Abstract
Even relatively simple species have evolved mechanisms to organize individual organisms into communities, such that the fitness of the group is greater than the fitness of isolated individuals. Within the fungal kingdom, the ability of many yeast species to organize into communities is crucial for their growth and survival, and this property has important impacts both on the economy and on human health. Over the last few years, studies of Saccharomyces cerevisiae have revealed several fundamental properties of yeast communities. First, strain-to-strain variation in the structures of these groups is attributable in part to variability in the expression and functions of adhesin proteins. Second, the extracellular matrix surrounding these communities can protect them from environmental stress and may also be important in cell signaling. Finally, diffusible signals between cells contribute to community organization so that different regions of a community express different genes and adopt different cell fates. These findings provide an arena in which to view fundamental mechanisms by which contacts and signals between individual organisms allow them to assemble into functional communities.
In many species, including our own, individual organisms assemble into communities to increase their overall fitness. Even in unicellular organisms like Saccharomyces cerevisiae, individual cells can organize themselves into a variety of types of multicellular aggregates. These biotic communities likely provide overall benefit to these yeast populations, for example by protecting organisms in the core of the structure from environmental stresses or by specializing functions to subpopulations within the community. Yeast communities also have broad relevance to human health and to industry. For example, biofilms formed by pathogenic yeasts on medical devices, such as catheters, are a major cause of the very high mortality rates of hospital-acquired fungal infections (reviewed in references 11, 43, and 44). Furthermore, surface film communities formed on food by spoilage yeasts may result in losses of billions of dollars annually (reviewed in reference 58). Yet the mechanisms underlying the organization of these communities are still poorly understood.
In the first section of this paper, I review types of yeast communities, focusing on the model yeast Saccharomyces cerevisiae, and briefly discuss broader aspects of two processes fundamental to yeast communities: cell-cell signaling and cell adhesion. In the second section, I discuss four aspects of yeast community organization highlighted by recent publications: boundary formation, cell adhesion, the extracellular matrix (ECM), and diffusible cell-cell signals. Much of this recent progress has focused on the cytological structures of these communities and on the identification of several genetic pathways required for this organization.
BACKGROUND. (I) THE MULTIFARIOUS YEAST COMMUNITY
Community types and variations.
Yeasts can grow as isolated cells when suspended in shaking cultures, but they can also group into an impressive array of types of communities, including flocs, flors, mats, colonies, and biofilms. These communities are found in natural habitats, in clinical settings, and in factories—indeed, wherever yeasts are found. Outside the laboratory, many yeast species are found in multispecies communities termed microbiomes, which can include other fungi or bacteria (reviewed in references 57 and 68).
Perhaps not surprisingly, there is no universal agreement on how some of these communities should be defined. For the purpose of this review, each type of community is defined operationally based on its structure and location relative to its food source, as follows. (i) Flocs are aggregations of cells that grow suspended in shaking liquid cultures. (ii) Flors, also called velum, are a thick layer of cells that form on the top surface of cultures. Flors are also easily visible to the eye. (iii) Colonies are compact structures with relatively small diameters that grow on agar plates. (iv) Mats grow specifically on moist plates that contain low concentrations of agar, and they are much wider and shallower than colonies. (v) Biofilms are structures that grow on and coat plastic or other hard surfaces submerged in a liquid nutrient source.
Not all of these yeast communities form in every strain of yeast. For example, many yeast strains used in the brewing and wine industries form either flocs (e.g., bottom-fermenting yeasts used to make lagers) or flors (e.g., top-fermenting yeasts used to make sherry wines). In general, strains that form flors have higher surface hydrophobicity than those that form flocs (12). Unlike these industrial yeasts, most laboratory yeasts form neither flors nor flocs. Flocs and flors are useful in fermentation yeasts to separate the yeast from the fermented product, whereas dispersed growth in cultures greatly simplifies most analyses of laboratory yeasts (36).
The structures of yeast communities also vary between species. For example, biofilms formed by S. cerevisiae differ dramatically from biofilms formed by Candida albicans. C. albicans is a commensal in healthy individuals but can become a serious pathogen in immunocompromised individuals, such as premature infants, transplant recipients, and HIV/AIDS patients. A number of cellular changes are required for virulence in C. albicans, including the dimorphic switch, as ovoid cells switch to form a mycelium composed of branched, thread-like cells (hyphae), which can invade the host tissue (reviewed in references 2 and 70). This dimorphic switch is also observed in mature C. albicans biofilms; these biofilms contain an underlying layer of ovoid cells covered by a thick mycelial layer embedded in the extracellular matrix (reviewed in references 5 and 14). In contrast, S. cerevisiae, which can also form biofilms that adhere tightly to plastic surfaces (51), has not been observed to form the bilayer structure characteristic of mature C. albicans biofilms. Unlike C. albicans, S. cerevisiae is seldom pathogenic (17, 45), but the relationship between mature biofilms and virulence may not be a simple one, since biofilms formed by several other pathogenic Candida species may also lack the mature structures formed by C. albicans (30).
Visibly organized communities.
One striking type of colony organization, which is visible even without magnification, is the “structured colony,” so termed because it contains striations on its surface (1, 66). These striations sometimes form a spoke-like pattern radiating from the center, a series of concentric rings, or a random distribution over the surface. In some strain backgrounds, such as 1278b, similar striations are observed on the surfaces of mats (51). For both mats and colonies, it has been proposed that these striations function as channels to move nutrients through the colony.
