Abstract
To understand the continuous problems that Escherichia coli O157:H7 causes as food pathogen, this study assessed global gene regulation in bacteria growing on meat. Since FlhD/FlhC of E. coli K-12 laboratory strains was previously established as a major control point in transducing signals from the environment to several cellular processes, this study compared the expression pattern of an E. coli O157:H7 parent strain to that of its isogenic flhC mutant. This was done with bacteria that had been grown on meat. Microarray experiments revealed 287 putative targets of FlhC. Real-time PCR was performed as an alternative estimate of transcription and confirmed microarray data for 13 out of 15 genes tested (87%). The confirmed genes are representative of cellular functions, such as central metabolism, cell division, biofilm formation, and pathogenicity. An additional 13 genes from the same cellular functions that had not been hypothesized as being regulated by FlhC by the microarray experiment were tested with real-time PCR and also exhibited higher expression levels in the flhC mutant than in the parent strain. Physiological experiments were performed and confirmed that FlhC reduced the cell division rate, the amount of biofilm biomass, and pathogenicity in a chicken embryo lethality model. Altogether, this study provides valuable insight into the complex regulatory network of the pathogen that enables its survival under various environmental conditions. This information may be used to develop strategies that could be used to reduce the number of cells or pathogenicity of E. coli O157:H7 on meat by interfering with the signal transduction pathways.
INTRODUCTION
Escherichia coli O157:H7, which belongs to the group enterohemorrhagic E. coli (EHEC), causes nearly 60,000 cases of human infection annually in the United States (17). Symptoms of infection range from bloody diarrhea to urogenital infections and can involve renal failure and meningitis. The severe forms of disease result from the bacterium's capability to produce Shiga toxin (53). Infection in cattle does not produce overt disease (e.g., diarrhea), but cattle can serve as a primary reservoir for humans (24). Transmission occurs via consumption of contaminated meat or meat products, drinking contaminated water, or personal contact (16, 35, 54).
Protection of consumers from infection by E. coli O157:H7 through meat occurs at various stages. At the production end, spraying the carcasses with a variety of chemicals, predominantly acids, is partially successful (5). At the consumer end, the Food Safety and Inspection Service (FSIS) recommends storage of meat at low temperature and thorough cooking before consumption. As a consequence of these measures, the number and frequency of outbreaks and meat recalls have declined steadily between 1994 and 2005 (FSIS). Still, recalls and outbreaks occur, and E. coli O157:H7 remains a safety hazard in the kitchen (1, 2).
Predominant among the reasons for the incomplete effectiveness of the acid sprays is the fact that bacteria develop resistance against any such antibacterial substances aimed at reducing their growth rate (11). In the case of acid resistance, this is particularly troublesome because bacteria now survive the acid barrier of the stomach (34, 46). As a novel research direction, impacting bacterial signal transduction may be less likely to induce resistance.
As one example of such a signal transduction pathway, early research by our lab has shown that extracellular serine, added to E. coli K-12 cells grown into mid-exponential phase on mixed amino acids, simultaneously increased the cell division rate and decreased the synthesis of flagella (40). The signaling transduction cascade involved formation of acetyl phosphate (41); phosphorylation of the response regulator of the osmoregulation two-component system, OmpR (37); and inhibition of flhDC expression by phosphorylated OmpR (49). The flhDC operon encodes the FlhD/FlhC transcriptional activator complex that was initially described as a flagellar regulator (4). Later research demonstrated that FlhD/FlhC was actually a global regulator, regulating a myriad of cellular processes in E. coli K-12 (38, 43), including cell division (39, 40). Gene regulation studies suggested a role for FlhD/FlhC in regulation of anaerobic respiration and sugar acid degradation via the methyl-accepting chemotactic protein Aer (43). Later on, a gene deletion study established an effect of Aer on colonization of the mouse intestine (21).
In a continuation of the gene regulation studies, Prüß and coworkers summarized a network of transcriptional regulation centered around FlhD/FlhC (42). The network included many genes encoding cell surface organelles that are characteristic of the different stages of biofilm formation, as well as their regulators. The study hypothesized the importance of the master regulator FlhD/FlhC in yet another important aspect of bacterial survival, biofilm formation. The effect of components of the network on biofilm formation was confirmed by a high-throughput study investigating the environmental and genetic factors that determine biofilm amounts (44).
In an attempt to investigate similar regulatory networks in pathogenic E. coli O157:H7, literature pertaining to individual regulation studies was reviewed, and a network was constructed (51). Two very intense centers of regulation were found. The first of these was the locus of enterocyte effacement (LEE), a chromosomally located pathogenicity island in E. coli O157:H7. The locus comprises many genes involved in attachment and type III secretion and the genes encoding intimin (eaeA) and its translocated receptor (tir) (36). The second center of intense regulation was observed around the flhDC operon. The network, however, lacked any indication of a regulatory connection between the LEE pathogenicity genes and FlhD/FlhC.
Understanding the importance of FlhD/FlhC as an integrator between environmental signals and cellular responses, this study investigated gene regulation by FlhD/FlhC in E. coli O157:H7 on meat. We carried out a series of microarray experiments, followed by real-time PCR analysis and comparison of the expression level of genes in an flhC mutant with its isogenic E. coli O157:H7 parent. This was done with E. coli growing on the surface of meat over a time period of 10 days at refrigeration temperature (10°C). This study provided the first evidence that FlhC regulates genes in E. coli O157:H7 grown on meat. The regulated genes that will be discussed in this study include genes involved in cell division, biofilm formation, and the LEE pathogenicity island. Phenotypes relating to cell division, biofilm formation, and pathogenicity paralleled gene regulation results. The implications of these findings in the context of the development of antimicrobials will be discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and storage conditions.
The bacterial strains used in the study are listed in Table 1. E. coli O157:H7 and its flhC isogenic mutant (14) were kindly provided by Scott A. Minnich (University of Idaho, Moscow). The mutant contains a 12-bp deletion in the open reading frame (ORF) of flhC which was confirmed by PCR and sequence analysis. Resistance toward streptomycin sulfate (1,000 μg/ml; Sigma-Aldrich, MO) was introduced into the parental strain (PS1) and the flhC mutant (PS2), using a previously established technique (6). PS1 was also made resistant toward nalidixic acid (Sigma-Aldrich) at a concentration of 50 μg/ml (PS1). PS2 was transformed with the FlhC-expressing plasmid pXL26 (33) to yield PS3. All strains were stored as freezer stocks at −80°C in 8% dimethyl sulfoxide (DMSO). The cultures were plated onto Luria-Bertani (LB) plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar) prior to use and incubated overnight at 37°C.
Table 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| ATCC 43894 | E. coli O157:H7 | American Type Culture Collection (14) |
| O157:H− | flhC | 12 |
| PS1 | E. coli O157:H7 Strr Nalr | This study |
| PS2 | E. coli O157:H− Strr | This study |
| PS3 | PS2 pXL26 | This study |
| Plasmid | ||
| pXL26 | pT7-7 flhC | 33 |
The designation “H−” refers to the lack of the H7 flagellin due to the flhC mutation, which renders the entire flagellar master regulator complex, FlhD/FlhC, dysfunctional.
