Abstract
The bacterial community in the sea surface microlayer (SML) (bacterioneuston) is exposed to unique physicochemical properties and stronger meteorological influences than the bacterial community in the underlying water (ULW) (bacterioplankton). Despite extensive research, however, the structuring factors of the bacterioneuston remain enigmatic. The aim of this study was to examine the effect of meteorological conditions on bacterioneuston and bacterioplankton community structures and to identify distinct, abundant, active bacterioneuston members. Nineteen bacterial assemblages from the SML and ULW of the southern Baltic Sea, sampled from 2006 to 2008, were compared. Single-strand conformation polymorphism (SSCP) fingerprints were analyzed to distinguish total (based on the 16S rRNA gene) and active (based on 16S rRNA) as well as nonattached and particle-attached bacterial assemblages. The nonattached communities of the SML and ULW were very similar overall (similarity: 47 to 99%; mean: 88%). As an exception, during low wind speeds and high radiation levels, the active bacterioneuston community increasingly differed from the active bacterioplankton community. In contrast, the particle-attached assemblages in the two compartments were generally less similar (similarity: 8 to 98%; mean: 62%), with a strong variability in the active communities that was solely related to wind speed. Both nonattached and particle-attached active members of the bacterioneuston, which were found exclusively in the SML, were related to environmental clones belonging to the Cyanobacteria, Bacteroidetes, and Alpha-, Beta-, and Gammaproteobacteria originally found in diverse habitats, but especially in water columns. These results suggest that bacterioneuston communities are strongly influenced by the ULW but that specific meteorological conditions favor the development of distinctive populations in the air-water interface.
INTRODUCTION
The uppermost level (top <1 mm) of virtually any body of water is defined as the (sea) surface microlayer (SML). Due to its location at the air-water interface, the SML influences exchange processes across this interface by forming a diffusive boundary and by virtue of its physicochemical properties (32). The bacterial community inhabiting the SML (bacterioneuston) is exposed to environmental conditions substantially different from those acting on the bacterioplankton in the underlying water (ULW). Thus, the question arises whether this unusual habitat determines a specific bacterial community that influences interfacial processes such as air-water gas fluxes.
One characteristic of the SML is the accumulation of diverse organic and inorganic compounds (25), and increased exoenzymatic activities have been measured in this layer (29, 35). Still, despite high concentrations of potential organic substrates, bacterial growth efficiencies in the SML are generally low (43), and the reduced bacterioneuston productivity suggests that the SML is a stressful habitat (36, 48). Conditions inhibitory to the bacterioneuston might include its increased exposure to organic pollutants, heavy metals, and UV radiation (34, 54).
Previous studies have reported contradictory data on differences in composition between bacterioneuston and bacterioplankton communities. A substantially lower diversity in the SML than in the ULW was determined based on 16S rRNA gene clone libraries from the North Sea (16). Furthermore, bacterioneuston communities of alpine lakes have been shown to be more similar to airborne than to planktonic populations (21). In contrast, there were no consistent differences between the SML and the ULW in 16S rRNA gene fingerprints from the Mediterranean Sea and the Atlantic Ocean (1, 36). Additionally, nearly all cultured bacterial strains isolated from the Mediterranean SML have been shown to be closely related to environmental clones from diverse marine habitats (1). The patterns of resistance to solar radiation of these isolates did not differ from those of their planktonic counterparts, indicating that adaptation to environmental stress factors is not a common trait among neustonic bacteria (3). However, the SML and the ULW are known to differ with respect to the relative abundances of bacterial groups incorporating leucine and the functional traits for trace gas metabolism (12, 23, 36), indicating that specific processes are driven by different active bacterial groups in the SML and the ULW.
At least three factors limit general comparisons between neustonic and planktonic communities. First, the various SML sampling techniques differ in their operating modes and therefore result in the collection of SML samples from layers of different thicknesses (24). Recent studies have highlighted this problem, emphasizing that SML studies carried out with different sampling methods must be compared with caution (2, 10, 48). Second, particulate organic material is consistently enriched in the SML (25), which might result in strong differences between particle-attached and nonattached bacterioneuston communities, as is known from estuaries (9). Particle-attached cells were found to be the dominant respiring fraction in the SML (20), where they show high abundances (4). However, distinctions between the particle-attached and nonattached bacterioneuston communities have only rarely been made, which limits generalizations. Third, meteorological conditions have a much stronger influence on the bacterioneuston than on the bacterioplankton community. For instance, high wind speeds cause increasing turbulence, and breaking waves disturb the SML. Conversely, decreasing wind speeds reduce surface mixing, and under highly calmed conditions, slicks are formed that can encompass large areas of coastal waters (44). Therefore, the variability of SML properties in response to changing meteorological conditions might at least partially explain the opposing results of bacterioneuston diversity reported so far.
The aim of this study was to obtain further insights into the patterns of bacterioneuston community structure and composition under various meteorological conditions. Accordingly, SML and ULW samples of the coastal Baltic Sea, one of the largest brackish basins of the world, were compared by single-strand conformation polymorphism (SSCP) fingerprint analysis of 16S rRNA and the 16S rRNA gene to account for the active and total communities, respectively. Additionally, nonattached and particle-attached bacterial communities were distinguished in order to obtain higher-resolution comparisons.
MATERIALS AND METHODS
Study site and sea surface microlayer sampling.
Samples from the SML were collected from the southern Baltic Sea, offshore of Warnemünde in proximity to position 54°19′N, 12°05′E, between July and September 2006, June and August 2007, and February and May 2008 (see Table S1 in the supplemental material). The glass-plate technique (19), which collects 7 to 10 ml of the upper 30 to 50 μm of the sea surface with each dip, was used to obtain 50 to 500 ml of the SML, sampled between 7 and 11 a.m. Control samples from the ULW were taken from a depth of 1 m with a 2-liter glass collection tube whose ends were closed by a drop-weight mechanism. Samples were kept in the cold and dark after sampling and were returned to the laboratory within 1 to 2 h, where they were processed for analysis. Meteorological data were recorded and provided by the meteorological station in Rostock-Warnemünde, located at the shore in proximity to the study site (see Table S1 in the supplemental material). Data for total organic carbon (TOC) and total nitrogen (TN) in the SML and the ULW were taken from previous studies (48, 49) (see also Table S1).