A less obvious type of colony organization, visible only under a microscope, forms when colonies are grown on agar medium containing limiting nitrogen (18). These colonies stop growing when they are still quite small and hence are termed “microcolonies.” Microcolonies initially grow as ovoid cells, but as nutrients become limiting, the ovoid cells at the edge of the microcolony undergo a dimorphic switch, i.e., they begin growing as pseudohyphae. Pseudohyphae are chains of elongated (i.e., filamentous) diploid cells. Thus, the dimorphic switch configures the organization of microcolonies such that ovoid cells are at the center and pseudohyphae at the periphery of a microcolony.
Cryptic communities.
Some types of colony organization are revealed by molecular rather than cytological analysis. One clear example of this type of organization occurs in colonies inoculated from a drop of liquid (here termed “spot colonies”) grown on glycerol medium. After 10 days of incubation, these colonies begin to cycle through alternating alkali and acid phases. These temporal phases are accompanied by periodic changes in the expression levels for hundreds of genes (40, 60). Spatial organization of these colonies occurs at the start of the first alkali phase; at this time, cells in the colony's center begin to undergo apoptosis, while cells at the colony's edge remain viable and continue to divide (35, 62).
Mats formed in most laboratory strains appear uniform, concealing a subtler form of organization in these communities, namely, that cells at the center of the mat adhere more tightly to the underlying agar than do cells at the periphery (52). This affinity difference could reflect differential gene expression between the two regions, since as mats develop, pH and glucose gradients form from the center to the edge. Similarly, some strains form colonies that invade the agar surface, and this invasive growth is typically detectable only after the main part of the colony has been washed from the agar surface (53).
Summary.
Yeast communities are organized in multiple ways, and this organization depends both on genotype and environment. In colonies, multiple kinds of organization have been discovered, including surface striations, localization of apoptosis, and positioning of pseudohyphae.
(II) CELL CONTACTS AND CELL ADHESION
Flocculins, the lynchpin of the yeast community.
Yeast communities are shaped in part by a family of adhesin proteins, which in S. cerevisiae are also termed “flocculins.” Yeast mutants lacking flocculins fail to form either flocs or flors (16), fail to form biofilms on plastic surfaces (49, 51), and fail to form either structured colonies (1) or mats (52). Thus, flocculins are required for most types of organization in yeast communities. Indeed, a major reason that many common S. cerevisiae laboratory strains, such as S288C, are unable to form flocs, flors, or structured colonies is because they do not express functional flocculin proteins (15, 33).
Collectively, the flocculin proteins have several biochemical functions in organizing yeast communities (reviewed in reference 64). First, flocculins mediate cell-cell adherence by binding to oligosaccharides on the surfaces of other cells. Flocculins are initially anchored to the cell wall by a glycosylphosphatidylinositol (GPI) anchor near the C terminus and require a lectin-like N-terminal domain to bind oligosaccharides on neighboring cells. Second, flocculins mediate the binding of cells to plastic and other surfaces. This binding occurs in part through a central domain of the flocculin that contains tandem repeats of a 10- to 20-amino-acid sequence (32). This repeat sequence is rich in Ser/Thr residues, is characterized by high levels of glycosylation, and may bind surfaces primarily through hydrophobic interactions (27). Third, flocculins are necessary to form pseudohyphae (34).
Flocculins organize yeast communities in part through cooperative association between cells. For example, an association between two cells that express Flo1p is stronger than an association between a Flo1p-expressing cell and a Flo1p-absent cell, probably because of reciprocal interactions (54). Thus, flocs formed in a mixed population of flo1Δ and FLO1+ strains contain disproportionately high levels of the FLO1 strain. If different regions of a yeast community express different flocculins, self-adhesion within each region may contribute to subdivisions within the community.
Summary.
Cell adhesion mediated by flocculins is critical to yeast community organization. One possibility for their required presence is that these proteins provide both a structural contact between cells and a signal that this contact is occurring.
(III) DIFFUSIBLE SIGNALS BETWEEN CELLS
Pheromone signals.
The exemplar of cell-to-cell communication in yeast is pheromone signaling (reviewed in reference 31). Pheromone signaling occurs during mating between haploids; in S. cerevisiae, these are haploids of the a and α mating types. Each mating type produces a peptide pheromone that induces a mating response in the opposite type. Pheromones activate the mating response through the Ste12p MAP kinase pathway—one of the most extensively studied and informative signal transduction pathways (reviewed in references 13 and 24).
Quorum-sensing signals.
Pheromones function as a signal between exactly two cells; in contrast, quorum-sensing signals coordinate the behavior of a large group of cells. In both bacterial and eukaryotic microbes, quorum sensing allows different cell behaviors at high versus low cell densities (see, for example, references 37 and 38). In yeasts, once a given cell density is reached, quorum sensing causes cells to undergo the dimorphic switch (reviewed in references 25 and 55).
Regulation of the dimorphic switch by cell-cell quorum signals has been studied most extensively in C. albicans (reviewed in reference 29). This switch is inhibited at low cell densities by secretion of farnesol (26, 39) and activated at high cell densities by secretion of tyrosol, an aromatic alcohol (10). These signals may be required to organize mature biofilms, and indeed, addition of farnesol to early biofilms prevents their maturation (50). The S. cerevisiae dimorphic switch is regulated by signals different from those used by C. albicans. For example, the S. cerevisiae switch is activated by phenylethanol and tryptophol, rather than tyrosol, and those two aromatic alcohols actually inhibit the C. albicans switch (9). These differences reflect that the dimorphic switch may have evolved as a host response in C. albicans and as a starvation response in S. cerevisiae.
