Abstract
Growing Dictyostelium cells secrete CfaD and AprA, two proteins that have been characterized as chalones. They exist within a high-molecular-weight complex that reversibly inhibits cell proliferation, but not growth, via cell surface receptors and a signaling pathway that includes G proteins. How the production of these two proteins is regulated is unknown. Dictyostelium cells possess three GCN2-type eukaryotic initiation factor 2 α subunit (eIF2α) kinases, proteins that phosphorylate the translational initiation factor eIF2α and possess a tRNA binding domain involved in their regulation. The Dictyostelium kinases have been shown to function during development in regulating several processes. We show here that expression of an unregulated, activated kinase domain greatly inhibits cell proliferation. The inhibitory effect on proliferation is not due to a general inhibition of translation. Instead, it is due to enhanced production of a secreted factor(s). Indeed, extracellular CfaD and AprA proteins, but not their mRNAs, are overproduced in cells expressing the activated kinase domain. The inhibition of proliferation is not seen when the activated kinase domain is expressed in cells lacking CfaD or AprA or in cells that contain a nonphosphorylatable eIF2α. We conclude that production of the chalones CfaD and AprA is translationally regulated by eIF2α phosphorylation. Both proteins are upregulated at the culmination of development, and this enhanced production is lacking in a strain that possesses a nonphosphorylatable eIF2α.
INTRODUCTION
The translational initiation factor eukaryotic initiation factor 2 (eIF2) functions to deliver initiator tRNAs to ribosomes that are assembling on mRNAs. The delivery, and thus the efficiency of translation, is regulated by phosphorylation of the α subunit of eIF2 (eIF2α) at serine 51 (11). While phosphorylation of eIF2α may decrease translation in a general manner, often it enhances the translation of specific mRNAs (5, 10, 16, 17, 19, 31) by a mechanism using short upstream open reading frames (18). There are four different types of eIF2α-specific kinases found in mammals, GCN2, PERK, HRI, and PKR (11). PERK is also found in all multicellular eukaryotes, while the GCN2 kinase is found in yeast and fungi, Dictyostelium, and many other eukaryotes (11). These kinases are activated by various cellular and environmental stimuli through the four types of regulatory domains that control the activity of the kinase domain. Translational regulation mediated by the four types of eIF2α kinases is involved in many cellular responses and processes, including the unfolded-protein response and endoplasmic reticulum stress (PERK); cell cycle progression and viral defense (PKR); iron/heme limitation (HRI); and a variety of stresses, including general nutrient limitation and amino acid starvation (GCN2) (11, 18). Among other recent studies of the functions associated with translational regulation by eIF2α phosphorylation are those demonstrating roles in controlling cell proliferation (4, 5, 20, 30, 39).
Dictyostelium cells have three initiation factor kinases, IfkA, IfkB, and IfkC, that are homologs of GCN2 and thus sense amino acid deprivation. Three other, uncharacterized genes have kinase domains similar to those of eIF2α kinases but lack regulatory domains similar to one of the four types described above. Studies on the expression of the ifkA, -B, and -C genes show that all three are expressed during growth and throughout development (14, 29). In situ hybridizations demonstrate a high degree of spatially restricted expression for each gene during development (29). Studies using disruptions of the ifk genes have indicated that IfkA functions in modulating the timing of aggregation and early gene expression, that IfkB functions in maintaining proper prestalk-specific gene expression, and that IfkA and IfkB function in maintaining proper cell-cell and cell-substrate adhesion and the equilibrium between different cell types for proper spatial patterning (14, 29).
Recent studies have characterized two proteins of Dictyostelium, CfaD and AprA, that act as chalones, i.e., endogenous proliferation inhibitors involved in reversibly regulating tissue size (7, 37). CfaD and AprA are secreted during growth and are part of an extracellular 150-kDa complex that represses proliferation but does not affect cell growth (1, 6). A receptor for AprA has been characterized (8), and the signaling pathway downstream of the chalone receptor involves G proteins (2) and a ROCO family kinase (26). While there is some understanding of the mechanism of action of CfaD and AprA in inhibiting proliferation, nothing is known about the control of their production. We demonstrate here that eIF2α phosphorylation regulates the production of the two chalone proteins.
MATERIALS AND METHODS
Generation of BS167.