Extraction of nucleic acids and fingerprint analysis.
Identical volumes of SML and ULW water samples from each station were filtered through 3-μm-pore-size Isopore filters (Millipore), which presumably retain the major particle-attached bacterial fraction (37). The filtrate was then filtered through 0.22-μm-pore-size Isopore filters (Millipore) to collect the nonattached bacterial fraction. In cases in which the SML water volume was less than the water volume in the corresponding ULW sample due to SML sample volume limitations, the concentrations of nucleic acids in the corresponding SML and ULW samples were adjusted to ensure the comparability of fingerprint analyses (see below). All filters were frozen rapidly in liquid nitrogen and were stored at −80°C. DNA and RNA were extracted from the material trapped on the filters using the phenol-chloroform extraction method of Weinbauer et al. (51). Briefly, after cell lysis, a pH-dependent procedure was carried out to extract DNA (phenol-chloroform at pH 7.5 to 8) or RNA (phenol-chloroform at pH 4.5 to 5). All extracts were washed with ethanol, and the nucleic acids were quantified spectrophotometrically using an ND-1000 spectrophotometer (NanoDrop Technologies).
For the analysis of the presumably more active community (16S rRNA fingerprints), RNA was reverse transcribed into cDNA. Coprecipitated DNA in 10-μl aliquots (containing 40 ng to 1 μg of total nucleic acid) was removed using DNase I (DNA-free kit; Ambion) according to the manufacturer's instructions. cDNA was synthesized from 20 ng of RNA by reverse transcriptase PCR (RT-PCR) using the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (31) and the iScript cDNA synthesis kit (Bio-Rad). Complete removal of DNA was tested by an additional RT-PCR without the reverse transcriptase enzyme. Successful cDNA synthesis was confirmed by a subsequent PCR, as described below.
For 16S rRNA and 16S rRNA gene SSCP fingerprint analyses of the bacterial communities, 10 ng of DNA extracts and 2 μl of cDNA amplicons were PCR amplified using primers Com1f (5′-CAGCAGCCGCGGTAATAC-3′) and Com2r-Ph (5′-CCGTCAATTCCTTTGAGTTT-3′) (46), which amplify Escherichia coli 16S rRNA gene positions 519 to 926, according to the protocol of Labrenz et al. (30). The annealing temperature was 50°C. Single-stranded DNA was generated and purified, and SSCP analysis was carried out as described by Schwieger and Tebbe (46). Agarose gel electrophoresis was performed to ensure complete exonuclease digestion of the PCR products and, if needed, was used to finally adjust template concentrations for subsequent SSCP analysis. Fifty to 250 ng of single-stranded DNA was loaded onto the SSCP gels, whereby the concentrations of the corresponding SML and ULW samples were always the same.
Cluster analysis.
Pairwise comparisons of band patterns from the SML and the ULW were carried out using digitized images and GelCompar II (Applied Maths NV) based on densitometric curves, i.e., comparing gray values of specific positions between two gel lanes. Background subtraction, least-square filtering, and optimization were carried out according to the manufacturers' instructions. The results of the comparisons are expressed as pairwise similarities, i.e., the percentage of similarity between the compositions of the bacterioneuston and bacterioplankton communities, based on the Pearson correlation coefficient (41). The pairwise similarity data from stations 16 to 19 (see Table S1 in the supplemental material) are those cited by Stolle et al. (49). The curve-based correlation was chosen because during band-based analysis (Jaccard coefficient) the optimization procedure was not as thorough as that in a curve-based correlation when tested on internal reference lanes. The values of the band-based pairwise similarities of the fingerprints were lower than those of curve-based analyses, but the values for the two procedures correlated with each other, implying that the trends evidenced by these analyses are comparable (see Table S3 in the supplemental material).
Sequence analysis.
In the SSCP gels, individual bands exclusively observed in the 16S rRNA fingerprints of the SML samples were excised and reamplified according to the method of Labrenz et al. (30). PCR products were purified using the NucleoSpin II extraction kit (Macherey-Nagel) as described by the manufacturer and were sequenced by Qiagen using primers Com1f and Com2r. The forward and reverse sequences of all bands were checked for accuracy using SeqMan software (DNAStar). The sizes of the sequences ranged from 242 to 399 bp. The phylogenetic affiliations of the partial 16S rRNA sequences were estimated by employing the basic local alignment search tool (BLAST) (5). Alignments were checked and the sequences taxonomically grouped using the ARB software package (33).
Statistical analysis.
The Wilcoxon test (for which the significance level was a P value of <0.05) was applied to test differences revealed by pairwise comparisons between the nonattached and particle-attached size fractions as well as between 16S rRNA and 16S rRNA gene fingerprints. Spearman's rank correlation (for which the significance level was a P value of <0.05) was applied to test the association between pairwise comparisons and meteorological conditions as well as concentrations of TOC and TN. Additionally, this association was further analyzed using the following polynomial, quadratic nonlinear regression: f(x) = y0 + a × x + b × x2.
Nucleotide sequence accession numbers.
For each operational taxonomic unit (OTU) determined in this study, a representative 16S rRNA sequence was deposited in the GenBank database. The sequences were assigned accession numbers HM193050 to HM193082 (Table 1).
Table 1.
