Abstract
Secondary metabolite (SM) production by fungi is hypothesized to provide some fitness attribute for the producing organisms. However, most SM clusters are “silent” when fungi are grown in traditional laboratory settings, and it is difficult to ascertain any function or activity of these SM cluster products. Recently, the creation of a chromatin remodeling mutant in Aspergillus nidulans induced activation of several cryptic SM gene clusters. Systematic testing of nine purified metabolites from this mutant identified an emodin derivate with efficacy against both human fungal pathogens (inhibiting both spore germination and hyphal growth) and several bacteria. The ability of catalase to diminish this antimicrobial activity implicates reactive oxygen species generation, specifically, the generation of hydrogen peroxide, as the mechanism of emodin hydroxyl activity.
INTRODUCTION
For decades, natural products produced by bacteria and fungi have been mined for use as antimicrobials. The easily accessible compounds (e.g., β-lactams, macrolides) were the first to be mined and patented for drug development. Recently, advances in genome sequencing have revealed an arsenal of untapped secondary metabolite (SM) gene clusters in both kingdoms (3, 25, 26). In many of these organisms the number of SM gene clusters (20 to 50) far exceeds the number of known metabolites, hence, the concept of “silent” SM gene clusters (7, 12, 14). The inaccessibility to these compounds in laboratory settings is hypothesized to be due to an inability to replicate the conditions that induce SM activation in nature.
In their ecological habitat, microbes are constantly being challenged, often by other microbes. These microbial interactions activate SM clusters that produce bioactive compounds that offer protection for the organism, niche security, and a myriad of other benefits (27). One case in point is the Aspergillus fumigatus gliotoxin gene cluster. Although most widely referred to for its role as a toxin in invasive aspergillosis (15), gliotoxin is a potent antifungal and likely helps secure A. fumigatus dominance in soil environments (1, 8, 16, 19, 21). A. fumigatus self protects from endogenous gliotoxin by expressing a gliotoxin reductase, GliT, also known to exhibit oxidase activity, encoded in the cluster (29, 31). Because the gliotoxin gene cluster is expressed under most laboratory conditions, this metabolite was identified decades ago (36). However, most antimicrobial gene clusters are silent in laboratory environments, presumably because there is little to no competition and therefore no need for protection. Until recently, the hurdle of silent gene cluster activation in laboratories has limited the mining resources for drug discovery and hindered any understanding of the ecological role of these SMs. With the advent of genome sequencing and recent advances in epigenetic modifications, formerly silent gene clusters can be activated, resulting in the production of novel compounds.
Epigenetic modification has been best illustrated in the model organism Aspergillus nidulans. The secondary metabolome of this fungus contains at least 25 SM gene clusters, many of which are characteristically silent in the laboratory environment, and therefore considerable numbers of its natural products are unknown (7, 12, 14). However, through chromosomal remodeling of A. nidulans, an array of novel bioactive compounds can be retrieved. For example, deletion of cclA, which encodes a protein (CclA) that is involved in histone 3 lysine 4 (H3K4) methylation, results in the activation of the several gene clusters yielding emodin derivatives, including monodictyophenone and the antiosteoporosis polyketides F9775A and F9775B, which are derived from orsellinic acid (5, 28).
At the same time that loss of CclA was found to induce these gene clusters, coculture of A. nidulans with Streptomyces hygroscopicus resulted in the induction of several cryptic SMs, including the F9775 metabolites (32). We hypothesized that the cclA deletant unveiled microbial SM induction pathways utilized by A. nidulans during interaction with other microbes. The purpose of the present study was to test this hypothesis by examining potential antimicrobial activities of compounds released by activated pathways against possible competitor microbes (Table 1). We found that a derivative of emodin, 2-hydroxyemodin (2-OH), effectively inhibits the growth of A. fumigatus, Aspergillus flavus, Candida albicans, Bacillus cereus, and Micrococcus luteus. We suggest that the process of SM cluster activation due to competition in nature can be mimicked through epigenetic modifications, resulting in an arsenal of novel compounds that can be characterized for their ecological roles and mined for new therapeutics.
Table 1.