Ammonium signals.
A third cell-cell signal in yeasts is ammonia/ammonium (reviewed in reference 42). For example, S. cerevisiae colonies produce and export ammonium during the alkaline phase (40, 60). A mutant (sok2Δ) that is defective in ammonium production is also defective in the subdivision of colonies into apoptotic and viable zones, suggesting that ammonium signals are required for this pattern (62). Ammonium is also a signal between colonies; it can diffuse from one colony to its neighbors on an agar plate, with the effect of synchronizing alkaline phases in neighboring colonies (41).
Summary.
The roles of cell-cell signals in yeast communities are still emerging, but the examples of quorum signaling and ammonium signaling suggest that cell-cell signals are essential for the association of yeast cells into communities.
RECENT PROGRESS
Several papers over the last 2 years revealed yeast community organization to be a promising research arena. These studies have determined that S. cerevisiae communities are: (i) subdivided into specialized regions with sharp boundaries, (ii) structurally diverse, in part due to variation in cell adhesion, (iii) surrounded by an extracellular matrix, and (iv) dependent on cell-cell signals for their organization.
(I) COMMUNITY BOUNDARIES
New approaches for examining yeast colony structure.
Two recent studies introduced new methods for investigating colony structure. In the first of these, the pattern of expression of a green fluorescent protein (GFP) fusion gene within colonies was monitored using two-photon excitation fluorescence microscopy (59). By this method, fusion gene expression in up to 10 to 20 cell widths can be visualized inside the colony. Colonies were viewed from above, below, and the side (after the colony was sliced down the middle), allowing a reconstruction of expression throughout the colony. In an alternative approach, colonies were embedded in plastic or frozen and then sectioned (47, 48). Embedded sections allowed the pattern of cell types (ovoid cells, pseudohyphae, and spores) within colonies to be determined (46, 48). Frozen sections (cryosections) allowed the pattern of expression of lacZ fusion genes within colonies to be visualized.
The above-described studies reveal that yeast colonies are far from being homogeneous. Instead, cells in separate regions of the colony express different genes and adopt different fates. Intriguingly, in both studies, these colony regions are separated by sharp boundaries.
Patterns of sporulated cells in colonies.
Patterns form in diploid colonies that first grow and then sporulate (48). Sporulated cells (asci) are easily distinguishable from nonsporulated cells, so the distribution of sporulating cells in these colonies is obvious from examining embedded sections (Fig. 1 A). After cell division ceases in these colonies, sporulation initiates in an internal layer of cells and in a second layer of cells at the agar surface. Over time, the internal layer of sporulated cells expands upward to include the top of the colony. Once sporulation in the colony ceases, the boundaries between sporulating and nonsporulating regions are very sharp (Fig. 1A). This same sporulation pattern forms in wild strains of S. cerevisiae and Saccharomyces paradoxus, as well as in several laboratory strain backgrounds, and wild strains form this same pattern on either fermentable or nonfermentable carbon sources and on either rich or synthetic nitrogen sources (46).
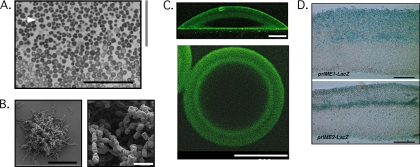
Fig. 1.
Patterns of differentiation and gene expression in colonies. (A) Section from central region of a 6-day-old wild yeast colony. The top of the image is closest to the top of the colony. The region of the section indicated by the gray bar has a high frequency of sporulation, whereas the underlying region contains no asci. A representative ascus is indicated by an arrow. Scale bar, 50 μm. (Reprinted from reference 46 with permission of the publisher.) (B) Pseudohyphal and meiotic differentiation on the surfaces of minicolonies. Scale bars, 100 μm (left panel) and 10 μm (right panel). (Reprinted from reference 69 with permission of the publisher.) (C) Expression of ATO1-GFP within a colony. The upper panel is a side view obtained after slicing a 10-day-old colony in half. Scale bar, 150 μm. The lower panel is viewed from the bottom of a 10-day-old colony. Scale bar, 500 μm. (Reprinted from reference 59 with permission of the publisher.) (D) Colony expression pattern of two meiotic genes. The upper panel is a colony containing the IME1 promoter fused to lacZ, and the lower panel is a colony containing the IME2 promoter fused to lacZ. (Reprinted from reference 48 with permission of the publisher.)
A different type of colony sporulation pattern forms in structures termed “minicolonies” (69). Minicolonies are related to biofilms in that they grow on a plastic surface submerged in liquid; however, unlike biofilms, minicolonies form limited structures rather than expanding to cover the entire surface. Interestingly, minicolonies grow initially as ovoid cells and then switch to pseudohyphal growth; finally, the pseudohyphae on the surface of the colony sporulate. Thus, minicolonies contain a high frequency of asci at their surfaces (Fig. 1B). A flo11Δ mutant, whose mutation prevents the dimorphic switch, inhibits sporulation in minicolonies, whereas disrupting the minicolonies leads to increased sporulation. Thus, the dimorphic switch may promote sporulation by dispersing cells from the tightly packed colony core.
