Abstract
A pilot-scale system was established to examine the feasibility of in situ U(VI) immobilization at a highly contaminated aquifer (U.S. DOE Integrated Field Research Challenge site, Oak Ridge, TN). Ethanol was injected intermittently as an electron donor to stimulate microbial U(VI) reduction, and U(VI) concentrations fell to below the Environmental Protection Agency drinking water standard (0.03 mg liter−1). Microbial communities from three monitoring wells were examined during active U(VI) reduction and maintenance phases with GeoChip, a high-density, comprehensive functional gene array. The overall microbial community structure exhibited a considerable shift over the remediation phases examined. GeoChip-based analysis revealed that Fe(III)-reducing bacterial (FeRB), nitrate-reducing bacterial (NRB), and sulfate-reducing bacterial (SRB) functional populations reached their highest levels during the active U(VI) reduction phase (days 137 to 370), in which denitrification and Fe(III) and sulfate reduction occurred sequentially. A gradual decrease in these functional populations occurred when reduction reactions stabilized, suggesting that these functional populations could play an important role in both active U(VI) reduction and maintenance of the stability of reduced U(IV). These results suggest that addition of electron donors stimulated the microbial community to create biogeochemical conditions favorable to U(VI) reduction and prevent the reduced U(IV) from reoxidation and that functional FeRB, SRB, and NRB populations within this system played key roles in this process.
INTRODUCTION
Uranium (U) is a relatively common contaminant, and remediation is of great importance because of the risk of transport off site. Bioremediation via microbial reduction of soluble U(VI) to insoluble U(IV) has been proposed (14). Microorganisms capable of U(VI) reduction include some sulfate-reducing bacteria (SRB) (15, 16, 26) and Fe(III)-reducing bacteria (FeRB) (17, 35), including Geobacter (24) and Shewanella spp. (19). Other microorganisms, including Clostridium sp. (7), Deinococcus radiodurans (8), and denitrifiers like Acidovorax spp. (22), have also been reported to reduce U(VI).
One promising strategy for improving U(VI) bioreduction is the addition of an electron donor (e.g., acetate, ethanol) to stimulate U-reducing microorganisms (2, 33). Experimental sites at the Integrated Field Research Challenge sites at Oak Ridge, TN (OR-IFRC), and Rifle, CO (Rifle-IFRC), have both shown long-term U(VI) reduction (33, 38) or sequestration (20). Studies have examined microbial communities during the reduction process, although these have primarily focused on phylogenetic information (5, 12, 25). However, little research on the functional gene diversity of microbial communities has been reported. Studies have examined the overall functional structure of communities during later phases of U(VI) bioremediation (28, 39), but none have studied the functional structure and dynamics of microbial communities during initiation and active phases of U(VI) bioremediation.
A major obstacle in monitoring complex microbial communities is that 99% of microorganisms have not yet been cultured (31). Therefore, to fully examine these communities, culture-independent methods like functional gene arrays (FGAs) (9, 34) are necessary. GeoChip 2.0 is a comprehensive FGA targeting ∼10,000 functional genes involved in the geochemical cycling of N, C, and S; metal reduction/resistance; and contaminant degradation (9). The GeoChip has been used to examine the microbial community functional structure at U(VI)-contaminated sites and has been shown to be a powerful tool for examining community changes (9, 28, 29, 33, 39).
A pilot-scale field test system, established at the OR-IFRC, has successfully shown the feasibility of in situ U(VI) bioremediation (18, 36–38). The current study was undertaken to examine functional gene changes in groundwater microbial communities during and after active bioreduction, with ethanol as an electron donor, using GeoChip. This study addressed how microbial community functional structures change over time with ethanol amendment and which environmental factors are important in shaping the microbial community's functional structure. Our results indicated that the functional community structure changed considerably over time in response to ethanol injections; additionally, different functional populations were responsible for initial U(VI) reduction and maintenance of reduced U(IV).
MATERIALS AND METHODS
Field treatment system.
A detailed description of the system used in this study is available elsewhere (18, 36). Briefly, the system was composed of two injection, two extraction, and three multilevel sampling (MLS) wells in a nested design (see Fig. S1 in the supplemental material). FW101-2 (13.7-m depth), FW102-2 (13.7 m), and FW102-3 (12.2 m) were selected for monitoring because of their hydraulic connection to the inner loop injection well (18, 36).
Groundwater sampling and analytical methods.
Groundwater (2 liters) was collected in sterile glass bottles using a peristaltic pump and kept on ice until delivered to the laboratory and then filtered (0.2 μm) to collect biomass. Filters were stored at −80°C until extraction. The use of groundwater allowed frequent sampling without disturbing well function. However, we could not obtain biological replicates. Detailed information on the source and quality of chemicals used and the methods for measuring geochemical variables was presented previously (36–38).
DNA extraction, amplification, labeling, and hybridization.
Community DNA was extracted using a freeze-grind method (40). DNA (100 ng) was amplified using the Templiphi kit (GE Healthcare, Piscataway, NJ) (33) and labeled with Cy-5 using random primers and Klenow (28). Labeled DNA was purified (QIAquick purification kit; Qiagen, Valencia, CA) and dried in a SpeedVac (45°C, 45 min; ThermoSavant, Waltham, MA).
Hybridizations were carried out using GeoChip 2.0 (9) overnight (14 to 16 h) at 50°C. Dried samples were suspended in hybridization buffer (50% formamide), and arrays were assembled and processed as described previously (33).
Microarray scanning and data processing.
Microarrays were scanned (ScanArray 5000) and digitally analyzed by quantifying pixel density (intensity) (ImaGene version 5.0). A signal-to-noise ratio [(signal mean − background mean)/background standard deviation] of ≥2 was considered a positive signal. A positive signal in at least 2 probes for a particular gene was required for a gene to be considered positive (9, 10). Outliers (P < 0.01) were removed. Data were normalized within and across samples using the mean-ratio approach as described previously (32).