Comparison of gene expression between the parental strain and the flhC mutant on the surface of meat. (i) Preparation and inoculation of the meat pieces.
Beef strip loin was purchased from a local supplier and transported to the lab within an hour of purchase. The meat was processed by trimming excess fat from the surface and surface sterilized by being dipped in boiling water for 30 s. The sterilized meat was retrimmed aseptically to remove the outer layer. The meat was then cut into uniform pieces of about 10 cm in length, 1 cm in width, and 1 cm in thickness.
For the inoculation of the meat pieces, overnight cultures of PS1 (parent E. coli) and PS2 (flhC mutant) in brain heart infusion broth (BHI; Becton Dickinson, LePont de Claix, France) were diluted to an optical density at 600 nm (OD600) of 0.1 in maximum recovery diluent (MRD; 0.85% peptone, 0.1% NaCl) and used as individual inocula for the meat pieces. The dilutions were done as previously reported (32), and the final inocula contained between 1 × 106 and 4 × 106 CFU/ml of bacteria. Equal numbers of meat pieces were dipped into the two separate inocula and placed in plastic trays. The trays were wrapped in cling film and incubated at 10°C for 10 days. Uninoculated meat pieces dipped in MRD were incubated as a negative control.
To enumerate background flora, uninoculated meat pieces from each test group were sampled on days 1, 3, 5, 7, and 10 postinoculation. Growth of the background flora that is characteristic for beef (32) was determined. Sampling involved homogenization of 20 g of meat sample in 80 ml of MRD, using a Stomacher 400 circulator (Seward, NY), followed by serial dilution and plating onto plates containing different media for enumeration. The population of Enterobacteriaceae on meat was established by using violet-red bile-glucose agar (VRBGA; Oxoid, Hampshire, United Kingdom), for Brocothrix by using streptomycin-thallous acetate-actidione agar (STAA; Oxoid) (18), for heterofermentative Lactobacillus by using all purpose Tween (APT) agar (BD) (15), and for Pseudomonas by using Pseudomonas (PS) agar (Oxoid). The growth of PS1 and PS2 within this background flora was established by inoculating meat pieces with PS1 and PS2. Samples were prepared and treated as described for the inoculated meat pieces. PS1 and PS2 were enumerated by plating onto sorbitol MacConkey agar (SMAC; BD), to which streptomycin was added at a concentration of 1,000 μg/ml (SMAC-strep). Additionally, the total bacterial count was determined for PS1- and PS2-inoculated, as well as uninoculated meat samples. This was done with standard plate count agar (PCA; BD). All enumeration experiments were performed twice. Representative data from one experiment are presented; the other sample yielded qualitatively similar results.
(ii) RNA isolation and cDNA synthesis.
The meat pieces inoculated with PS1 and PS2 were homogenized separately after an incubation period of 10 days as described above. The eluents from the homogenized samples were collected in sterile bottles. Stop solution (5% phenol in ethanol) was added at a ratio of 1:10 to inhibit further bacterial growth and degradation of mRNA. The sample was then pelleted at 1,000 × g for 10 min to sediment the remaining meat debris. The resultant supernatant was subjected to high-speed centrifugation at 7,000 × g for 10 min to pellet bacterial cells. The bacterial pellets hence obtained from the two inocula were flash frozen over a mixture of dry ice and acetone and stored at −80°C for a maximum of 10 days. The pellets were thawed as required for RNA isolation.
RNA for microarray experiments was isolated from pellets of three independently grown bacterial cultures per strain. For the first real-time PCR experiment, RNA samples were used that were produced for the microarray experiment. Since one of the experiments did not yield enough RNA, an additional RNA sample was produced from each strain for the real-time PCR. Isolation of RNA was done by the previously established hot phenol-sodium dodecyl sulfate method (12). Briefly, one extraction each with phenol, phenol-chloroform, and chloroform was performed, followed by isopropyl alcohol precipitation. RNA was treated with DNase I twice and passed through RNeasy minicolumns (Qiagen, Los Angeles, MD) for a final cleanup. RNA was reverse transcribed to cDNA, using a previously validated protocol by Patrick O. Brown (http://cmgm.Stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm). For real-time PCR, cDNA was prepared with unlabeled deoxynucleoside triphosphates (dNTPs). cDNA was stored at −20°C. For the microarray experiment, RNA was reverse transcribed with amino-allyl-labeled dUTP. The cDNA from the parent strain was labeled with Alexa Fluor 555 (Invitrogen, CA), and the cDNA from the mutant was labeled with Alexa Fluor 647 (Invitrogen). The labeled cDNA samples were vacuum dried individually in a Vacufuge (Eppendorf, NY) and stored at −20°C.
(iii) Microarray experiment.
For the microarray experiment, the E. coli O157 OciChip from Ocimum Biosolutions (Hyderabad, AP, India) was used. The chip consists of 6,336 spots and covers the genomes of three strains, E. coli K-12 MG1655, E. coli O157:H7 EDL933, and E. coli O157:H7 VT2-Sakai. Each spot is comprised of a 50-mer oligonucleotide, which is representative of one protein-coding open reading frame. The three strains used for the design of the microarray chips share a backbone of roughly 3,500 genes.
Prior to the hybridization, two differentially labeled cDNA samples (derived from the two bacterial strains) were resuspended separately in 60 μl of the hybridization buffer (Ocimum Biosolutions) and preheated at 42°C for 10 min. The two suspensions were then combined to form the hybridization mixture. The hybridization mixture was heated for 3 min at 95°C, cooled on ice, and briefly centrifuged. This mixture was then loaded onto the slides in a Hyb4 hybridization station (Genomic Solutions, Ann Arbor, MI). After hybridization at 42°C overnight, the slides were washed for 5 min each in wash buffer 1 (2× SSC, 0.1% SDS) and wash buffer 2 (1× SSC) and then washed for 1 min in wash buffer 3 (0.1× SSC). (SSC is 0.15 M NaCl plus 0.015 M sodium citrate.) A total of five such hybridizations were performed from the three independently grown bacterial cultures. All slides were scanned with a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA) at 635 and 532 nm. The voltage of the photomultiplier tubes was adjusted such that maximal signal could be obtained with minimal loss due to bleaching of the dye.
Analysis of the individual slides was done using the GenePrix Pro 6.0 software package from Axon (Molecular Devices). Median intensities were used after background subtraction. Pixel values for the parental strain were divided by those for the mutant to obtain expression ratios. Data were imported into Microsoft Excel. Ratio-based normalization was performed across the five arrays, using the average expression ratios for each array. We considered genes to be expressed higher in the parent strain than in the mutant when the expression ratio was higher than 2 and to be expressed higher in the mutant than in the parent strain when the expression ratio was lower than 0.5. This is consistent with current literature (3, 29). Genes that were lacking more than two data points out of the five replicate experiments were excluded from the analysis. Statistical testing was completed using the z-test (http://www.mathworks.com/help/toolbox/stats/f13914.html), which evaluated whether the means of the expression ratios are statistically different from a set number. In our case, this set number is 1, indicating genes that are not regulated. Genes that had a P value of ≤0.05 were considered putative targets of FlhC and further investigated by real-time PCR. The Results and Discussion sections will focus on a selection of these genes.