To replace the endogenous eIF2α gene with a copy carrying a mutation in codon 51 that results in an S51A mutated protein product, a knock-in construct was made as follows. (In Dictyostelium eIF2α, the residue equivalent to the canonical S51 phosphorylation site of yeast GCN2 [10] is S53. Here, we refer to this residue generically as S51, given the prevalent and general use of the yeast numbering to identify the residue phosphorylated by eIF2α kinases.) A 1,875-bp fragment of the promoter and the entire coding region was amplified from genomic DNA using primers if2-5 (5′) and if2-2 (3′) (Table 1) and cloned into the pGEM T-easy vector (Promega). Mutagenesis of the S51 codon was carried out as described previously (32) employing the if2-4 oligonucleotide (Table 1), which alters the serine codon to one encoding alanine. A blasticidin resistance gene cassette (bsr) (35) was attached at the end of the coding region. A 1,350-bp fragment of the coding region amplified from genomic DNA with if2-1 and if2-2 primers (Table 1) was cloned to the opposite side of the bsr cassette, resulting in the knock-in construct pif2α-25. The second coding region also possessed the S51A mutation made as described above. pif2α-25 was digested to release the eIF2α-bsr DNA, and this was transformed into Ax4 cells using electroporation (33). Genomic DNA was isolated from blasticidin-resistant clones possessing insertions at the endogenous eIF2α gene (determined by PCR screening) and used as a template in a PCR to amplify a 1,000-bp fragment spanning the proximal promoter and coding region past the S51 codon. PCR products were sequenced to identify clones in which double-crossover points occurred, so as to replace the S51 codon with the A51 codon. In such clones, there is an intact eIF2α promoter and a coding region possessing the S51A mutation. Independent isolates gave the same phenotypes, and one such strain was named BS167 and was used for the experiments described in this paper.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| if2-4 | 5′ ATAGATCTGATACGACGACGTGCAATTTC 3′ |
| if2-5 | 5′ AGGGCCCACTGGTTAATGCTGTTAAAATTG 3′ |
| if2-2 | 5′ GGATCCTTATTTTCTGCTCGATTTCTTTTCC 3′ |
| if2-1 | 5′ GGATCCTAAAAATGGTATTCGAAAATGATTG 3′ |
| ifkA-9 | 5′ GGATCCGGAGGTTCAATGTTTAGATATCATTCTAGA 3′ |
| ifkA-10 | 5′ CTCGAGTTAAAACAATTTCTCCATCAA 3′ |
| ifkA-13 | 5′ GGAGTATTGTTCAAAGGGAACCTTGAAAAC 3′ |
| ifkA-14 | 5′ AAAACCAGCCAATATTTTAATTGATAATG 3′ |
| AprA-5 | 5′ GAACCACTTTGGTTAGCAAG 3′ |
| AprA-3 | 5′ CCCATTAAAGAGTGTTCAG 3′ |
| CfaD-5 | 5′ GTAGTCACCCAGATGTTTAC 3′ |
| CfaD-3 | 5′ ACCAAAAGTCCAACATGAAC 3′ |
Generation of GyrB/kinase domain fusions.
To generate an inducible, active kinase domain, the ifkA kinase domain was fused to the GyrB domain of the Escherichia coli gyrase. This GyrB domain contains an ATPase active site that can be inhibited by coumermycin, causing dimerization of the domain and any fused polypeptides (15, 36). A 1,896-bp fragment of the ifkA coding region was amplified from genomic DNA using oligonucleotides ifkA-9 and ifkA-10 (Table 1), with flanking BamHI and XhoI sites derived from the ends of the primers. This represents amino acids 881 to 1514, which align with the kinase domain of yeast GCN2. The fragment was cloned into the pGEM T-easy vector. Activating mutations based on studies with yeast GCN2 mutations (28) were made as described above employing primers ifkA-13 and ifkA-14 (Table 1). The double-mutated kinase domain possessed a K1258G and an F1320L change. An EcoRI/BamHI 690-bp fragment from pC939 (36) and coding for residues 1 to 220 of the E. coli GyrB protein (its dimerization domain) was fused to the actin 15 promoter in the plasmid Exp4(+) (13). The BamHI/XhoI kinase domain fragments were subsequently fused to the 3′ end of the GyrB coding region, resulting in pifkA-6 (wild-type kinase domain) and pifkA-14 (double-mutant kinase domain). Each plasmid was transformed into various strains using electroporation. In each instance, at least two independent populations of transformed cells were used in independent experiments.
Cell growth and conditioned medium.
Cells were grown in HL-5 medium (9) at 21°C. The titers of the cells were determined by direct counting with a hemocytometer. When present, coumermycin was added to a final concentration of 1 μg/ml from a stock of 5-mg/ml coumermycin in DMSO (dimethyl sulfoxide). For collection of conditioned medium, cultures were started at 5 × 105 cells per ml and grown for 48 h or to the desired cell titers. Coumermycin was added for 12, 24, or 48 h, and the cells were removed by centrifugation for 4 min at 5,000 × g. The supernatant was removed and spun for 5 min at 9,000 × g to ensure removal of all cells. The supernatant from the second spin was either used immediately or frozen at −80°C.
Protein synthesis measurements.
Cell cultures at a titer of 3 × 106 cells per ml were split into two subcultures, one with and one without coumermycin at 1 μg/ml. The titers of cells were determined at 4 and 24 h after addition to monitor the effect, if any, of coumermycin on proliferation. To minimize any manipulation-induced stress on the cells that might affect translation, 5 ml of cells after 4 h of growth with or without coumermycin treatment was removed and placed in a new flask. After being shaken for 5 min, 25 μl of 10 mCi/ml Tran35S-label (MP Biomedicals) was added to each shaking flask. At 0, 5, 10, 15, and 20 min; a 1-ml sample was removed; and the cells were rapidly pelleted (15 s), washed with ice-cold PDF (22 mM potassium phosphate, pH 6.5, 20 mM KCl, 5 mM Mg2Cl) containing unlabeled methionine, and taken up in 75 μl of protein lysis buffer (200 mM NaPO4, 20 mM KCl, 5 mM MgCl2, 5% [wt/vol] sucrose, 0.5% NP-40 at pH 7.5). Samples were incubated at room temperature for 5 min and clarified by centrifugation for 5 min at 8,000 × g. The supernatant was collected and stored at −80°C.
