Abstract
Microbial communities host unparalleled taxonomic diversity. Adequate characterization of environmental and host-associated samples remains a challenge for microbiologists, despite the advent of 16S rRNA gene sequencing. In order to increase the depth of sampling for diverse bacterial communities, we developed a method for sequencing and assembling millions of paired-end reads from the 16S rRNA gene (spanning the V3 region; ∼200 nucleotides) by using an Illumina genome analyzer. To confirm reproducibility and to identify a suitable computational pipeline for data analysis, sequence libraries were prepared in duplicate for both a defined mixture of DNAs from known cultured bacterial isolates (>1 million postassembly sequences) and an Arctic tundra soil sample (>6 million postassembly sequences). The Illumina 16S rRNA gene libraries represent a substantial increase in number of sequences over all extant next-generation sequencing approaches (e.g., 454 pyrosequencing), while the assembly of paired-end 125-base reads offers a methodological advantage by incorporating an initial quality control step for each 16S rRNA gene sequence. This method incorporates indexed primers to enable the characterization of multiple microbial communities in a single flow cell lane, may be modified readily to target other variable regions or genes, and demonstrates unprecedented and economical access to DNAs from organisms that exist at low relative abundances.
INTRODUCTION
The composition, organization, and spatial distribution of environmental microbial communities are still poorly understood. Enormous progress in method development has begun to enable the study of alpha, beta, and gamma diversity, but a substantial limitation remains: the coverage of most sequencing methods remains insufficient to analyze single samples comprehensively or to conduct field-scale comparisons of the microbial diversity in most environments. Methodology is still required to provide (i) high sample throughput, (ii) information on the microbial species (or phylotypes) present at both high and low relative abundances, and (iii) affordability for the average research laboratory. Although comprehensive metagenomic analysis could eventually be used for microbial community profiling (sampling both abundant and rare populations), this is not yet feasible for most environmental samples due to enormous computational and sequencing limitations. Instead, an alternative community profiling approach involves surveying distributions of the small subunit rRNA gene due to its ubiquity across all domains of life (16S rRNA in the Bacteria and Archaea and 18S rRNA in the Eukarya) (27). Additionally, the 16S rRNA gene provides valuable phylogenetic information (6) for comparison to database collections. For many years, the use of Sanger sequencing to sequence collected 16S rRNA genes from environmental samples has revealed that sample sizes, and thus coverage, afforded by Sanger sequencing have been insufficient to adequately describe and compare microbial communities (6, 24). The advent of serial analysis of ribosomal sequence tags (SARST) (17, 26, 33) and 454 pyrosequencing provided a major advance by enabling the collection of thousands of sequences from multiple samples. These approaches have provided a new window into the diversity and composition of microbial communities (11, 25, 31), increased sample throughput by use of indexing (1, 10, 19), and sparked interest in elucidating the members of the rare biosphere, which are microorganisms that exist at low relative abundances (23, 28, 31). To further reduce the costs of sequencing, the Illumina platform was recently used to generate data sets of unprecedented size (3, 8, 20) that surpass 454 pyrosequencing data sets by over an order of magnitude in number of sequences per unit cost (30). Initial Illumina-based methods for sequencing 16S rRNA genes have been limited by ≤101-base sequence reads (3, 8, 12, 20, 34) and/or an inability to leverage the paired-end approach that would allow for assembly of reads and reduced sequencing errors (3, 4, 20).
Here we present a novel application-ready method for generating multimillion-sequence data sets at a fraction of the cost of Sanger or 454 pyrosequencing. Without factoring sample preparation costs, Illumina is currently ∼50- and ∼12,000-fold less expensive per sequenced megabase than pyrosequencing (i.e., 454 pyrosequencing) and Sanger sequencing, respectively (S. Pereira, The Centre for Applied Genomics, Toronto, Ontario, Canada, personal communication). This method leverages the paired-end Illumina sequencing platforms (i.e., GAIIx genome analyzer, Hiseq 2000 platform, and MiSeq genome analyzer) to assemble ∼200-base hypervariable region 3 (V3) amplicons with individual forward and reverse read lengths of 125 nucleotides each. We demonstrate with replicate defined community and Arctic tundra (AT) libraries that 16S rRNA gene sequencing with the Illumina sequencing platform enables rapid, affordable, reproducible, and comprehensive assessments and comparisons of the taxonomic diversity present in complex microbial communities and provides unprecedented access to organisms present at low relative abundances.