(iv) Real-time PCR.
Unlabeled cDNA samples from PS1 and PS2 grown on meat were used for this part of the experiment and tested in three replicate reactions. The primer couples used to quantify the selected genes were designed using BeaconDesign 3.0 (Premier Biosoft International, CA) and are listed in Table S1 in the supplemental material.
The real-time PCR was performed with iQ SYBR green supermix (Bio-Rad, Hercules, CA). The reaction mixture contained 10 to 100 ng cDNA from one of the two strains, 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 0.625 U iTaq DNA polymerase, 0.2 mM each dNTP, 3 mM MgCl2, SYBR green I, 10 nM fluorescein, and 100 nM each primer. The PCR was performed with the Bio-Rad iQ5 system using the following cycling conditions: 95°C for 3 min, followed by 50 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The reactions were concluded at 72°C for 10 min.
Melting curves for each reaction were obtained and yielded single peaks for all primer couples. The absence of genomic DNA was determined by subjecting the original RNA samples to the procedure described above. Additionally, dilutions of cDNA samples were analyzed as controls. For all samples, a 2-fold dilution yielded a difference in the threshold cycle (CT) of 1.
To normalize real-time PCR data, various techniques are routinely employed (58). For our experimental purposes, we used the 2−ΔΔCT technique after normalization with two housekeeping genes. The genes ompX and gapA were selected as housekeeping genes after screening a total of five genes for the apparent lack of regulation by FlhC under the conditions of our experiment. The average difference in CT was determined for ompX and gapA (ΔCT house). Differential threshold crossings (ΔCT gene) were calculated for each tested gene by subtracting the CT of the mutant strain from that of the parent. ΔCT gene values were normalized by subtracting ΔCT house from ΔCT gene to yield ΔΔCT. Expression ratios were then calculated as 2−ΔΔCT. This was done for each replicate. For each gene, the average of the expression ratios was determined across the replicate experiments. The standard error is indicated.
Comparison of phenotypes between the parental strain and the flhC mutant. (i) Biofilm and cell division.
The estimations of biofilm amounts and bacterial cell numbers (indicative of cell division) were done in a combined experiment with 96-well plates. Three independent overnight cultures (biological replicates) of PS1 (parent), PS2 (flhC mutant), and PS3 (PS2 transformed with FlhC-expressing plasmid pXL26) were diluted 1:100 in beef broth (0.3% beef extract, 0.5% peptone). One hundred microliters of the diluted cultures was dispensed into each well of a 96-well plate in such a way that five technical replicates of each of the three biological replicates were achieved. The plate was incubated at 10°C for 10 days. On day 10, the supernatant from each well was carefully aspirated, serially diluted, and plated on SMAC-strep (BD) agar plates to obtain cell counts. Cell counts were reported as CFU/ml. The biofilms that formed in each well were then quantified by using the previously established ATP assay (50). Briefly, biofilms were washed twice with phosphate-buffered saline (PBS) in order to remove any residual medium components. The biofilms were air dried and quantified with 100 μl of BacTiter Glo reagent (Promega, WI). The biofilms were incubated with the reagent for 10 min at room temperature, and the bioluminescence was recorded with a TD 20/20 luminometer (Turner Design, CA). The bioluminescence was reported as relative lux units (RLU). For both cell division and biofilm amounts, we performed a Student's t test across all 15 data points.
(ii) Electron microscopy of biofilms.
Biofilms were processed for scanning electron microscopy as described previously (50). Biofilms from PS1 and PS2 were grown for 10 days at 10°C in beef broth separately on glass coverslips (Assistant, Niderau, Germany), air dried, and fixed in glutaraldehyde. The fixed biofilm was subjected to graded alcohol dehydration and coated with gold-palladium. Images were obtained with a JEOL JSM-6490LV (JOEL, Ltd., Tokyo, Japan) scanning electron microscope at a 1,000× magnification.
(iii) Chicken ELA.
For the chicken embryo lethality assay (ELA), overnight cultures of PS1 and PS2 grown in beef broth were centrifuged and washed twice with PBS. The bacterial pellets were resuspended in PBS and diluted to a range of 100 to 300 CFU/ml. One hundred microliters of this dilution was used as the inoculum.
Pathogen-free chicken embryos (Sunrise Farms, Inc., NY) were incubated in a Sportsman incubator, model 1502 (G.Q.F. Manufacturing Co., Savannah, GA), at 37°C for 12 days. The humidity level of the incubator was maintained at 86%. On the twelfth day, the allantoic fluids of the embryonated eggs were inoculated with the bacterial suspensions, each containing 10 to 30 bacteria. Each strain was tested in four independent experiments in test groups of 15 embryos, totaling 60 embryos per bacterial strain. The previously described (19, 52) avian E. coli strains V1 and A4 served as positive and negative controls, respectively. Additional negative control groups of either uninoculated embryos or embryos inoculated with PBS were also processed. All embryos were candled once a day for 3 consecutive days, and the number of dead embryos on each day was recorded. The lethality was reported as the percentage of dead embryos across the total of 60 embryos. This was done for each strain and the uninoculated and PBS-inoculated controls.
Due to the discontinuous nature of percentage data and the fact that these data are not normally distributed, percent lethality was subjected to logistic regression analysis. Odds were calculated for experimental strains as p/(1 − p), with p being the percentage lethality determined by ELA. Odds ratios across multiple experiments were determined, using the PS1 strain as baseline reference strain for the logistic regression. Lethality by the tested strains was also compared using the Duncan's multiple-group comparison, a test that determines the statistical significance of differences in lethality between the test groups. All statistical analysis was performed on SAS v 9.1 (SAS Institute, Inc., Cary, NC).
Microarray data accession number.
The results from the complete microarray experiment are deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE28836.
RESULTS
FlhC-mediated gene regulation in E. coli O157:H7 growing on the surface of meat was investigated with microarray experiments. The putative target genes were confirmed by real-time PCR and grouped according to their cellular functions. Additional genes from the same functions were tested that the microarray had not hypothesized as putative targets. Three different phenotypes were determined and correlated with the observed gene regulation. The following sections report the experimental and statistical results of the outlined experimental procedures.
Determination of bacterial flora on meat.
