Abstract
Antigenic variation in the human malaria parasite Plasmodium falciparum depends on the transcriptional regulation of the var gene family. In each individual parasite, mRNA is expressed exclusively from 1 var gene out of ∼60, while the rest of the genes are transcriptionally silenced. Both modifications to chromatin structure and DNA regulatory elements associated with each var gene have been implicated in the organization and maintenance of the silent state. Whether silencing is established at the level of entire chromosomal regions via heterochromatin spreading or at the level of individual var promoters through the action of a silencing element within each var intron has been debated. Here, we consider both possibilities, using clonal parasite lines carrying chromosomally integrated transgenes. We confirm a previous finding that the loss of an adjacent var intron results in var promoter activation and further show that transcriptional activation of a var promoter within a cluster does not affect the transcriptional activity of neighboring var promoters. Our results provide more evidence for the hypothesis that var genes are primarily silenced at the level of an individual gene, rather than by heterochromatin spreading. We also tested the intrinsic directionality of an intron's silencing effect on upstream or downstream var promoters. We found that an intron is capable of silencing in either direction and that, once established, a var promoter-intron pair is stably maintained through many generations, suggesting a possible role in epigenetic memory. This study provides insights into the regulation of endogenous var gene clusters.
INTRODUCTION
The malaria parasite Plasmodium falciparum is capable of maintaining lengthy infections of its human host, thus contributing to more efficient transmission from one individual to another. This process is dependent upon antigenic variation—the process by which the organism changes the proteins displayed on the surfaces of infected red blood cells (RBCs) in order to evade the host immune response. Malaria parasites invade host RBCs and modify them by transporting many proteins to the RBC membrane. One of these proteins is the primary antigenic molecule PfEMP1 (P. falciparum erythrocyte membrane protein 1), which extends through the red cell membrane to the extracellular surface. PfEMP1 makes infected erythrocytes cytoadherent to each other and to vascular endothelia and leads to the sequestration of parasitized cells in the capillary beds. The ensuing hemostasis, hemorrhage, and inflammation are responsible for many of the often fatal clinical symptoms. Thus, PfEMP1 is not only the major antigenic determinant, it is also the most important virulence factor of P. falciparum infections (12, 41).
Different forms of PfEMP1 are highly variable and are encoded by the multicopy var gene family, which consists of ∼60 members per haploid genome (20). Each variant can have different adhesive properties, which account for sequestration within different tissues and the associated clinical syndromes, such as cerebral and placental malaria (32, 35). Each variant is also sufficiently antigenically distinct to render antibodies specific against one PfEMP1 ineffective against another. Expressing only one var gene per parasite and periodically changing the variant that is expressed allows a parasite population to evade the antibody response and establish consecutive waves of parasitemia during the course of an infection. Efficient transmission of the parasite to subsequent hosts depends on the establishment of a persistent infection, which in turn depends on the strict control of each member of the var gene family—from ensuring that only a single var gene is expressed at a time, to maintaining the rest of the repertoire in a silent state, to regulating the switch rate (12, 41).
Research over the past 15 years has identified several key features of var gene regulation. As has been recently proposed (12), different layers of var regulation are likely at play: (i) DNA control elements and the regulatory proteins that bind them, (ii) histone modifications and epigenetic memory, and (iii) subnuclear positioning. The DNA control elements that have been implicated in the control of var gene expression are located in and around each var gene and include an upstream promoter responsible for mRNA production, as well as a conserved intron that also possesses promoter activity that results in expression of noncoding RNAs of unknown function (5, 8). While two examples of parasites that actively transcribed two var genes simultaneously have recently been reported (3, 22), this is thought to be a rare situation, and most investigations have determined that at any one time in an individual parasite, only one var gene is expressed as mRNA, while all other var promoters within the repertoire are transcriptionally silent (6, 13, 40, 44). Changes in gene expression do not correlate with changes in DNA sequence or chromosomal position, and specific transcription factor binding is unlikely to be responsible for monoallelic expression. There is, however, ample evidence that epigenetic mechanisms, including chromatin restructuring and subnuclear positioning, are important in regulating var gene expression. Silent and active var genes each have a distinct chromatin structure with characteristic histone marks (7, 10, 18, 29). Of particular interest, trimethylated H3K9, a modification typical of silent chromatin in other organisms, appears to be specific to multicopy gene families in P. falciparum (30). The nuclear periphery likely houses silent and active gene expression compartments that are associated with var gene silencing and activation, respectively (14, 34, 38, 44).
Histone modification and nuclear positioning are likely to be only one component, or consequence, of a pathway that governs var gene silencing and monoallelic expression. Other elements of such a mechanism, including the initiating DNA control elements and effector molecules, are poorly characterized. The conserved var introns are important regulatory elements, according to various studies that used both episomal and chromosomally integrated transgenic constructs. Early work relied on transient-transfection experiments, which identified the intron's own promoter activity as a requirement for upstream var promoter silencing (5, 19). This silencing was specific to var promoters and was S-phase dependent, a feature typical of mechanisms that involve chromatin modifications (8). The presence or absence of an adjacent intron influences not only the transcriptional state of the associated upstream var promoter, but also that promoter's participation in the allelic-exclusion pathway. The intron appears to be necessary for the upstream promoter to be included, or “counted,” as a member of the repertoire (13, 14). Additional evidence for the importance of var introns is that the endogenous var1csa gene, a var pseudogene often lacking the intron and exon 2, is the only var gene promoter that is not silenced and not counted for allelic exclusion (27, 46). This endogenous example is consistent with data from transgenic studies of the intron's role in var gene regulation.