Patterns of gene expression during the alkaline phase of colony development.
Several recent studies clearly establish that different regions of yeast communities express different genes. In one striking example, spot colonies express an ATO1-GFP fusion gene (59) in different regions of the colony at different stages of colony development. ATO1 encodes a transporter that exports ammonium from the cell, and prior to extensive ammonium production, colonies express ATO1-GFP only in a narrow layer of cells at the top surface of the colony. As ammonium production increases in the colony, a second layer of cells also begins to express this gene (Fig. 1C). Still later, this second layer becomes the primary site of ATO1-GFP expression.
Localization of expression patterns within specific regions of the colony is likely to be a general property of colonies. For example, when proteins were isolated from either the outer zone or central zone of spot colonies, the outer zone of the colony preferentially expressed respiration and peroxisome enzymes (e.g., Cit3p, Icl2p, and Cta1p), whereas the central zone expressed stress defense proteins (e.g., Ctt1p and Sod1p) (61).
Overlapping expression patterns for meiotic regulators.
Not only are genes expressed in sharply defined regions within colonies, but different genes can be expressed in different regions (48). For example, colonies displaying colony sporulation patterns express two sporulation genes: IME1 (a transcription factor) and IME2 (a protein kinase) in different patterns (Fig. 1D). Initially, colonies express IME1-lacZ at their top surface, and over time the layer of cells that express this gene expands downward to the center of the colony. At this time, IME2-lacZ, which depends on IME1 for its transcription, is induced specifically in a narrow band of cells in the center of the colony near the bottom of the band of cells that express IME1-lacZ.
Thus, the first cells that express both IME1 and IME2, and hence initiate sporulation, are within an internal layer of the colony.
Summary.
New methods reveal sharply defined expression patterns within colonies, and in some cases, these expression patterns localize particular cell types to specific regions of the colony. In colonies containing sporulated cells, localization of these cells to the top of the colony may promote the dissemination of these environmentally resistant spores to new locations. In colonies entering the alkaline phase of growth, localization of apoptosis to the central region of the colonies may release nutrients that are subsequently utilized for growth at the colony edge.
(II) COMMUNITY DIVERSITY AND FLOCCULIN DIVERSITY
Several types of S. cerevisiae communities display remarkably diverse organizations, and a unifying theme for much of this diversity is the role played by the flocculin family of proteins.
Community diversity.
Structured colonies display a particularly striking range of morphologies from very “lacy” structures (Fig. 2 A) to colonies that appear “mountainous” (Fig. 2B) (22). Even the same strain can have very different morphologies when grown on different media (Fig. 2C and D). This morphological variation may reflect allelic differences, particularly in genes that sense nutrient status and/or regulate filamentation (22). The region of the colony that invades the agar also displays variability; colonies in one strain background (Σ1278b) invade the agar in a single bubble-like structure at the center of the colony, whereas colonies in another strain background (SK1) invade the agar across a much larger region of the agar-colony interface (46). Similarly, S. cerevisiae strains used in wine production differ greatly in the sizes and structures of the flors that they form (73).
Fig. 2.
Dependence of colony structure on strain background and growth conditions. (A) YJM311 (a clinical isolate of S. cerevisiae) (reprinted from reference 22 with permission of the publisher); (B) PMY348 (reprinted from reference 22 with permission of the publisher); (C) SH561 (SK1 background) grown on yeast extract-peptone-dextrose (YPD); (D) SH561 grown on YP-glycerol. Scale bars, 1 mm.
FLO functional variability.
In many cases, strain differences in community organization have a molecular basis in the several types of variability characteristic of flocculins (reviewed in reference 64). The first level of variability reflects functional differences between FLO genes. A well-established example of these differences is in the sugar-binding specificities of particular flocculins; e.g., some flocculins bind primarily mannose, whereas others bind glucose and mannose equally (reviewed in reference 19). More recently, comparing strains each expressing a different flocculin demonstrates that some flocculins, (e.g., Flo11p), are sufficient for invasive growth and flors but not for flocculation or adhesion to plastic surfaces. In contrast, other flocculins (e.g., Flo1p) show the opposite specificity (20, 21, 63). Different strains express different flocculins, so the functional diversity of the flocculin gene family likely underlies much of the strain-dependent structural diversity of S. cerevisiae communities.
FLO allele diversity.
A second type of flocculin variability, allele diversity, is illustrated by variation in the numbers of central-domain tandem repeats in different alleles. Progressively deleting tandem repeats within FL01 causes corresponding decreases in adherence to plastic and flocculation (65). Furthermore, when FLO11 alleles from 20 flor-producing wild strains were compared, they contained from 11 to 78 tandem repeats of a 12-amino-acid sequence, and in general, alleles with greater numbers of repeats led to more massive flors (73). Furthermore, FLO11 expression levels varied in these strains, with expression above a threshold level necessary to produce more-massive flors. Thus, flor mass depends both on the repeat number in a particular FLO11 allele and on its expression levels (73).
FLO epigenetic variability.