Residual bacterial spoilage flora on surface-sterilized meat was determined prior to the microarray experiments. This uninoculated control piece of meat had a total bacterial count of 8.7 × 1010 CFU/g (Table 2). The bacterial counts obtained from each selective medium (1.25 × 1010 for Brocothrix, 5 × 1010 for heterofermentative Lactobacillus, and 1.25 × 1010 for Pseudomonas) added up to approximately this number. The ability of our inocula (PS1 and PS2) to grow within this bacterial community was investigated in an independent experiment. Meat pieces were inoculated with PS1 and PS2 and tested for E. coli O157:H7. The total bacterial count of the meat sample inoculated with PS1 was 1.25 × 1011 CFU/g; meat inoculated with PS2 had a total bacterial count of 1.37 × 1011 CFU/g. Addition of the background flora count obtained from the control group (8.7 × 1010 CFU/g) to the PS1 (1.25 × 1010 CFU/g) and PS2 (5 × 1010 CFU/g) counts yielded the approximate total bacterial count for the respective group.
Table 2.
Bacterial counts on the beef surface
| Tested bacterial species | Bacterial count (CFU/g of meat) |
||
|---|---|---|---|
| PS1 (parent) | PS2 (flhC mutant) | Control | |
| Naturally occurring bacteria on beef | |||
| Total bacterial count | 1.25 × 1011 | 1.37 × 1011 | 8.7 × 1010 |
| Enterobacteriaceae | NDa | ND | 0 |
| Brocothrix | ND | ND | 1.25 × 1010 |
| Heterofermentative Lactobacillus | ND | ND | 5 × 1010 |
| Pseudomonas | ND | ND | 1.25 × 1010 |
| Inoculated bacteria | |||
| E. coli O157:H7 | 1.25 × 1010 | 5 × 1010 | 0 |
ND, not done.
In E. coli O157:H7 growing on the surface of meat, FlhC regulates genes responsible for cell division, biofilm formation, and pathogenicity.
The previous experiment had established that our inocula (PS1 and PS2) were able to grow on meat and represented 10 to 50% of the total bacterial flora. Determination of genes that were regulated by FlhC in E. coli O157:H7 growing on meat was done next. The following section outlines the results obtained from the microarray experiments and the real-time PCR.
The microarray experiment hypothesized 287 genes as being regulated by FlhC.
PS1 and PS2 were grown on meat, and differentially labeled cDNA samples were obtained as described in Materials and Methods. mRNA levels for 287 genes were at least 2-fold higher in the flhC mutant (PS2) than in the parental strain (PS1). The putative target genes were then divided into functional groups, based on information provided by the manufacturer of the microarray chips. This information is summarized in Table S2 in the supplemental material. This included 125 genes belonging to the central metabolism, 10 genes that were virulence and LEE pathogenicity island related, and seven genes that were related to cell division. Additionally, 35 of the regulated genes encoded transporters. Two biofilm related genes were also identified as a putative target of FlhC. The remaining putative FlhC targets were associated with DNA synthesis and transcription/translation. Table 3 lists a selection of relevant genes belonging to six functional groups. Representative genes from each of these groups were selected and subjected to real-time PCR analysis for confirmation.
Table 3.
Microarray analysis of PS1 and PS2 grown on the surface of beef
| ORF (total no. of genes)a | Geneb | Protein functionc | Expression ratiod | z-test P valuee |
|---|---|---|---|---|
| Metabolism (125) | ||||
| b3236 | mdh* | Malate dehydrogenase | 0.15 | 5.5 × 10−10 |
| ECS0753 | sucC | Succinate dehydrogenase | 0.17 | 2.6 × 10−5 |
| Z0877 | sdhA* | Succinate dehydrogenase | 0.28 | 7.7 × 10−4 |
| b0430 | cyoA* | Cytochrome o-oxidase | 0.25 | 0.0032 |
| b0432 | cyoC | Cytochrome o-oxidase | 0.14 | 1.27 × 10−4 |
| Transport (34) | ||||
| Z1276 | ompF* | Outer membrane protein F | 0.47 | 0.0098 |
| Z3473 | ompC* | Outer membrane protein C | 0.13 | 2.74 × 10−5 |
| ECS1744 | oppB* | Oligopeptide permease | 0.38 | 0.0009 |
| Z3570 | hisQ | Histidine permease | 0.39 | 0.0187 |
| ECS5330 | osmY | Hyperosmotically inducible periplasmic protein | 0.20 | 6.1 × 10−4 |
| b4123 | dcuB | Anaerobic dicarboxylate transporter | 0.29 | 0.0548 |
| Cell division (7) | ||||
| b0039 | ftsW | Membrane protein involved in shape determination | 0.085 | 1.93 × 10−9 |
| ECS0975 | ftsK* | Cell division | 0.14 | 8.06 × 10−9 |
| Z0105 | ftsZ* | Tubulin-like GTP binding protein | 0.43 | 0.0032 |
| Z4333 | ftsY* | Cell division membrane protein | 0.33 | 1.6 × 10−4 |
| b0924 | mukB | Kinesin-like cell division protein | 0.093 | 7.7 × 10−7 |
| Z5336 | minC* | Inhibits ftsZ ring formation | 0.30 | 0.057 |
| ECS1670 | chpB | Probable growth inhibitor | 0.40 | 0.0062 |
| LEE pathogenicity island (10) | ||||
| Z5119 | escN* | Type III secretion system | 0.35 | 0.0047 |
| Z5112 | tir* | Translocated intimin receptor | 0.10 | 4.03 × 10−9 |
| Biofilm (2) | ||||
| b0196 | rcsF* | Regulator in colanic acid synthesis | 0.38 | 0.0025 |
| b2058 | wcaB* | Putative transferase | 0.44 | 0.0087 |
| Miscellaneous | ||||
| b0438 | clpX* | ATP-dependent specificity component of ClpX/ClpP complex | 0.23 | 6.17 × 10−5 |
Open reading frames (ORF) are taken from E. coli K-12 MG1655 (7), E. coli O157:H7 EDL933 (36), and E. coli O157:H7 Sakai (20).
Asterisks indicate genes that were selected for real-time PCR.
Protein functions were determined with the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg).
The expression ratio (or fold difference) was determined by dividing the pixel values of the parent strain by those of the flhC mutant for each experiment. The average over five experiments is presented.
z-tests were performed on the expression ratios obtained, and ratios having a P value of ≤0.05 were selected.
Real-time PCR confirmed microarray data.