To separate labeled proteins from amino acids, 15 μl of the above-mentioned samples was spotted on squares of Whatman chromatography paper and submerged in ice-cold 10% trichloroacetic acid (TCA) for 15 min. The filters were transferred to boiling 5% TCA for 10 min and then once again transferred to ice-cold 5% TCA for 10 min. The filters were washed once with ethanol, followed by an ether wash. The filters were dried, and the radioactivity was determined by scintillation counting.
Protein isolation and Western blotting.
Conditioned medium obtained as described above was concentrated 10-fold using Amicon Ultracel-10K filtration units, and 20-μl samples were electrophoresed on NuPage Bis-Tris gels (Invitrogen). Western analysis was carried out using either anti-Countin antibodies at a dilution of 1:500, anti-AprA at 1:1,000, or anti-CfaD at 1:3,000. The antibodies were generously provided by Richard Gomer.
For isolation of extracellular protein during development, the method described by Bakthavatsalam and Gomer (3) was used. After removal of bacteria by differential centrifugation, 2 × 107 cells, harvested from growth on bacteria, were placed on type 353102 1-μm-pore-size polyethylene terephthalate membrane six-well format cell culture inserts (Becton Dickinson). The liquid was removed after the cells had settled, and 0.45 ml of PDF was added to the wells so that the liquid just touched the membrane. At various times poststarvation, the conditioned buffer was removed from a well and stored at −80°C. After collection at all time points, the samples were lyophilized and reconstituted to 0.06 ml, and 15 μl of each sample was electrophoresed and subjected to Western analysis.
RT-PCR.
RNA was isolated using Trizol (Invitrogen). Reverse transcription (RT)-PCR was carried out as described previously (25) using either cfaD- or aprA-specific oligonucleotides (Table 1). Oligonucleotides specific for the H7 gene were used as an internal control (40).
Spore viability analysis.
Spore-filled sori from Ax4 and BS167 cells growing on bacteria were picked and resuspended in PDF. After two washes in PDF, the titer of the spores was determined, and they were resuspended in 10 mM EDTA, pH 7.5, 0.1% NP-40. The samples were incubated at 42°C for 45 min, followed by two washes with PDF. The spores were resuspended with Klebsiella pneumoniae and plated at 100 spores per plate onto SM plates (34). Colonies, derived from individual spores, were counted after incubation for several days at 21°C.
RESULTS
Ectopic activation of Ifks inhibits proliferation.
eIF2α kinases require dimerization for activation (27, 36). In order to examine the functions of the initiation factor kinases in Dictyostelium, the kinase domains of the three kinases were fused to the chemically inducible dimerization domain of the GyrB protein of E. coli. The drug coumermycin induces dimerization of the GyrB protein and, thus, polypeptides fused to the dimerization domain (15, 36). Fusions were made using the normal kinase domain coding regions, as well as coding regions that had been mutated in residues demonstrated in yeast GCN2 to be activating mutations of the kinase activity (28). Initial characterizations indicated that ectopic activation of the kinase domains during growth inhibited proliferation. The extent of inhibition was dependent upon which of the three kinases was used, the presence or absence of activating mutations, and the presence or absence of coumermycin, presumably reflecting differences in the intrinsic activities of the kinase domains separated from their regulatory domains. The IfkA fusions were chosen for more detailed examination.
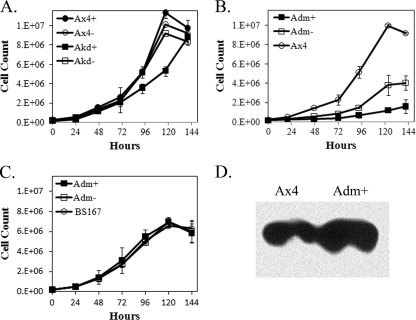
Ax4 cells transformed with pifkA-6 (GyrB fused to the IfkA kinase domain, referred to here as Akd) resulted in no effect on proliferation relative to untransformed Ax4 cells unless coumermycin was added to the culture (Fig. 1A). Coumermycin addition resulted in a slight but reproducible decrease in the proliferation rate. In contrast, cells transformed with pifkA-14 (GyrB fused to the IfkA kinase domain possessing two activating mutations, referred to here as Adm, for double mutant of IfkA) resulted in severe inhibition of proliferation, even without coumermycin (Fig. 1B). Addition of coumermycin resulted in a further reduction in proliferation, and most often addition resulted in only 2 to 4 cell doublings, followed by little to no further proliferation. The enhanced effect on proliferation by the Adm fusion protein relative to the unmutated Akd fusion indicates that the activating mutations that were patterned on the yeast GCN2 protein do activate the Dictyostelium Ifks.
Fig. 1.