A model for intron-mediated silencing has emerged from a few recent studies. Frank et al. provided evidence suggesting that there is a strict one-to-one pairing requirement between var promoters and var introns in order for silencing to occur and that each var intron can silence only a single var promoter (17). Their experimental approach took advantage of the parasite's ability to maintain stably transfected episomes as multicopy concatemers (36). Intramolecular homologous recombination between promoter and intron cassettes within the large concatemers resulted in smaller episomes containing different combinations of the following cassettes: (i) a var promoter and intron adjacent to one another, (ii) a var promoter cassette without an intron, and (iii) a single var promoter with two intron cassettes. Whether these concatemers were carried episomally or integrated into a chromosome, it was always true that all var promoters in the concatemer were silent when the number of introns was at least equal to the number of promoters. In other words, when each var promoter was paired with an intron, all the var promoters were silent. In contrast, in every concatemer in which the var promoters outnumbered the introns, i.e., there were unpaired promoters, at least one of the var promoters was actively expressed (17). This pairing requirement was later supported by the work of Dzikowski et al., who additionally showed that unpaired var promoters are incapable of being epigenetically silenced (14). These lines of evidence have led to the “promoter-intron pairing model” of var gene regulation. In this model, the default state of an upstream var promoter is active, and both silencing and recognition by the mutually exclusive expression pathway require the presence of a var intron in cis.
Other studies have suggested alternative or additional models for var gene silencing. For examples, Voss et al. proposed a model in which the default state of an upstream var promoter is silent, with transcriptional activation occurring as a result of nuclear repositioning and changes in the local chromatin environment, regardless of the presence of an intron. They suggested a limited, supportive role for the intron, which they found to enhance silencing of one subtype of upstream var promoter (44). In a subsequent study, parasites were stably transfected with an episome that carried two var promoters and no introns, and one of the var promoters was forced to be active by drug selection. In this context, the neighboring var promoter was also constitutively active, even without selection (45). This result is consistent with the pairing hypothesis, since no introns were present in cis, and thus, both var promoters would be predicted to be active, and it is inconsistent with a model in which the default state of an unselected var promoter is silent. However, Voss et al. proposed an alternative interpretation in which the unselected var promoter was activated by default through the “spreading” of open chromatin along the episome from the adjacent var promoter that was selected for activation. They suggest that there must be boundary elements present within endogenous var gene clusters that separate individual genes and thereby limit the spread of open chromatin (44, 45). It is generally recognized that both DNA elements and chromatin structure play roles in var gene regulation. However, the degree to which var promoter-intron pairing or chromatin spreading influences the silencing of var genes has been debated (11, 14, 41, 45) and deserves further investigation. Is it the interaction of DNA elements that is the primary determinant of a silent state that is then maintained by chromatin modifications, or is the spreading of heterochromatin alone enough to silence a var promoter, even in the absence of the regulatory element in the var intron?
In this study, we set out to differentiate between the “pairing” and “chromatin-spreading” models of var gene silencing. These two models are not necessarily mutually exclusive, and both mechanisms may be at work, even as part of the same silencing pathway. However, these models do differ in what is considered to be the first level of the silencing mechanism, the influence of either the var intron or heterchromatin spreading. In the chromatin-spreading model, the var intron is dispensable, but the pairing model contends that the var intron is a necessary component of silencing. Analysis of transgenic parasite clones allowed us to test both hypotheses and to explore the intrinsic directionality of the var intron's silencing effect. Our results support the previously described var promoter-intron pairing hypothesis and indicate that var introns have the ability to silence var promoters located either upstream or downstream. Once established, var promoter-intron pairs appear to be stable through many cell generations, suggesting a possible role for these interactions in epigenetic memory. This work addresses a controversial issue in the field of var gene regulation and lays some groundwork for more detailed study of the var gene silencing mechanism.
MATERIALS AND METHODS
DNA constructs.
pVBH was described previously (14). It consists of a var promoter driving expression of the blasticidin S deaminase (BSD) gene, terminated by the 3′ untranslated region (UTR) of the P. falciparum hrp2 gene, in a pGEM backbone. pVRHIDH was created from pVLHIDH (5) and pVRH was created from pVLH (8) by replacing the firefly luciferase reporter genes with Renilla luciferase.