MATERIALS AND METHODS
Sample collection and DNA isolation.
A composite Arctic tundra soil sample was prepared from a pristine site in Alert, Nunavut, Canada. The same soil sample was used previously for 16S rRNA gene tag sequencing using the SARST technique (26). For a defined community, six bacterial strains were chosen as controls: Escherichia coli (ATCC 11303), Pseudomonas aeruginosa (ATCC 10145), Bacillus subtilis (ATCC 6633), Flexibacter canadensis (ATCC 29591), Methylococcus capsulatus Bath (ATCC 33009), and Paracoccus denitrificans (ATCC 17741). These organisms were chosen to provide wide coverage of phyla and rRNA operon copy numbers. Genomic DNAs were extracted from soil and log-phase bacterial cultures by use of a FastDNA spin kit for soil (MP Biomedicals) according to the manufacturer's instructions. Soil DNA was extracted in triplicate, and the extracts were subsequently pooled. Ten nanograms of each pure culture template DNA was combined prior to PCR in order to eliminate possible bias associated with DNA extraction.
Illumina library generation.
The V3 region of the 16S rRNA gene was amplified using modified 341F and 518R primers (22) (see Table S1 in the supplemental material). In addition to V3-specific priming regions, these primers are complementary to Illumina forward, reverse, and multiplex sequencing primers (with the reverse primer also containing a 6-bp index allowing for multiplexing). All custom primers were synthesized and purified by polyacrylamide gel electrophoresis (PAGE; IDT, Coralville, IA). Three PCR amplifications were carried out for each sample, using 50-μl reaction mixtures. Each reaction mixture contained 25 pmol of each primer, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, and 1 U Phusion Taq polymerase (Finnzyme, Finland). The PCR conditions involved an initial denaturation step at 95°C for 5 min followed by 20 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min and ended with an extension step at 72°C for 7 min in a DNA Engine thermocycler (Bio-Rad, Mississauga, Ontario, Canada). Following separation of products from primers and primer dimers by electrophoresis on a 2% agarose gel, PCR products of the correct size were recovered using a QIAquick gel extraction kit (Qiagen, Mississauga, Ontario, Canada). For each library, triplicate soil PCR products with unique indexes were mixed in equal nanogram quantities, quantified on a NanoDrop ND2000 spectrophotometer (Thermo Scientific, Wilmington, DE), and sent to Illumina (Hayward, CA) for 125-nucleotide paired-end multiplex sequencing. The Alert DNA was included in a larger proportion than the defined community. Together, the Alert libraries accounted for approximately 75% of the total DNA sent for sequencing in a single lane; other samples unrelated to this study occupied the balance (∼25%) of the template mixture. The library was clonally amplified on a cluster generation station using Illumina, version 4, cluster generation reagents to achieve a target density of approximately 150,000 clusters per tile in a single channel of a flow cell. The resulting library was then sequenced on a GAIIx genome analyzer using Illumina, version 4.0, sequencing reagents, generating paired reads of 125 bases. After sequencing was complete, image analysis, base calling, and error estimation were performed using Illumina Analysis Pipeline (version 2.6).
Clone libraries.
Both soil and pure culture genomic DNAs were used as templates with primers 27f and 1492r (18), targeting the full-length bacterial 16S rRNA gene. PCR amplifications were performed in 25-μl volumes with the concentrations of reagents and reaction conditions described for Illumina library generation, with the exception of the extension step, which was extended to 1.5 min to accommodate the longer amplicon. Reaction products were cloned into the TOPO vector (Invitrogen, Burlington, Ontario, Canada) according to the manufacturer's instructions. Ninety-five positive clones were selected from each library (either soil or pure culture library) and sequenced with Sanger technology (Beckman Coulter Genomic Services, Danvers, MA).
Initial quality filtering.
Using a custom algorithm (PANDAseq [see the supplemental material]), Illumina reads were binned according to index sequence. Overlapping regions within paired-end reads were then aligned to generate “contigs.” If a mismatch was discovered, the paired-end sequences involved in the assembly were discarded. All sequences with ambiguous base calls were also discarded.
Bioinformatic analysis.
All sequences (Illumina- and Sanger-based) were assigned taxonomic affiliations based on naïve Bayesian classification (RDP classifier) (32) with an assignment cutoff of 0.5. Additionally, assembled contigs and Sanger clone library sequences were used as input for modified single-linkage clustering using CD-HIT (21). Good's coverage estimate (9) was calculated for each of the resulting libraries to estimate the sequence coverage of the composite Alert library (AT).