Strains used in this study
| Organism | Name | Source of strain |
|---|---|---|
| Aspergillus fumigatus | WT (Af293) | Nancy Keller laboratory |
| Aspergillus fumigatus | AF41 | David Denning laboratory |
| Aspergillus fumigatus | AF72 | David Denning laboratory |
| Aspergillus fumigatus | F14532 | David Denning laboratory |
| Aspergillus fumigatus | F14403 | David Denning laboratory |
| Aspergillus fumigatus | F16216 | David Denning laboratory |
| Aspergillus fumigatus | F11628 | David Denning laboratory |
| Aspergillus fumigatus | F12776 | David Denning laboratory |
| Aspergillus fumigatus | F13747 | David Denning laboratory |
| Aspergillus flavus | WT (NRRL3357) | Nancy Keller laboratory |
| Aspergillus nidulans | WT (RJW64; veA1) | Nancy Keller laboratory |
| Aspergillus nidulans | cclAΔ (RAAS74.29; pyroA4ΔcclA::pyroA veA1) | Nancy Keller laboratory |
| Aspergillus nidulans | cclAΔ mdpGΔ (LO2149; pyrG89 pyroA4ΔnkuA::argB riboB2ΔstcJ::AfriboBΔcclA::AfpyroA ΔmdpG::AfpyrG veA1) | Berl Oakley laboratory |
| Aspergillus nidulans | alcA(p):mdpE [LO2333; pyrG89 pyroA4 ΔnkuA::argB riboB2 ΔstcJ::AfriboB ΔmdpE::AfpyrG-alcA(p)::mdpE veA1] | Berl Oakley laboratory |
| Pseudomonas aeruginosa | PAO1 | Nancy Keller laboratory |
| Bacillus cereus | U85 | Nancy Keller laboratory |
| Micrococcus luteus | M. luteus | Nancy Keller laboratory |
MATERIALS AND METHODS
Microbial growth conditions.
Aspergillus, Candida, Pseudomonas, Bacillus, and Micrococcus strains (Table 1) were maintained as frozen glycerol stocks at −80°C and grown at 37°C on glucose minimal medium (GMM) or yeast extract-peptone-dextrose medium (YPD).
Preparation of test organisms.
Aspergillus spp. were point inoculated onto GMM and incubated at 37°C for 3 to 5 days. Spore suspensions were prepared as follows: 7 ml of 0.01% Tween–water was added to each plate, and then spores were isolated by gently scraping the surface of each plate with a sterile plastic spreader. The spore suspensions were quantified using a hemocytometer. Candida albicans and bacterial species were grown on YPD agar plates at 37°C for 24 h. Immediately prior to use, cells were removed from the plate with a sterile applicator, suspended in sterile phosphate-buffered saline, and counted using a hemocytometer.
Morphology.
For macroscopic phenotype analysis, 104 spores in 10 μl Tween-water were point inoculated onto GMM with supplements or lactose minimal medium (LMM) with cyclopentanone and supplements for induction of the alcA promoter. Cultures were grown at 37°C for 4 days. For microscopic analysis, spores were added to 25 ml of GMM with supplements to a final concentration of 106 spores/ml. Cultures were grown for 20 h at 37°C shaking at 250 rpm. Cultures were then shifted either to fresh GMM plus supplements or LMM plus cyclopentanone and supplements and grown at 37°C shaking at 250 rpm for an additional 4 h. Microscopic hyphal morphology was then viewed at 400× magnification.
Inhibition analysis.
To determine the effect of coculturing Aspergillus spp. strains on growth, 5 μl of a 104 spores/ml suspension of A. nidulans wild type, cclAΔ, or cclAΔ mdpGΔ strains was point inoculated on GMM plates with supplements and A. fumigatus wild-type (WT) strain AF293 or A. nidulans WT. The alcA(p)::mdpE inoculations were made on LMM with cyclopentanone and supplements for induction of the alcA promoter. Plates were incubated for 3 days at 37°C, and radial growth was assessed visually for signs of growth inhibition at 3 to 5 days postinoculation. Images shown in the figure insets were obtained with a dissecting microscope at 50× magnification.
Preparation of crude extracts and purified metabolites.
A. nidulans wild-type and cclAΔ strains were inoculated on GMM agar and incubated at 37°C for 4 to 5 days, and then mycelia and supporting agar were excised from the plates with a sterile blade as described by Bok et al. (5). In general, 10 to 20 plates, or 20 ml of agar, were extracted. Briefly, the mycelia and agar were homogenized for 10 s in a blender containing 250 ml of methanol. An additional 500 ml of methanol was added to the slurry and was stirred slowly overnight at room temperature. The slurry was filtered through Miracloth and three layers of Whatman filter paper and using a vacuum apparatus to remove agar and mycelia. The filtrate was then evaporated using a Rotavapor R-210. The evaporated material was suspended in 200 ml of a 1:1 mixture of water and ethyl acetate and separated using a 250-ml separatory funnel, and the aqueous and organic fractions were collected and evaporated using the Rotavapor. These crude fractions were stored in sealed glass vials at −80°C. Purified metabolites were collected as described by Bok et al. (5).