To create the larger plasmid pDual-var, the ampicillin resistance cassette of pVRH was replaced with a kanamycin resistance cassette. pVRH(kan) and pVLHIDH(amp) were each linearized with ApaI. Linearized pVRH(kan) was treated with calf intestinal phosphatase (New England Biolabs). The two linearized plasmids were ligated to each other using a Rapid Ligation Kit (Promega). XL-10 Gold (Stratagene) chemically competent Escherichia coli was transformed with the ligation mixture, grown for 1 h in 250 μl of NZYM medium (39), and plated on kanamycin agar plates. The colonies were then replica plated on ampicillin agar plates. The two surviving colonies were grown in ampicillin plus kanamycin medium and screened for the presence of the 17-kb pVRH+VLHIDH (pDual-var) plasmid by restriction digestions/agarose gel electrophoresis and automated sequencing. One colony was found to carry the correct plasmid. To obtain larger quantities of the plasmid, the bacteria were always grown in medium containing both kanamycin and ampicillin. Restriction digestions and sequencing revealed that this large plasmid was stable in E. coli.
All constructs, in their final preparations, were verified using restriction digestions/gel electrophoresis and automated sequencing prior to their use in experiments.
Parasite culture.
P. falciparum parasites (the 3D7 line or its transgenic derivatives) were cultured using standard procedures as described by Trager and Jensen (43). The transgenic line E5D10 was created by Frank et al. (17). Parasites were grown at 5% hematocrit in RPMI 1640 medium, 0.5% AlbuMax II (Invitrogen), 0.25% sodium bicarbonate, and 0.1 mg/ml gentamicin. Cultures were incubated at 37°C in an atmosphere of 5% oxygen, 5% carbon dioxide, and 90% nitrogen.
Stable transfection, selection of integrants, and parasite cloning.
Parasites were transfected, as previously described (9), by utilizing their ability to spontaneously take up DNA from “plasmid-loaded” red blood cells. DNA loading was done by electroporation of RBCs. RBCs (175 μl, packed) were washed in incomplete Cytomix (120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 10 mM K2HPO4-KH2PO4, 25 mM HEPES, pH 7.6) and combined with the appropriate amount of plasmid DNA and enough incomplete Cytomix to reach a final volume of 400 μl. This mixture was transferred to 0.2-cm cuvettes (Bio-Rad), which were then chilled on ice, and electroporated by using a Bio-Rad Gene Pulser II and standard conditions of 0.31 kV and 975-F capacitance.
For stable transfections of pVBH, 100 μg of DNA was used per electroporation cuvette, and two cuvettes of loaded RBCs were used for one 5-ml parasite culture. DNA-loaded red cells were washed and taken up in 4.5 ml culture medium. Schizont stage parasites (E5D10 line [17]) were purified using the Percoll-sorbitol method (1, 5), washed in culture medium, and added to culture flasks containing the DNA-loaded red cells. Two days (one P. falciparum generation) after the initial DNA loading and parasite invasion of red cells, the DNA-loading step was repeated, and the loaded RBCs were added to the parasite culture. Two days after the second DNA loading, 2 μg/ml blasticidin was added to the culture to select for parasites stably carrying the pVBH construct. Upon addition of the drug, most parasites in the culture died within two generations. After 10 days of drug pressure, parasites were detected using Giemsa-stained smears of the cultures.
To confirm that the parasites seen on the smear were stably transformed with the pVBH construct, PCR, plasmid rescue, and Southern blotting were employed. PCR was performed on genomic DNA (gDNA) extracted from the transformed culture, using primers for the bsd gene. All PCRs were carried out on a PTC-2000 Peltier thermal cycler using Taq polymerase (Invitrogen) under the following conditions: 95°C for 5 min, followed by 37 cycles of 94°C for 30 s, 52°C for 40 s, 68°C for 30 s, and a final extension step of 68°C for 10 min. The reaction products were analyzed by gel electrophoresis and automated sequencing.
For plasmid rescue, 1 μg of genomic DNA was used to transform XL-10 Gold (Stratagene) E. coli. The growth of colonies on ampicillin agar plates suggested that pVBH plasmid DNA was present in the gDNA preparation. Plasmid DNA from five of these colonies was isolated and sequenced to confirm that the parasites in the transformed culture were indeed carrying the correct sequence of pVBH. Southern blotting confirmed that the pVBH was episomal and that the plasmid had not yet integrated into the genome.
To select for chromosomal integration, the E5D10 line, now stably transformed with pVBH, was cycled on and off blasticidin drug pressure (off drug for 3 weeks, on drug for 1 week) until the culture appeared to survive drug addition without significant death of the parasites, as seen on smears. Confirmation of genomic integration was done using plasmid rescue to quantify the episomal load, PCR for presence of the bsd gene, and Southern blotting.
Clonal cultures of pVBH/E5D10 integrants were generated by limiting dilution using 96-well microtiter plates (24). The bulk culture was diluted to 50 parasites per 20 ml, which was divided on the plate into 200 μl per well. The medium was changed on days 7, 14, 21, and 23. Wells were screened for the presence of parasites on day 25. Twenty-three of 96 wells were positive for parasites. Of these, eight were selected for further genotypic and phenotypic analyses.
Transient transfection.
For transient transfections, the amount of each construct used in the experiment was adjusted based on molecular weight to ensure equal molar amounts of each were used. The specific amounts were as follows: pVRH, 448 μg; pVRHIDH, 656 μg; pDual-var, 1,176 μg; pVLHIDH, 720 μg; and pVLH, 520 μg. Transient transfections of each construct were done using eight cuvettes, which were combined into 20-ml cultures. The media of these cultures were changed once or twice a day for 3 days, and on the fourth day (after two generations), schizonts from these cultures were purified on Percoll-sorbitol gradients and split into 3 or 4 5-ml cultures, each containing fresh (unloaded) RBCs at 5% hematocrit. These cultures were then assayed for Renilla or firefly luciferase activity 24 h after the gradient-purified schizonts were added to the culture, ensuring that the parasites would be assayed at ring stage, when the var promoter is most active (27). Giemsa-stained smears of these cultures were also made in order to calculate parasitemia.