Nucleotide sequence accession numbers.
All Illumina sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under accession number SRA024100. Sanger sequences for the Alert and control libraries were submitted to GenBank under accession numbers JF508183 to JF508359.
RESULTS
Development of Illumina for 16S rRNA gene sequence analysis.
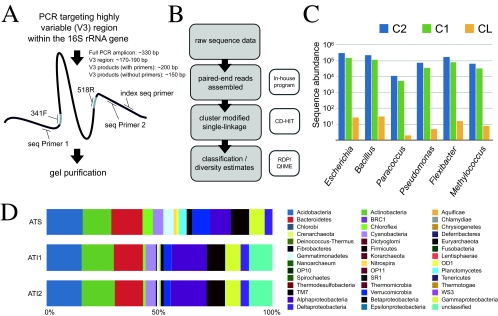
The V3 region of the 16S rRNA gene was selected for this method because of its taxonomic resolution (14), conserved flanking regions (22), and length (8) (∼170 to 190 nucleotides) (Fig. 1A), which is compatible with paired-end 125-base read assembly (Fig. 1B). Complete variable region assembly, by virtue of overlapping 3′-end sequences, reduces sequencing errors and generates data sets that are compatible with established computational analysis pipelines (e.g., QIIME) (2). Custom primers (see Table S1 in the supplemental material) contain regions specific to the Illumina flow cell, unique indexing to allow for multiplexing of samples, and regions complementary to the conserved portions of the bacterial 16S rRNA gene flanking the V3 region. Additional error-correcting indexes (indexes 13 to 84) were designed using the Barcrawl software package (7). The Bacteria-specific primers are identical to those used for the initial application of denaturing gradient gel electrophoresis (DGGE) for microbial community analysis (22). The use of a single low-cycle-number PCR step and subsequent gel purification (Fig. 1A) greatly decreased hands-on library preparation time compared to that for previous sequencing approaches. To validate this method, we analyzed a defined mixture of genomic DNAs from six microorganisms as a control library (C) and an Arctic tundra soil from Alert, Nunavut, Canada (AT), which was previously analyzed by SARST (25). Technical replicates of each sample (C1/C2 and AT1/AT2) were performed to confirm the reproducibility of this technique. Paired-end reads were assembled by aligning the 3′ ends of forward and reverse reads. This assembly step provided additional quality control in the lower-quality 3′ region of each read (Fig. 2), given that Phred scores are additive in the overlapping region. The average assembly overlap was 66 ± 11 bases, and the average postassembly sequence length of our libraries was 150 ± 11 bases (without sequenced primers). This overlap resulted in 2-fold coverage across a substantial portion of each sequence in our libraries. Paired-end reads that did not assemble as contigs were discarded because they possessed sequencing errors (presenting as mismatches between the complementary ends of the two reads). This greatly decreased the number of artifactual sequences used in downstream analyses, with almost 50% of sequences omitted from subsequent analysis for replicate control and Alert tundra libraries (Table 1).
Fig. 1.
Overview of the Illumina 16S rRNA gene sequencing method and generated library data. (A) The schema indicates a PCR (20 cycles) and gel purification of ∼330-base PCR products, including the conserved 16S rRNA gene primer-binding region. (B) Informatics pipeline for generating clusters and taxonomic affiliations. (C) Resulting taxonomic affiliations for the replicate control libraries (C1 and C2) and the Sanger sequencing-based library (CL). (D) Taxonomic affiliations for the Alert tundra duplicate libraries (AT1 and AT2) and the Sanger sequencing-based library (ATS).
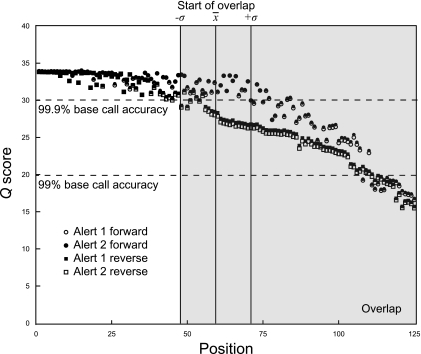
Fig. 2.