A third type of flocculin variability, epigenetic variation, allows genetically identical cells in identical environments to express dramatically different levels of flocculins. For example, in a single culture, FLO11 can be fully repressed in some cells and highly induced in others (23). One mechanism for this biphasic expression is a “toggle switch” involving two long noncoding RNAs transcribed from opposing strands in the FLO11 regulatory region (6). These two RNAs are reciprocally interfering so that in a given cell, only one of the two is expressed. Furthermore, transcription of one of these RNAs extends into the FLO11 transcription start site and hence inhibits its transcription. Which of these two RNAs is expressed depends in part on competitive binding at the promoter between the Flo8p transcriptional activator and the Sfl1p transcriptional repressor (6), and expression of one or the other RNA may “lock” the promoter into a metastable “on” or “off” state. Chromatin structure, in particular, histone acetylation, is also critical for the epigenetic regulation of FLO11; for example, both the Hda1p and Rpd3Lp histone deacetylases influence the switch between on and off transcriptional states (6, 23).
In microcolonies containing FLO11-GFP, the fusion gene is expressed in only a subset of the cells in the colony, including both elongated and ovoid cells (56), though preferential expression of FLO11 in daughter cells may ensure that this gene is expressed in cells undergoing the dimorphic switch (71). The role of flocculins in cooperative cell association (see above) could be related to their expression in only a subpopulation within a community. It will be interesting to discover whether the two epigenetic states of FLO11 are equally distributed throughout a yeast community or limited to one region.
Summary.
The structures of S. cerevisiae communities differ among strains, and these differences are attributable in part to flocculin variability, including functional, allele, and epigenetic variability.
(III) ECM AS SHIELD AND SIGNAL
In metazoans, the extracellular matrix (ECM), which consists primarily of polysaccharides and proteins, surrounds and anchors cells in tissue and is essential for many types of cell signaling. Some microorganisms, including C. albicans, produce an extensive ECM, which is thought to help organize them into communities. The recent discoveries of ECM in S. cerevisiae communities have implicated both structural and regulatory roles for this matrix in yeast community organization.
Protecting the community.
Several studies establish that yeast communities are both covered and protected by a layer of ECM. When flocs formed from a strain expressing FLO1 were examined by transmission electron microscopy, an outer layer of ECM was observed to surround the floc (Fig. 3 A); in contrast, flocs that do not express FLO1 lack this outer layer (3). The ECM layer, which consists mainly of hexose polysaccharides, may protect the flocs against environmental toxins, since only flocs containing an ECM layer were able to exclude large molecules from penetrating the floc.
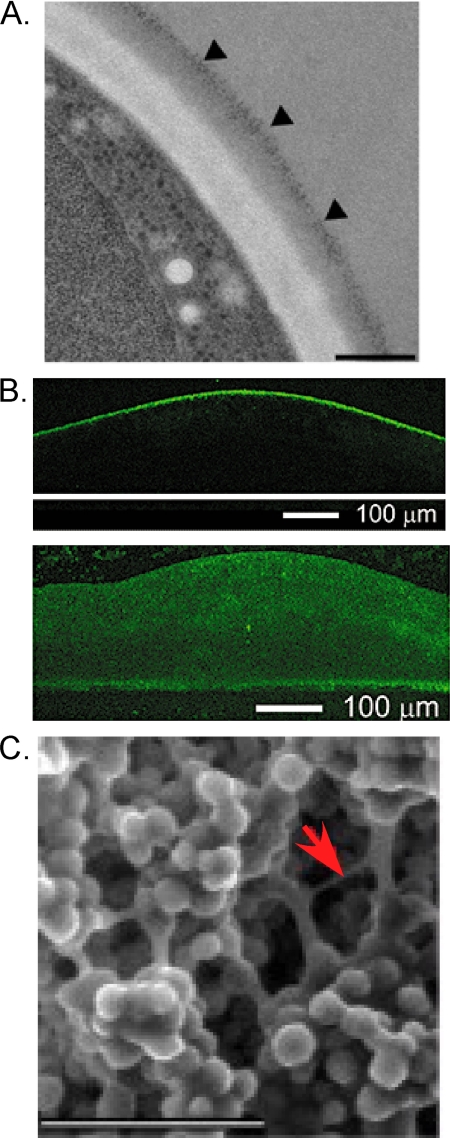
Fig. 3.
Roles of extracellular matrix (ECM) in community organization. (A) Flocs expressing FLO1 are coated with extracellular matrix, as viewed by transmission electron microscopy. Arrowheads indicate a gray staining region on the surface of a floc. (Reprinted from reference 3 with permission of the publisher.) (B) Tight attachments at the colony surface. (Top) ConA-Alexa Fluor staining of the top of a colony; (bottom) ConA-Alexa Fluor staining of a sliced colony from the side. (Reprinted from reference 59 with permission of the publisher.) (C) Scanning electron micrograph of a fluffy colony. A representative bridge of ECM is indicated by the arrow. Scale bar, 20 μm. (Reprinted from reference 56 with permission of the publisher.)
Smooth yeast colonies do not form detectable ECM but instead form an alternative type of protective layer, namely, a thin layer of tightly connected cells on the surface of the colony (59). Dyes such as concanavalin A (ConA) fail to penetrate the colony from the top but can easily penetrate into colonies that have been sliced open, suggesting that this “skin” protects the colony from its environment (Fig. 3B). Consistent with this view, only the intact colony is resistant to the lethal effect of ethanol. The protective nature of this layer can be disrupted by either proteases or polysaccharide-degrading enzymes that attack the cell wall, suggesting that tight contacts between cell walls are essential for this structure.
ECM and signaling.