Luciferase assays.
For stably transformed and chromosomally integrated lines, parasites were synchronized by the Percoll-sorbitol method. Schizonts were isolated from 20-ml cultures using a 70%-40% Percoll-sorbitol gradient and were used to inoculate a 20-ml culture at 5% hematocrit. Luciferase activity was measured 24 h after purified schizonts were added to the culture. Synchronization was confirmed by light microscopy, which revealed that nearly 100% of the parasites were in ring stage. Parasitemia was counted for 1,000 red cells per culture. Parasites were obtained from 200 μl of culture by centrifugation, and the cells were lysed by the addition of 100 μl of Glo Lysis Buffer. One hundred microliters of Bright-Glo luciferase reagent was added to the lysate. Luciferase activity was measured immediately in a TD-20/20 luminometer. Luciferase activity is expressed as luminescence units per 1% ring stage parasitemia. The transgenic parasite line E4 (17) was used as a positive control for luciferase activity. The luciferase activity of each clonal line was determined in at least three independent experiments.
For reporter gene assay of transiently transfected cultures, the reporter gene activities of entire 5-ml cultures were measured. Equal molar amounts of each plasmid were transfected into synchronized, cultured parasites, and assays were performed in triplicate. Parasites were obtained from 5-ml cultures by mixing the centrifuged red cells with 500 μl phosphate-buffered saline (PBS) and 20 μl 10% saponin and washing them three times in 1,000 μl of PBS. Parasite pellets were taken up in 100 μl of either Glo Lysis Buffer (for the firefly luciferase assay) or Renilla Assay Lysis Buffer (Promega). Either Bright-Glo luciferase reagent or Renilla assay reagent (100 μl) was added to the lysate. Luminescence was measured and adjusted as described above.
Genomic DNA extraction.
Infected red blood cells from a 2-ml culture were centrifuged at 6,000 rpm. The supernatant was discarded, and the pellet was resuspended in 2 ml of phosphate-buffered saline and 75 μl 10% saponin. The mixture was divided into two 1.5-ml centrifuge tubes, and the parasite pellets were spun and washed twice in 750 μl of PBS. The pellet was then taken up in 400 μl Tris-sodium chloride-EDTA buffer, along with 80 μl of 10% SDS and 40 μl of 6 M NaClO4. This suspension was placed on a rocker at room temperature overnight, and the genomic DNA was extracted with phenol-chloroform the following morning. The final aqueous phase was ethanol precipitated and resuspended in 35 μl of sterile distilled H2O. After removal of contaminating RNA by digestion with RNase, the final DNA concentration was determined by absorbance at 260 nm.
Southern blots and determination of integration sites.
The presence of episomes and the arrangements of integrated constructs in various parasites cultures were determined by Southern blotting. Southern blots were performed according to standard protocols (39). Genomic DNA was extracted from parasites, digested to completion using restriction endonucleases, and subjected to gel electrophoresis using 0.8% agarose in Tris-boric acid-EDTA buffer. The gels were treated with 0.25 N HCl to improve the transfer of large (>15-kb) fragments. The DNA was transferred to high-bond nylon membranes (Amersham) by capillary action after alkaline denaturation. The DNA was cross-linked to the membrane in a UV cross-linker. DNA probe labeling, detection, and stripping/reprobing were done using DIG-High Prime kits (Roche) according to the manufacturer's protocols.
RNA extraction and real-time quantitative PCR (qPCR).
RNA was extracted from synchronized ring stage parasites 16 to 18 h postinvasion. RNA extraction was performed with the Trizol LS Reagent (Invitrogen) as previously described (26). RNA was purified using RNeasy MiniElute columns (Qiagen) according to the manufacturer's protocol. The isolated RNA was then treated with DNase I (Invitrogen) to degrade contaminating gDNA. cDNA synthesis was performed with Superscript II RNase H reverse transcriptase (Invitrogen) with random primers (Invitrogen), as described by the manufacturer. Total RNA (800 ng) was used for each cDNA synthesis reaction. A control reaction without reverse transcriptase was performed with identical amounts of template.
Real-time quantitative PCR was performed using either cDNA or genomic DNA as a template. To quantify luciferase expression or the genomic copy number, we used the primers 5′-GCTGGGCGTTAATCAGAGAG-3′ and 5′-GTGTTCGTCTTCGTCCCAGT-3′. To quantify blasticidin S deaminase expression or the genomic copy number, we used the primers 5′-TTTGTCTCAAGAAGAATCCA-3′ and 5′-TCCCCCAGTAAAATGATATAC-3.′. Primers 5′-AAGTAGCAGGTCATCGTGGTT-3′ and 5′-TTCGGCACATTCTTCCATAA-3′ were used for the control housekeeping gene, the seryl tRNA synthetase gene, which is thought to be expressed at similar levels in all parasites. All primer pairs had similar amplification efficiencies, as determined by standard curves from real-time measurements of 10-fold dilutions of linearized plasmid DNA.