Data analysis.
Hierarchical cluster analysis was performed using the pairwise average-linkage algorithm in CLUSTER and visualized using TREEVIEW. Statistical analyses were performed using R (v.2.2.0; http://www.r-project.org/) or Canoco (Version 4.5; Biometris-Plant Research International, Netherlands).
The microarray data presented are available at http://ieg.ou.edu/4download/.
RESULTS
Geochemical changes after ethanol biostimulation.
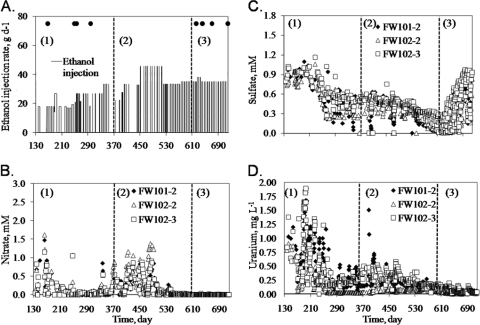
Changes in geochemistry between days 130 and 719 are illustrated in Fig. 1. Ethanol was injected weekly, except for days 142 to 163, 354 to 374, and 415 to 441 (Fig. 1A). The test period can be divided into three U(VI) reduction phases: active reduction (days 137 to 304), continuous reduction (days 304 to 621), and maintenance (days 622 to 720). Samples for GeoChip analysis were taken during active-reduction and maintenance phases (Fig. 1A).
Fig. 1.
Geochemical measurements in MLS wells (days 135 to 720). Ethanol injection and groundwater sampling dates (A) and concentrations of nitrate (B), sulfate (C), and uranium (D) are shown. Black closed circles indicate time points at which samples were taken: 1, active U(VI) reduction phase (days 137 to 304); 2, continuous-reduction phase (days 304 to 621); 3, maintenance phase (days 622 to 720).
After preconditioning (36), nitrate concentrations were ∼1.0 to 1.5 mM (Fig. 1B). Nitrate decreased rapidly as ethanol was injected and was <0.1 mM on day 304. There was a rebound in the absence of ethanol injection due to a change in the groundwater nitrate level and a release of nitrate from the sediment matrix. In the maintenance phase, nitrate was low (<0.05 mM). Sulfate concentrations were relatively stable until day 201, when sulfate decreased from 0.9 to 1.2 to 0.3 mM (Fig. 1C) and increased after sulfite, added on day 638 to remove dissolved oxygen (DO), penetrated the inner loop.
Sulfide was detected after day 201 and increased to 1.5 mg liter−1 during active reduction and to 12 mg liter−1 during the maintenance phase in FW101-2 (data not shown). Sulfide concentrations were relatively low in FW102-2 and FW102-3 (<0.1 mg liter−1) during active reduction but increased to 3 and 11 mg liter−1, respectively, in the maintenance phase. Dissolved Fe increased from ≤0.1 to 1.0 to 2.0 mg liter−1 after day 220 (data not shown), indicating that Fe(III) reduction was occurring. After day 184, U concentrations started to decrease but rebounded when ethanol injections stopped due to a recirculation of U-containing groundwater. U levels decreased to below the EPA maximum contaminant level (MCL; 0.03 mg liter−1) after day 615 in FW102-2 and then in FW101-2 and FW102-3 (Fig. 1D). In the maintenance phase, U concentrations remained stable at ∼0.03 mg liter−1. Based on X-ray absorption near edge structure (XANES) analysis, U(IV) in FW101-2 sediment increased to 35% (of the total U) on day 535 (38). Higher U(IV) levels (74 to 82%) were observed later in FW101-2, FW102-2, and FW102-3 (38).
Detrended correspondence analysis (DCA) of geochemical variables indicated a change in geochemistry over time, corresponding to the bioremediation phase (see Fig. S2b in the supplemental material). By day 255, environmental conditions in FW102-2 had begun to shift and appeared to be relatively stable by day 622. FW101-2 and FW102-3 reached relative stability by day 670.
Functional gene diversity.
Functional gene richness, indicated by the total number of genes detected, remained relatively low during early stages of remediation. In FW102-2, the total functional population increased by day 248, peaked on day 269, rebounded by day 642 when DO control started, and decreased again at day 719 when ethanol injections stopped (Table 1). Similar trends were observed for all wells (see Tables S2 and S3 in the supplemental material).
Table 1.
Percentages of unique and overlapping genes detected and diversity indices from each time point in well FW102-2
| Sampling day or index | % of unique and overlapping genes detecteda or diversity index on sampling day: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 163 (923)b | 166 (564) | 170 (390) | 184 (359) | 248 (1,204) | 255 (1,223) | 269 (2,350) | 278 (685) | 298 (652) | 323 (791) | 622 (676) | 642 (1,318) | 670 (1,915) | 719 (1,301) | |
| 163 | 20 | 16.6 | 18.2 | 13.7 | 24.4 | 24.6 | 21.1 | 19.5 | 21.7 | 19.1 | 23.4 | 24.8 | 24.3 | 18.5 |
| 166 | 6.2 | 36.5 | 38.6 | 19 | 33.8 | 17.4 | 34.6 | 17.9 | 25.9 | 23.3 | 21.9 | 15.9 | 23.8 | |
| 170 | 4.1 | 41.3 | 19.3 | 25.3 | 12.6 | 30.6 | 19.1 | 23.5 | 22.9 | 19.9 | 14.5 | 16.1 | ||
| 184 | 1.7 | 15.5 | 22.3 | 12.7 | 31.3 | 15.9 | 24.5 | 19.9 | 17.7 | 12.5 | 17.2 | |||
| 248 | 1.3 | 43.6 | 43.8 | 34.3 | 35.2 | 38.9 | 29.6 | 45.1 | 41.9 | 34.1 | ||||
| 255 | 1.1 | 40 | 46.9 | 36.3 | 49.7 | 37.1 | 48 | 39.2 | 44.4 | |||||
| 269 | 13.9 | 26.5 | 22.8 | 30.4 | 21.4 | 41.7 | 53.2 | 42 | ||||||
| 278 | 0.3 | 35 | 54.9 | 37.2 | 38.3 | 27.8 | 34.3 | |||||||
| 298 | 1.8 | 38.3 | 36.9 | 37.1 | 27.3 | 24.5 | ||||||||
| 323 | 0.6 | 34.3 | 42.1 | 34 | 37.8 | |||||||||
| 622 | 3.2 | 40.9 | 26.2 | 24.1 | ||||||||||
| 642 | 4.1 | 47.8 | 40.3 | |||||||||||
| 670 | 11.4 | 40.5 | ||||||||||||
| 719 | 6.7 | |||||||||||||
| Shannon | 6.648 | 6.141 | 5.682 | 5.495 | 6.852 | 6.874 | 7.523 | 6.319 | 6.31 | 6.523 | 6.34 | 6.993 | 7.395 | 7.04 |
| Simpson | 643.7 | 391.6 | 210.4 | 156.1 | 715.2 | 767.5 | 1,447.8 | 455.7 | 451.6 | 578.6 | 475 | 891.3 | 1,372.5 | 1,006.6 |
| Simpson E | 0.697 | 0.694 | 0.539 | 0.435 | 0.594 | 0.628 | 0.616 | 0.665 | 0.693 | 0.731 | 0.703 | 0.676 | 0.717 | 0.774 |
Values for unique genes are bold, and those for overlapping genes are italic.