Effects on proliferation by ectopic expression of active kinase domains. Cultures were started at 1.5 × 105 cells per ml, and the cells were counted every 24 h. At least three independent experiments were performed for each strain. The error bars indicate SEM. (A) Ax4 cells and Ax4 cells transformed with pifkA-6 (Akd, wild-type IfkA kinase domain fused to GyrB). +, coumermycin, −, no coumermycin. For clarity, the error bars for only the Ax4+ and Akd+ cells are shown; the differences seen at 93 and 117 h are significant (P < 0.05; Student's t test). (B) Ax4 cells and Ax4 cells transformed with pifkA-14 (Adm, double-mutant kinase domain fused to GyrB). The differences between Ax4 and Adm− and Ax4 and Adm+ are significant at 20 h and all subsequent time points (P < 0.01). The differences between Adm− and Adm+ are significant at 47 h and all subsequent time points (P < 0.05). (C) BS167 cells (possessing S51A eIF2α) and BS167 cells transformed with pifkA-14 (Adm). There were no significant differences at any time points. (D) Western analysis of extracellular Countin produced in Ax4 cells or Ax4 cells transformed with pifkA-14 (Adm). Cultures started at 5 × 105 cells per ml were grown to 3 × 106 cells per ml, followed by the addition of coumermycin for 12, 24, or 48 h. Cells were removed from the medium, and the medium was used in the Western analysis. Shown are the results from 24 h plus coumermycin conditioned medium.
To demonstrate that the effect on proliferation was due to eIF2α phosphorylation by the kinase domain fusions, the plasmids were transformed into BS167. This strain is a knock-in strain that possesses an eIF2α gene in which the serine codon corresponding to the phosphorylation site has been mutated to an alanine codon. No effect on cell proliferation was seen in BS167 cells transformed with the kinase fusion plasmids, with or without coumermycin addition (Fig. 1C). This indirect method of demonstrating that dependence of the inhibitory effect on proliferation was due to phosphorylation of eIF2α was necessitated by the fact that no phospho-eIF2α-specific antibodies currently exist for the Dictyostelium protein. Further confirmation that the kinase domain fusions phosphorylate eIF2α was obtained by demonstrating that the protein Countin was induced by the presence of the fusion proteins (Fig. 1D). It was previously shown that eIF2α phosphorylation by IfkA enhances the production of Countin (14).
Ectopic activation of Ifks does not result in general inhibition of translation.
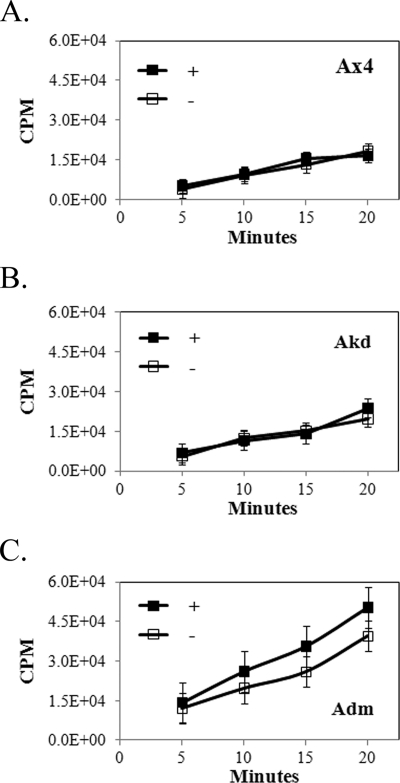
The severe inhibitory effect on proliferation by the expression of the activated kinase domain of IfkA might be due to a general shutdown or severe reduction in protein synthesis as a result of substantial phosphorylation of eIF2α. Such an effect is seen in yeast when an active PKR is overexpressed (36). The relative rates of protein synthesis in the presence and absence of coumermycin were examined in Ax4 cells and Ax4 cells transformed with the Akd and Adm constructs. Cells were counted for a 24-h period to confirm the effects on proliferation shown in Fig. 1. The cells were taken from the culture medium (with or without coumermycin) and incubated with radioactive methionine. Incorporation levels were measured over a 20-min period. No significant differences were found in the presence versus absence of coumermycin, importantly, even in cells expressing the Adm fusion kinase domain (Fig. 2). Overall incorporation varied somewhat from strain to strain, and the presence of the double-mutant kinase domain repeatedly gave higher incorporation, a finding that remains unexplained. Silver staining of gels of total protein isolated from Ax4 cells expressing or not expressing the Adm fusion protein in the presence of coumermycin indicated no significant or reproducible differences in the total protein patterns.
Fig. 2.
Effect of ectopic expression of active kinase domains on overall protein synthesis. The incorporation of radioactive methionine (CPM) in Ax4 cells (A), Ax4 cells transformed with pifkA-6 (B), and Ax4 cells transformed with pifkA-14 (C) in the presence (+) or absence (−) of coumermycin is shown. Three independent trials were performed. The error bars indicate SEM. No significant differences were found with or without coumermycin in any of the strains (Student's t test).
Ectopic activation of Ifks inhibits proliferation through a secreted factor.
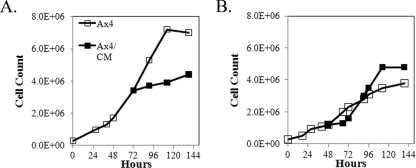
To determine if the inhibitory effect on proliferation was cell autonomous or due to a secreted factor, conditioned medium was obtained from Ax4 cells expressing the Adm fusion kinase domain and growing in medium containing coumermycin for 12, 24, or 48 h. Untransformed Ax4 cells were grown in normal medium to a titer of ca. 3 × 106 cells per ml. The culture was split into two groups of cells, and both were pelleted. One was resuspended at the initial titer in Adm conditioned medium, while the other was resuspended at the initial titer in the same medium in which the cells were previously growing. As seen in Fig. 3A, conditioned medium inhibited cell proliferation. The maximal extent of inhibition was found using conditioned medium obtained after 24 h of growth in the presence of coumermycin. The effect was reversible, as Adm cells growing with coumermycin that were resuspended in fresh medium showed a burst of proliferation prior to slowing to the initial low rate (Fig. 3B).