16S rRNA gene similarities of SSCP bands identified exclusively in the sea surface microlayer
| Taxon and OTUa | Station(s)b | Fractionc | Relative band intensity (%) | No. of sequences | Closest phylogenetic relatived |
Phylogenetic affiliatione | GenBank accession no. (this study) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Accession no. | 16S rRNA sequence similarity (%) | Habitat | |||||||
| Alphaproteobacteria | ||||||||||
| BaSML1 | 6 | N | 3.9 | 1 | Uncultured bacterium clone 1C227571 | EU799901 | 98 | Marine harbor | Rhodospirillales | HM193051 |
| BaSML2 | 9 | N | 2.2 | 1 | Uncultured alphaproteobacterium clone CB51E03 | EF471665 | 100 | Coastal water | Rhodobacteraceae | HM193053 |
| BaSML3 | 9 | P | 3.6 | 1 | Uncultured alphaproteobacterium clone CB51E03 | EF471665 | 98 | Coastal water | Rhodobacteraceae | HM193073 |
| BaSML4 | 11 | P | 0.9 | 1 | Uncultured alphaproteobacterium clone 131718 | AY922222 | 99 | Marine microbial mat | Rhodobacteraceae | HM193064 |
| BaSML5 | 4 | N | 3.3 | 2 | Alphaproteobacterium MPCa6SU_Cu07 isolate | EF414048 | 100 | Sponge associated | Sphingomonadaceae | HM193058 |
| Betaproteobacteria | ||||||||||
| BaSML6 | 17 | N | 3.5 | 1 | Uncultured bacterium clone TBb-04 | DQ791365 | 98 | Geothermally heated soil | Burkholderiales | HM193061 |
| BaSML7 | 15 | P | 0.5 | 1 | Uncultured Pelomonas sp. clone B6 | EF125953 | 99 | Biofilm reactor | Burkholderiales | HM193071 |
| BaSML8 | 7, 8 | N | 3.5 | 2 | Uncultured betaproteobacterium isolate, SSCP band 77-7-15 | GU088518 | 100 | Tropical lagoon water | Burkholderiales | HM193052 |
| Gammaproteobacteria | ||||||||||
| BaSML9 | 4 | N | 3.6 | 2 | Rheinheimera sp. JSM 083085 | FJ527418 | 100 | Forest soil | Chromatiaceae | HM193056 |
| BaSML10 | 9 | N/P | 2.8 | 2 | Rheinheimera sp. 3006 | AM110966 | 99 | Deep-sea sediment | Chromatiaceae | HM193072 |
| BaSML11 | 4 | P | 3.4 | 1 | Rheinheimera sp. enrichment culture clone AM15 | FJ516460 | 99 | Enrichment culture | Chromatiaceae | HM193081 |
| BaSML12 | 19 | P | 7.8 | 1 | Uncultured bacterium clone CCSD_DF2700_B7 | DQ128248 | 100 | Drilling fluid | Chromatiaceae | HM193082 |
| BaSML13 | 1 | P | 7.7 | 1 | Buchnera aphidicola strain 5A | CP001161 | 99 | Endosymbiont (insect) | Enterobacteriaceae | HM193076 |
| BaSML14 | 2 | P | 6.3 | 3 | Buchnera aphidicola | M63248 | 99 | Endosymbiont (insect) | Enterobacteriaceae | HM193078 |
| BaSML15 | 5 | N | 0.8 | 1 | Uncultured bacterium clone C7p3_ML_171 | FJ353210 | 98 | Lake water | Halothiobacillaceae | HM193050 |
| BaSML16 | 1 | P | 1.8 | 1 | Eubacterium from Cecidotrioza sozanica | AF286124 | 97 | Associated with insects | Coxiellaceae | HM193075 |
| BaSML17 | 1, 17 | P | 3.2 | 2 | Uncultured gammaproteobacterium clone SR-O-03-32 | AM905315 | 98 | Beach sediment | Moraxellaceae | HM193077 |
| BaSML18 | 17 | N | 0.3 | 1 | Shewanella sp. enrichment culture clone XB | GU001720 | 98 | Sediment of gas plant | Shewanellaceae | HM193059 |
| BaSML19 | 17 | N | 0.7 | 2 | Shewanella frigidimarina NCIMB400 | Y13699 | 95 | S. frigidimarina | Shewanellaceae | HM193060 |
| BaSML20 | 17 | N | 12.8 | 4 | Shewanella sp. Lac1 | GQ327990 | 99 | Intestines of gastropoda | Shewanellaceae | HM193062 |
| Bacteroidetes | ||||||||||
| BaSML21 | 3 | P | 0.4 | 1 | Uncultured bacterium clone C6S16_ML_382 | FJ352992 | 95 | Lake water | Cryomorphaceae | HM193079 |
| BaSML22 | 3 | P | 2.8 | 2 | Uncultured Bacteroidetes bacterium clone PI_4z12e | AY580718 | 97 | Coastal water | Cryomorphaceae | HM193080 |
| BaSML23 | 4 | N | 1.3 | 1 | Uncultured bacterium clone S23_1172 | EF573073 | 98 | Marine water | Flavobacteriaceae | HM193057 |
| BaSML24 | 17 | N | 3.4 | 1 | Uncultured Flavobacterium isolate, DGGE gel band | EU878141 | 100 | Baltic Sea mesocosms | Flavobacteriaceae | HM193063 |
| BaSML25 | 11 | P | 0.4 | 1 | Uncultured Haliscomenobacter sp. clone XZXXH27 | EU703445 | 100 | Lake water | Saprospiraceae | HM193068 |
| Cyanobacteria | ||||||||||
| BaSML26 | 12 | N | 9.8 | 2 | Anabaena sp. BIR361 | EF568895 | 99 | Baltic Sea | Halospirulina | HM193054 |
| BaSML27 | 14, 17, 18 | N/P | 13.8 | 4 | Uncultured cyanobacterium clone B1-06-24L | FM212461 | 99 | Macrophyte associated | Halospirulina | HM193070 |
| BaSML28 | 17 | P | 3.8 | 1 | Anabaena sp. BIR358 | EF568894 | 99 | Baltic Sea | Halospirulina | HM193065 |
| BaSML29 | 17 | P | 7.4 | 1 | Uncultured Coelosphaerium sp. clone SC1-1 | EF638722 | 97 | Coastal water | Halospirulina | HM193066 |
| BaSML30 | 9, 10 | P | 6.0 | 2 | Nodularia spumigena strain NSLA01 | AF268009 | 99 | Nodularia spumigena | Halospirulina | HM193074 |
| BaSML31 | 17 | P | 1.9 | 1 | Uncultured organism clone Lc2yS22_ML_277 | FJ355041 | 99 | Lake water | Oscillatoria | HM193067 |
| BaSML32 | 13 | P | 1.6 | 2 | Uncultured eukaryote chloroplast gene | AB469950 | 99 | Cow manure compost | Oscillatoria | HM193069 |
| Other: BaSML33 | 2 | N | 2.4 | 1 | Uncultured bacterium clone mek61a11 | AY537289 | 94 | Unclassified | HM193055 | |
OTUs are grouped according to their phylogeny. The relative abundances of the OTUs are shown in Fig. S1 in the supplemental material.
