying that MAT activity was no longer the only rate-limiting factor in SAM synthesis. Moreover, an increase in MAT activity from 1.05 to 1.33 U/mg of protein led to a moderate decrease in biomass. From these results, 1.05 U/mg of protein may thus be an optimum MAT activity for SAM accumulation. Beyond this point, although elevating MAT expression had a slightly positive effect on specific SAM production, it showed an adverse effect on growth (Fig. 6), thereby resulting in decreased volumetric SAM production (Table 3).
Fig. 6.
Impacts of specific MAT activity on specific SAM production (closed circles) and cell growth (open triangles) in recombinant P. pastoris. Experiments were carried out in shake flask cultures for 60 h in BMGY medium. Samples for measuring MAT activity, SAM production, and biomass were taken at the end of the fermentation. Error bars present the standard deviations for three independent experiments.
DISCUSSION
In the current study, from nearly 30,000 random promoter mutants, 33 mutants were chosen to form a functional PGAP library for modulating steady-state gene expression in P. pastoris. This collection of constitutive promoters exhibits a 296-fold range of yEGFP fluorescence (Fig. 2A). Several extraordinarily strong variants were obtained, with the strongest showing a nearly 20-fold increase of specific fluorescence and a 48-fold increase in mRNA level compared to that of the wild-type PGAP. Seven representative members were then rigorously characterized to confirm that the chosen promoters drove stable monovariate yEGFP expression in shake flask culture by flow cytometry (Fig. 2B), and the yEGFP expression was transcriptionally controlled by sequence-specific PGAP mutants (Fig. 3). Two additional reporters (lacZ and ds56) were also used to verify the broader applicability of the results obtained with the reporter yEGFP. Both enzyme activity (Fig. 4A) and mRNA transcript (Fig. 5A) levels were regulated by the selected promoters. The relative enzyme activities (Fig. 4B and C) and mRNA amounts (Fig. 5B and C) of these two reporters showed a linear correlation with yEGFP expression level. When under the control of the selected promoters, the transcript levels of egfp, lacZ, and ds56 varied with the same orders of magnitude, spanning 955-, 463-, and 711-fold ranges, respectively. Therefore, the constitutive promoter library that we constructed exhibits a broad dynamic range and presents a tool for fine-tuning of gene expression in P. pastoris, which behaves similarly regardless of the gene being regulated.
When the representative promoter variants were used to control the expression of three reporter genes, the relative mRNA transcript levels spanned a wider range than did the protein or enzyme levels, and the increase in protein expression did not totally reflect the increase in message levels (Table 3). The mRNA levels of egfp, lacZ, and ds56 spanned 955-, 463-, and 711-fold ranges, respectively. However, the concomitant levels of yEGFP fluorescence, Gal, and MAT enzyme activity spanned only 115-, 370-, and 57-fold ranges, respectively. Such a limited correlation between mRNA and protein/enzyme activity levels may be due to complex posttrans-criptional processes such as transcript (de)stabilization, translation, posttranslational modifications, protein degradation that determines and modulates the quality and quantity of expressed proteins, and protein folding efficiency (42). Protein concentrations may be only 20% to 40% attributable to altered mRNA levels (6, 38). Furthermore, analyses integrating mRNA and protein profiling in cells have suggested that approximately 60% to 80% discordance exists between mRNA abundance and protein abundance (33).