A different role for the ECM in yeast communities was indicated by scanning electron microscopy of two other types of yeast communities. Flors were found to contain bridges of material linking cells together (73), and similar bridge-like structures were present in structured (or “fluffy”) colonies (Fig. 3C) though absent in smooth colonies (56). These bridge-like structures may act as a conduit for cell-to-cell signals. The high water content of ECM surrounding colonies may also allow more rapid diffusion of nutrients and signal molecules along the colony surface (56).
Mats are encased in a fluid-like ECM that may regulate the transition from a relatively slow-growing colony to a rapidly expanding mat (28). This transition involves the processing and release of Flo11p and the mucin Msb2p into the surrounding ECM. Interestingly, released Flo11p increases the rate of mat expansion but inhibits agar invasion and binding of cells to polystyrene. Thus, released Flo11p inhibits cell adhesion, which is opposite to its function when it is anchored to the cell surface (28).
Summary.
Now that ECM has been discovered in colonies, flocs, and mats, it is clear that ECM has a role both in protecting the community from environmental stress and in cell signaling and cell adhesion. Future studies may reveal the mechanisms by which ECM is generated and regulated and the role of ECM in shaping yeast communities.
(IV) CELL-CELL SIGNALS: DIFFERENT SIGNALS FOR DIFFERENT COMMUNITIES
Several recent studies have revealed cell-to-cell signaling as a hallmark of yeast community organization. Below, I discuss three of these signals—alkali, reactive oxygen species (ROS), and ammonia—and their corresponding signaling pathways.
Alkaline pH and the Rim101p pathway.
The sharp boundaries between colony regions described above (Community Boundaries) suggest involvement of cell-cell signals in colony patterning. The signals regulating sporulation patterns in colonies were investigated using chimeric colonies, i.e., spot colonies composed of a mixture of a “reporter strain” containing an IME2-lacZ fusion gene and a signal strain lacking this fusion (48). By comparing levels of reporter gene expression in chimeric colonies containing wild-type or mutant signal strains, the role of alkali signals, sensed through the Rim101 pathway, in regulating colony sporulation was established (48). Specifically, this pathway is required for the wave of sporulation that extends upward over time from the middle to the top of the colony. As a result, increasing the pH of the agar medium advances the timing of this wave. Conversely, in rim101Δ colonies, unlike in wild-type colonies, the initial narrow layer of sporulation fails to expand.
Ammonia and ROS as signals that localize apoptosis in colonies.
Recent work has begun to reveal how ammonium signals subdivide colonies into apoptotic and viable zones (61). In particular, cells isolated from the edge of the colony produce higher levels of ammonium than cells isolated from the center. This difference may help to differentiate gene expression in the center of the colony from expression at the edge. In particular, Cta1p (mitochondrial/peroxisomal catalase A), a reactive oxygen species (ROS) scavenger, is induced to higher levels at the edge than in the center. Thus, ammonium production at the colony edge may specifically induce genes that limit cell death in this region.
A second signal shaping the organization of 10- to 20-day-old spot colonies may be ROS. ROS is a cell-cell signal in both mammals and plants (4, 72), but it is not known whether ROS is an extracellular signal in yeast. However, cells in the center of 23- to 30-day-old colonies produce higher levels of cellular ROS, such as H2O2, than cells at the margin (7). Mutant colonies defective in the ROS-scavenging enzymes Sod2p (mitochondrial superoxide dismutase) or Ctt1p (cytosolic catalase) are defective both in ammonia production and in localization of dying cells to the colony center (7, 8). Thus, production of extracellular ROS at the colony center may reinforce ammonium production at the edge, further distinguishing cellular fates in the two regions.
Ammonia as a trigger for the dimorphic switch.
Remarkably, two microcolonies that grow in close proximity undergo the dimorphic switch along their opposing faces (67). As a result, pseudohyphae grow from each colony toward the other, and eventually the two colonies link together. This coordinated dimorphic transition requires ammonium signals; in addition, ammonium may actually inhibit the dimorphic switch in older colonies (67). Thus, ammonium can inhibit apoptosis and stimulate or inhibit the dimorphic switch, depending on the concentration of ammonium and the stage of colony development.
Summary.
Only a few signals have been implicated in yeast community organization; it is likely that many others remain to be identified. Studies of yeasts and other simple eukaryotes may reveal some of the earliest mechanisms of communication between individuals.
CONCLUSIONS
Research on S. cerevisiae communities has begun to identify the components that organize and shape these communities, including cell-cell signal pathways, adhesins, and extracellular matrix (Fig. 4). This rapid progress highlights the potential over the next few years to discover mechanisms by which these communities are subdivided into functional regions. These studies may help elucidate how species evolved to increase overall fitness through the formation of communities.
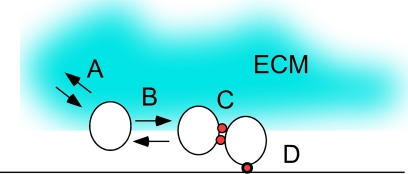
Fig. 4.
Summary of interactions affecting yeast community organization. (A) Cell-ECM interactions. Mucins, such as Msb2p, and flocculins, such as Flo11p, that are secreted from cells help to establish the S. cerevisiae extracellular matrix (ECM) and may also signal other cells through this matrix. (B) Diffusible signals between cells. Alkaline pH sensed through the Rim101 pathway, ammonia/ammonium produced by the Ato transporters, and perhaps also reactive oxygen species are all diffusible signals which contribute to the organization of yeast into communities. (C) Cell-cell contacts. Flocculins are cell surface proteins required for contacts between cells. (D) Cell-surface contacts. Flocculins also required for contacts between cells and between cells and surfaces.