The value in parentheses is the total number of genes detected.
Gene overlap was also determined, and a large percentage (12.5 to 54.9%) of the genes detected was shared among the time points in FW102-2 (Table 1). The percentages of overlapping genes from time points which are close together are higher than those of genes which are more distant. For example, day 255 had 43.6% of the genes in common with day 248 genes but only 24.6% in common with the day 163 genes. About 20% of the genes detected from day 163 were unique to that time point (Table 1). The microbial community dynamics in FW102-3 and FW101-2 (see Tables S2 and S3 in the supplemental material) were similar to those of FW102-2, although functional population peaks occurred sooner (day 212, FW102-3; day 255, FW101-2). The percentage of unique genes detected on day 166 from FW102-3 (3.6%) and FW101-2 (6.3%), were much lower than in FW102-2 and consistent with the more rapid functional population changes observed in these wells. Over 60% of the genes detected were shared by all 3 wells, 58 to 63% were shared by at least 2, and <20% were unique to a single well (Table 2).
Table 2.
Gene overlap among sampling wells
| Sample | No. (%) of overlapping genesa |
||
|---|---|---|---|
| FW102-2 (3,419)b | FW102-3 (3,431)b | FW101-2 (2,744)b | |
| FW102-2 | 630 (18.73) | 2,656 (63.33) | 2,260 (57.90) |
| FW102-3 | 449 (13.35) | 2,413 (64.14) | |
| FW101-2 | 2,127(63.23) | 158 (4.70) | |
Numbers and percentages in italics are unique gene numbers and percentages. Numbers and percentages in bold are overlapped gene number and percentage overlapped by three wells. Numbers and percentages not italic or bold are gene numbers and percentages overlapped by two of the three sampling wells.
Total number of genes detected.
Cluster analysis of all functional genes.
Approximately 3,000 functional genes in 11 categories were detected by GeoChip 2.0. Hierarchical cluster analysis of all of the genes detected was performed (see Fig. S3 in the supplemental material). A total of 35 different groups were observed which could be separated into two main divisions. Genes in the first division (groups 1 to 5 [35% of the genes detected]), were generally shared by all samples. Genes in group 1 (11%) were most abundant in FW102-2 on days 248 and 255, in FW102-3 on day 212, and in FW101-2 on day 255. Genes in group 2 (4%) peaked in FW102-3 by days 191 to 255, in FW101-2 at days 166 to 298, and at day 269 in FW102-2 and seemed dominant in FW101-2 and FW102-3. The genes in group 3 (7%) were most abundant in FW102-2 (day 191) and FW102-3 (day 248) and then decreased gradually. Group 4 (11%) genes were detected primarily in FW102-3, with a peak at day 191. Group 5 (3%) was small, but its members were abundant across all samples. The second division includes the remaining 30 groups (75%) and contains genes which dominated in individual wells or at specific times. Most of these groups have only one or two peaks, with the remaining sample points being at or near zero.
The relative abundance of cytochrome c and nitrification genes was higher within the common/shared genes (division 1) than in the unique genes (division 2), while the relative abundance of organic contaminant and carbon degradation genes was lower in the unique genes (see Fig. S4 in the supplemental material). For instance, in FW102-2 on day 255, division 1 cytochrome genes accounted for 8% of the genes; nitrification genes accounted for 14%. However, genes from division 2 contained only 5% cytochrome genes and 6% nitrification genes. In contrast, division 1 contained 34% organic contaminant degradation genes and 7% carbon degradation genes while division 2 contained 44% organic contaminant degradation genes and 10% carbon degradation genes. A complete list of the genes detected within each division and group is in Table S1 in the supplemental material.
Relationships among the microbial communities.
DCA was used to examine overall functional structure changes in the microbial communities (see Fig. S2a in the supplemental material). In the DCA ordination plot, samples that are more similar cluster together. The DCA of all detected genes from all three wells did not produce any clear clustering based on bioremediation phase (see Fig. S2a in the supplemental material). However, there was a large shift in overall community structure from the beginning of this study to the end, 507 days later. Generally, the communities were arranged by time along axis 1 and by well along axis 2.