Extract testing.
Crude extracts and purified metabolites were assayed for antifungal activities against various strains of A. fumigatus and A. nidulans, as well as A. flavus, Pseudomonas aeruginosa, B. cereus, M. luteus, and C. albicans, by using a 96-well plate format as previously described (24). Briefly, spores, yeast, or bacteria (1 × 103/well) were added to triplicate wells containing 200 μl of RPMI with morpholinepropanesulfonic acid (MOPS) and various concentrations of crude extracts or purified metabolites from the cclAΔ strain. Plates were incubated for 24 to 48 h, and MICs were determined by visual examination of wells. In preliminary experiments, we determined that there was no solvent-related inhibitory activity against the test species from the evaporated organic fraction of the crude extracts prior to medium addition. Similarly, the addition of dimethyl sulfoxide (DMSO) at comparable concentrations as observed with the addition of purified metabolites had no inhibitory activity against the fungi tested. For quantification of germinated spores, three biological replicates and two technical replicates of 100 spores were counted for each treatment. Spores were considered germinated if an apparent germ tube was seen emerging from the spores.
Catalase inhibition assay.
To determine if the observed 2-OH antifungal activity was due to generation of reactive oxygen species, catalase (Sigma-Aldrich, St. Louis, MO) was added at various concentrations to test wells containing 2-OH. Neither catalase nor high concentrations of the protein bovine serum albumin exhibited antimicrobial activity against the fungi tested.
RESULTS
Competition growth assays.
Our laboratory previously showed that the loss of CclA, which is involved in H3K4 methylation, turns on cryptic secondary metabolite gene clusters in A. nidulans. This resulted in the production of several silent SMs, including monodictyophenone, emodin and emodin derivatives, and the antiosteoporosis polyketides F9775A and F9775B (5). The expansive armamentarium of SMs produced by fungi is thought to provide an adaptive advantage, allowing them to compete with other microbiota and colonize diverse ecological niches. We hypothesized that the cryptic SM gene clusters (e.g., the monodictyophenone cluster emodin derivatives) turned on in the A. nidulans cclAΔ strain might produce SMs with antimicrobial activities against other fungi and bacteria.
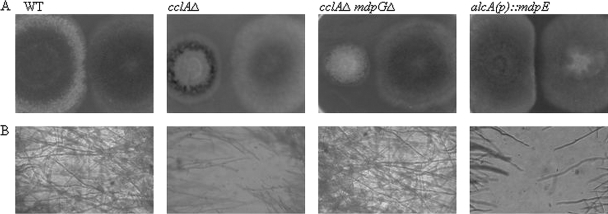
To investigate this hypothesis we cocultured the wild type and three A. nidulans mutants [cclAΔ upregulated in monodictyophenone and other SM clusters, alcA(p)::mdpE upregulated in just the monodictyophenone cluster, and cclAΔ mdpGΔ unable to express metabolites in the monodictyophenone cluster but upregulated in other SMs] with wild-type A. fumigatus on GMM agar medium (WT, cclAΔ, and cclAΔ mdpGΔ) or LMM agar with cyclopentanone [alcA(p)::mdpE]). Interestingly, we observed that the radial growth of A. fumigatus mycelia did not extend into regions of the plate containing strains where the monodictyophenone cluster was expressed [cclAΔ and alcA(p)::mdpE] (Fig. 1). In contrast, the mycelia of wild-type A. nidulans and A. fumigatus strains overlapped with no signs of inhibitory activity. Inhibition by the cclAΔ strain was abrogated in a strain lacking the backbone PKS for this cluster (cclAΔ mdpGΔ). Similar results were observed with the mutant series against wild-type A. nidulans (data not shown). These results indicated that upregulation of the monodictyophenone gene cluster produced SMs with inhibitory activities against wild-type strains of A. fumigatus and A. nidulans.
Fig. 1.
A. nidulans cclAΔ inhibits the growth of A. fumigatus strains on GMM agar. Both 2-OH-overproducing strains [cclAΔ and alcA(p)::mdpE] prevented hyphal intercalation of the radial growth of both A. nidulans (data not shown) and A. fumigatus wild-type strains. In contrast, the mycelia of wild-type A. nidulans and A. fumigatus strains overlapped with no signs of inhibitory activity. The cclAΔ mdpGΔ strain, which failed to produce 2-OH, showed interactions similar to wild type. All strains were grown on GMM with the exception of the confrontation between A. nidulans alcA(p)::mdpE and A. fumigatus, which was conducted on LMM with cyclopentanone.