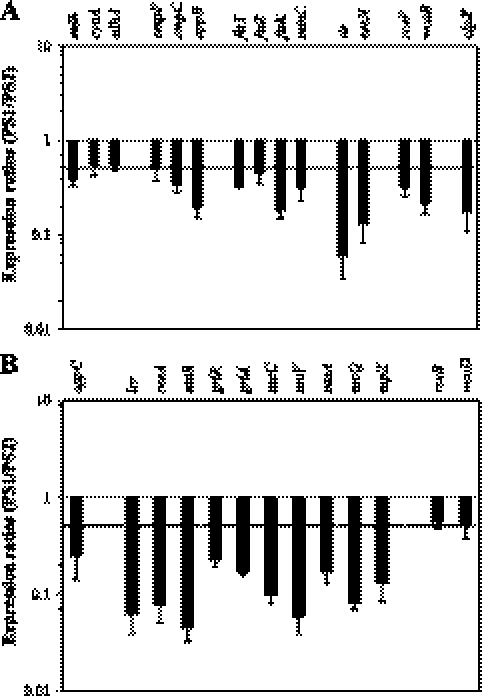
Real-time PCR was carried out to confirm putative FlhC targets obtained from the microarray experiments. A total of 15 representative genes were selected from the functional groups listed in Table 3. The genes included the metabolic genes mdh, cyoA, and sdhA; transporter genes ompF, ompC, and oppB; cell division genes ftsY, ftsZ, ftsK, and minC; LEE pathogenicity genes tir and escN; biofilm-related genes rcsF and wcaB; and a protease-encoding gene, clpX. The results from the real-time PCR are presented in Fig. 1A. Of the 15 genes tested, 13 (87%) could be confirmed. The regulation of cyoA and sdhA was borderline, using 2-fold as a cutoff for regulated genes. Interestingly, the two genes (tir and escN) that are involved in virulence and located on the LEE pathogenicity island were the most highly regulated genes (10- to 30-fold). Genes from the remaining functional groups were between 2- and 5-fold inhibited by FlhC.
Fig. 1.
Real-time PCR. The regulation by FlhC of selected genes was tested with real-time PCR. The experiment was done three times each with cDNA from two independently grown bacterial cultures. The average was determined across all six data points. Standard errors are indicated. Panel A contains the genes that were hypothesized as being regulated by FlhC in the microarray experiment. Fifteen of the genes that were found to be regulated by FlhC in the microarray experiment were subjected to real-time PCR. The different functional groups are separated by blank spaces on the x axis. Panel B contains additional genes associated with transport, the LEE pathogenicity island, and biofilm formation. These were selected based on their location and functional similarity to the genes in panel A.
Based on this observation, 13 additional genes were selected for real-time PCR analysis (Fig. 1B). These genes were either from the same operon or in the same functional group as the 15 genes that were previously tested (Fig. 1A) but either were not spotted on the microarray chip or were missing data points. With respect to the LEE pathogenicity island, we selected genes from each of the five LEE operons and the glrR and grlA regulatory genes that are located between LEE 1 and LEE 2. One additional gene was selected from the transport group, oppC. For the biofilm genes, we selected one other rcs gene and one other gene from the colanic acid operon, wca. The results from the real-time PCR with these additional genes are presented in Fig. 1B. All of the additionally tested genes were determined as being regulated by FlhC. Within the functional groups, regulation was similar to that shown in Fig. 1A. The LEE genes were highly regulated (10- to 30-fold); the two biofilm genes showed a borderline regulation of 2-fold. Altogether, 26 genes were determined as being inhibited by FlhC. These genes represent six functional groups, of which three will be investigated further.
A comparison of phenotypes between the parental strain and the flhC mutant indicates differences in cell division, biofilm formation, and lethality.
The microarray and the real-time PCR indicated genes involved in cell division, biofilm formation, and pathogenicity of E. coli O157:H7 as being targets of FlhC regulation. The following experiments were done to investigate the effect of the above confirmed regulation at the phenotypic level.
Comparison of cell division and biofilm formation between the parental strain and the flhC mutant.
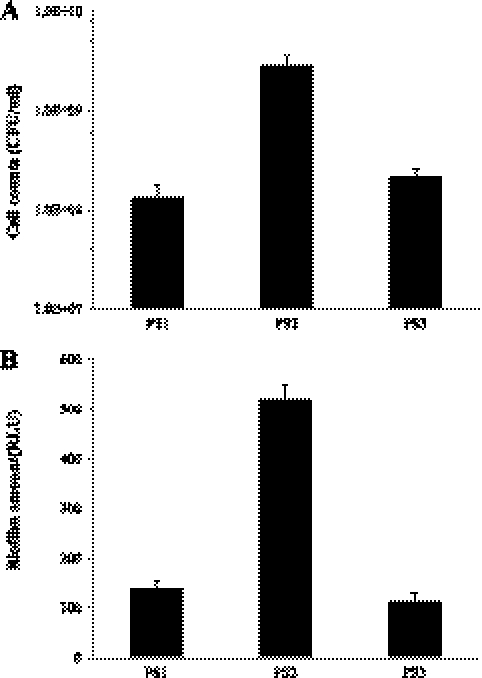
Overnight cultures of PS1, PS2, and PS3 were grown in 96-well plates for 10 days as described earlier. Bacterial cell numbers and biofilm amounts were determined in each well. The results for the cell count experiment are represented in Fig. 2A. At day 10, PS2 had divided to a cell density of 3 × 109 CFU/ml, which is 20 times higher than the parental PS1 strain (with a P value from the t test of 2.9 × 10−2), indicating an inhibitory effect of FlhC on the cell division rate. Complementation of PS2 with pXL26 (PS3) restored the phenotype of the mutant to almost that of the parental strain.
Fig. 2.
Cell division and biofilm formation. PS1, PS2, and PS3 were grown in beef broth on 96-well microtiter plates at 10°C for 10 days. The growth medium was carefully aspirated; serial dilutions were plated on SMAC-strep agar plates to obtain cell counts. The cell counts were indicative of cell division. The results are reported as CFU/ml (A). Biofilms were washed twice with PBS and quantified with the ATP assay. Biofilm amounts are given as RLU (B). The experiment was done with five replicates from each of three independently grown bacterial cultures.
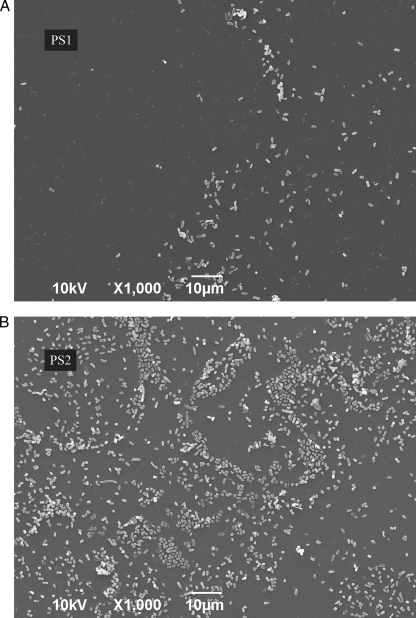
After removal of the supernatants from each well, the biofilm that was formed at the bottom of each well was carefully processed and subjected to ATP assay. The ATP assay is an indirect measure of biomass and is indicative of the number of cells that form the biofilm. The results from this experiment are represented in Fig. 2B. At day 10, PS2 had formed approximately 5 times more biofilm than the PS1 strain (with a P value from the t test of 3.2 × 10−15), implying that a loss in FlhC was beneficial for biofilm. Furthermore, PS2 complemented with pXL26 (PS3), showed about a 5-fold reduction in the ability of PS2 to form biofilm, restoring the phenotype of the flhC mutant to that of the parental strain. The electron micrographs of the biofilm formed by PS1 and PS2 (Fig. 3) at day 10 showed visual differences in the number of bacterial cells that had attached to the surface of the glass slips, corroborating the quantitative differences observed.