The presence of dsr, cytochrome c, and denitrification genes were used as indicators of SRB, FeRB, and NRB, respectively. The presence of denitrification genes indicates the growth of NRB in the subsurface. While there is no clear evidence that NRB play a role in enzymatic U(VI) reduction under field conditions; NRB are essential for the removal of nitrate to create the low-redox conditions favorable for U(VI) reduction and subsequently prevent nitrate-dependent U(IV) oxidation (3, 6, 33). FeRB and SRB activity favors direct (enzymatic) and indirect (abiotic) U(VI) reduction. Since these bacterial groups are often involved in U(VI) reduction (30), DCA analysis was performed with these genes and the results indicated similar community changes (see Fig. S5 in the supplemental material). SRB, represented by dsr genes, and metal-reducing bacteria, represented by cytochrome c genes, ordered fairly well by time along axis 1 (Fig. S5a and b). NRB, represented by nitrate/nitrite-reducing genes, clustered more closely together than the sulfate- or metal-reducing bacterial communities (Fig. S5c).
Relative abundances of functional gene categories.
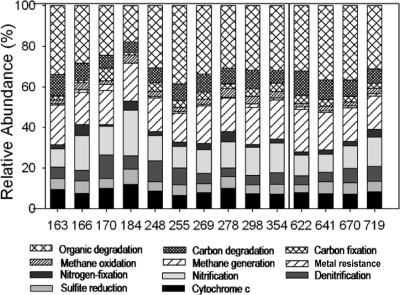
The relative abundances of functional gene categories at different time points were compared (Fig. 2; see Fig. S6 in the supplemental material). A list of the genes covered under each category can be found in the supplementary data to reference 9. Most of the changes to the community structure occurred during active U(VI) reduction (days 163 to 255, Fig. 2), for example, organic degradation genes accounted for 33.9% of the genes detected on day 163, 17.9% of those detected on day 184, and 38.5% of those detected on day 255. But this same category (organic degradation genes) accounted for 30.8 to 36.6% from day 269 to day 719. Assuming that these genes represent the background functional population (i.e., those microorganisms which are not directly involved in bioreduction), decreases in these functional populations suggest that other functional populations more directly involved in bioreduction are increasing. This relative decrease in the background functional population was most obvious on day 184; however, the peak in the abundance of potential bioreducing functional populations did not occur until day 255 (Fig. 3; see Fig. S7 in the supplemental material; discussed below). The relative abundance of functional gene categories involved in U(IV) reduction or indicative of U(VI)-reducing functional populations (i.e., dsr, cytochrome c, and denitrification genes) increased during the active-reduction phase, decreased, and then reached relative stability. For example, cytochrome genes increased from 9.4% on day 163 to 12.0% on day 184 and then remained relatively stable at 7 to 8% by day 298 (Fig. 3). Nitrogen-cycling genes displayed the same trend. Both nitrification and denitrification genes increased from day 163, reaching peaks on days 184 (22.5%) and 170 (11.5%), respectively, and then declined and remained relatively stable by day 255. The nitrogen fixation genes also increased during the active-reduction phase. Sulfate-reducing (dsr) genes did not show much of a change during this period, remaining at ∼5%. Methane generation genes increased from day 163 to day 170 (1.4% to 3.1%) and then decreased and remained at ∼1%. Methane oxidation genes maintained a relatively higher level (∼2 to 4%). In contrast, contaminant and carbon degradation genes decreased from day 163 (33.9%, 10.7%) to day 184 (17.9%, 5.0%) and then rebounded on day 248.
Fig. 2.
Relative abundances of all functional gene categories detected. The total number of genes detected at each time point was used to calculate the relative abundance of each gene category from FW102-2. Numbers along the x axis are operational days. Days to the left of the black line are in the active U(VI) reduction phase, while those to the right are in the maintenance phase.
Fig. 3.
Abundance of metal-reducing functional populations in FW102-2. The relative abundance of dsr, nitrate/nitrite reduction, and cytochrome c genes was determined using the total signal intensity of each gene category. Days to the left of the black line are in the active U(VI) reduction phase, while those to the right are in the maintenance phase.
The microbial communities in FW102-3 and FW101-2 did not show as obvious a pattern as that observed in FW102-2; however, the carbon and contaminant degradation genes did increase in FW102-3 from day 166 to day 191, suggesting that changes in environmental conditions, which affected the community structure, may have occurred earlier in FW102-3. Similarly, contaminant degradation genes also increased from day 166 (29.9%) to day 255 (32.9%) in FW101-2.
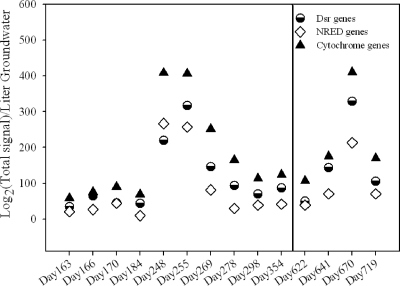
Changes in denitrification and metal and sulfate reduction genes.
The functional gene dynamics of metal-, sulfate-, and nitrate/nitrite-reducing bacteria were examined based on signal intensity (Fig. 3; see Fig. S7 in the supplemental material). The relative abundance of all functional gene categories is based on the number of genes detected; however, to make differences more visible, the signal intensities for metal-, sulfate-, and nitrate/nitrite-reducing genes are presented alone in these figures. In all wells, there was a peak in the abundance of all three functional categories between days 200 and 260, concomitant with a decrease in sulfate and U(VI) concentration (Fig. 1C and D). The peak in FW102-2 (days 248 to 255), corresponded to a period of rapid U(VI) reduction in that well (Fig. 3 and 1D). The functional populations began to decrease almost immediately following the peak. FW101-2 peaked at day 255 (Fig. S7a), and FW102-3 peaked at days 191 to 212, the earliest of all wells (Fig. S7b).