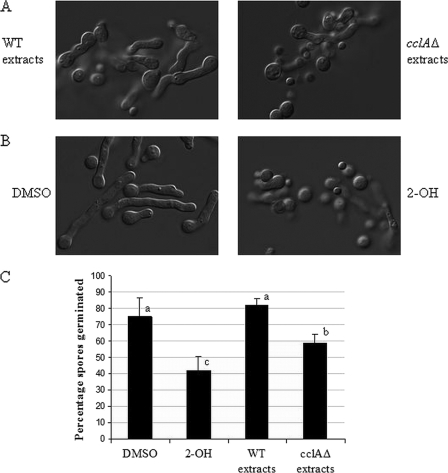
Next, crude extracts were prepared from A. nidulans cclAΔ and wild-type strains grown on GMM agar at 37°C. The aqueous and organic fractions of the crude extracts were then assessed for antifungal activities against A. fumigatus wild-type AF293 in a 96-well microtiter plate-based assay. A. fumigatus spores (1 × 103 CFU/well) were added to wells containing various concentrations of the crude extracts, and germination was assessed microscopically over time to evaluate inhibitory activity. Although aqueous extracts had no effect on fungal growth (data not shown), we did observe that organic extracts from A. nidulans cclAΔ, but not A. nidulans wild type, possessed potent antifungal activity and largely inhibited the germination of A. fumigatus spores (Fig. 2A and C). This result was consistent with our hypothesis that an SM(s) with antifungal activity was produced as a result of the loss of CclA.
Fig. 2.
AF293 germination and germling morphology are affected by cclAΔ but not wild-type extracts. (A) Germination of spores is inhibited by addition of organic extracts from cclAΔ strains, but not from WT strains. These effects included decreased percentages of germination and decreased conidial swelling. (B) Addition of purified 2-OH reconstitutes these effects. DMSO is the control solvent for 2-OH. (C) Quantification of the percentage of germinated spores for each treatment. Letters (a to c) indicate differences with statistical significance (P < 0.05) according to the Tukey-Kramer multiple comparison test.
Antimicrobial activities of purified compounds from A. nidulans cclAΔ.
We previously identified several organically soluble SMs produced by the A. nidulans cclAΔ mutant strain, including desacetylaustin, dehydroaustinol, citreorosein, emodic acid, 2-OH, 2-aminoemodin, emodin, and the polyketides F9775A and F9775B (5). We speculated that one or more of these SMs might mediate the inhibitory activities observed with the crude extracts. First, each of the compounds was purified and assessed for antifungal activity against A. fumigatus spores in a 96-well microtiter plate assay at concentrations ranging from 9 μM to 567 μM. We observed that of the nine compounds tested, only 2-OH exhibited significant antifungal activity against A. fumigatus, and 2-OH activity reconstituted the inhibition of germination effects seen with treatment with cclAΔ extracts (Fig. 2B and C).
To further investigate whether the antifungal activity exhibited by 2-OH was specific for A. fumigatus, or if 2-OH exhibited broader antimicrobial activity, we assessed 2-OH activity against three fungi (A. fumigatus, A. flavus, and C. albicans) and three bacteria (P. aeruginosa, B. cereus, and M. luteus). We observed that 2-OH exhibited robust antimicrobial activities against both aspergilli, C. albicans, and two of the three bacterial species tested (Table 2). A. flavus (MIC100, 142 to 284 μM) was more resistant than A. fumigatus (MIC100, 35 to 71 μM), which was more resistant than C. albicans (MIC100, 9 to 18 μM). 2-OH was also quite effective against B. cereus (MIC100, ≤9 μM) and M. luteus (MIC100, ≤9 μM), while P. aeruginosa showed significantly higher resistance to 2-OH (MIC, 284 to 567 μM) (Table 2).
Table 2.
2-OH exhibits inhibitory activities against M. luteus, B. cereus, P. aeruginosa, A. fumigatus, A. flavus, and C. albicans at various concentrations
| 2-OH concn (μM) | Growth of species following the indicated treatmenta |
|||||
|---|---|---|---|---|---|---|
| M. luteus | B. cereus | P. aeruginosa | A. fumigatus | A. flavus | C. albicans | |
| 567 | − | − | − | − | − | − |
| 284 | − | − | + | − | − | − |
| 142 | − | − | + | − | + | − |
| 71 | − | − | + | − | + | − |
| 35 | − | − | + | + | + | − |
| 18 | − | − | + | + | + | − |
| 9 | − | − | + | + | + | + |
| Untreated control | + | + | + | + | + | + |
The symbols + and − denote growth and no growth, respectively. Growth was assessed by microscopic visualization of wells 48 h posttreatment.