Fig. 3.
Scanning electron micrographs of biofilms formed by PS1 (A) and PS2 (B) on separate glass slides after 10 days of incubation at 10°C. Micrographs were taken at a magnification of ×1,000. One representative image is presented per strain.
Comparison of the lethality of PS1 and PS2 with the ELA.
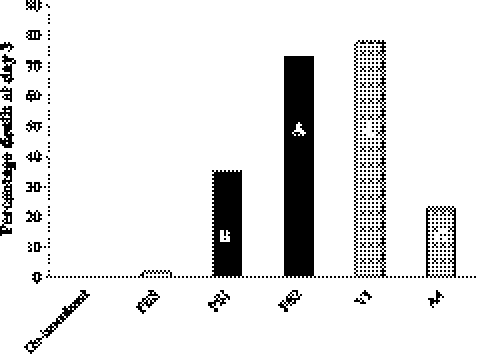
The ELA was performed to test whether the observed regulation of the LEE pathogenicity genes correlated with changes in bacterial phenotypes. The assay involved inoculation of 10 to 30 CFU of each bacterial strain into the allantoic fluid of embryonated eggs. Avian E. coli strains V1 and A4 were used as positive and negative controls, respectively. Additionally, embryos were inoculated with PBS, and uninoculated embryos were maintained as negative controls. The eggs were candled daily for 3 consecutive days postinoculation, and the number of dead embryos was noted. Figure 4 represents the percentage of dead embryos for all strains tested, as well as the uninoculated and PBS controls on day 3. Seventy-three percent of the embryos inoculated with PS2 were reported dead by the end of day 3, exhibiting a lethality similar to that of the positive control strain, V1 (78%). In agreement with this, Duncan's multiple-group comparison sorted these two groups into one group, labeled A in Fig. 4. The PS1 strain was less lethal than PS2 and V1, and only 35% of the embryos that had been inoculated with PS1 were reported dead. This strain formed group B in Duncan's test. The odds ratio between PS1 and PS2 was 5.107, indicating that an embryo was roughly 5 times more likely to die if inoculated with the flhC mutant strain than with the parental E. coli. This lethality was comparable to that of the negative control strain, A4 (23%), although the two strains each formed their own Duncan's group (groups B and C, respectively). There was no lethality in the uninoculated embryos, and one embryo inoculated with PBS was reported dead.
Fig. 4.
Lethality in the ELA. Test groups of 15 embryos were inoculated with 10 to 30 bacteria of PS1, PS2, V1, and A4. Additional test groups included uninoculated and PBS-inoculated embryos. The experiment was done four times. The percentage of dead embryos at day 3 postinoculation was determined for each test group and calculated over all 60 embryos. Negative controls are shown as white bars, E. coli O157:H7 strains as black bars, and avian E. coli control strains as gray bars. Capital letters within the bars indicate the grouping that resulted from the Duncan's multiple-range test, which was done for embryos only if they were inoculated with bacterial strains.
DISCUSSION
In the experiments described above, the effect of FlhC, one subunit of the global regulator complex FlhD/FlhC, on the expression level of various genes in E. coli O157:H7 was investigated in bacteria that had been grown on the surface of meat. The hypothesis was that FlhD/FlhC may play a crucial role in regulating genes of the LEE pathogenicity island. In addition to the genes in the LEE pathogenicity island, many other genes were hypothesized as putative FlhC targets. These genes were grouped into cellular functions and shall be discussed in the following sections. Interestingly, flagellar genes did not show up as induced by FlhC in the microarray. It is possible that the expression level of flhC on the nutrient-rich meat is not high enough to induce flagellar genes but is high enough to repress cell division, biofilm, and pathogenicity genes.
FlhC negatively regulates cell division in E. coli O157:H7.
In the present study, it was demonstrated that FlhC negatively regulated seven cell division genes when E. coli O157:H7 was grown on meat at 10°C. The quantitative real-time PCR experiments confirmed the microarray analysis, and in phenotypic experiments carried out subsequently, it was observed that the flhC mutant divided to 20 times higher cell numbers than its parental strain. The flhC phenotype could be restored back to the parental phenotype by complementation with the FlhC-expressing plasmid pXL26.
An earlier study by Prüß and coworkers had described a cell division effect in several E. coli K-12 strains (40). However, this study was exclusively phenotypic, and the corresponding regulation of the cell division genes was never demonstrated. Intriguingly, regulation of the cell division rate in the E. coli K-12 strains appeared to be mediated by the other subunit of the FlhD/FlhC complex, FlhD. The disparity between E. coli K-12 and E. coli O157:H7 regarding regulation (or lack thereof) by FlhC may be attributed to the different functional properties of the FlhD and FlhC subunits within the heterohexameric complex (56). The FlhC subunit is capable of binding to DNA alone, while FlhD can only bind to DNA in the presence of FlhC (13). However, binding of FlhC alone does not activate transcription; binding of FlhD to FlhC is a required step for transcription activation (10). We currently believe in a working model of regulation by FlhD and/or FlhC that includes potential substitute regulators that may bind within the same region of the target promoters as FlhD/FlhC. The model is in agreement with the observation that the four promoters that appeared regulated by FlhD and not FlhC in E. coli K-12 (43) were subject to complex regulation by numerous global regulators and the fact that E. coli O157:H7 has about 1,000 genes in excess of the K-12 lab strains (20). We believe that regulation of the FlhD- and/or FlhC-regulated promoters may be determined by the ratio of multiple competing regulators, each of whose expression is in itself affected by numerous environmental conditions. In E. coli O157:H7, the study was restricted to an flhC mutant only. The possibility that expression of the FlhC-regulated genes is affected by FlhD also cannot be excluded and warrants further study.
FlhC negatively regulates biofilm formation in E. coli O157:H7.
Microarray and real-time PCR data demonstrated that FlhC downregulates the expression of several biofilm-related genes, such as wcaB, wcaD, rcsF, and rcsB. In agreement with this, flhC mutants formed larger amounts of biofilm than the parent E. coli strain. Introduction of flhC into the mutant strain with plasmid pXL26 resulted in a complemented strain with a phenotype similar to that of its parental type. The increased biofilm amount of the flhC mutant was supported by electron microscopy (Fig. 3). Altogether, this study indicates that FlhC may be an inhibitor of biofilm formation, at least under the conditions of our experiment, which included long-term growth at low temperature on the surface of meat.