See Table S1 in the supplemental material.
N, nonattached fraction; P, particle-attached fraction; N/P, OTUs found in both size fractions.
According to alignment to the NCBI database. DGGE, denaturing gradient gel electrophoresis.
According to alignment to the ARB database.
RESULTS
Comparison of bacterioneuston and bacterioplankton community structures.
Nineteen samples of the SML in the southern Baltic Sea were taken from 2006 to 2008, during spring and late summer (see Table S1 in the supplemental material), and the community structure of the bacterioneuston was compared to that of the bacterioplankton in the ULW. Their pairwise similarities for the nonattached size fraction were 46.7 to 96.9% (mean, 84.7%) in the active communities and 66.3 to 98.7% (mean, 91.8%) in the total communities (Fig. 1). The corresponding values for the particle-attached size fraction were 25.4 to 97.8% (mean, 62.8%) and 7.8 to 96.6% (mean, 60.8%), respectively (Fig. 1). The differences between the nonattached and particle-attached size fractions for both the active and total communities were significant (P, <0.01 by the Wilcoxon test; n = 19) (Fig. 1). Also, the differences between the active and total communities in the nonattached size fraction were significant but at a lower probability (P, 0.011 by the Wilcoxon test; n = 19) (Fig. 1).
Fig. 1.
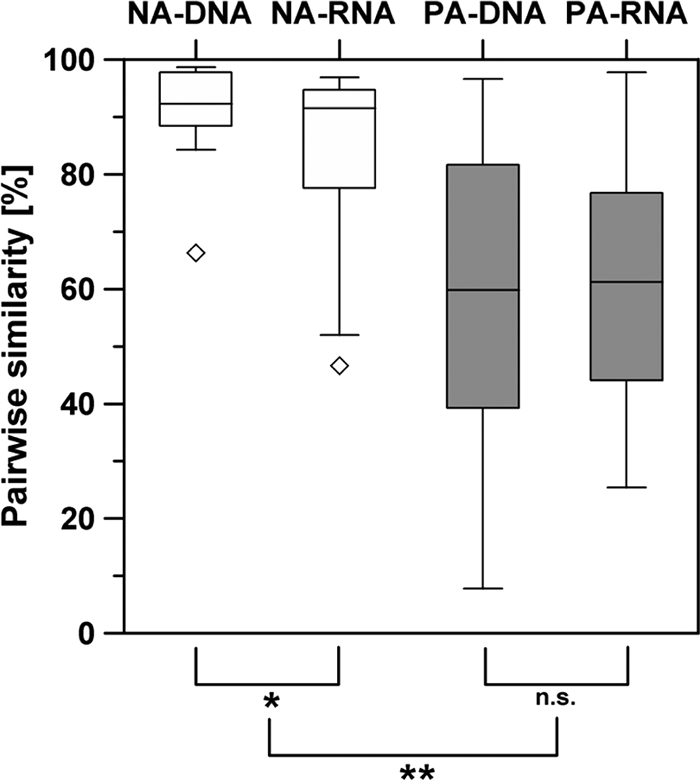
Differences in pairwise similarities between bacterioneuston and bacterioplankton communities of 16S rRNA (active community) (RNA) and 16S rRNA gene (total community) (DNA) fingerprints of the nonattached (NA) and particle-attached (PA) communities (n = 19). Levels of significance by the Wilcoxon test are indicated as follows: *, P < 0.05; **, P < 0.01; n.s. (not significant), P > 0.05.
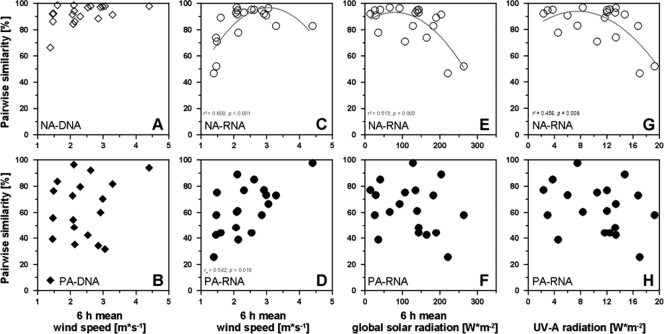
To determine whether meteorological conditions were responsible for the differences between the bacterioneuston and bacterioplankton community compositions, values for the pairwise similarities were plotted against wind speed, global solar radiation, and UV-A radiation during sampling, and against mean wind speed and mean global solar radiation 6 h prior to sampling (see Table S2 in the supplemental material). Spearman's rank correlation showed no relation between any of these parameters and the differences among the bacterial communities, except between the mean wind speed 6 h prior to sampling and the active communities of the particle-attached size fraction (Fig. 2D). Additionally, the relation between the pairwise similarities and the accumulation of TOC and TN was significant only for the total communities in the particle-attached size fraction (see Table S2 in the supplemental material). Despite this overall lack of correlation, the similarities among the active communities in the nonattached size fraction tended to decrease at the lowest wind speeds and at the highest radiation levels (Fig. 2C, E, and G). Nonlinear regression was applied to further elucidate these effects and showed an association with mean wind speed 6 h prior to sampling (Fig. 2C), mean global solar radiation 6 h prior to sampling (Fig. 2E), and UV-A radiation during sampling (Fig. 2G). In contrast, nonlinear regression did not reveal any influence of meteorological conditions on the total communities of the nonattached size fraction or on the changing patterns of all fingerprints within the particle-attached size fraction (data not shown). In summary, wind speed history was positively related to pairwise similarities in the active communities of both size fractions, and most meteorological parameters were related to the decreasing similarities in the active communities of the nonattached size fraction.