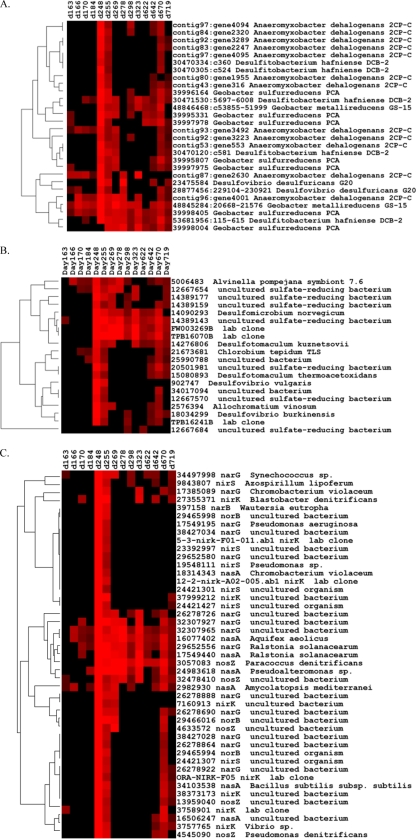
Cytochrome genes similar to those from Geobacter metallireducens (48845284), G. sulfurreducens (39998405), and Anaeromyxobacter dehalogenans were detected in FW102-2 at all time points during the active-reduction phase (Fig. 4A). Genes similar to several Geobacter genes were still detected afterwards, as were genes similar to those from Desulfitobacterium hafniense (53681956, 30471530). Similar patterns were observed in FW101-2 and FW102-3 (see Fig. S8a and S9a in the supplemental material).
Fig. 4.
Hierarchical cluster analysis of functional genes in FW102-2. Cluster analysis was done for cytochrome c (A), dsr (B), and nitrate/nitrite reduction (C) genes. Results were generated in CLUSTER and visualized using TREEVIEW. Red indicates signal intensities above the background, while black indicates signal intensities below the background. Brighter red indicates higher signal intensities.
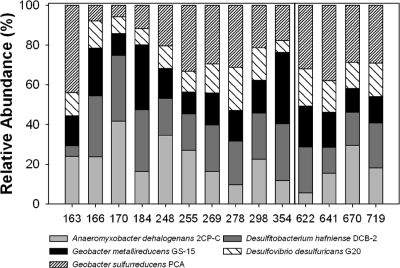
Since this category of genes appeared to be important in metal reduction, the relative abundance of cytochrome c genes derived from six microorganisms that were detected in high abundance in all wells and known to reduce U(VI) (Fig. 4A; see Fig. S8a and S9a in the supplemental material) was calculated (Fig. 5). Those genes from G. sulfurreducens-like strains were in the highest abundance (43.9%) at day 163 in FW102-2, but their abundance decreased to 7.87% on day 166 and remained low until day 255. While this sudden decrease in relative abundance may seem surprising, this most likely indicates a rapid increase in the abundance of G. metallireducens- and D. hafniense-like strains as a result of ethanol addition. Rapid response to ethanol addition has been observed previously (28). The functional population then remained relatively stable at ∼30%. During the period of rapid U(VI) reduction, A. dehalogenans-, D. hafniense-, and G. metallireducens-like strains were the dominant cytochrome-containing strains detected. Afterwards, in the maintenance phase, G. sulfurreducens-like strains appeared in greater abundance, suggesting a shift in the metal-reducing functional population during these two phases.
Fig. 5.
Relative abundances of U(VI)-reducing microorganisms in FW102-2. Relative abundances of known U(VI)-reducing microorganisms detected in high abundance in all MLS wells and time points were calculated based on the total signal intensities of cytochrome c genes. Days to the left of the black line are in the active U(VI) reduction phase, while those to the right are in the maintenance phase.
Few sulfate-reducing or denitrifying genes were detected in FW102-2 until day 248 (Fig. 3 and 4B). After day 255, dsr genes from organisms like Desulfomicrobium norvegicum (14090293), an uncultured SRB (14389143), a symbiont of Alvinella pompeiana (5006483), and Desfulfotomaculum kuznetsovii (14276806) dominated. More dsr genes were detected in the active-reduction phase in FW101-2 (see Fig. S7a and S8b in the supplemental material).
Denitrification genes that were detected at most time points included narG derived from an uncultured bacterium (32307965, 26278726), Ralstonia solanacearum (29652556) and Synechococcus sp. (34497998); nasA derived from Aquifex aeolicus (16077402), R. solanacearum (17549440), Pseudoalteromonas sp. (24983618), and Amycolatopsis mediterranei (2982930); and nosZ derived from an uncultured bacterium (32478410). Similar patterns were observed with FW102-3 (see Fig. S9c in the supplemental material).
Relationship of environmental conditions to the microbial community.
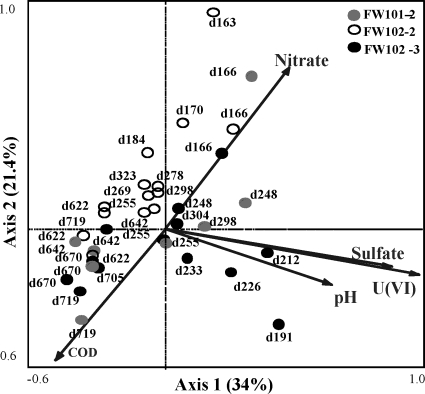
To examine the relationship between microbial community structure and geochemistry, canonical correspondence analyses (CCA) were performed (Fig. 6). The CCA was used to correlate environmental variables with the functional community structure and determine which environmental variables are most important in determining that structure. An initial analysis was performed using all available geochemical variables [chemical oxygen demand (COD), sulfate, nitrate, pH, nitrate, U(VI), sulfide, Fe], and five environmental variables were selected based on variance inflation factors (VIF) and Monte Carlo permutation P values: COD (VIF = 1.41, P = 0.008), U(VI) (VIF = 5.32; P = 0.010), pH (VIF = 2.51, P = 0.318), nitrate (VIF = 1.71, P = 0.068), and sulfate (VIF = 3.22, P = 0.598). Although pH and sulfate were not significant in an integrated model, when tested individually, P values were ≤0.05 (pH = 0.058, sulfate = 0.002). The small angles between the sulfate, pH, and U(VI) vectors indicate a strong correlation between these variables; however, VIFs for pH and sulfate were each <5, suggesting little correlation. The specified model was significant (first axis, P = 0.038; all axes, P = 0.002) and explained 10.61% of the variation (sum of all canonical eigenvalues, 0.782). When samples were separated by phase, the percentages of variation explained were 15.0 and 21.8% for the active and maintenance phases, respectively (data not shown). U(VI), pH, and sulfate showed a strong positive correlation with the first axis and a negative correlation with the second axis. COD showed a negative correlation with both the first and second axes, while nitrate showed a strong positive correlation. The microbial communities in FW102-2 and FW101-2 were affected primarily by nitrate and COD, while FW102-3 was affected by pH, sulfate, and U(VI).