A previous paper from the Prüß lab had summarized a network of regulation in E. coli K-12 that included the FlhD/FlhC complex and many genes that contribute to biofilm formation (42). The study hypothesized that this FlhD/FlhC-centered network may be involved in the regulation of biofilm formation, a hypothesis that was confirmed in a subsequent study (44). The present study expands on all of those previous experiments, demonstrating a negative effect of FlhD/FlhC itself on biofilm formation. The study constitutes one more example in a long row of contradictory reports on the role of FlhD/FlhC and motility during the formation of biofilm, supporting those reports that consider motility a disadvantage. On the other hand, our data are in contradiction to a report on Yersinia pseudotuberculosis, in which a mutation in flhDC was detrimental for biofilm formation in both biotic and abiotic surfaces (57). All of these contradictions may be explained by previous observations where motility had varied effects on Bacillus cereus biofilm formation, depending on the surface on which biofilm forms (23). The precise condition under which the biofilms form undoubtedly has an effect on the observed gene regulation under each of these conditions. It is also possible that flagella constitute an advantage early during biofilm development and a disadvantage at later time points. This hypothesis would be in agreement with our previous study (44) and the present one, in which the bacteria were collected from the meat samples after 10 days of incubation.
FlhC negatively regulates pathogenicity in E. coli O157:H7.
The relationship between motility and pathogenicity has long been investigated in various bacterial species and led to contradictory results. While flagella and motility have certainly been recognized as a virulence factor in many bacterial species, there are also examples where a nonmotile species of a genus is more pathogenic than its motile counterpart. One such example is the genus Yersinia: the nonmotile species Yersinia pestis is one of the most lethal bacteria on earth, while the motile species Yersinia enterocolitica is only a moderate pathogen. At a molecular level, three nonflagellar genes, yplA (48), inv (60), and virF (8), were described as part of the flagellar regulon and activated by the sigma factor of the regulon, FliA. As an apparent contradiction, temperature regulation of many Y. enterocolitica flagellar (27, 28) and late virulence (8) genes appeared to follow an inverse pattern. This was mediated by temperature-dependent expression of fliA (22).
The present study sheds new light into this apparent controversy; it was demonstrated that E. coli O157:H7 FlhC negatively regulates genes belonging to the LEE pathogenicity island. These genes were, in fact, the most highly regulated genes in our experiments. In an attempt to investigate the extent of this regulation, additional genes from the LEE pathogenicity island were selected and subjected to real-time PCR analysis that had not been postulated as FlhC regulated in the microarray experiment. One hundred percent of the selected LEE genes were confirmed as being negatively regulated by FlhC, indicating the possibility of FlhC regulating the entire LEE pathogenicity island. These data are consistent with Y. enterocolitica, where flagella and seven plasmid-encoded virulence genes were inversely regulated by FliA (22). They are also in agreement with observations made during colonization of the intestine, where the lack of flagella was discussed as a selective advantage (31). In environments where there is abundant nutrition available, motility would not be required to move to more nutritious environments. Therefore, the bacteria may be able to afford shutting down motility and turning on genes that aid colonization and pathogenicity. The surface of meat may just constitute one such nutrient-rich environment.
Future directions.
E. coli O157:H7 has been a constant concern to the food industry. Various antimicrobial sprays and treatment methods have been employed with partial success to actively reduce the bacterial contamination from beef carcasses and food products (9, 25, 26, 30, 45, 47, 55, 59). This study observed that three vital cellular processes, cell division, biofilm formation, and pathogenicity, were all downregulated by FlhC. Such an observation opens endless possibilities of simultaneously limiting these three cellular processes, via increase of FlhD/FlhC expression. For example, in previous studies, we were able to decrease FlhD/FlhC expression and increase the cell division rate by the addition to the bacterial growth medium of serine (40) and other carbon sources (37) that can get converted to acetate. In the future, we will identify carbon sources that will decrease the cell division rate, as well as biofilm formation and pathogenicity, by increasing the expression of FlhC. Ultimately, one of these carbon sources could be used as a new control measure to protect consumers from infection by E. coli O157:H7.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott A. Minnich (University of Idaho, Moscow) for providing the E. coli O157:H7 strains, Curt Doetkott (North Dakota State University [NDSU]) for statistical analysis of the data, Jayma Moore (NDSU) for electron microscopy, and Penelope Gibbs (NDSU) for helping with the ELA.
The work was funded by the North State Board of Agricultural Research and Education, the ND Beef Commission, and a Leap Lab Renovation grant from ADVANCE/FORWARD and ND EPSCoR.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Anonymous 2010. Surveillance for foodborne disease outbreaks—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 59:973–979 [PubMed] [Google Scholar]
- 2. Anonymous 2010. Two multistate outbreaks of Shiga toxin-producing Escherichia coli infections linked to beef from a single slaughter facility—United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 59:557–560 [PubMed] [Google Scholar]
- 3. Barbosa T. M., Levy S. B. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartlett D. H., Frantz B. B., Matsumura P. 1988. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J. Bacteriol. 170:1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berry E. D., Cutter C. N. 2000. Effects of acid adaptation of Escherichia coli O157:H7 on efficacy of acetic acid spray washes to decontaminate beef carcass tissue. Appl. Environ. Microbiol. 66:1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackburn C. D., Davies A. R. 1994. Development of antibiotic-resistant strains for the enumeration of foodborne pathogenic bacteria in stored foods. Int. J. Food Microbiol. 24:125–136 [DOI] [PubMed] [Google Scholar]
- 7. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 8. Bleves S., Marenne M., Detry G., Cornelis G. R. 2002. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 184:3214–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bosilevac J. M., et al. 2004. Protocol for evaluating the efficacy of cetylpyridinium chloride as a beef hide intervention. J. Food Prot. 67:303–309 [DOI] [PubMed] [Google Scholar]
- 10. Campos A., Matsumura P. 2001. Extensive alanine scanning reveals protein-protein and protein-DNA interaction surfaces in the global regulator FlhD from Escherichia coli. Mol. Microbiol. 39:581–594 [DOI] [PubMed] [Google Scholar]
- 11. Castanie-Cornet M., Penfound T. A., Smith D., Elliott J. F., Foster J. W. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chuang S. E., Daniels D. L., Blattner F. R. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Claret L., Hughes C. 2002. Interaction of the atypical prokaryotic transcription activator FlhD2C2 with early promoters of the flagellar gene hierarchy. J. Mol. Biol. 321:185–199 [DOI] [PubMed] [Google Scholar]