ACKNOWLEDGMENTS
This research was supported by the NIH (grant R15GM094770).
I am grateful to Alex Idnurm (UMKC) for comments on the manuscript and to Z. Palkova (Charles University), P. Magwene (Duke University), and A. Beauvais (Institut Pasteur) for allowing their micrographs to be reproduced in this review.
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1. Barrales R. R., Jimenez J., Ibeas J. I. 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 178:145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastidas R. J., Heitman J. 2009. Trimorphic stepping stones pave the way to fungal virulence. Proc. Natl. Acad. Sci. U. S. A. 106:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beauvais A., Loussert C., Prevost M. C., Verstrepen K., Latge J. P. 2009. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 9:411–419 [DOI] [PubMed] [Google Scholar]
- 4. Bedard K., Krause K. H. 2007. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87:245–313 [DOI] [PubMed] [Google Scholar]
- 5. Blankenship J. R., Mitchell A. P. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9:588–594 [DOI] [PubMed] [Google Scholar]
- 6. Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R. 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 106:18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cap M., Vachova L., Palkova Z. 2009. Yeast colony survival depends on metabolic adaptation and cell differentiation rather than on stress defense. J. Biol. Chem. 284:32572–32581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cap M., Vachova L., Palkova Z. 2010. How to survive within a yeast colony?: change metabolism or cope with stress? Commun. Integr. Biol. 3:198–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H., Fink G. R. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20:1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen H., Fujita M., Feng Q., Clardy J., Fink G. R. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 101:5048–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crump J. A., Collignon P. J. 2000. Intravascular catheter-associated infections. Eur. J. Clin. Microbiol. Infect. Dis. 19:1–8 [DOI] [PubMed] [Google Scholar]
- 12. Dengis P. B., Rouxhet P. G. 1997. Surface properties of top- and bottom-fermenting yeast. Yeast 13:931–943 [DOI] [PubMed] [Google Scholar]
- 13. Dohlman H. G., Slessareva J. E. 2006. Pheromone signaling pathways in yeast. Sci. STKE 2006:cm6. [DOI] [PubMed] [Google Scholar]
- 14. Douglas L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36 [DOI] [PubMed] [Google Scholar]
- 15. Fichtner L., Schulze F., Braus G. H. 2007. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288C. Mol. Microbiol. 66:1276–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fidalgo M., Barrales R. R., Ibeas J. I., Jimenez J. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. U. S. A. 103:11228–11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Floch M. H. 2003. Saccharomyces: is it a probiotic or a pathogen and what is the significance of an elevated anti-S. cerevisiae antibody? J. Clin. Gastroenterol. 36:5–6 [DOI] [PubMed] [Google Scholar]
- 18. Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68:1077–1090 [DOI] [PubMed] [Google Scholar]
- 19. Goossens K., Willaert R. 2010. Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 32:1571–1585 [DOI] [PubMed] [Google Scholar]
- 20. Govender P., Bester M., Bauer F. F. 2010. FLO gene-dependent phenotypes in industrial wine yeast strains. Appl. Microbiol. Biotechnol. 86:931–945 [DOI] [PubMed] [Google Scholar]
- 21. Govender P., Domingo J. L., Bester M. C., Pretorius I. S., Bauer F. F. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:6041–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Granek J. A., Magwene P. M. 2010. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 6:e1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Halme A., Bumgarner S., Styles C., Fink G. R. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405–415 [DOI] [PubMed] [Google Scholar]
- 24. Herskowitz I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80:187–197 [DOI] [PubMed] [Google Scholar]
- 25. Hogan D. A. 2006. Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot. Cell 5:613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hornby J. M., et al. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang S., Choi H. 2005. Effect of surface hydrophobicity on the adhesion of S. cerevisiae onto modified surfaces by poly(styrene-ran-sulfonic acid) random copolymers. Colloids Surf. B Biointerfaces 46:70–77 [DOI] [PubMed] [Google Scholar]
- 28. Karunanithi S., et al. 2010. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr. Biol. 20:1389–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kruppa M. 2009. Quorum sensing and Candida albicans. Mycoses 52:1–10 [DOI] [PubMed] [Google Scholar]
- 30. Kuhn D. M., Chandra J., Mukherjee P. K., Ghannoum M. A. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurjan J. 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27:147–179 [DOI] [PubMed] [Google Scholar]
- 32. Li F., Palecek S. P. 2005. Identification of Candida albicans genes that induce Saccharomyces cerevisiae cell adhesion and morphogenesis. Biotechnol. Prog. 21:1601–1609 [DOI] [PubMed] [Google Scholar]
- 33. Liu H., Styles C. A., Fink G. R. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144:967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lo W. S., Dranginis A. M. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meunier J. R., Choder M. 1999. Saccharomyces cerevisiae colony growth and ageing: biphasic growth accompanied by changes in gene expression. Yeast 15:1159–1169 [DOI] [PubMed] [Google Scholar]