Fig. 6.
Canonical correspondence analysis for FW101-2, FW102-2, and FW102-3. Analysis was done using all of the functional genes detected (symbols) and environmental variables (arrows) from FW101-2, FW102-2, and FW102-3. Environmental variables were chosen by automatic selection significance calculated from individual CCA results and variance inflation factors calculated during CCA. Numbers indicate operational days sampled.
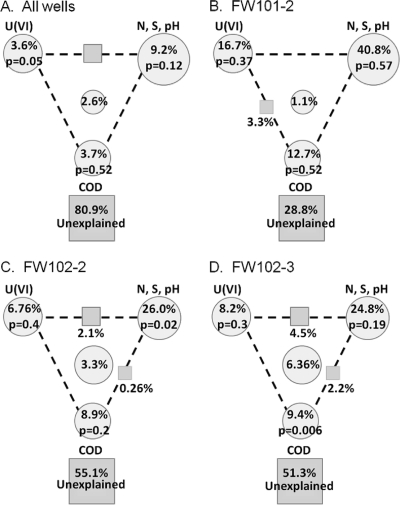
Variance partitioning analyses (VPA) were performed for each well (Fig. 7) to better understand how the environmental variables were affecting the microbial communities. The variance was partitioned into three categories: COD, U(VI), and other geochemical components (nitrate, sulfate, and pH). For FW102-2, 44.9% of the variance could be explained by the environmental variables, with COD, U(VI), and other geochemistry components explaining 8.9%, 6.7%, and 26%, respectively. Although COD only explained a small portion of the variance, there was little covariance between it and the other two variables (0.22% and 0.26%, respectively), indicating that COD was an independent factor. Similar partitioning results were obtained for FW102-3. The model was not significant for FW101-2. Much of the variance in the individual wells (45 to 70%) could be explained. Similar results were obtained when only metal-reducing functional populations (i.e., SRB, NRB, FeRB) were examined.
Fig. 7.
Variance partitioning of environmental variables analyzed by CCA. The diagram represents the relative effect of each variable upon the functional community in all wells (A), FW101-2 (B), FW102-2 (C), and FW102-3 (D). The circles represent the effects of individual variables by partitioning out the effects of the other variables. The squares between the circles represent the combined effect of the circles on either side of the square. The square at the bottom of each panel represents the effect that could not be explained by any of the variables tested. Variables used in CCA were used for the VPA. P values shown were generated during partial CCA.
DISCUSSION
Biostimulation of microbial communities.
Successful bioremediation strategies depend greatly on the microorganisms present at the target site and appropriate geochemical conditions. Previous studies have shown the presence of U(VI)-reducing microorganisms at the OR-IFRC (1, 4, 5, 21, 22), indicating that bioremediation could be a successful approach. To ensure adequate microbial growth, the test area was preconditioned to be geochemically favorable for microbial activity (37). Our results demonstrated that appropriate microbial communities were stimulated by ethanol injection, resulting in geochemical changes, including Fe(III), sulfate, and U(VI) reduction (Fig. 1).
Studies at the Rifle-IFRC demonstrated that acetate injection resulted in Fe(III)-reducing conditions and stimulation of Geobacter spp., resulting in removal of aqueous U(VI) (2). After 50 days, the system shifted to sulfate-reducing conditions and U(VI) levels began to rise. A second experiment indicated that a switch from a U(VI)-reducing, Geobacter-dominated community to a Firmicutes-dominated community, which may sequester U(VI) via sorption (20). In contrast, our system was fed intermittently with ethanol, resulting in long-term U(VI) reduction (<0.03 mg liter−1 for several months) (28, 37, 38). XANES analysis confirmed that U(VI) had been partially reduced to U(IV) by day 258 (37, 38). The different results observed at the Rifle- and OR-IFRC sites may be due to several factors, including dissimilar starting communities and the electron donor sources used (i.e., acetate versus ethanol), which stimulated different community members.
U(VI)-reducing microorganisms have been found in both sediment and groundwater at the OR-IFRC. Cardenas et al. (5) reported that (in order of decreasing importance) Desulfovibrio, Geobacter, Anaeromyxobacter, Desulfosporosinus, and Acidovorax were likely the most important sediment microorganisms involved in U(VI) reduction based on the frequency observed in clone libraries. Hwang et al. (12) detected similar groundwater microorganisms, with Desulfovibrio and Geobacter predominating, during the active U(VI) reduction phase and afterwards. These previous studies used the 16S rRNA gene to detect microbial populations. Similarly, results from the current study suggest that Anaeromyxobacter, Geobacter, and Desulfovibrio were abundant based on the amount of cytochrome genes detected. Having diverse functional populations may be important in maintaining long-term U(VI) reduction. Injection of ethanol as an electron donor may stimulate a more diverse, sustainable U(VI)-reducing community than that of acetate.
Functional categories related to U(VI) reduction.