For the three reporter genes tested, although protein expression increased with relative mRNA transcript level, this correlation varied with the gene and was not always linear. In the reporter Gal, a high correlation between mRNA and enzyme activity was observed. Compared with the other two reporters, higher Gal activity was obtained with the strong promoters G1, G2, and G3 (Table 3). The fact that Gal activity levels tended to be much higher in some of the strains could be partly attributed to the longer half-life of Gal, more than 20 h (3), compared with 7 h for yEGFP (27) and 2 h for MAT (44). When yEGFP was driven by the strong promoters (G1 and G2), the considerably large increase in mRNA level (up to 48-fold) resulted in only a moderate increase of specific fluorescence (up to 20-fold). A possible reason, as suggested by Lenassi Zupan et al. (23), may be the high-level expression of fluorescent GFP in P. pastoris, which leads to the formation of fluorescent GFP particles unavailable for fluorescence measurements. Remarkably, even though the transcriptional levels of all three genes were of the same magnitude, the enzymatic activity of MAT was extraordinarily low and tended to stay constant as the mRNA transcript levels increased to the maximum values observed. Such a huge gap between enzyme activity and mRNA level has also been reported by Lu and Jeffries (25). When they adjusted the enzyme activity of transaldolase (TAL1), transketolase (TKL1), and pyruvate kinase (PYK1) by tuning the promoter strength, the enzyme activity of PYK1 was significantly lower than those of the other two genes. In the present study, the extraordinarily low MAT activity may suggest that dimerization of the protein, which is important for MAT activity (9), maintenance of the cellular redox potential (26), and SAM inhibition (32), plays important roles in the modulation of MAT activity. In summary, the expression of these three reporters was regulated at several stages, and the huge variance in mRNA level exerted by the different promoters was attenuated at the protein/enzyme activity level by various degrees for each reporter.
In our previous study to improve SAM accumulation in P. pastoris by overexpression of recombinant MAT gene ds56, an increase in enzyme activity (2.8-fold) did not result in an equal increase in SAM production (about 1.0-fold) (17). Now, enabled by the collection of constitutive promoter mutants, we analyzed the effect of MAT enzyme activity on SAM accumulation, and the optimum MAT activity was identified at around 1.05 U/mg of protein, which maximized volumetric SAM production. When the MAT activity increased from 1.05 to 1.33 U/mg of protein, only a slight increase in specific SAM production was obtained. This result implies that MAT activity is not the rate-limiting factor for SAM synthesis. To enhance SAM production further, other limiting steps, such as elevating ATP level through pulsed feeding of glycerol (19), optimizing the l-Met feeding strategy (18), or blocking the transsulfuration pathway through cystathionine by disrupting the cystathionine synthase (16), should be targeted. In addition, a negative influence on the cell growth was observed when the specific SAM production exceeded 73 mg/g (dry cell weight [DCW]) (Fig. 6). An increased level of SAM has deleterious effects on yeast, particularly when the excess SAM cannot be sequestered in the vacuole (10). The growth inhibition associated with SAM hyperaccumulation could be explained by the transient G1 cell cycle delay caused by the excess SAM in the cytosol (28). The net ATP loss during SAM synthesis could also exert inhibitory action on cell growth.
In conclusion, the library of mutant constitutive promoters constitutes a valuable addition to the genetic toolbox for fine-tuning of gene expression in P. pastoris. This library contains several extremely strong constitutive promoters and spans, at the transcription level, a <0.1- to >45-fold range of the native PGAP activity. Therefore, the constitutive promoter collection provided here could be an attractive alternative to the strong methanol-inductive promoter AOX1 for tailoring a production strategy specific to the needs of the individual protein, particularly when the heterologous proteins are not toxic to the host. With a simplified fermentation process, the hazards and costs associated with the storage and delivery of a large volume of methanol can also be eliminated. In addition, in metabolic pathway analysis and optimization, the library has several advantages over existing inducible expression systems. One advantage is the precise control of the steady-state expression of a target gene at the single-cell level and the ability to determine the optimal level for product yield or cell growth, as highlighted by our investigation of SAM production. Another advantage is the option to modulate the expression of many genes in the same strain simultaneously to different extents. The third advantage is that the length of the PGAP mutants is only half of that of PAOX, which facilitates a more efficient PCR-based generation of mutant libraries. Finally, as P. pastoris PGAP has been used to express the hepatitis B virus surface antigen in S. cerevisiae (40) and appears to have a broad host range, the PGAP library might be applicable to other yeast species, an idea which should be tested in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the National Natural Science Foundation of China (20976050) and the National Basic Research Program (973 Program 2007CB714306) for their financial support of this research.