All reactions were performed in triplicate. The reactions were performed at a final primer concentration of 0.25 μM using Bio-Rad ITAQ Sybr Supermix in 20-μl reaction volumes on an ABI Prism 7900HT real-time PCR machine. The ΔCT for each individual primer pair was determined by subtracting the threshold cycle (CT) value from the CT value of the control seryl tRNA synthetase gene (Applied Biosystems user bulletin 2). ΔCTs were then converted to relative copy numbers by the formula 2ΔCt. Expression or the genomic copy number was normalized to the amount of control seryl tRNA synthetase gene in order to ensure comparison of equal amounts of template across samples.
RESULTS
Chromosomal integration of active, unpaired var promoters into a cluster of silent var promoters.
To test the pairing and chromatin-spreading hypotheses, we utilized a genetically modified parasite line (E5D10) created by Frank et al. (17) in which a tandem array of var promoter-intron cassettes had been integrated into an endogenous var locus (gene PFB1055c on chromosome 2) in the 3D7 parasite line. Due to the repetitive nature of the tandem array of reporter gene cassettes, it was not possible to directly determine changes in chromatin modifications at individual cassettes within the concatemer using typical techniques, like chromatin immunoprecipitation; therefore, instead of assaying chromatin spreading directly, we determined the expression state of promoters using reporter gene assays. Since substantial work has recently been published regarding the chromatin structure found at active and silent var promoters, changes in reporter gene expression likely reflect alterations in chromatin structure. In E5D10, the transgenic var promoters drive expression of firefly luciferase, and the numbers of var promoters and introns are equal (Fig. 1 A). All the transgenic var promoters are silent in this arrangement, and thus, the parasites express very low levels of luciferase. When assayed after several months of continuous culture, the luciferase cassettes remained silent, indicating that the phenotype is stable. We therefore asked how the introduction of a constitutively active var promoter not linked to an intron or other active promoter into this silent cluster would affect the expression profiles of other var promoters within the array.
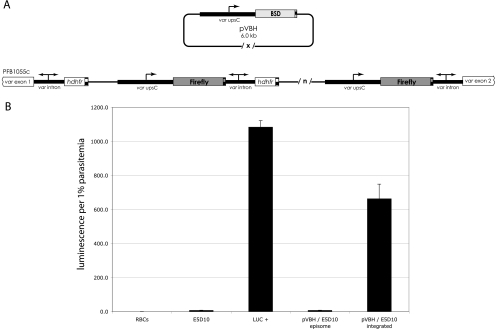
Fig. 1.
Stable transfection of pVBH into the E5D10 line. (A) Schematic of the E5D10 integration site and diagram of pVBH. The ends of the original pVLHIDH integration at the PFB1055c intron are shown. Transgenic var promoters are labeled var upsC and marked with a single arrow. The var introns, which have bidirectional promoter activity, are indicated by double arrows. Each coding sequence is terminated by the 3′ UTR (striped boxes) of the P. falciparum hrp2 gene. Multiple copies of pVLHIDH cassettes are indicated by /n/. Multiple copies of pVBH are maintained as episomes in large, tandem concatemers, indicated by /x/. (B) Luciferase expression following transfection of pVBH into the E5D10 line. As controls, uninfected RBCs were assayed, in addition to LUC+, a transgenic parasite line carrying several chromosomally integrated copies of transcriptionally active var promoters driving luciferase expression. Luciferase activities of pVBH/E5D10 cultures before and after chromosomal integration of pVBH are shown. The error bars indicate standard deviations.
The plasmid construct pVBH features a var promoter driving expression of the blasticidin S deaminase resistance gene (bsd) and contains no var introns (Fig. 1A). This var promoter, of the upsC subtype from the Dd2 strain, is identical to the var promoter in the integrated array in E5D10 and does not have a homologue elsewhere in the 3D7 genome. Previous work demonstrated that in pVBH, the promoter is constitutively active and is not recognized by the mechanism that limits expression to a single var gene (11, 14). We transfected pVBH into the E5D10 line and used blasticidin pressure to select for parasites that stably carried the episome. Plasmid rescue and sequencing of rescued plasmids confirmed that these parasites were indeed carrying pVBH episomes, and Southern blotting of genomic DNA extracted from the newly transformed culture found that chromosomal integration of the episomes was initially undetectable. This culture was then cycled on and off blasticidin to select for parasites in which pVBH had integrated into the genome. Integration was indicated by an absence of colonies upon plasmid rescue and confirmed by Southern blotting. Given the sequence homology to the original (pVLHIDH) integrated tandem array, the pVBH episome was most likely to integrate into the original transgene concatemer by homologous recombination, although integration at the endogenous hrp2 locus was also possible. Furthermore, pVBH itself was likely to integrate as a multicopy concatemer.