Fig. 3.
Inhibition of proliferation by ectopic expression of the active kinase domain is due to a secreted factor(s). (A) Conditioned medium was obtained by starting cultures of Ax4 cells transformed with pifkA-14 at 5 × 105 cells per ml and growing them to 3 × 106 cells per ml. Coumermycin was added for 24 h, and the culture medium was cleared of cells. Conditioned medium was added to half of the cells from a growing Ax4 culture to give the same titer as the other half of the culture, and the cell count was examined for another 72 h. The result shown is one of two trials, both of which resulted in inhibition of proliferation. (B) Reversibility of the inhibition of proliferation by coumermycin. Adm cells were grown in the absence of coumermycin (open squares). At 48 h, coumermycin was added to half of the culture (solid squares), resulting in inhibition of proliferation. After a further 24 h, the cells were transferred to fresh medium without coumermycin, resulting in enhanced proliferation.
Ectopic activation of Ifks induces CfaD and AprA production.
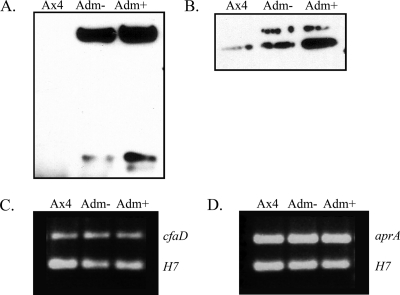
Previous studies characterized two chalones in Dictyostelium, AprA and CfaD, that are secreted during growth and that reversibly inhibit proliferation (1, 6, 8). The production of these two proteins was examined in Ax4 cells transformed with the Adm plasmid. The conditioned medium after growth with or without coumermycin was collected and used in Western analysis. Extracellular AprA and CfaD were found at much higher levels when the cells were expressing the Adm fusion protein (Fig. 4A and B). The levels were even higher when coumermycin was present in the culture medium.
Fig. 4.
Western analysis of extracellular CfaD and AprA, and RT-PCR on their mRNAs during ectopic expression of the active kinase domain. Conditioned medium was collected from Ax4 cells and Ax4 cells transformed with pifkA-14 in the presence (+) or absence (−) of coumermycin. (A) The 58.5- and 27-kDa forms of CfaD detected using anti-CfaD antiserum. (B) The 60-kDa band of AprA detected using anti-AprA antiserum. The faint upper band was sometimes seen and may represent a modified version of AprA. RNA was isolated from the indicated cells in the presence (+) or absence (−) of coumermycin. (C) RT-PCR was carried out with primers specific for cfaD (upper band) and for the H7 gene (lower band) as an internal control. (D) RT-PCR was carried out with primers specific for aprA (upper band) and for the H7 gene (lower band) as an internal control.
To examine aprA and cfaD mRNA levels, RT-PCR was carried out on RNA isolated from Ax4 cells and Ax4 cells transformed with the Adm plasmid growing in the presence or absence of coumermycin. In contrast to the greatly enhanced production of the chalone proteins, the corresponding mRNA levels were unchanged in cells expressing the Adm fusion protein (Fig. 4C and D), and they remained unchanged when coumermycin was included in the medium.
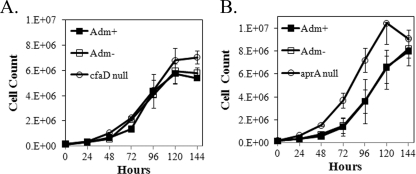
These results suggest that the inhibition of proliferation caused by the presence of the activated IfkA kinase domain is due at least in part to enhanced production of extracellular AprA and CfaD. To confirm involvement in the inhibition by the two chalones, the Adm plasmid was transformed into the null strains of aprA and cfaD. Proliferation of the transformants was compared to that of the respective untransformed null strains in the presence and absence of coumermycin. Expression of the Adm protein had no effect on proliferation for cfaD-null cells with or without the presence of coumermycin (Fig. 5A). A somewhat lower rate of proliferation was seen in aprA-null cells when they expressed the Adm fusion protein, but the cells did reach a titer at saturation similar to that of the untransformed cells (Fig. 5A). The inhibitory effect is much less pronounced than that seen in Ax4 cells (compare Fig. 1B). Importantly, the inhibition was not enhanced by the presence of coumermycin. That little to no difference in proliferation was found in the chalone-null strains suggests the inhibitory effect is mostly attributable to the overproduction of extracellular AprA and CfaD when the IfkA kinase domain is ectopically active in growing cells that contain the corresponding genes.
Fig. 5.