We thank James M. Cregg for his generous gift of the plasmid pSAOH5.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Alper H., Fischer C., Nevoigt E., Stephanopoulos G. 2005. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U. S. A. 102:12678–12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andersen H. W., Pedersen M. B., Hammer K., Jensen P. R. 2001. Lactate dehydrogenase has no control on lactate production but has a strong negative control on formate production in Lactococcus lactis. Eur. J. Biochem. 268:6379–6389 [DOI] [PubMed] [Google Scholar]
- 3. Bachmair A., Finley D., Varshavsky A. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179–186 [DOI] [PubMed] [Google Scholar]
- 4. Bakke I., et al. 2009. Random mutagenesis of the Pm promoter as a powerful strategy for improvement of recombinant-gene expression. Appl. Environ. Microbiol. 75:2002–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bottiglieri T., Hyland K., Reynolds E. H. 1994. The clinical potential of ademetionine (S-adenosylmethionine) in neurological disorders. Drugs 48:137–152 [DOI] [PubMed] [Google Scholar]
- 6. Brockmann R., Beyer A., Heinisch J. J., Wilhelm T. 2007. Posttranscriptional expression regulation: what determines translation rates? PLoS Comput. Biol. 3:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cadwell R. C., Joyce G. F. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28–33 [DOI] [PubMed] [Google Scholar]
- 8. Cederbaum A. I. 2010. Hepatoprotective effects of S-adenosyl-l-methionine against alcohol- and cytochrome P450 2E1-induced liver injury. World J. Gastroenterol. 16:1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chamberlin M. E., Ubagai T., Pao V. Y., Pearlstein R. A., Yang Chou J. 2000. Structural requirements for catalysis and dimerization of human methionine adenosyltransferase I/III. Arch. Biochem. Biophys. 373:56–62 [DOI] [PubMed] [Google Scholar]
- 10. Chan S. Y., Appling D. R. 2003. Regulation of S-adenosylmethionine levels in Saccharomyces cerevisiae. J. Biol. Chem. 278:43051–43059 [DOI] [PubMed] [Google Scholar]
- 11. Cregg J. M., Cereghino J. L., Shi J. Y., Higgins D. R. 2000. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 16:23–52 [DOI] [PubMed] [Google Scholar]
- 12. De Mey M., Maertens J., Lequeux G. J., Soetaert W. K., Vandamme E. J. 2007. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellis T., Wang X., Collins J. J. 2009. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 27:465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasser B., Maurer M., Gach J., Kunert R., Mattanovich D. 2006. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol. Bioeng. 94:353–361 [DOI] [PubMed] [Google Scholar]
- 15. Hartner F. S., et al. 2008. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 36:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He J. Y., Deng J. J., Zheng Y. H., Gu J. 2006. A synergistic effect on the production of S-adenosyl-l-methionine in Pichia pastoris by knocking in of S-adenosyl-l-methionine synthase and knocking out of cystathionine-beta synthase. J. Biotechnol. 126:519–527 [DOI] [PubMed] [Google Scholar]
- 17. Hu H., et al. 2009. DNA shuffling of methionine adenosyltransferase gene leads to improved S-adenosyl-l-methionine production in Pichia pastoris. J. Biotechnol. 141:97–103 [DOI] [PubMed] [Google Scholar]
- 18. Hu H., et al. 2009. Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Appl. Microbiol. Biotechnol. 83:1105–1114 [DOI] [PubMed] [Google Scholar]
- 19. Hu X. Q., et al. 2008. Effects of different glycerol feeding strategies on S-adenosyl-l-methionine biosynthesis by PGAP-driven Pichia pastoris overexpressing methionine adenosyltransferase. J. Biotechnol. 137:44–49 [DOI] [PubMed] [Google Scholar]
- 20. Jensen P. R., Hammer K. 1998. Artificial promoters for metabolic optimization. Biotechnol. Bioeng. 58:191–195 [PubMed] [Google Scholar]
- 21. Khlebnikov A., Risa O., Skaug T., Carrier T. A., Keasling J. D. 2000. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 182:7029–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koebmann B. J., Andersen H. W., Solem C., Jensen P. R. 2002. Experimental determination of control of glycolysis in Lactococcus lactis. Antonie Van Leeuwenhoek 82:237–248 [PubMed] [Google Scholar]