If it is true that one var intron can silence only one var promoter, as postulated by the pairing model proposed by Frank et al., then the introduction of additional, unpaired var promoters into the original concatemer could potentially uncouple a previously paired var promoter from the adjacent intron and result in the expression of luciferase. Likewise, under the chromatin-spreading model proposed by Voss et al., introduction of constitutively active promoters into the cluster could result in the opening of chromatin to allow the transcriptional machinery to access neighboring, previously silent, var promoters driving luciferase. Figure 1B shows the luciferase activity of the episomal pVBH/E5D10 culture and of the bulk culture after selection for integration of the plasmid. Prior to integration of pVBH, luciferase expression was similar to that of the nontransformed line E5D10; all promoters in the original transgene array were silent. However, once chromosomal integration of an unpaired/active promoter(s) occurred, the culture expressed robust luciferase activity, suggesting that previously silent var promoters within the original transgenic cluster had become activated. This result is consistent with both the intron-pairing model and the chromatin-spreading model.
Phenotype and genotype analyses of pVBH/E5D10 clones.
It was likely that the pVBH-integrated culture used for the assays shown in Fig. 1B was a heterogeneous population in which several distinct independent integration events had occurred. To analyze individual integration events, clonal populations of parasites were isolated from the bulk integrated culture of pVBH/E5D10 by limiting dilution. Each clonal line was analyzed for luciferase expression, and six clones, displaying a range of luminescence, were selected for further analysis.
Two of the clones (D5 and D6) expressed very low levels of luciferase activity, similar to that of the E5D10 background line, while the other four (G5, G7, H2, and H4) expressed varying levels, all well above that observed in the parent line. These levels were constant regardless of maintenance on blasticidin (data not shown), indicating the stability of the phenotypes. Parasite growth rates were constant regardless of the presence of blasticidin in the media, suggesting that at least one var promoter driving bsd expression was active in all of these clones. The clones displaying higher luciferase expression than the E5D10 background indicate that at least one previously silent var promoter in the original concatemer also must have been activated as a result of pVBH integration.
Southern blotting was used to determine whether the luciferase expression phenotypes of the clones correlated with the integration site of pVBH and the specific arrangement of transgenic var promoters and introns. An extensive series of Southern blots revealed that all of the clones had integrated pVBH into the original VLHIDH concatemer and allowed us to determine the specific site of integration for each clone (only representative blots are shown in the figures). Southern blots specific for the endogenous hrp2 locus, the only other part of the genome that was homologous to sequences on the pVBH construct, showed that none of the clones had integrations at that site (data not shown).
Analysis of a single promoter integration supports the var-pairing hypothesis.
One clone, D5, had a single integrated copy of pVBH (Fig. 2 A and B), while all other clones (H2, H4, G5, G7, and D6 [Fig. 3 ]) had multiple copies. This clone had a luciferase-silent phenotype, similar to the E5D10 background (Fig. 2C), indicating that the transgenic var cluster in D5 consisted of one active var promoter expressing bsd, while all the other promoters within the array remained silent. Active transcription of the bsd gene was confirmed by real-time reverse transcription (RT)-PCR, which detected bsd RNA at a level similar to that of the var gene previously shown to be active in this parasite line (PFA0010c), while the var gene at the original site of integration (PFB1055c) remained silent (Fig. 2D). This clonal line displayed a stable luciferase phenotype regardless of drug pressure and grew continuously upon addition of blasticidin. Both observations suggest that the silent or active state of the var promoters was stably inherited. The genotype and phenotype displayed by clone D5 are consistent with the pairing hypothesis: one unpaired promoter (driving bsd) is constitutively active, and the surrounding var promoters that are paired with introns are silent. However, analysis of clone D5 does not support the chromatin-spreading model, which predicts that the simple insertion of an active promoter within this transgenic cluster would result in activation of nearby var promoters, as well.
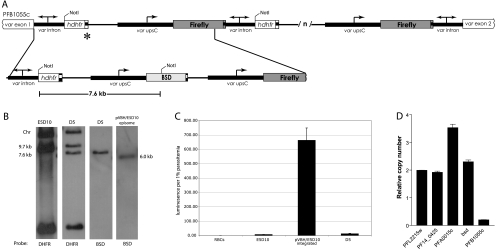
Fig. 2.
Single-copy pVBH integration. (A) Schematic of the pVBH integration site at the original pVLHIDH concatemer. A single copy of pVBH integrated at the hrp2 terminator at the 5′ end of the original pVLHIDH concatemer (marked with an asterisk). (B) Southern blots. gDNA from the D5 clone of pVBH/E5D10 shows the same bands as the original E5D10 line after digestion with NotI, with the addition of a 7.6-kb band (see panel A) that hybridizes to both dhfr and bsd probes. The absence of the 6-kb plasmid unit, seen in the blot of DNA from the pVBH/E5D10 culture prior to chromosomal integration, indicates that D5 contains only one integrated copy of pVBH. (C) Luciferase expression from uninfected cells (RBCs), the original E5D10 parasites (E5D10), the uncloned bulk culture of E5D10 after integration of pVBH, and the D5 clone. The error bars indicate standard deviations. (D) Quantitative RT-PCR of cDNA obtained from ring stage D5 parasites. Levels of expression are shown for two control genes, the actin (PFL2215w) and fructose biphosphate aldolase (PF14_0425) genes; the dominantly expressed var gene (PFA0015c); the blasticidin S deaminase (bsd) gene; and the var gene at the site of integration (PFB1055c).