- 36. Mortimer R. K., Johnston J. R. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics 113:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng W. L., Bassler B. L. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Novick R. P., Geisinger E. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 39. Oh K. B., Miyazawa H., Naito T., Matsuoka H. 2001. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 98:4664–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palkova Z., et al. 2002. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol. Biol. Cell 13:3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palkova Z., et al. 1997. Ammonia mediates communication between yeast colonies. Nature 390:532–536 [DOI] [PubMed] [Google Scholar]
- 42. Palkova Z., Vachova L. 2003. Ammonia signaling in yeast colony formation. Int. Rev. Cytol. 225:229–272 [DOI] [PubMed] [Google Scholar]
- 43. Perlroth J., Choi B., Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 44. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piarroux R., Millon L., Bardonnet K., Vagner O., Koenig H. 1999. Are live saccharomyces yeasts harmful to patients? Lancet 353:1851–1852 [DOI] [PubMed] [Google Scholar]
- 46. Piccirillo S., Honigberg S. M. 2010. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res. Microbiol. 161:390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Piccirillo S., Honigberg S. M. Yeast colony embedding method. J. Vis. Exp., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Piccirillo S., White M. G., Murphy J. C., Law D. J., Honigberg S. M. 2010. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics 184:707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Purevdorj-Gage B., Orr M. E., Stoodley P., Sheehan K. B., Hyman L. E. 2007. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 7:372–379 [DOI] [PubMed] [Google Scholar]
- 50. Ramage G., Saville S. P., Wickes B. L., Lopez-Ribot J. L. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reynolds T. B., Fink G. R. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878–881 [DOI] [PubMed] [Google Scholar]
- 52. Reynolds T. B., Jansen A., Peng X., Fink G. R. 2008. Mat formation in Saccharomyces cerevisiae requires nutrient and pH gradients. Eukaryot. Cell 7:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roberts R. L., Fink G. R. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974–2985 [DOI] [PubMed] [Google Scholar]
- 54. Smukalla S., et al. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135:726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sprague G. F., Jr., Winans S. C. 2006. Eukaryotes learn how to count: quorum sensing by yeast. Genes Dev. 20:1045–1049 [DOI] [PubMed] [Google Scholar]
- 56. St'ovicek V., Vachova L., Kuthan M., Palkova Z. 2010. General factors important for the formation of structured biofilm-like yeast colonies. Fungal Genet. Biol. 47:1012–1022 [DOI] [PubMed] [Google Scholar]
- 57. Straight P. D., Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118 [DOI] [PubMed] [Google Scholar]
- 58. Stratford M. 2006. Food and beverage spoilage yeasts, p. 335–380 In Querol A., Fleet G. (ed.), Yeasts in food and beverages. Springer-Verlag, Berlin, Germany: [Google Scholar]
- 59. Vachova L., et al. 2009. Architecture of developing multicellular yeast colony: spatio-temporal expression of Ato1p ammonium exporter. Environ. Microbiol. 11:1866–1877 [DOI] [PubMed] [Google Scholar]
- 60. Vachova L., et al. 2004. Sok2p transcription factor is involved in adaptive program relevant for long term survival of Saccharomyces cerevisiae colonies. J. Biol. Chem. 279:37973–37981 [DOI] [PubMed] [Google Scholar]
- 61. Vachova L., Kucerova H., Devaux F., Ulehlova M., Palkova Z. 2009. Metabolic diversification of cells during the development of yeast colonies. Environ. Microbiol. 11:494–504 [DOI] [PubMed] [Google Scholar]
- 62. Vachova L., Palkova Z. 2005. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169:711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van Mulders S. E., et al. 2009. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9:178–190 [DOI] [PubMed] [Google Scholar]
- 64. Verstrepen K. J., Fink G. R. 2009. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu. Rev. Genet. 43:1–24 [DOI] [PubMed] [Google Scholar]
- 65. Verstrepen K. J., Jansen A., Lewitter F., Fink G. R. 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37:986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vopalenska I., Hulkova M., Janderova B., Palkova Z. 2005. The morphology of Saccharomyces cerevisiae colonies is affected by cell adhesion and the budding pattern. Res. Microbiol. 156:921–931 [DOI] [PubMed] [Google Scholar]
- 67. Vopalenska I., St'ovicek V., Janderova B., Vachova L., Palkova Z. 2010. Role of distinct dimorphic transitions in territory colonizing and formation of yeast colony architecture. Environ. Microbiol. 12:264–277 [DOI] [PubMed] [Google Scholar]
- 68. Walker J. J., Pace N. R. 2007. Endolithic microbial ecosystems. Annu. Rev. Microbiol. 61:331–347 [DOI] [PubMed] [Google Scholar]
- 69. White M. G., et al. 2011. Flo11p adhesin required for meiotic differentiation in S. cerevisiae minicolonies grown on plastic surfaces. FEMS Yeast Res. 11:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wilson D., et al. 2009. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 9:688–700 [DOI] [PubMed] [Google Scholar]
- 71. Wolf J. J., et al. 2010. Feed-forward regulation of a cell fate determinant by an RNA-binding protein generates asymmetry in yeast. Genetics 185:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wong H. L., Shimamoto K. 2009. Sending ROS on a bullet train. Sci. Signal. 2:pe60. [DOI] [PubMed] [Google Scholar]
- 73. Zara G., Zara S., Pinna C., Marceddu S., Budroni M. 2009. FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae. Microbiology 155:3838–3846 [DOI] [PubMed] [Google Scholar]