Fig. 2.
Relationship of meteorological conditions (mean wind speed 6 h prior to sampling [A to D], mean global solar radiation 6 h prior to sampling [E and F], and UV-A radiation [G and H]) with the pairwise similarities between bacterioneuston and bacterioplankton community structures in the 16S rRNA gene (DNA) (A and B) and 16S rRNA (RNA) (C to H) fingerprints. Results for the nonattached (NA) (upper row of panels) and particle-attached (PA) (lower row of panels) communities are shown. Significant correlations (rs, Spearman correlation coefficient) and nonlinear regression (r2) are indicated (gray lines). p, level of significance.
Identification of members of the bacterioneuston community.
An additional aim of this study was to identify specific, active bacterioneuston members by analyzing SSCP bands detectable solely in the SML 16S rRNA fingerprints. No unique SSCP bands were identified in the SML fingerprints from 6 out of 19 stations; however, in the remaining 13 stations, 133 SSCP bands, ranging from 1 to 22 bands per sample, were found exclusively in the SML fingerprints. Of these, 52 bands belonged to the nonattached and 81 to the particle-attached size fraction. Sufficient sequencing results were obtained for 38% of all sequences (n = 51). Based on their relative band intensities (Table 1), these sequences accounted for an average of 65% (±28%) (n = 13) of all SML-specific SSCP bands at each station. Notably, the highest number of SML-specific sequences (n = 15) per sample was retrieved from station 17 (Table 1; see also Table S1 in the supplemental material), where a dense surface film (slick) had been observed during sampling (49).
Only 3 out of the 51 sequences showed a <97% 16S rRNA gene sequence similarity to environmental clones in the NCBI database (Table 1). Sequences related to the identical environmental clone and with >99% sequence similarity to each other were grouped into an operational taxonomic unit (OTU). The 33 OTUs thus identified and their closest environmental clones are shown in Table 1. These clones were originally found in several habitats, such as different water columns, microbial mats, and soils, or were associated with metazoans. Active members of the SML included those belonging to the phyla Cyanobacteria and Bacteroidetes as well as to the alpha, beta, and gamma subgroups of the phylum Proteobacteria, with similar distributions in the two size fractions and a dominance of Gammaproteobacteria and Cyanobacteria (see also Fig. S1 in the supplemental material). Overall, within each phylogenetic group, several orders and families were identified, except in the Betaproteobacteria, in which identification was limited to members of Burkholderiales (Table 1). While several phylogenetic affiliations, e.g., Chromatiaceae, were detected in both size fractions, others were present exclusively in either the nonattached (e.g., Shewanellaceae) or the particle-attached (e.g., Enterobacteriaceae) size fraction (Table 1). However, only two OTUs were common to both size fractions; the remaining OTUs were found exclusively in either the nonattached or the particle-attached fingerprints. Most OTUs belonged to Gammaproteobacteria (n = 12) or to Cyanobacteria (n = 7) (Table 1).
DISCUSSION
The aim of the present study was to better understand the influence of various meteorological conditions on the development of a particular bacterial community in the sea surface microlayer of the southern Baltic Sea. Thus, low wind speed history was found to have the strongest differentiation effect on active bacterioneuston assemblages (as determined by 16S rRNA analysis), followed by global solar radiation and UV radiation. In contrast, the total communities (as determined by 16S rRNA gene analysis) were not influenced by any of the meteorological conditions investigated. Phylogenetically, active members of the bacterioneuston belonged to Cyanobacteria, Bacteroidetes, and the alpha, beta, and gamma subgroups of Proteobacteria. Their 16S rRNA sequences were related mainly to environmental clones from soils and different water columns, and thus, they most likely did not form distinct bacterioneuston assemblages.
Comparison of bacterioneuston and bacterioplankton community compositions.
Opposing results have been reported regarding the differences between SML and ULW bacterial communities. For instance, 16S rRNA gene-based analyses showed bacterioneuston diversity patterns strongly distinguishable from those of bacterioplankton when the SML was sampled with membrane filters (12, 16) but not when a screen sampler was used (1, 36). This discrepancy was caused partly by the different SML sampling techniques (2, 10), demonstrating the importance of sample collection. The most important considerations for obtaining high-quality SML samples are minimal dilution with bulk water, a lack of bias due to sampler selectivity, collection of a large sample volume within a reasonable sampling time, and easy handling of the samplers (17, 24). The glass-plate sampling device used in our study was chosen because (i) its layer thickness is in the range of 50 μm, as recommended in previous reports (10, 55), (ii) larger amounts of water could be sampled for further analyses, and (iii) previous studies demonstrated that this method shows no selectivity in the analysis of bacterial community composition (48).
Pairwise comparisons of bacterial community structures of the SML and ULW, as carried out in this and other studies, allow assessment of the extent of bacterioneuston and bacterioplankton differentiation. Whereas previous studies generally compared total bacterial communities, we sought to achieve a higher-resolution comparison by distinguishing between nonattached and particle-attached communities as well as between active and total communities. In our study it became obvious that the differences between the SML and ULW exhibited by particle-attached assemblages were much more pronounced than those for the nonattached assemblages, in which the two compartments were highly similar. This is in accordance with the notion that particulate material is generally inhabited by specific bacterial assemblages that are distinguishable from those of the surrounding bulk water and that differ depending on the particle type (47). These observations underline the importance of distinguishing different size fractions as well as active and total communities in order to obtain more-profound comparisons of bacterioneuston and bacterioplankton diversity analyses.
Influence of meteorological conditions.