- 14. Dobbin H. S., Hovde C. J., Williams C. J., Minnich S. A. 2006. The Escherichia coli O157 flagellar regulatory gene flhC and not the flagellin gene fliC impacts colonization of cattle. Infect. Immun. 74:2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evans J. B., Niven C. F. 1951. Nutrition of the heterofermentative Lactobacilli that cause greening of cured meat products. J. Bacteriol. 62:599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fratamico P. M., Smith L. J., Buchanan R. L. 2002. Escherichia coli, p. 79–101 In Cliver D. O., Riemann H. P., Thippareddi H. (ed.), Foodborne diseases, 2nd ed Academic Press, London, United Kingdom [Google Scholar]
- 17. Frenzen P. 2007. An online cost calculator for estimating the economic cost of illness due to Shiga toxin-producing E. coli (STEC) O157 infections. Economic Information Bulletin 28. USDA Economic Research Service, Washington, DC [Google Scholar]
- 18. Gardner G. A. 1985. Streptomycin-thallous acetate-actidione (STAA) agar: a medium for the selective enumeration of Brochothrix thermosphacta. Int. J. Food Microbiol. 2:69–70 [Google Scholar]
- 19. Gibbs P. S., Maurer J. J., Nolan L. K., Wooley R. E. 2003. Prediction of chicken embryo lethality with the avian Escherichia coli traits complement resistance, colicin V production, and presence of the increased serum survival gene cluster (iss). Avian Dis. 47:370–379 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi T., et al. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 21. Horne S. M., Mattson K. R., Prüß B. M. 2009. An Escherichia coli aer mutant exhibits a reduced ability to colonize the streptomycin-treated mouse large intestine. Antonie Van Leeuwenhoek 95:149–158 [DOI] [PubMed] [Google Scholar]
- 22. Horne S. M., Prüß B. M. 2006. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch. Microbiol. 185:115–126 [DOI] [PubMed] [Google Scholar]
- 23. Houry A., Briandet R., Aymerich S., Gohar M. 2010. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156:1009–1018 [DOI] [PubMed] [Google Scholar]
- 24. Hussein H. S., Sakuma T. 2005. Shiga toxin-producing Escherichia coli: pre- and postharvest control measures to ensure safety of dairy cattle products. J. Food Prot. 68:199–207 [DOI] [PubMed] [Google Scholar]
- 25. Kalchayanand N., et al. 2009. Effectiveness of 1,3-dibromo-5,5 dimethylhydantoin on reduction of Escherichia coli O157:H7− and Salmonella-inoculated fresh meat. J. Food Prot. 72:151–156 [DOI] [PubMed] [Google Scholar]
- 26. Kalchayanand N., et al. 2008. Evaluation of various antimicrobial interventions for the reduction of Escherichia coli O157:H7 on bovine heads during processing. J. Food Prot. 71:621–624 [DOI] [PubMed] [Google Scholar]
- 27. Kapatral V., Minnich S. A. 1995. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol. Microbiol. 17:49–56 [DOI] [PubMed] [Google Scholar]
- 28. Kapatral V., Olson J. W., Pepe J. C., Miller V. L., Minnich S. A. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061–1071 [DOI] [PubMed] [Google Scholar]
- 29. Kapatral V., et al. 2004. Gene array analysis of Yersinia enterocolitica FlhD and FlhC: regulation of enzymes affecting synthesis and degradation of carbamoylphosphate. Microbiology 150:2289–2300 [DOI] [PubMed] [Google Scholar]
- 30. Laury A. M., et al. 2009. Validation of a lactic acid- and citric acid-based antimicrobial product for the reduction of Escherichia coli O157:H7 and Salmonella on beef tips and whole chicken carcasses. J. Food Prot. 72:2208–2211 [DOI] [PubMed] [Google Scholar]
- 31. Leatham M. P., et al. 2005. Mouse intestine selects non-motile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Q., Logue C. M. 2005. The growth and survival of Escherichia coli O157:H7 on minced bison and pieces of bison meat stored at 5 and 10°C. Food Microbiol. 22:415–421 [Google Scholar]
- 33. Liu X., Matsumura P. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mead P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng J., Doyle M. P., Zhao T., Zhao S. 2001. Enterohemorrhagic Escherichia coli, p. 193–213 In Doyle M. P., Beuchat L. R., Montville T. J. (ed.), Food microbiology: fundamentals and frontiers, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 36. Perna N. T., et al. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prüß B. M. 1998. Acetyl phosphate and the phosphorylation of OmpR are involved in the regulation of the cell division rate in Escherichia coli. Arch. Microbiol. 170:141–146 [DOI] [PubMed] [Google Scholar]
- 38. Prüß B. M., Liu X., Hendrickson W., Matsumura P. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91–97 [DOI] [PubMed] [Google Scholar]
- 39. Prüß B. M., Markovic D., Matsumura P. 1997. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J. Bacteriol. 179:3818–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prüß B. M., Matsumura P. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prüß B. M., Wolfe A. J. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973–984 [DOI] [PubMed] [Google Scholar]
- 42. Prüß B. M., Besemann C., Denton A., Wolfe A. J. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prüß B. M., et al. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prüß B. M., et al. 2010. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 192:715–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raftari M., et al. 2009. Effect of organic acids on Escherichia coli O157:H7 and Staphylococcus aureus contaminated meat. Open Microbiol. J. 3:121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Russell J. B., Diez-Gonzalez F., Jarvis G. N. 2000. Potential effect of cattle diets on the transmission of pathogenic Escherichia coli to humans. Microbes Infect. 2:45–53 [DOI] [PubMed] [Google Scholar]
- 47. Sawyer J. E., et al. 2008. Effect of xylitol on adhesion of Salmonella typhimurium and Escherichia coli O157:H7 to beef carcass surfaces. J. Food Prot. 71:405–410 [DOI] [PubMed] [Google Scholar]
- 48. Schmiel D. H., Young G. M., Miller V. L. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shin S., Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sule P., et al. 2009. A combination of assays reveals biomass differences in biofilms formed by Escherichia coli mutants. Lett. Appl. Microbiol. 49:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sule P., Prüß B. M. 2009. Regulation of virulence genes in Escherichia coli O157:H7, p. 43–59 In Pandalai S. G. (ed.), Recent research and developments in microbiology. Research Signpost, Trivandrum, India [Google Scholar]
- 52. Townsend M. K., et al. 2008. Pleiotropic phenotypes of a Yersinia enterocolitica flhD mutant include reduced lethality in a chicken embryo model. BMC Microbiol. 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tzipori S., et al. 1987. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect. Immun. 55:3117–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Varma J. K., et al. 2003. An outbreak of Escherichia coli O157 infection following exposure to a contaminated building. JAMA 290:2709–2712 [DOI] [PubMed] [Google Scholar]
- 55. Vurma M., Pandit R. B., Sastry S. K., Yousef A. E. 2009. Inactivation of Escherichia coli O157:H7 and natural microbiota on spinach leaves using gaseous ozone during vacuum cooling and simulated transportation. J. Food Prot. 72:1538–1546 [DOI] [PubMed] [Google Scholar]
- 56. Wang S., Fleming R. T., Westbrook E. M., Matsumura P., McKay D. B. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798–808 [DOI] [PubMed] [Google Scholar]
- 57. Wang Y., et al. 2007. The flhDC gene affects motility and biofilm formation in Yersinia pseudotuberculosis. Sci. China Ser. C Life Sci. 50:814–821 [DOI] [PubMed] [Google Scholar]
- 58. Wong M. L., Medrano J. F. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39:75–85 [DOI] [PubMed] [Google Scholar]
- 59. Yoder S. F., et al. 2010. Investigation of water washes suitable for very small meat plants to reduce pathogens on beef surfaces. J. Food Prot. 73:907–915 [DOI] [PubMed] [Google Scholar]
- 60. Young G. M., Badger J. L., Miller V. L. 2000. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect. Immun. 68:4323–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.