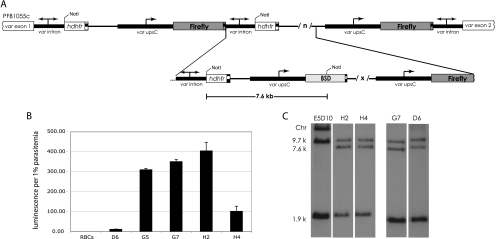
Fig. 3.
Multiple-copy pVBH integration. (A) Schematic of pVBH integration into the original pVLHIDH concatemer. Multiple copies (/x/) of pVBH integrated into the 3′ region of the original pVLHIDH concatemer. (B) Luciferase expression from uninfected cells (RBCs) and the clones D6, G5, G7, H2, and H4. The error bars indicate standard deviations. (C) Representative Southern blots showing four genotypically indistinguishable clones. gDNA digested with NotI and probed with hdhfr show a loss of the large chromosomal band (Chr), indicating integration after the last hdhfr in the original concatemer. The 7.6-kb band also hybridizes to a bsd probe (not shown).
Analysis of clones with multiple unpaired promoters reveals the variability of var promoter-intron pairing.
The pairing hypothesis predicts that integrating a single unpaired var promoter into the original transgenic array and selecting for that promoter to be active through drug pressure would not disrupt any previously formed promoter-intron pairs, and thus, luciferase expression should remain extremely low. This is consistent with the phenotype displayed by the D5 clone described above. However, the pairing hypothesis also predicts that adding multiple unpaired promoters (integration of a pVBH concatemer) to the original array would allow spontaneous activation of previously silent var promoters. Specifically, since all the clones are blasticidin resistant, we know that at least one var promoter driving bsd must be active in each clone. However, if multiple copies of pVBH were integrated, then some of these newly integrated unpaired promoters would be free to pair with the introns in the original concatemer and be silenced. In turn, if var promoter-intron pairing is strictly one to one, a previously silent luciferase-driving var promoter would then become transcriptionally active, resulting in parasites that actively express luciferase. In short, there would no longer be enough introns for each promoter to be paired. Further, if var promoter-intron pairs form at random, the pairing hypothesis predicts that variable levels of luciferase activities would be observed in parasites that contained multiple integrated copies of pVBH, ranging from near zero (all introns remain paired with promoters driving luciferase) to high (several introns become paired with promoters driving bsd expression). This is precisely what was observed in the clones derived from the bulk VBH/E5D10 culture (Fig. 3A). Figure 3B shows that, of the clones with multiple integrated copies of pVBH, four express luciferase (G5, G7, H2, and H4) and one does not (D6).
It was possible that activation of previously silent var promoters within the transgenic cluster was dependent on the exact site of integration and the resulting arrangement of the various cassettes. To determine if luciferase expression correlated with the specific integration site of pVBH within the original VLHIDH concatemer, several Southern blots utilizing a number of different restriction enzyme digestions were used to resolve the arrangement of the array in each clone (one such blot is shown in Fig. 3C). On the other hand, genotypic analysis of these six clones revealed that the var promoter-silencing phenomenon within the transgenic cluster does not correlate with the integration site of the unpaired promoter(s). In fact, four of the clones (D6, G7, H2, and H4) were shown to be genotypically identical on all Southern blots (Fig. 3C), yet they displayed a wide range of luciferase expression. The G5 clone, which actively expressed luciferase, had integrated pVBH at a site distinct from those of the integrations found in the other clones (data not shown). In addition, quantitative real-time PCR of gDNA extracted from these clones showed that all contained approximately 4 bsd cassettes, indicating that the level of luciferase expression was not correlated with the copy number of the inserted concatemer. We hypothesize that, when multiple copies of pVBH integrated into the transgene array, one or more of the unpaired var promoters from the pVBH plasmid were able to pair with introns found within the cluster, thereby “unpairing” previously silent var promoters, resulting in luciferase expression. The promoter-intron pairs appear to initially form in a random manner, resulting in variable levels of expression. However, once formed, the pairs appear to be stable, resulting in stable luciferase expression phenotypes for each clone.
The var intron-mediated silencing effect is bidirectional.