In general, differences between active bacterioneuston and bacterioplankton community compositions in both size fractions were highest during low wind speeds (Fig. 2), suggesting a shift in the composition of active bacteria in the SML under calm conditions. In a previous study, a correlation between wind speed history and changing SML properties was demonstrated (36). Also in the present study, only wind speed history, expressed as the mean wind speed 6 h prior to sampling and not the actual wind speed during sampling, was related to the differences between the bacterioneuston and bacterioplankton fingerprints. This probably reflected the fact that the reduction of turbulent surface mixing induced by low wind speeds is time delayed. In the presence of less wind-induced surface mixing, the sea surface calms. This may in turn promote the development of a distinct, active bacterioneuston community. The development of a distinct total-bacterioneuston community most likely requires more than 6 h of low turbulence in the SML. Similar suggestions have been made regarding estuarine bacterioneuston assemblages, since they are exposed to strong hydrodynamics (45), and might explain previous observations of highly distinctive bacterioneuston assemblages after a few days in mesocosm experiments and during slick formation (13, 49). In addition, alpine lakes, which are exposed to less wind stress than marine systems, contain unique archaeal assemblages in the air-water interface and ULW (6), and even specific neustonic bacteria, such as Nevskia ramosa, may live in limnic habitats (39).
Compared to wind speed history, global solar radiation and UV-A radiation had only minor impacts on the differences between the active bacterioneuston and bacterioplankton communities, implying low structuring effects on bacterioneuston assemblages in the present study. This has also been shown for the near-surface bacterioplankton composition of the North Sea, which was only slightly influenced by changing levels of UV radiation (52). However, in the active communities of the nonattached size fraction examined in the present study, the strongest differences occurred in response to the highest radiation levels. This suggests that increasing exposure to UV-induced stress alters the community composition of nonattached, active bacterioneuston members, which again is favored during low wind conditions. In contrast, variability in the particle-attached fingerprints was not related to changing levels of radiation, probably due to the sheltering from UV exposure conferred by particle association (22, 53).
Interestingly, none of the meteorological conditions correlated with changes in the total bacterioneuston and bacterioplankton assemblages, at least based on our methodological approach. A previous study likewise detected no association between wind speed and differences between total bacterioneuston and bacterioplankton communities in coastal Mediterranean waters (1), indicating that the high spatial and temporal variability in the SML is also caused by other factors. The dynamic patterns of the similarities between the SML and ULW in the total, particle-attached communities were found to be related to TOC and TN accumulation in the SML (see Table S2 in the supplemental material). The SML is generally characterized by strong enrichment of organic and inorganic substances, especially of the particulate fraction (25). Under very calm conditions, e.g., in visible surface films, the particulate fraction increasingly contributes to the total-organic-matter pool (49). Thus, the changing enrichment of particulate organic matter may account even better for the differences in community composition that were already shown for bacterial abundances and productivity (49). Further evidence of large differences between the community compositions of the SML and the ULW following increases in the amount of organic matter comes from mesocosm experiments, in which autotrophic production resulted in high concentrations of transparent exopolymeric particles in the SML (10, 11).
Identification of active bacteria in the SML.
To identify active bacterioneuston members of the southern Baltic Sea, SML-specific SSCP bands from 16S rRNA fingerprints of the particle-attached and nonattached size fractions were sequenced. This approach allowed us to obtain sequences of most of the abundant (as revealed by their relative band intensities) and potentially active members of the bacterioneuston. Mostly, these sequences were closely related to 16S rRNA sequences in the NCBI database, covering diverse habitats such as marine, brackish, and limnic waters, as well as soils and metazoans. This supports previous findings showing that unknown taxa are not widespread among bacterioneuston communities (1, 21). Compared to those in the other samples investigated, an unusually high number of OTUs (eight, belonging to several phylogenetic groups) (Table 1) were detected in a slick sample from station 17 (49), supporting the conclusions from the fingerprint analyses that highly calmed conditions promote distinctive bacterioneuston populations.
Most of the OTUs phylogenetically located within the Cyanobacteria were detected in the particle-attached size fraction and were related predominantly to sequences from planktonic habitats (Table 1), e.g., Nodularia spumigena, which is highly abundant in Baltic Sea surface blooms (40). Several proteobacterial OTUs of the Rhodobacteriaceae, Burkholderiales, Chromatiaceae, Moraxellaceae, and Shewanellaceae were originally assigned to sediments and microbial mats; interestingly enough, in this study most of these OTUs were allocated to the particle-attached size fraction. Organisms from the sediment-water interface and particle-attached organisms might have specific requirements to inhabit the air-water interface, where conditions might be comparable to those of solid substrata (28). However, since the water depth at the sampling site in the southern Baltic Sea is shallow (∼10 m), sediment turbulence and thus passive transport of these OTUs into the SML, rather than specific colonization within this layer, could also have occurred.
A large proportion of the OTUs from the nonattached and particle-attached size fractions showed the highest similarities to planktonic members of limnic, brackish, and marine waters. Among these, all Bacteroidetes were related to bacterioplankton sequences. Generally, Bacteroidetes are widespread among bacterioneuston populations (1, 12, 36), most likely the result of their high potential for degrading complex polymeric substances (27). Interestingly, Flavobacteriaceae were detected only in the nonattached size fraction. One such OTU was closely related to Polaribacter filamentus, which is known to contain gas vacuoles (18). This finding suggests that some members of the bacterioneuston actively regulate the duration of their stay in the SML.
The dominance of Gammaproteobacteria in our samples confirms previous studies in which culture-independent (16, 36) and culture-dependent (14, 15, 50) methods were used to examine marine bacterioneuston assemblages. The ability of Gammaproteobacteria to respond quickly to nutrient enrichment may account for their predominance in the SML (38, 42). Members of the Gammaproteobacteria and Bacteroidetes from the Mediterranean bacterioneuston contributed strongly to strains with a high resistance to solar radiation (3). Leucine uptake in the Atlantic SML was also dominated by these groups, further indicating their strong presence in bacterioneuston assemblages (36). Notably, the Chromatiaceae-related sequences within the Gammaproteobacteria detected in the present study clustered closely together with Rheinheimera baltica and Rheinheimera perlucida, isolated from Baltic Sea surface waters (7, 8). The high contribution of organisms related to Rheinheimera spp. further suggests that the dominant active members of the bacterioneuston in the Baltic Sea do not specifically inhabit the SML.