2-OH mechanism of action.
To begin the investigation of potential mechanisms of action, we assessed the inhibitory activities of 2-OH against a collection of A. fumigatus strains with known antifungal drug resistance to voriconazole, itraconazole, or posaconazole. The rationale for this approach was that if a strain with a known phenotype (and corresponding genotype) exhibited resistance to 2-OH, then we could gain insights into the mechanism by which 2-OH kills A. fumigatus. All of the A. fumigatus drug-resistant strains assessed exhibited susceptibility to 2-OH that was comparable to wild-type A. fumigatus (data not shown). This suggested that the mechanism of activity was novel and independent of defined pathways known to be involved in resistance to azole drugs.
Emodin (1,3,8-trihydrozy-6-methylanthraquinone) is a free radical generator that, when used as a therapeutic agent, can cause immunosuppression, vasorelaxation, and hypolipidemia (37). The production of free radicals, specifically, hydrogen peroxide, is thought to contribute to the immunosuppressive activities of emodin (17). 2-OH has also been shown to specifically produce hydrogen peroxide and not superoxide (18). Therefore, we hypothesized that hydrogen peroxide might account for the fungicidal activity of 2-OH against A. fumigatus. To test this hypothesis, we assessed whether catalase, which catalyzes the reduction of hydrogen peroxide to water and oxygen, could diminish the inhibitory activity of 2-OH against A. fumigatus. In agreement with our hypothesis, we observed that catalase did effectively abrogate the inhibitory activity of 2-OH against A. fumigatus (Table 3). The complete loss of antifungal activity in the presence of catalase suggested that the inhibitory activity of 2-OH was dependent on the production of hydrogen peroxide. Interestingly, challenge of A. fumigatus with 2-OH did not result in increased endogenous catalase activity (data not shown), in accordance with the sensitivity to this metabolite.
Table 3.
Catalase abrogates the inhibitory activity of 2-OHa
| Additive (amt or concn) |
Growthb | |||
|---|---|---|---|---|
| Catalase (U) | 2-OH (μM) | BSA (μg) | H2O2 (mM) | |
| 0 | 0 | 370 | 0 | + |
| 10,000 | 0 | 0 | 0 | + |
| 0 | 60 | 0 | 0 | − |
| 5,000 | 60 | 0 | 0 | + |
| 0 | 60 | 370 | 0 | − |
| 0 | 0 | 0 | 0.5 | + |
| 0 | 0 | 0 | 1 | + |
| 0 | 0 | 0 | 1.5 | − |
| 0 | 0 | 0 | 2 | − |
| 5,000 | 0 | 0 | 2 | + |
AF293 spores (1 × 103 total CFU) were added to wells with the indicated additives. Addition of 60 μM 2-OH resulted in growth inhibition of spores similar to that caused by addition of 1.5 mM H2O2. Bovine catalase at a concentration of 5,000 units/well was able to eliminate the growth inhibition by 2-OH and H2O2.
The symbols + and − denote growth and no growth, respectively. Growth was assessed by microscopic visualization of wells 48 h posttreatment.
Lack of self-resistance activity.
Frequently, fungal SM gene clusters include a gene encoding a protein that imparts a resistance mechanism for the fungus against the respective SM produced by the cluster. For instance, gliT in the gliotoxin gene cluster encodes a reductase that detoxifies gliotoxin in order to protect A. fumigatus against endogenous gliotoxin (29, 31). We were interested to see if a cclAΔ strain (overproducing 2-OH), a cclAΔ strain, mdpGΔ strain (unable to produce 2-OH or other metabolites in this pathway due to deletion of mpdG encoding the monodictyophenone cluster polyketide synthase [10]), or an alcA(p)::mdpE strain (overproducing 2-OH due to overexpression of mdpE encoding the monodictyophenone pathway-specific transcription factor [10]) showed variances in susceptibilities to 2-OH. Table 4 shows that there was little difference in 2-OH sensitivity between these three A. nidulans strains and the wild type. The slight increase in sensitivity of cclAΔ may be suggestive of the endogenous overproduction of 2-OH in this strain, although this does not appear to be the case for the alcA(p)::mdpE strain. Regardless, this suggests that the monodictyophenone gene cluster does not include a gene that encodes resistance to 2-OH-mediated growth inhibition.
Table 4.