Quality (Q) scores for all 125-base sequence reads. The Q score is an integer mapping of P, the probability that the corresponding base call is incorrect, with higher Q scores indicating lower error rates. The magnitude of sequence overlap for each assembled read was characterized, and the mean (x̄) and standard deviation (±σ) were plotted relative to sequence length. The region of potential read overlap as presented does not explicitly calculate the additive Q score at each position, as the range of overlap varied due to the large range of V3 lengths.
Table 1.
Counts of paired-end rRNA gene sequences obtained from the Illumina flow cell (preassembly) and following assembly (postassembly) for the replicate libraries included in this study
| Library | No. of sequences |
Remaining sequences (%) | |
|---|---|---|---|
| Preassembly | Postassembly | ||
| AT1 | 7,570,249 | 4,073,963 | 53.8 |
| AT2 | 4,371,453 | 2,396,331 | 54.8 |
| C1 | 716,366 | 464,045 | 64.8 |
| C2 | 1,350,602 | 842,585 | 62.4 |
Defined community clustering and error rates.
To generate taxonomic profiles of the samples included in this study, the assembled sequence data were assigned to taxonomic groups by use of the naïve Bayesian classifier v.2.1 (32) from the Ribosomal Database Project (RDP) (5). A six-organism defined community was constructed for control purposes by mixing equal proportions of extracted genomic DNAs from six bacterial species for the generation of both Illumina and Sanger libraries. The resulting read counts generated by the Illumina method were at least four orders of magnitude higher than the counts of the corresponding sequences generated using a clone library (Fig. 1C); the costs of generating the Illumina C1 and C2 replicate libraries (>1 million sequences in total) and the Sanger control library (CL; 95 sequences) were roughly equivalent at the time of sequencing.
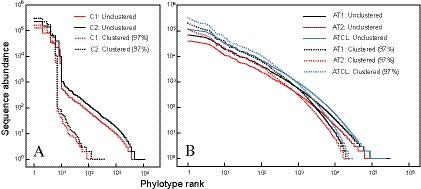
Huse and coworkers (13) reported that a single-linkage preclustering step followed by average linkage clustering at 3% gave a more accurate operational taxonomic unit (OTU) characterization in pyrosequencing data sets and minimally affected the presence and distribution of microbial taxa. Given the large size of our Illumina libraries, linkage clustering would be too computationally intensive. Instead, we used CD-HIT (21) to cluster our control and Arctic tundra data sets at an equivalent of 97% sequence identity (Fig. 3A). Applying such clustering to the C1 and C2 libraries revealed V3 region sequences from the six microorganisms that were well above background noise (Fig. 3A). Clustering the same C1 and C2 libraries at 95% identity had a minimal effect on further reducing low-abundance sequences (data not shown). The 97% clustering step with the assembled control library sequences provided a measure of the total effect of sequencing and PCR errors on the resulting libraries. Clustering of control libraries at 97% identity increased the counts of sequences binned within expected phylotypes by 18%, suggesting that approximately one in every five ∼200-base sequences (including 16S rRNA gene primers) contained at least one error (∼0.1% error, including PCR error). For a comparison, the error rate of Sanger sequencing can be as low as 0.001% (excluding PCR errors), with raw error rates for pyrosequencing (excluding PCR errors) ranging from 1 to 1.5% (30). However, these single-nucleotide errors had little effect on classification of the sequences due to the clustering step. Additionally, sequences were detected in the Illumina libraries (C1 and C2) that were not seen in the Sanger libraries (CL) and which did not cluster within expected V3 regions of the defined communities. These errors did not appear to be caused by PCR error or chimeras because they were confidently affiliated with 16S rRNA gene sequences from known organisms (see Table S2 in the supplemental material). Instead, these sequences likely resulted from the coextraction of DNA from the bacterial growth medium or from low-level contamination of reagents used for PCR, an effect also observed in a recent pyrosequencing study (13). If associated with the bacterial growth medium, these contaminating sequences would not affect results obtained for environmental samples.
Fig. 3.
Rank-abundance curves for duplicate control libraries (A) and Alert Arctic tundra libraries (B). The data shown are the raw data and also the data clustered using CD-HIT at a cutoff of 97%. Note that the Alert Illumina library was considered as separate replicates (AT1 and AT2) and also as a composite library (ATCL), which represents the combined replicates.
Clustering and characterization of Arctic tundra libraries.