Ectopic expression of the active kinase domain in strains that lack either CfaD or AprA. The cfaD- and aprA-null strains were transformed with pifkA-14. Cell proliferation was followed in the presence (+) and absence (−) of coumermycin. Three independent experiments were performed. The error bars indicate SEM. The difference seen at the 144-h time point between cfaD-null cells and cfaD-null cells expressing Adm was significant (P < 0.05; Student's t test). In panel B, differences between transformed (both with and without coumermycin) and untransformed aprA-null cells at the 48-h through the 120-h time points were significant (P < 0.05).
Lack of increase in chalone production during development in BS167 cells.
Enhanced production of extracellular AprA and CfaD by ectopic phosphorylation of eIF2α inhibits cell proliferation. While these results demonstrate a competency for regulating chalone production by eIF2α phosphorylation, they do not reveal when such regulation might normally come into play during growth and/or development of Dictyostelium cells. One possible scenario might be to regulate cell proliferation during development. Development occurs during amino acid starvation, which should be sensed by the Ifks, and previous work demonstrated that IfkA phosphorylates eIF2α during development (14). Whether and how much cell division occurs during development is somewhat controversial. Recent experiments indicate that a low and decreasing percentage of cells divide during the first 9 h of development, followed by a burst of division during the next 3 h while mounds form tips (24). The number of cells undergoing division subsequently drops rapidly to very low numbers during the remaining stages of finger formation and culmination (24). We wondered if chalone production functions at these times to influence proliferation during development.
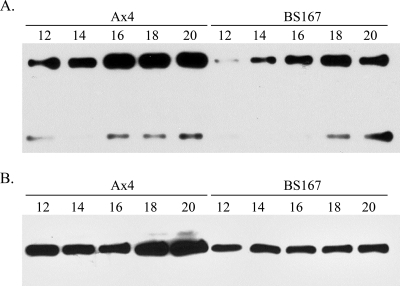
Ax4 and BS167 cells were plated for development in a manner that facilitated the isolation of secreted proteins (3). Several time points were examined by Western analysis for CfaD and AprA production. Little to no protein could be reliably detected during the first half of development. Both proteins were detected once mounds had formed (10 to 12 h poststarvation) and at subsequent times. In developing Ax4 cells, CfaD levels increased from 12 to 20 h poststarvation (Fig. 6A), during which time tipped mounds transitioned to fingers that then underwent culmination. This corresponds to the time at which Muramoto and Chubb found decreasing numbers of dividing cells (24). In developing BS167 cells, which possess an eIF2α that cannot be phosphorylated (S51A mutation), there was much less CfaD protein present at 12 h, and only a modest increase occurred, so that the level at 20 h was roughly equivalent to that at 12 h in Ax4 cells (Fig. 6A). Similar results were found for AprA. Increased production with time of development was seen in Ax4 cells, with little to no increase found in the BS167 strain, and the levels at 20 h in BS167 cells were less than those at 12 h in Ax4 cells (Fig. 6B).
Fig. 6.
Western analysis of extracellular CfaD and AprA during development in the Ax4 and BS167 strains. Cells growing in the presence of bacteria were harvested and plated for development in a manner that allows easy recovery of secreted proteins. Samples were collected at the indicated times (in hours poststarvation). After concentration, the samples were subjected to Western analysis. (A) The 58.5- and 27-kDa forms of CfaD detected using anti-CfaD antiserum. (B) The 60-kDa band of AprA detected using anti-AprA antiserum.
BS167 spores are not viable.
One characteristic of the aprA- and cfaD-null strains is the production of spores with reduced viability—about 44% that of wild type spores for cfaD-null cells (1) and 18% of wild-type viability for aprA-null cells (6). It was thus of interest to examine spore viability in the BS167 strain. After growth on bacteria and sporulation, Ax4 and BS167 spores were collected and assayed for viability; 43% ± 2% of Ax4 spores were viable, while only 3% ± 2% of BS167 spores were viable (± standard error of the mean [SEM]; n = 4). Morphologically, the BS167 spores were atypical in that they were round instead of oval, as is found for wild-type spores.
DISCUSSION
Expression of the unregulated and active kinase domain of IfkA severely inhibited proliferation of Dictyostelium cells. Inhibition of proliferation was due to phosphorylation of eIF2α, as demonstrated by a lack of inhibition in cells possessing an eIF2α that could not be phosphorylated (S51A mutation) and by the increased production of Countin, a protein previously demonstrated to be upregulated by a shift of its mRNA onto ribosomes upon eIF2α phosphorylation (14). No decrease in overall protein synthesis was detected. Thus, the inhibitory effect was not due to a gross repression of translation, suggesting instead a specific effect on activation of one or more mRNAs that produce proteins that inhibit proliferation.
Conditioned medium obtained from the kinase domain-expressing cells was found to inhibit the proliferation of nonexpressing cells. Greatly enhanced production of extracellular AprA and CfaD was found in the conditioned medium. Extracellular CfaD is found in several sizes, with the 60- and 27-kDa polypeptides typically being the prevalent forms (1). Enhanced production of both the 60- and 27-kDa forms was found in cells expressing the activated IfkA kinase domain. While there was enhanced protein production for both chalones, the mRNA levels for aprA and cfaD essentially remained unchanged. This is consistent with an enhanced rate of translation of the two mRNAs upon phosphorylation of eIF2α. Both genes have several small open reading frames upstream of the respective coding regions; such open reading frames are hallmarks of mRNAs upregulated by eIF2α phosphorylation (11). Direct examination of the polysome profiles of aprA and cfaD mRNAs will need to be carried out to confirm enhanced translation brought about by eIF2α phosphorylation.