A. nidulans wild-type, cclAΔ, cclAΔ mdpGΔ, and alcA(p)::mdpE strains exhibit similar patterns of susceptibility to 2-OHa
| 2-OH treatment (μM) | Growth of strain in RPMI with indicated 2-OH concn |
Growth of alcA::mdpE in RPMI + 30 mM cyclopentanone | |||
|---|---|---|---|---|---|
| WT | cclAΔ | cclAΔ mdpGΔ | alcA::mdpE | ||
| 567 | − | − | − | − | − |
| 284 | − | − | − | − | − |
| 142 | + | − | + | − | − |
| 71 | + | + | + | + | + |
| 35 | + | + | + | + | + |
| 18 | + | + | + | + | + |
| 9 | + | + | + | + | + |
| Untreated control | + | + | + | + | + |
The patterns of susceptibility suggest that the monodictyophenone gene cluster does not include a resistance-encoding gene to protect the fungus from the metabolites produced by this pathway. The symbols + and − denote growth and no growth, respectively. Growth was assessed by microscopic visualization of wells 48 h post treatment.
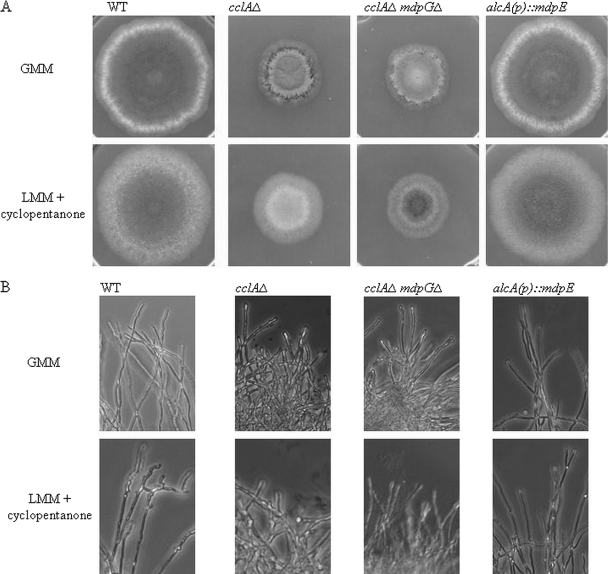
We also speculated that the reduced rate of growth and/or altered morphology of all three A. nidulans mutants might give insight into SM function in A. nidulans. Macroscopic and microscopic examination of all three strains compared to the wild type revealed a dominant impact of cclAΔ on diameter growth that was not alleviated in the double cclAΔ mpdGΔ strain (Fig. 3). However, loss of monodictyophenone cluster expression in cclAΔ did result in an increase in altered colony morphology. Because induction of the alcA(p)::mdpE allele led to decreased/delayed conidial production, it is possible that one or more of the monodictyophenone cluster-generated metabolites has a negative impact on conidiophore development.
Fig. 3.
Morphology abberancies in A. nidulans mutants. (A) Colony morphology varies between strains. cclAΔ leads to a decrease in colony diameter, likely independent of production of emodin derivatives, as the cclAΔ mdpGΔ strain devoid of emodin derivative production also was inhibited in diameter. However, the decreased conidiation observed in both the cclAΔ and alcA(p)::mdpE strains appeared to be associated with the increased expression of the monodictyophenone cluster. (B) Microscopic examination of hyphal morphology differs among strains but is not dependent upon production of emodin derivatives.
DISCUSSION
Historically, the vast majority of antimicrobial compounds have either been natural products or derived from natural products. These include a diverse spectrum of drugs, including antibiotics, antifungals, antiparasitics, lipid control agents, immunosuppressants, and anticancer drugs (20). In the last 2 decades there has been a shift in pharmaceutical research that has emphasized high-throughput screening of large libraries of synthetically produced compounds. This shift in focus away from natural products occurred in part because of the difficulty of finding new novel natural products. For example, members of the aspergilli are known to be rich sources of bioactive metabolites, but there remains a discontinuity between the vast number of SM gene clusters and the relatively few compounds described for the genus, leaving the true SM potential of these fungi unknown (7, 12, 14).
The disproportionate cluster to SM ratio of Aspergillus species is due largely to the inability to promote the expression of SM gene clusters in the laboratory. However, recent genomic mining of A. nidulans has uncovered several techniques to stimulate formerly silent clusters (4, 5, 6, 9, 11, 12, 22, 30, 32, 34). Epigenetic tools, in particular (e.g., inactivation of genes encoding histone-modifying enzymes or use of histone deacetylase inhibitors), have tremendous power as they activate tracks of chromatin otherwise subject to regulation by unknown environmental ligands (5, 15, 33).