Conclusions.
The identification of active bacterioneuston members and the high similarity between the nonattached communities of the SML and ULW in the Baltic Sea together support previous findings that the bacterioneuston is strongly influenced by the underlying bacterioplankton (26). Increasing differences between the active, nonattached communities were associated with low wind speed and high radiation levels, indicating that in a stable SML the profile of the active bacterioneuston community changes in response to specific conditions, e.g., enhanced radiation exposure. In contrast, the difference in the particle-attached community structure between the bacterioneuston and the bacterioplankton was found to be highly variable for both the total and the active communities and only weakly linked to wind speed, probably reflecting the importance of particle dynamics in this unusual habitat. Thus, a combinatory influence of wind stress, radiation exposure, and particulate organic material accumulation seems to differentiate the community structure of bacterioneuston from that of bacterioplankton, especially in the active communities. It is thus plausible that under these conditions, air-water exchange processes might be specifically influenced by the bacterioneuston; however, the significance still needs to be resolved.
Supplementary Material
ACKNOWLEDGMENTS
We thank U. Hehl and M. Poetzsch for excellent assistance during sampling. Analyses of total organic carbon and nitrogen by K. Nagel are also highly appreciated. Furthermore, we thank A. Eggert and C. Wagner for very helpful comments and advice during statistical analyses, and we thank two anonymous reviewers for very helpful comments.
C.S. was financed by the German Leibniz Society (WGL-PAKT project FILGAS) and the Federal Ministry of Education and Research (SOPRAN Phase II).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Agogué H., et al. 2005. A survey on bacteria inhabiting the sea surface microlayer of coastal ecosystems. FEMS Microbiol. Ecol. 54:269–280 [DOI] [PubMed] [Google Scholar]
- 2. Agogué H., et al. 2004. Comparison of samplers for the biological characterization of the sea surface microlayer. Limnol. Oceanogr. Methods 2:213–225 [Google Scholar]
- 3. Agogué H., Joux F., Obernosterer I., Lebaron P. 2005. Resistance of marine bacterioneuston to solar radiation. Appl. Environ. Microbiol. 71:5282–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aller J. Y., Kuznetsova M. R., Jahns C. J., Kemp P. F. 2005. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. J. Aerosol Sci. 36:801–812 [Google Scholar]
- 5. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Auguet J., Casamayor E. O. 2008. A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ. Microbiol. 10:1080–1086 [DOI] [PubMed] [Google Scholar]
- 7. Brettar I., Christen R., Hofle M. G. 2002. Rheinheimera baltica gen. nov., sp. nov., a blue-coloured bacterium isolated from the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 52:1851–1857 [DOI] [PubMed] [Google Scholar]
- 8. Brettar I., Christen R., Hofle M. G. 2006. Rheinheimera perlucida sp. nov., a marine bacterium of the Gammaproteobacteria isolated from surface water of the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 56:2177–2183 [DOI] [PubMed] [Google Scholar]
- 9. Crump B. C., Armbrust E. V., Baross J. A. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cunliffe M., et al. 2009. Comparison and validation of sampling strategies for the molecular microbial analysis of surface microlayers. Aquat. Microb. Ecol. 57:69–77 [Google Scholar]
- 11. Cunliffe M., et al. 2009. Dissolved organic carbon and bacterial populations in the gelatinous surface microlayer of a Norwegian fjord mesocosm. FEMS Microbiol. Lett. 299:248–254 [DOI] [PubMed] [Google Scholar]
- 12. Cunliffe M., et al. 2008. Phylogenetic and functional gene analysis of the bacterial and archaeal communities associated with the surface microlayer of an estuary. ISME J. 2:776–789 [DOI] [PubMed] [Google Scholar]
- 13. Cunliffe M., et al. 2009. Comparison of bacterioneuston and bacterioplankton dynamics during a phytoplankton bloom in a fjord mesocosm. Appl. Environ. Microbiol. 75:7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donderski W., Walczak M., Mudryk Z., Kobyliński M. 1999. Neustonic bacteria number, biomass and taxonomy. Pol. J. Environ. Stud. 8:137–141 [Google Scholar]
- 15. Fehon W. C., Oliver J. D. 1979. Taxonomy and distribution of surface microlayer bacteria from two estuarine sites. Estuaries 2:194–197 [Google Scholar]
- 16. Franklin M. P., et al. 2005. Bacterial diversity in the bacterioneuston (sea surface microlayer): the bacterioneuston through the looking glass. Environ. Microbiol. 7:723–736 [DOI] [PubMed] [Google Scholar]
- 17. Garrett W. D., Duce R. A. 1980. Surface microlayer samplers, p. 471–490 In Dobson F., Hasse L., Davis R. (ed.), Air-sea interaction. Plenum, New York, NY [Google Scholar]
- 18. Gosink J. J., Woese C. R., Staley J. T. 1998. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48:223–235 [DOI] [PubMed] [Google Scholar]
- 19. Harvey R. W., Burzell L. A. 1972. A simple microlayer method for small samples. Limnol. Oceanogr. 17:156–157 [Google Scholar]
- 20. Harvey R. W., Young L. Y. 1980. Enumeration of particle-bound and unattached respiring bacteria in the salt marsh environment. Appl. Environ. Microbiol. 40:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hervas A., Casamayor E. O. 2009. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol. Ecol. 67:219–228 [DOI] [PubMed] [Google Scholar]
- 22. Hess-Erga O. K., Kihle Attramadal K. J., Vadstein O. 2008. Biotic and abiotic particles protect marine heterotrophic bacteria during UV and ozone disinfection. Aquat. Biol. 4:147–154 [Google Scholar]