The duplicate Arctic tundra libraries displayed high degrees of similarity, based on a comparison of phylum representation, to one another (AT1 to AT2; r = 0.999) and to a small Sanger-sequenced Arctic tundra clone library (ATS; r = 0.950) (Fig. 1D). Representational differences between the two sequencing approaches were likely due to primer bias, because different primers were used for the construction of each library. The similarity of the V3 sequencing data and the Sanger-based clone library was much higher by overall phylum distribution than those between these libraries and additional data sets generated from the same sample by V1 region sequencing (17 to 55 bases in length) with either SARST (25) or Illumina-based approaches (unpublished data; see Fig. S1 in the supplemental material).
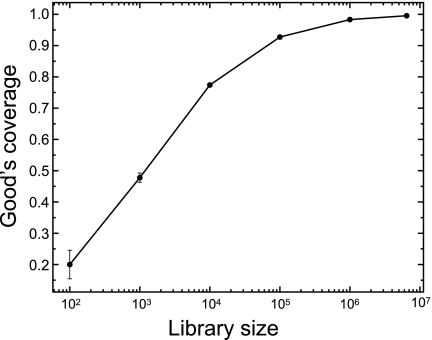
Clustering at 97% similarity reduced the proportion of singleton sequences from 5.9% (unclustered) to 0.17% (clustered) of all sequences in the combined AT libraries (Fig. 3B), and high-abundance phylotypes increased disproportionately to low-abundance phylotypes when AT1 was combined with AT2 to form ATCL. Calculated Good's nonparametric coverage estimates for the combined AT Illumina data set increased from 0.962 to 0.996 for unclustered and clustered libraries, respectively. In contrast, Good's coverage estimate for the full-length 16S rRNA gene Sanger sequencing-based clone library clustered at 97% identity (87 clones) was only 0.207. Good's coverage estimate was also used to assess the effect of library size on coverage with increasing subsamples of the combined AT data set. The Good's coverage estimates were >0.95 with >1 million sequences sampled. Additionally, once millions of sequences were sampled, Chao1 richness estimates began to reach an asymptote (see Fig. S3A in the supplemental material). This illustrates that multimillion-sequence libraries (generated using the Illumina sequencing method) result in high estimates of completeness of sampling (Fig. 4). Replicate Illumina sequence libraries for the Alert tundra DNA sample (AT1 and AT2) were highly similar, with the majority of sequences (99.57%) corresponding to clusters detected in both replicates (see Fig. S2, inset) and with a high Bray-Curtis similarity value (0.96), especially when the 50 most abundant phylotypes were considered (0.99) (see Fig. S3B). Rank-abundance curves for the most abundant phylotypes were nearly identical in distribution (see Fig. S3B). Although there were several clusters unique to one of the replicate libraries, these were composed largely of clusters represented by single sequences (see Fig. S2).
Fig. 4.
Effect of library size on phylotype coverage. Randomly subsampled libraries were drawn in triplicate from combined AT libraries and used to calculate Good's coverage estimates. Averages for triplicates were plotted with standard deviations.
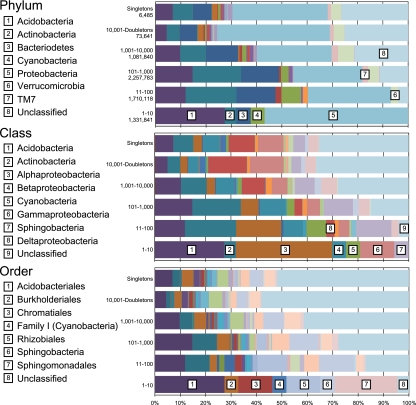
Taxonomic classification of 97% sequence identity clusters demonstrated a distinct taxonomic shift in comparing predominant to low-abundance clusters or ranks (Fig. 5). The 10 most abundant ranks accounted for 20.6% of all sequences and belonged to the Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and Proteobacteria (see Tables S3, S4, and S5 in the supplemental material). Except for Cyanobacteria, which were largely absent in the lower ranks and singletons, these phyla remained predominant throughout the low-abundance ranks. There was an increase in the number of phyla present in low-abundance ranks, with a maximum of 28 phyla represented by the 10,001-doubleton abundance rank. There was also a notable increase in the proportion of Verrucomicrobia sequences in midrange abundance ranks (11 to 100 and 101 to 1,000 most abundant), as these were absent in the high-abundance ranks (1 to 10). Furthermore, sequences affiliated with TM7 were predominant only in low-abundance ranks. The proportion of sequences assigned as unclassified (i.e., weakly classified or not classified at all) increased from 0% among the high-abundance ranks (1 to 10) to approximately 25% of all rare sequence cluster ranks (see Table S3). Future work will aim to separate errors from genuine diversity, confirming that low-abundance sequences are not simply accumulated artifacts from increased sequencing intensity, as suggested by Kunin et al. (16).