We suggest that at least one microbial activation pathway proceeds through H3K4 modification. The A. nidulans cclAΔ mutant, defective in H3K4 methylation (5), inhibited growth of wild-type A. nidulans and the pathogen A. fumigatus. We hypothesized that this inhibitory property might be related to altered SM production in cclAΔ. This was confirmed when crude extracts from A. nidulans cclAΔ strains, but not extracts from wild-type strains, exhibited antifungal activities against A. flavus and A. fumigatus, fungi with soil niches similar to that of A. nidulans. This activity appeared quite broad as crude extract and also showed antibiosis against C. albicans, as well as the bacteria P. aeruginosa, B. cereus, and M. luteus. Based on our studies with the purified compounds from two of the cclAΔ-induced clusters, the inhibitory activity of crude extracts is mediated primarily by 2-OH. This was surprising, because neither emodin nor 2-aminoemodin exhibited any detectable antimicrobial activity at low molarities in our assays, despite other investigators' reports of antibacterial properties of emodin (2, 35).
We utilized strains of A. fumigatus with known drug resistance genotypes in an effort to identify molecular pathways susceptible to 2-OH activity. This pathogenic fungus in particular is increasingly resistant to common antifungals, with some scientists citing possible resistance acquisition due to overuse of agricultural fungicides (23). Since all of the A. fumigatus drug-resistant strains exhibited equivalent susceptibilities, we conclude that the mechanism of action is independent of ergosterol biosynthesis and drug efflux pump activity. The ability of catalase to abrogate 2-OH activity points toward the production of reactive oxygen intermediates as the primary mechanism of inhibitory activity. This is consistent with previous studies that have reported 2-OH is a free radical generator (18). The absence of emodin-mediated inhibitory activity suggests that the level of reactive oxygen species produced may be below that threshold required for antifungal activity. However, the addition of the hydroxyl group to emodin is likely to result in a compound with increased potential of producing reactive oxygen species. This may account for why 2-OH is more inhibitory than emodin. Previous results support an increased reactivity of 2-OH over emodin (13).
Microbes often exhibit self-protection from their own antimicrobials. As mentioned earlier, the gliotoxin-producing species A. fumigatus is tolerant of gliotoxin but shows enhanced sensitivity to this antifungal when one of the genes in the gliotoxin gene cluster, gliT, is deleted (29, 31). GliT encodes a gliotoxin reductase that reduces the oxidized form of gliotoxin, thus inhibiting glutathione depletion in the fungal cell. An examination of the monodictyophenone gene cluster did not identify a protein with possible 2-OH protection activity (10), and our examination of sensitivity of the cclAΔ, cclAΔ mdpGΔ, and alcA(p)::mdpE strains supported a lack of specific 2-OH protection as encoded by the cluster. However, morphological examination of the three mutants did suggest that one or more of the monodictyophenone cluster-generated SMs negatively impacts conidiophore formation in the producing organisms.
Here, we have presented 2-OH as an example of the antimicrobial possibilities possessed within silent SM gene clusters. We suggest that pathways activated during niche securement and microbial interactions can be turned on in the absence of these interactions through chromatin remodeling. The procedures outlined in this paper present a methodology that can be used to activate these clusters with subsequent production of novel compounds that can be explored for their ecological niche, antimicrobial activity, and new drug therapies.
ACKNOWLEDGMENTS
We thank David Denning for providing us with drug-resistant A. fumigatus strains.