Microorganisms play vital roles directly or indirectly in the reduction of various metals. Some SRB (15, 16, 26) and FeRB (17, 35) can enzymatically reduce U(VI). In general, the bioreduced products of FeRB and SRB (e.g., sulfide or ferrous compounds) can reduce U(VI) under certain geochemical conditions (e.g., pH 6.0 to 7.0, low bicarbonate). The OR-IFRC site was manipulated to reach these ideal conditions. In this study, we detected functional genes which indicate the presence of SRB, FeRB, and NRB (dsr, cytochrome, and nitrate/nitrate reductase, respectively) which may be involved in U(VI) reduction. Cytochrome genes represented on the GeoChip were selected only from microorganisms known to be involved in U(VI) reduction so that this gene category could reasonably be used to represent these functional populations. While dsr, cytochrome, and nitrate/nitrite reductases were used to represent specific microbial functional populations, the presence of these genes does not provide absolute proof that these functional populations are involved in metal reduction. Additional experiments using mRNA are required to measure their activity.
As described previously, there is no clear evidence that NRB play a key role in enzymatic U(VI) reduction under field conditions but NRB are essential for the removal of nitrate to create the low-redox conditions favorable for U(VI) reduction and prevent nitrate-dependent oxidation of U(IV) (3, 6, 33). Although not all SRB or FeRB are involved in enzymatic U(VI) reduction, their activity could favor abiotic U(VI) reduction in situ. However, some categories important to U(VI) reduction will not be detected if probes do not exist on the array. The peaks of the SRB, FeRB, and NRB functional populations occurred as U concentrations decreased in the active-reduction phase, suggesting the importance of these categories, which could be involved directly in U(VI) reduction or indirectly by creating geochemical conditions favorable for U(VI) reduction. Additionally, these functional genes remained at higher levels throughout the maintenance phase. This is in contrast to the results from the Rifle-IFRC, which indicated a decrease in FeRB as SRB increased (2).
Functional population changes during active U(VI) reduction.
As described above, significant functional population change occurred during the initial active-reduction phase, resulting in an increase in U(VI) reduction related NRB, FeRB, and SRB. Strains with cytochrome genes similar to those derived from A. dehalogenans, G. sulfurreducens, G. metallireducens, D. vulgaris, and D. hafniense were the main constituents of the metal-reducing bacteria detected in this study (Fig. 5). The relative abundances of these species fluctuated over time, indicating growth after ethanol injection. During the active-reduction phase, when U(VI) levels began to decrease (Fig. 1), the relative abundance of G. metallireducens-like cytochrome c genes began to increase at the same time as overall cytochrome c genes began to increase (Fig. 5). A similar increase in A. dehalogenans- and D. hafniense-like cytochrome c genes was also observed. These results suggest that Anaeromyxobacter- and Geobacter-like strains may play an important role in U(VI) reduction and maintenance of reduced U(IV). All of the these strains have been shown to reduce U(IV) (references 13 and 30 and references therein). Previous studies at the OR-IFRC have suggested that these genera are likely important in U(VI) reduction at this site (5, 12). Geobacter spp. have been shown to be important in rapid U(VI) reduction at other sites (2, 11). The increase in gene number may also be due to the introduction of ethanol into the system. The number of Anaeromyxobacter spp. has been shown to increase in the groundwater during periods of biostimulation with ethanol (27). Further work is required to confirm the roles of individual species in U(VI) reduction.
The microbial communities in the current study responded to the bioremediation treatment used (i.e., ethanol injection). There was a gradual shift in the community structure as U(VI) concentrations decreased, indicating that the communities were able to adjust to the changing conditions and resulting in very different communities during the active-reduction and maintenance phases. We also observed a functional shift with more reduction-related genes at increased abundance detected during the active-reduction phase. Functional gene richness in all three wells increased until around day 255 and then dropped, most likely due to lower ethanol concentrations, an increase in DO, and depletion of other nutrients. A rebound occurred when DO was controlled, but a drop occurred again when the ethanol injection stopped. Our continuous studies at the OR-IFRC site (days 746 to 992) indicated further that ethanol (as COD) was a major driver in the system and had a strong influence on community structure (28). DO had less of an effect, but community changes were observed during a period of increased DO (28).
Effects of environmental conditions on functional community structure.
COD, nitrate, pH, sulfate, and U(VI) appeared to be the most important environmental parameters determining the groundwater community structure during the active-reduction and maintenance phases (days 137 to 720). Similarly, sulfate, pH, and mean travel time (which caused a COD gradient) were the most important parameters for sediment communities on day 774 (39). After reoxidation by DO (days 713 to 992), COD, temperature, sulfate, and U(VI) were found to be the most important parameters (28). In this study, environmental conditions appeared to have a greater impact after day 255, when U(VI) concentrations had decreased. The amount of variation unexplained in this study (∼30 to 55%) is comparable to that observed in other contaminated aquifers (35 to 45%) (28, 29) or more diverse soil systems (50 to 80%) (23, 41).
This study characterized changes in microbial community structure during the startup of an in situ biostimulation system for U(VI) reduction. U(VI) concentrations below the EPA MCL were achieved by reduction of U(VI) to U(IV), and changes in the community structure occurred as the U(VI) levels decreased. FeRB, SRB, and NRB functional populations appeared to be key to the success of U(VI) reduction via enzymatic reduction or provided a geochemically suitable environment for reduction to occur. Community changes during the active-reduction and maintenance phases suggest that different functional populations are responsible for initial U(VI) reduction and subsequent maintenance of the reduced U(IV). These results provide a clearer picture of microbial community dynamics during U(VI) reduction and indicate that with the appropriate biostimulation strategy, specific microbial functional populations can be stimulated and maintained in situ in order to achieve a low U level in contaminated aquifers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chuanmin Ruan and Kenneth Lowe for help with analytical work.