Fig. 5.
Taxonomic affiliations at the levels of phylum, class, and order for consecutive abundance ranks of sequence data clustered at 97% with CD-HIT. Predominant taxa are represented in the bottom row, and singletons are at the top for each taxonomic level. Full details of RDP affiliations are summarized in Tables S3, S4, and S5 in the supplemental material.
Patterns of rank-specific taxonomic distributions of phyla were also present in class and order classifications (Fig. 5; see Tables S4 and S5 in the supplemental material). In each case, the abundant clusters maintained their predominance in lower-abundance ranks (with the notable exception of cyanobacterial sequences), with taxonomic diversity increasing in lower-abundance ranks. Notably, in class and order classifications, the presence of clusters labeled unclassified increased incrementally from <5% (ranks 1 to 10) to ∼50% (singletons) for both classifications. The increase in unclassified sequences was larger for low-abundance clusters within a taxonomic level and increased with depth of classification (i.e., the majority of sequences were successfully classified to the phylum level, while even some highly abundant sequences were not successfully classified to the ordinal level).
DISCUSSION
Here we demonstrate improvements in both sampling depth and sequence quality by use of an inexpensive and rapid sequencing methodology. An advantage of this technique over current high-throughput methods is the assembly of paired-end reads, which greatly reduces the number of erroneous sequences included in downstream analyses. Importantly, as the read lengths for the Illumina platform increase (currently at ∼125 bases), so too will the quality of the libraries generated with this technique. Additionally, the use of index sequences enables many samples to be sequenced in parallel. We have tested 24 indexed primers in our laboratory (data not shown), and additional index sequences have been provided that can further increase sample throughput (see Table S1 in the supplemental material). Further improvements to this method can be introduced, such as the addition of a highly diverse series of bases adjacent to the forward sequencing primer binding area (see Table S1). This addition improves Illumina base calling because the algorithm identifies clusters optimally on the flow cell when maximum nucleotide diversity is present across the first four bases sequenced in the forward read. In addition, the long oligonucleotide primers used here were purified commercially by PAGE for an additional cost (IDT, Coralville, IA). Future research will determine if standard desalting of primers will be sufficient to generate Illumina data sets, which would reduce the start-up cost for this new technology.
With the increase in recovered sequences, there is a corresponding increase in artifactual sequences. The capacity of the Illumina platform to generate enormous data sets is undoubtedly an advantage; however, if low-abundance phenotype discovery and accurate measurements of alpha diversity are desired, errors must be managed effectively. Otherwise, community characterization is only useful at a coarse level. In this study, assembly was accomplished by the use of overlapping paired-end reads, and a modified single-linkage clustering protocol was applied at 97% sequence identity. Future work will identify effective clustering algorithms that adequately reduce data sets to the expected phylotype diversity, as shown recently for 454 pyrosequencing data (13), and that are scalable to sequence libraries possessing many millions of sequences and hundreds (or thousands) of samples. Additionally, problems resulting from the sensitivity of the technology (e.g., sequencing of low-abundance sequence contamination in laboratory growth medium) would be bypassed by multiplex PCR amplifications directly from environmental samples as outlined in this protocol.