The project was supported by grant PO1GM084077 from the National Institute of General Medical Sciences to N.P.K, B.R.O, and C.C.C.W. and, in part, by NIH grant R01 AI065728-S1 to S.S.G.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Aliaa R. E.-S. 2008. Control of root-rot diseases of Phaseolus vulgaris using gliotoxin. Malays J. 4:40–43 [Google Scholar]
- 2. Beattie K. D., et al. 2010. Antibacterial metabolites from Australian macrofungi from the genus Cortinarius. Phytochemistry 71:948–955 [DOI] [PubMed] [Google Scholar]
- 3. Bentley S. D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 4. Bergmann S., et al. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213–217 [DOI] [PubMed] [Google Scholar]
- 5. Bok J. W., et al. 2009. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5:462–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bok J. W., et al. 2006. Genomic mining for Aspergillus natural products. Chem. Biol. 13:31–37 [DOI] [PubMed] [Google Scholar]
- 7. Brakhage A. A., et al. 2008. Activation of fungal silent gene clusters: a new avenue to drug discovery. Prog. Drug Res. 66:1–12 [DOI] [PubMed] [Google Scholar]
- 8. Brian P. W., Hemming H. G. 1945. Gliotoxin, a fungistatic metabolic product of Trichoderma viride. Ann. Appl. Biol. 32:214–224 [DOI] [PubMed] [Google Scholar]
- 9. Chiang Y. M., Lee K. H., Sanchez J. F., Keller N. P., Wang C. C. 2009. Unlocking fungal cryptic natural products. Nat. Prod. Commun. 4:1505–1510 [PMC free article] [PubMed] [Google Scholar]
- 10. Chiang Y. M., et al. 2010. Characterization of the Aspergillus nidulans monodictyophenone gene cluster. Appl. Environ. Microbiol. 76:2067–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiang Y. M., et al. 2009. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 131:2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiang Y. M., et al. 2008. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem. Biol. 15:527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi J. S., Chung H. Y., Jung H. A., Park H. J., Yokozawa T. 2000. Comparative evaluation of antioxidant potential of alaternin (2-hydroxyemodin) and emodin. J. Agric. Food Chem. 48:6347–6351 [DOI] [PubMed] [Google Scholar]
- 14. Cichewicz R. H. 2010. Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep. 27:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dagenais T. R., Keller N. P. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22:447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howell C. R., Stipanovic R. D. 1995. Mechanisms in the biocontrol of Rhizoctonia solani-induced cotton seedling disease by Gliocladium virens: antibiosis. Phytopathology 85:469–472 [Google Scholar]
- 17. Huang H. C., et al. 1992. Immunosuppressive effect of emodin, a free radical generator. Eur. J. Pharmacol. 211:359–364 [DOI] [PubMed] [Google Scholar]
- 18. Kodama M., et al. 1987. Generation of free radical and hydrogen peroxide from 2-hydroxyemodin, a direct-acting mutagen, and DNA strand breaks by active oxygen. Toxicol. Lett. 37:149–156 [DOI] [PubMed] [Google Scholar]
- 19. Larkin R. R., Fravel D. R. 1998. Efficiency of various fungal and bacterial biocontrol organisms for control of fusarium wilt of tomato. Plant Dis. 82:1022–1028 [DOI] [PubMed] [Google Scholar]
- 20. Li J. W., Vederas J. C. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165 [DOI] [PubMed] [Google Scholar]
- 21. Lorito M., Leterbauer C., Hayes C. K., Woo S. L., Harman G. E. 1994. Synergistic combination of cell wall degrading enzymes and different antifungal compounds enhances inhibition of spore germination. Microbiology (London) 140:623–629 [DOI] [PubMed] [Google Scholar]
- 22. Losada L., Ajayi O., Frisvad J. C., Yu J., Nierman W. C. 2009. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 47(Suppl. 1):S88–S96 [DOI] [PubMed] [Google Scholar]
- 23. Meneau I., Sanglard D. 2005. Azole and fungicide resistance in clinical and environmental Aspergillus fumigatus isolates. Med. Mycol. 43(Suppl. 1):S307–S311 [DOI] [PubMed] [Google Scholar]
- 24. National Committee for Clinical Laboratory Standards 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 25. Nierman W. C., et al. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156 [DOI] [PubMed] [Google Scholar]
- 26. Omura S., et al. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 98:12215–12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rohlfs M., Albert M., Keller N. P., Kempken F. 2007. Secondary chemicals protect mould from fungivory. Biol. Lett. 3:523–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanchez J. F., et al. 2010. Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol. BioSyst. 6:587–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scharf D. H., et al. 2010. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc. 132:10136–10141 [DOI] [PubMed] [Google Scholar]
- 30. Scherlach K., Hertweck C. 2006. Discovery of aspoquinolones A-D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Org. Biomol. Chem. 4:3517–3520 [DOI] [PubMed] [Google Scholar]
- 31. Schrettl M., et al. 2010. Self-protection against gliotoxin: a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schroeckh V., et al. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 106:14558–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shwab E. K., et al. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 9:1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Szewczyk E., et al. 2008. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 74:7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., et al. 2010. Microcalorimetric assay on the antimicrobial property of five hydroxyanthraquinone derivatives in rhubarb (Rheum palmatum L.) to Bifidobacterium adolescentis. Phytomedicine 17:684–689 [DOI] [PubMed] [Google Scholar]
- 36. Weindling R., Emerson O. H. 1936. The isolation of a toxic substance from the culture filtrate of Trichoderma. Phytopathology 26:1068–1069 [Google Scholar]
- 37. Wu Y., et al. 2007. Emodin-mediated protection from acute myocardial infarction via inhibition of inflammation and apoptosis in local ischemic myocardium. Life Sci. 81:1332–1338 [DOI] [PubMed] [Google Scholar]