AprA and CfaD are chalones that are found in a 150-kDa complex that inhibits proliferation, but not growth, of Dictyostelium cells (1, 6, 8). The presence of both is required for the inhibition of proliferation, even though it appears there may be separate receptors for the two proteins (8). Exogenously added CfaD does not decrease the proliferation rate of aprA-null cells (1), and likewise, added AprA does not decrease the proliferation rate of cfaD-null cells (8). This predicts that if the inhibitory effect of expressing the activated IfkA kinase domain is mostly due to the enhanced production of CfaD and AprA, then the effect should not be seen in either of the null strains. Indeed, expressing the activated kinase domain in aprA-null cells had no effect on proliferation. Expression in the cfaD-null cells did give a reduction in proliferation, but one that was not nearly as dramatic as that seen in Ax4 cells and that was not abrogated further by the addition of coumermycin. While most of the reduction may thus be attributable to the enhanced production of CfaD in the Adm-expressing cells, the slight but significant reduction seen may indicate the involvement of an additional, unknown factor(s).
The inhibition of proliferation by the ectopically expressed IfkA kinase domain is an artificial situation, as the phosphorylation of eIF2α is not under normal regulation. Nonetheless, it points to a capability of controlling chalone production by eIF2α phosphorylation. Previous work demonstrated that eIF2α phosphorylation occurs during the developmental program of Dictyostelium (14, 29). Recent studies on cell division during development demonstrated a low level of cell division occurring during early development, followed by a sizable increase in the number of cells undergoing division during tipped-mound formation (9 to 12 h poststarvation) (24). The number of cells undergoing division drops substantially at the culmination of development, at which time prestalk and prespore tissues are being formed.
Examination of the production of AprA and CfaD from 12 to 20 h poststarvation revealed an increase in the extracellular levels of both chalones in wild-type cells. This increase coincides with the time when the number of cells undergoing division drops from the highest numbers to the lowest numbers found during the developmental program (24). The increase in AprA and CfaD levels was not seen in developing cells that possessed the S51A eIF2α that could not be phosphorylated. In this strain, the levels of both proteins at 20 h were comparable to or less than those found at 12 h in wild-type cells. These results suggest that AprA and CfaD play a role in inhibiting cell division during culmination that is mediated by eIF2α phosphorylation, enhancing the production of the chalones during the later stages of development. The lack of inhibition of division during culmination thus predicted in the BS167 and the aprA- and cfaD-null strains might contribute to spores from these strains being nonviable. Testing this postulated function of the two chalones will require examining cell division during development in the null strains for these two genes and in the BS167 strain. Regulation of cell numbers during the stages of development when prestalk and prespore tissues are being formed would certainly substantiate the naming of AprA and CfaD as chalones.
Amino acid deprivation plays a major role in the Dictyostelium developmental program (21–23). That the Ifks in Dictyostelium are involved in chalone production and hence in regulating cell proliferation is perhaps not surprising, as these GCN2-type kinases possess a regulatory domain that senses and responds to amino acid starvation (12, 38). In mammals, kinases that phosphorylate eIF2α have been found to affect cell proliferation by several mechanisms (4, 5, 20, 30, 39). The finding that in Dictyostelium chalone production is controlled at least in part by eIF2α phosphorylation opens up the possibility that eIF2α kinases may function similarly in regulating chalones, such as myostatin, in mammals.
ACKNOWLEDGMENTS
We thank Richard Gomer for generously providing strains and antibodies, Tom Dever for providing the gene encoding the GyrB dimerization domain, the Dictyostelium Stock Center for providing strains, and Leigh Singleton for assistance with the figures.
The work was supported in part by NSF IOS 0639836.
Footnotes
Published ahead of print on 28 January 2011.