- 23. Lenassi Zupan A., Trobec S., Gaberc-Porekar V., Menart V. 2004. High expression of green fluorescent protein in Pichia pastoris leads to formation of fluorescent particles. J. Biotechnol. 109:115–122 [DOI] [PubMed] [Google Scholar]
- 24. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Lu C., Jeffries T. 2007. Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 73:6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Markham G. D., Satishchandran C. 1988. Identification of the reactive sulfhydryl groups of S-adenosylmethionine synthetase. J. Biol. Chem. 263:8666–8670 [PubMed] [Google Scholar]
- 27. Mateus C., Avery S. V. 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 16:1313–1323 [DOI] [PubMed] [Google Scholar]
- 28. Mizunuma M., Miyamura K., Hirata D., Yokoyama H., Miyakawa T. 2004. Involvement of S-adenosylmethionine in G1 cell-cycle regulation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:6086–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nevoigt E., et al. 2007. Engineering promoter regulation. Biotechnol. Bioeng. 96:550–558 [DOI] [PubMed] [Google Scholar]
- 30. Nevoigt E., et al. 2006. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 72:5266–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reguera R. M., et al. 2002. Cloning expression and characterization of methionine adenosyltransferase in Leishmania infantum promastigotes. J. Biol. Chem. 277:3158–3167 [DOI] [PubMed] [Google Scholar]
- 33. Seliger B., et al. 2009. Combined analysis of transcriptome and proteome data as a tool for the identification of candidate biomarkers in renal cell carcinoma. Proteomics 9:1567–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siegele D. A., Hu J. C. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. U. S. A. 94:8168–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solem C., Jensen P. R. 2002. Modulation of gene expression made easy. Appl. Environ. Microbiol. 68:2397–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Solem C., Koebmann B. J., Jensen P. R. 2003. Glyceraldehyde-3-phosphate dehydrogenase has no control over glycolytic flux in Lactococcus lactis MG1363. J. Bacteriol. 185:1564–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Surribas A., Resina D., Ferrer P., Valero F. 2007. Rivoflavin may interfere with on-line monitoring of secreted green fluorescence protein fusion proteins in Pichia pastoris. Microb. Cell Fact. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tian Q., et al. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3:960–969 [DOI] [PubMed] [Google Scholar]
- 39. Tschopp J. F., Brust P. F., Cregg J. M., Stillman C. A., Gingeras T. R. 1987. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. Nucleic Acids Res. 15:3859–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vellanki R. N., Komaravelli N., Tatineni R., Mangamoori L. N. 2007. Expression of hepatitis B surface antigen in Saccharomyces cerevisiae utilizing glyceraldeyhyde-3-phosphate dehydrogenase promoter of Pichia pastoris. Biotechnol. Lett. 29:313–318 [DOI] [PubMed] [Google Scholar]
- 41. Waterham H. R., Digan M. E., Koutz P. J., Lair S. V., Cregg J. M. 1997. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37–44 [DOI] [PubMed] [Google Scholar]
- 42. Waters K. M., Pounds J. G., Thrall B. D. 2006. Data merging for integrated microarray and proteomic analysis. Brief. Funct. Genomic. Proteomic. 5:261–272 [DOI] [PubMed] [Google Scholar]
- 43. Williams A. L., Girard C., Jui D., Sabina A., Katz D. L. 2005. S-adenosylmethionine (SAMe) as treatment for depression: a systematic review. Clin. Invest. Med. 28:132–139 [PubMed] [Google Scholar]
- 44. Yoon G. S., et al. 2006. Characterization of S-adenosylmethionine synthetase from Streptomyces avermitilis NRRL8165 and its effect on antibiotic production. Enzyme Microb. Technol. 39:466–473 [Google Scholar]
- 45. Zhang A. L., et al. 2009. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol. Biol. Rep. 36:1611–1619 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.