This research was supported by The U.S. DOE under the Environmental Remediation Science Program and in part by the U.S. Department of Energy through ENIGMA under contract DE-AC02-05CH11231 with the Lawrence Berkeley National Laboratory and by the Oklahoma Center for the Advancement of Science and Technology under the Oklahoma Applied Research Support Program.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Amos B. K., et al. 2007. Detection and quantification of Geobacter lovleyi strain SZ: implications for bioremediation at tetrachloroethene- and uranium-impacted sites. Appl. Environ. Microbiol. 73:6898–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson R. T., et al. 2003. Stimulating the in-situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beller H. R. 2005. Anaerobic, nitrate-dependent oxidation of U(IV) oxide minerals by the chemolithoautotrophic bacterium Thiobacillus denitrificans. Appl. Environ. Microbiol. 71:2170–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brodie E. L., et al. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cardenas E., et al. 2008. Microbial communities in contaminated sediments associated with bioremediation of uranium to submicromolar levels. Appl. Environ. Microbiol. 74:3718–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Finneran K., Housewright M. E., Lovely D. R. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 4:510–516 [DOI] [PubMed] [Google Scholar]
- 7. Francis A. J., Dodge C. J., Lu F., Halada G. P., Clayton C. R. 1994. XPS and XANES studies of uranium reduction by Clostridium sp. Environ. Sci. Technol. 28:636–639 [DOI] [PubMed] [Google Scholar]
- 8. Fredrickson J. K., Kostandarithes H. M., Li S. W., Plymale A. E., Daly M. J. 2000. Reduction of Fe(III) Cr(VI), and Te(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 66:2006–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He Z., et al. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67–77 [DOI] [PubMed] [Google Scholar]
- 10. He Z., Wu L. Y., Li X. Y., Fields M. W., Zhou J. Z. 2005. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol. 71:3753–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holmes D. E., Finneran K. T., O'Neil R. A., Lovley D. R. 2002. Enrichment of Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hwang C., et al. 2009. Bacterial community succession during in-situ uranium bioremediation: spatial similarities along controlled flow paths. ISME J. 3:47–64 [DOI] [PubMed] [Google Scholar]
- 13. Leigh M. B., et al. 2005. Understanding the bioremediative potential of FRC microbial communities. Abstr. DOE Natural and Accelerated Bioremediation Research (NABIR) PI Meeting, Warrenton, VA http://esd.lbl.gov/research/projects/ersp/generalinfo/pi_meetings/PI_mtg_05/05_PI_Mtg_pdf/posters05/Tiedje_poster05nabir.pdf [Google Scholar]
- 14. Lovley D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Indust. Microbiol. 14:85–93 [DOI] [PubMed] [Google Scholar]
- 15. Lovley D. R., Phillips E. J. P. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lovley D. R., Phillips E. J. P. 1994. Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome. Appl. Environ. Microbiol. 60:726–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lovley D. R., Phillips E. J. P., Groby Y. A., Landa E. R. 1991. Microbial reduction of uranium. Nature 350:413–416 [Google Scholar]
- 18. Luo J., et al. 2006. A nested-cell approach for in situ remediation. Ground Water 44:266–274 [DOI] [PubMed] [Google Scholar]
- 19. Marshall M. J., et al. 2006. c-type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4:e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. N′Guessan A. L., Vrionis H. A., Resch C. T., Long P. E., Lovley D. R. 2008. Sustained removal of uranium from contaminated groundwater following stimulation of dissimilatory metal reduction. Environ. Sci. Technol. 42:2999–3004 [DOI] [PubMed] [Google Scholar]
- 21. North N. N., et al. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nyman J. L., et al. 2006. Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation 17:303–316 [DOI] [PubMed] [Google Scholar]
- 23. Ramette A., Tiedje J. M. 2007. Multiscale responses of microbial life in spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. U. S. A. 104:2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shelobolina E. S., et al. 2007. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 7:16–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spain A. M., et al. 2007. Identification and isolation of a Castellaniella species important during biostimulation of an acidic nitrate- and uranium-contaminated aquifer. Appl. Environ. Microbiol. 73:4892–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tebo B. M., Obraztsova A. Y. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193–198 [Google Scholar]
- 27. Thomas S. H., Padilla-Crespo E., Jardine P. M., Sanford R. A., Löffler F. E. 2009. Diversity and distribution of Anaeromyxobacter strains in a uranium-contaminated subsurface environment with a nonuniform groundwater flow. Appl. Environ. Microbiol. 75:3679–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Nostrand J. D., et al. 2009. GeoChip-based analysis of functional microbial communities during the reoxidation of a bioreduced uranium-contaminated aquifer. Environ. Microbiol. 11:2611–2626 [DOI] [PubMed] [Google Scholar]
- 29. Waldron P. J., et al. 2009. Functional gene array-based analysis of microbial community structure in groundwaters with a gradient of contaminant levels. Environ. Sci. Technol. 43:3529–3534 [DOI] [PubMed] [Google Scholar]
- 30. Wall J. D., Krumholtz L. R. 2006. Uranium reduction. Annu. Rev. Microbiol. 60:149–166 [DOI] [PubMed] [Google Scholar]
- 31. Whitman W. B., Coleman D. C., Wiebe W. J. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu L., Kellogg L., Devol A. H., Tiedje J. M., Zhou J. 2008. Microarray-based characterization of microbial community functional structure and heterogeneity in marine sediments from the Gulf of Mexico. Appl. Environ. Microbiol. 74:4516–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu L., Liu X., Schadt C. W., Zhou J. 2006. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl. Environ. Microbiol. 72:4931–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu L., et al. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Q., Sanford R. A., Löffler F. E. 2006. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl. Environ. Microbiol. 72:3608–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu W. M., et al. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer 1: conditioning of a treatment zone. Environ. Sci. Technol. 40:3978–3985 [DOI] [PubMed] [Google Scholar]
- 37. Wu W. M., et al. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 2: U(VI) reduction and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986–3995 [DOI] [PubMed] [Google Scholar]
- 38. Wu W. M., et al. 2007. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ. Sci. Technol. 41:5716–5723 [DOI] [PubMed] [Google Scholar]
- 39. Xu M., et al. 2010. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 4:1060–1070 [DOI] [PubMed] [Google Scholar]
- 40. Zhou J., Bruns M. A., Tiedje J. M. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou J., Kang S., Schadt C. W., Garten C. T., Jr 2008. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U. S. A. 105:7768–7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.