REFERENCES
- 1. Bakthavatsalam D., et al. 2008. The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 121:2473–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakthavatsalam D., Choe J. M., Hanson N. E., Gomer R. H. 2009. A Dictyostelium chalone uses G proteins to regulate proliferation. BMC Biol. 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bakthavatsalam D., Gomer R. H. 2010. The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics 10:2556–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baltzis D., et al. 2007. The eIF2alpha kinases PERK and PKR activate glycogen synthase kinase 3 to promote the proteasomal degradation of p53. J. Biol. Chem. 282:31675–31687 [DOI] [PubMed] [Google Scholar]
- 5. Brewer J. W., Diehl J. A. 2000. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. U. S. A. 97:12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brock D. A., Gomer R. H. 2005. A secreted factor represses cell proliferation in Dictyostelium. Development 132:4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bullough W. S. 1962. The control of mitotic activity in adult mammalian tissues. Biol. Rev. 37:307–342 [DOI] [PubMed] [Google Scholar]
- 8. Choe J. M., Bakthavatsalam D., Phillips J. E., Gomer R. H. 2009. Dictyostelium cells bind a secreted autocrine factor that represses cell proliferation. BMC Biochem. 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cocucci S., Sussman M. 1970. RNA in cytoplasmic and nuclear fractions of cellular slime mold amebae. J. Cell Biol. 45:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dever T., et al. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585–596 [DOI] [PubMed] [Google Scholar]
- 11. Dever T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545–556 [DOI] [PubMed] [Google Scholar]
- 12. Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. 2000. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6:269–279 [DOI] [PubMed] [Google Scholar]
- 13. Dynes J. L., et al. 1994. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 8:948–958 [DOI] [PubMed] [Google Scholar]
- 14. Fang R., Xiong Y., Singleton C. K. 2003. IfkA, a preumptive eIFalpha kinase of Dictyostelium, is required for proper timing of asggregation and regulation of mound size. BMC Dev. Biol. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Farrar M. A., Albero L., Perlmutter R. M. 1996. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 383:178–181 [DOI] [PubMed] [Google Scholar]
- 16. Fernandez J., Yaman I., Sarnow P., Snider M., Hatzoglou M. 2002. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J. Biol. Chem. 277:19198–19205 [DOI] [PubMed] [Google Scholar]
- 17. Harding H., et al. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 18. Hinnenbusch A. 1996. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2, p. 199–244In Hershey J., Mathews M., Sonenberg N. (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Kaufman R., Davies M., Pathak V., Hershey J. 1989. The phosphorylation of eukaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9:946–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kazemi S., et al. 2007. A novel function of eIF2alpha kinases as inducers of the phosphoinositide-3 kinase signaling pathway. Mol. Biol. Cell 18:3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margolskee J. P., Froshauer S., Skrinska R., Lodish H. 1980. The effects of cell density and starvation on early developmental events in Dictyostelium discoideum. Dev. Biol. 74:409–421 [DOI] [PubMed] [Google Scholar]
- 22. Marin F. T. 1976. Regulation of development in Dictyostelium discoideum. Dev. Biol. 48:110–117 [DOI] [PubMed] [Google Scholar]
- 23. Marin F. T. 1977. Regulation of development in Dictyostelium discoideum. Dev. Biol. 60:389–395 [DOI] [PubMed] [Google Scholar]
- 24. Muramoto T., Chubb J. R. 2008. Live imaging of the Dictyostelium cell cycle reveals widespread S phase during development, a G2 bias in spore differentiation and a premitotic checkpoint. Development 135:1647–1657 [DOI] [PubMed] [Google Scholar]
- 25. Pekovich S. R., Martin P. R., Singleton C. K. 1998. Thiamine deficiency decreases steady-state mRNA levels for transketolase and pyruvate dehydrogenase but not for α-ketoglutarate dehydrogenase in three human cell types. J. Nutr. 128:683–687 [DOI] [PubMed] [Google Scholar]
- 26. Phillips J. E., Gomer R. H. 2010. The ROCO kinase QkgA is necessary for proliferation inhibition by autocrine signals in Dictyostelium discoideum. Eukaryot. Cell 9:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu H., Garcia-Barrio M. T., Hinnebusch A. G. 1998. Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol. 18:2697–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu H., Hu C., Dong J., Hinnebusch A. G. 2002. Mutations that bypass tRNA binding activate the intrinsically defective kinase domain in GCN2. Genes Dev. 16:1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rai M., Xiong Y. H., Singleton C. K. 2006. Disruption of the ifkA and ifkB genes results in altered cell adhesion, morphological defects and a propensity to form pre-stalk O cells during development of Dictyostelium. Differentiation 74:583–595 [DOI] [PubMed] [Google Scholar]
- 30. Raven J. F., et al. 2008. PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2alpha phosphorylation. J. Biol. Chem. 283:3097–3108 [DOI] [PubMed] [Google Scholar]
- 31. Roussou I., Thireos G., Hauge B. 1988. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN2 transcriptional activator and the GCN2 protein kinase. Mol. Cell. Biol. 8:2132–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singleton C. K., Wang J. J.-L., Shen L., Martin P. R. 1996. Conserved residues are functionally distinct within transketolases of different species. Biochemistry 35:15865–15869 [DOI] [PubMed] [Google Scholar]
- 33. Singleton C. K., Zinda M. J., Mykytka B., Yang P. 1998. The histidine kinase dhkC regulates the choice between migrating slugs and terminal differentiation in Dictyostelium discoideum. Dev. Biol. 203:345–357 [DOI] [PubMed] [Google Scholar]
- 34. Sussman M. 1966. Biochemical and genetic methods in the study of cellular slime mold development, p. 397–410In Prescott D. (ed.), Methods of cell physiology. Academic Press, New York, NY [Google Scholar]
- 35. Sutoh K. A. 1993. A transformation vector for Dictyostelium discoideum with a new selectable marker Bsr. Plasmid 30:150–154 [DOI] [PubMed] [Google Scholar]
- 36. Ung T. L., Cao C., Lu J., Ozato K., Dever T. E. 2001. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 20:3728–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weiss P., Kavanau J. L. 1957. A model of growth and growth control in mathematical terms. J. Gen. Physiol. 41:1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wek R., Jackson B., Hinnebusch A. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availablity. Proc. Natl. Acad. Sci. U. S. A. 86:4579–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye J., et al. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29:2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Q. 1995. Studies of H7 gene function and regulation of its expression by a bidirectional promoter in Dictyostelium discoideum. Ph.D. thesis. Vanderbilt University, Nashville, TN [Google Scholar]