- 23. Hörtnagl P., Pérez M. T., Sommaruga R. 2010. Living at the border: a community and single-cell assessment of lake bacterioneuston activity. Limnol. Oceanogr. 55:1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huehnerfuss H. 1981. On the problem of sea surface film sampling: a comparison of 21 microlayer-, 2 multilayer-, and 4 selected subsurface-samplers. Part 2. Sonderdr. Meerestechnik 12:170–173 [Google Scholar]
- 25. Hunter K. A. 1997. Chemistry of the sea-surface microlayer, p. 287–320 In Liss P. S., Duce R. A. (ed.), The sea-surface and global change. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 26. Joux F., et al. 2006. Microbial community structure in the sea surface microlayer at two contrasting coastal sites in the northwestern Mediterranean Sea. Aquat. Microb. Ecol. 42:91–104 [Google Scholar]
- 27. Kirchman D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91–100 [DOI] [PubMed] [Google Scholar]
- 28. Kjelleberg S. 1985. Mechanisms of bacterial adhesion at gas-liquid interfaces, p. 163–194 In Savage D. C., Fletcher M. (ed.), Bacterial adhesion. Plenum, New York, NY [Google Scholar]
- 29. Kuznetsova M., Lee C. 2001. Enhanced extracellular enzymatic peptide hydrolysis in the sea-surface microlayer. Mar. Chem. 73:319–332 [Google Scholar]
- 30. Labrenz M., Jost G., Jürgens K. 2007. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat. Microb. Ecol. 46:177–190 [Google Scholar]
- 31. Lane D. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 32. Liss P. S., Duce R. A. 1997. The sea surface and global change. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 33. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maki J. S. 1993. The air-water interface as an extreme environment, p. 409–439 In Ford T. E. (ed.), Aquatic microbiology: an ecological approach. Blackwell Scientific Publications, Boston, MA [Google Scholar]
- 35. Münster U., Heikkinen E., Knulst J. 1997. Nutrient composition, microbial biomass and activity at the air-water interface of small boreal forest. Hydrobiologia 363:261–270 [Google Scholar]
- 36. Obernosterer I., et al. 2008. Biochemical characteristics and bacterial community structure of the sea surface microlayer in the South Pacific Ocean. Biogeosciences 5:693–705 [Google Scholar]
- 37. Paerl H. W. 1975. Microbial attachment to particles in marine and freshwater ecosystems. Microb. Ecol. 2:73–83 [DOI] [PubMed] [Google Scholar]
- 38. Pinhassi J., Berman T. 2003. Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pladdies T., Babenzien H. D., Cypionka H. 2004. Distribution of Nevskia ramosa and other rosette-forming neustonic bacteria. Microb. Ecol. 47:218–223 [DOI] [PubMed] [Google Scholar]
- 40. Ploug H. 2008. Cyanobacterial surface blooms formed by Aphanizomenon sp. and Nodularia spumigena in the Baltic Sea: small-scale fluxes, pH, and oxygen microenvironments. Limnol. Oceanogr. 53:914–921 [Google Scholar]
- 41. Rademaker J. L. W., Louws F. J., Rossbach U., Vinuesa P., de Bruijn F. J. 1999. Computer-assisted pattern analysis of molecular fingerprints and database construction, p. 1–33 In Akkermans A. D. L., van Elsas J. D., de Bruijn F. J. (ed.), Molecular microbial ecology manual 7.1.3. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 42. Reche I., et al. 2009. Effect of Saharan dust inputs on bacterial activity and community composition in Mediterranean lakes and reservoirs. Limnol. Oceanogr. 54:869–879 [Google Scholar]
- 43. Reinthaler T., Sintes E., Herndl G. J. 2008. Dissolved organic matter and bacterial production and respiration in the sea-surface microlayer of the open Atlantic and the western Mediterranean Sea. Limnol. Oceanogr. 53:122–136 [Google Scholar]
- 44. Romano J.-C. 1996. Sea-surface slick occurrence in the open sea (Mediterranean, Red Sea, Indian Ocean) in relation to wind speed. Deep Sea Res. Part I 43:411–423 [Google Scholar]
- 45. Santos A. L., et al. 2009. Short-term variability of abundance, diversity and activity of estuarine bacterioneuston and bacterioplankton. J. Plankton Res. 31:1545–1555 [Google Scholar]
- 46. Schwieger F., Tebbe C. C. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Simon M., Grossart H. P., Schweitzer B., Ploug H. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28:175–211 [Google Scholar]
- 48. Stolle C., Nagel K., Labrenz M., Jürgens K. 2009. Bacterial activity in the sea-surface microlayer: in situ investigations in the Baltic Sea and the influence of sampling devices. Aquat. Microb. Ecol. 58:67–78 [Google Scholar]
- 49. Stolle C., Nagel K., Labrenz M., Jürgens K. 2010. Succession of the sea-surface microlayer in the coastal Baltic Sea under natural and experimentally induced low-wind conditions. Biogeosciences 7:2975–2988 [Google Scholar]
- 50. Tsyban A. V. 1971. Marine bacterioneuston. J. Oceanogr. Soc. Jpn. 27:56–66 [Google Scholar]
- 51. Weinbauer M. G., Fritz I., Wenderoth D. F., Höfle M. G. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winter C., Moeseneder M. M., Herndl G. J. 2001. Impact of UV radiation on bacterioplankton community composition. Appl. Environ. Microbiol. 67:665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu Y., Clevenger T., Deng B. 2005. Impacts of goethite particles on UV disinfection of drinking water. Appl. Environ. Microbiol. 71:4140–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wurl O., Obbard J. P. 2004. A review of pollutants in the sea-surface microlayer (SML): a unique habitat for marine organisms. Mar. Pollut. Bull. 48:1016–1030 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Z., Liu L., Liu C., Cai W. 2003. Studies on the sea surface microlayer. II. The layer of sudden change of physical and chemical properties. J. Colloid Interface Sci. 264:148–159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.