Regardless of sequencing artifacts, advances in sequencing technologies are paralleled by increased magnitudes of phylotype diversity surveyed from microbial communities. Although a small number of sequences may be sufficient to detect underlying patterns differentiating highly divergent communities (15), larger data sets are required to identify more subtle responses to environmental factors among less predominant populations and for increased sequence coverage of the rare biosphere (13, 31). Rare microbial taxa likely represent microorganisms that (i) are adapted to life at low relative abundance, (ii) have not been discovered previously, and (iii) possess abundance distributions with important correlates to measured physicochemical parameters. In this study, the Illumina sequencing platform provided access to low-abundance phylotypes from soil with adequate coverage (Fig. 4) and combined library sizes greater than those reported previously (3, 4, 29). The main limitation of recent iterations of the Illumina platform has been the reduced taxonomic resolution of short sequence reads (3, 8, 20). With the introduction of 125-base paired-end reads reported here, this sequencing methodology can now span the taxonomically informative V3 variable region of the 16S rRNA gene and will soon generate two-fold coverage of complete PCR amplicons as sequence length continues to increase. Note that the V3 region chosen here was selected because the primers used are the same as those used for DGGE of bacterial communities (22) and that this region is longer (∼170 to 190 bases) than the V6 region, which was sequenced elsewhere (∼105 to 120 bases) (8). Although base-calling accuracy decreases markedly toward the 3′ end, the sequence read overlap of 66 ± 11 nucleotides (ATCL library) greatly increased data quality in this region (Fig. 2). The primers and adaptors are modular, so this sequencing methodology can readily be modified to target other genes or regions of interest. This versatile, affordable, and powerful methodology greatly increases the depth at which low-abundance organisms can now be probed, as noted by high Good's coverage estimates (Fig. 4), high levels of similarity between replicates (see Fig. S3 in the supplemental material), and the number of unclassified or unique taxa in low-abundance groups (Fig. 5), suggesting that we are now able to comprehensively and reproducibly characterize and compare abundant and rare populations across multiple samples derived from complex microbial communities.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), a catalyst grant from the Canadian Institutes of Health Research (CIHR), and funds from the Canadian Foundation for Innovation (CFI).
We thank Abizar Lakdawalla, Keith Moon, Christian Haudenschild, and Grace Murray of Illumina, Inc., for providing sequencing support for this study. Andre Masella is thanked for technical assistance with Illumina data analysis.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Andersson A. F., et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caporaso J. G., et al. 2010. QIIME allows analysis of high-throughput community sequence data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caporaso J. G., et al. 15 March 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Claesson M. J., et al. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cole J. R., et al. 2008. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Curtis T. P., et al. 2006. What is the extent of prokaryotic diversity? Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2023–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Frank D. N. 2009. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics 10:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gloor G. B., et al. 2010. Microbiome profiling by Illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5:e15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Good I. J. 1952. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 10. Hamady M., Walker J. J., Harris J. K., Gold N. J., Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huber J. A., et al. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100 [DOI] [PubMed] [Google Scholar]
- 12. Hummelen R., et al. 2010. Deep sequencing of the vaginal microbiota of women with HIV. PLoS One 5:e12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huse S. M., Welch D. M., Morrison H. G., Sogin M. L. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huse S. M., et al. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuczynski J., et al. 2010. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat. Methods 7:813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12:118–123 [DOI] [PubMed] [Google Scholar]
- 17. Kysela D. T., Palacios C., Sogin M. L. 2005. Serial analysis of V6 ribosomal sequence tags (SARST-V6): a method for efficient, high-throughput analysis of microbial community composition. Environ. Microbiol. 7:356–364 [DOI] [PubMed] [Google Scholar]
- 18. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. Wiley and Sons Ltd., Chichester, United Kingdom [Google Scholar]
- 19. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lazarevic V., et al. 2009. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Methods 79:266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W., Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 22. Muyzer G., De Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neufeld J. D., Li J., Mohn W. W. 2008. Scratching the surface of the rare biosphere with ribosomal sequence tag primers. FEMS Microbiol. Lett. 283:146–153 [DOI] [PubMed] [Google Scholar]
- 24. Neufeld J. D., Mohn W. W. 2005. Assessment of microbial phylogenetic diversity based on environmental nucleic acids, p. 219–260 In Stackebrandt E. (ed.), Molecular identification, systematics, and population structure of prokaryotes. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 25. Neufeld J. D., Mohn W. W. 2005. Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl. Environ. Microbiol. 71:5710–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neufeld J. D., Yu Z., Lam W., Mohn W. W. 2004. Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities. Environ. Microbiol. 6:131–144 [DOI] [PubMed] [Google Scholar]
- 27. Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337–365 [DOI] [PubMed] [Google Scholar]
- 28. Pedrós-Alió C. 2007. Dipping into the rare biosphere. Science 315:192–193 [DOI] [PubMed] [Google Scholar]
- 29. Rousk J., et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 30. Shendure J., Hanlee J. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 31. Sogin M. L., et al. 2006. Microbial diversity in the deep sea and underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Z., Yu M., Morrison M. 2006. Improved serial analysis of V1 ribosomal sequence tags (SARST-V1) provides a rapid, comprehensive, sequence-based characterization of bacterial diversity and community composition. Environ. Microbiol. 8:603–611 [DOI] [PubMed] [Google Scholar]
- 34. Zhou H. W., et al. 21 October 2010. BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J. 5:741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.