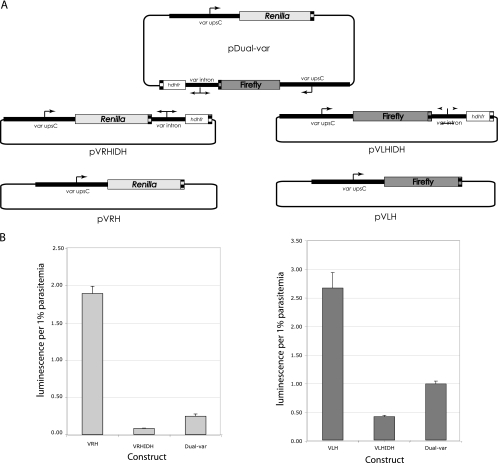
If random intron-promoter pairing is indeed responsible for the variable phenotypes observed in the clones described above, then introns must be able to pair equally efficiently with var promoters when located in either an upstream or a downstream position. To test this hypothesis, we studied intron silencing in a more controlled setting by creating a large (17-kb) dual-var plasmid that had an intron roughly equidistant between two identical var promoters, each driving a different luciferase reporter gene (Fig. 4 A). This construct is similar to one previously described by Voss et al. (45) that also contained two var promoters, but no intron, and in which both var promoters remained active. This construct was then transiently transfected into the wild-type 3D7 parasite line and assayed for the activity of each reporter gene. We used transient, and not stable, transfection for this experiment in order to avoid the concatemerization, recombination, and shuffling of cassettes that frequently occur when episomes are stably maintained by parasites (17). In addition, by using luciferase expression instead of a drug resistance marker, we were able to assess the default state of each promoter in the absence of selection. In the context of a transiently transfected episome, while complete silencing is not achieved, an intron can strongly repress transcription from a paired var promoter (Fig. 4B, pVLHIDH and pVRHIDH). If the single intron on the pDual-var plasmid has an inherent propensity to interact with a var promoter in one direction, then one reporter gene will be highly repressed while the other will be preferentially expressed. However, if the intron is capable of repressing expression of either promoter at random, but not both simultaneously, then the measured activity of each reporter gene on the dual-var plasmid will be at an intermediate level between that of a paired, repressed promoter (pVLHIDH and pVRHIDH) and that of one that is unpaired and fully active (pVLH and pVRH). The data in Fig. 4B are consistent with the second model and show that each reporter gene on the dual-var plasmid is expressed, but not at the high levels expressed by the same promoters in constructs in which they are isolated and constitutively active. Data from transient transfections should be interpreted with caution, since these experiments involve many aspects that can be difficult to control, for example, DNA uptake efficiency and stability. However, the idea that an intron can interact with a var promoter in either direction but in a strictly one-to-one manner was also suggested by the work of Frank et al. (17) using constructs that were stably integrated into the chromosome. Thus, at least in the context of the plasmid-derived arrangements described here, this seems to be a consistent property of intron-promoter interactions.
Fig. 4.
Transient transfection of pDual-var. (A) Schematic of transfected plasmids. (B) Renilla (left) and firefly (right) luciferase expression from parasites transfected with each plasmid shown in panel A. Equal molar amounts of each plasmid were transfected into cultured 3D7 parasites, and assays were performed in triplicate. The experiment was repeated three times independently, and a representative experiment is shown. The error bars indicate standard deviations.
DISCUSSION
In this report, we tested two previously proposed models of var gene silencing. Both are based on data gathered from experiments using transgenes, and they both address the intrinsic properties of var upstream promoters and introns. The intron-pairing model contends that an upstream var promoter is only capable of being silenced when it is in cis with a var intron, a regulatory element which perhaps directs the establishment of silent chromatin in the 5′ region of the var gene. The chromatin-spreading model posits that the var intron is not a necessary factor for silencing and that the transcriptional states of neighboring genes can influence the activity of a var promoter either through the spreading of an open chromatin structure that allows active transcription or, conversely, through similar spreading of heterochromatin. It then follows from this model that larger-scale chromosome and nuclear organization, and not the presence of the regulatory element within a var intron, would primarily determine the transcriptional status of var genes. Analysis of transgenic parasites generated in the present study allowed us to test each hypothesis. Our results are consistent with the pairing model, but not the chromatin-spreading model. However, this is not to say that chromatin modifications, and the possibility of the spreading of chromatin structure along a chromosome, play no role in var gene regulation. Rather, at least in the context of the artificial constructs utilized in the experimental design described in this paper, the primary determinants of var promoter activity are the DNA regulatory elements found in var upstream promoters and introns.
Several studies, including this one, support the idea that DNA regulatory elements represent the first level of var gene regulation. Previous work has shown that both the silencing phenomenon and recognition by the pathway that controls mutually exclusive expression are contingent upon the presence of a var intron in the vicinity of a var upstream promoter. In the present study, the integration of an active var promoter into a transgenic cluster of paired, silent promoters resulted in the single unpaired promoter being active while surrounded by var promoters that remained silent. Similar to the results of Frank et al. and Dzikowski et al. (14, 17), we also found that expression of endogenous var genes was not affected by the presence of a transcriptionally active, unpaired promoter, further supporting the notion that such promoters are not “counted” as part of the family. Our results therefore indicate that there is fine-scale control of var promoters in a closely spaced cluster and that each var promoter is silenced or activated individually. In addition, coordinated expression of the entire gene family relies on the interactions of both intronic and upstream regulatory elements for proper recognition of each gene.
Although the cis DNA elements appear to be the primary determinants of silencing, the surrounding chromatin structure is important and may be the next level of regulation that influences the maintenance of that silent state from generation to generation. For example, there is evidence that subgroups of var genes located in different parts of the chromosomes are also regulated differently. var promoters are classified into four main groups based on their conserved sequences (25, 28). These groups are differentially regulated by two different paralogs of a histone deacetylase called PfSir2 (10, 18, 42). Compared to centrally located var genes, var genes located in heterochromatic subtelomeric regions are more likely to be maintained in the silent state and are more likely to be switched off once activated (16). This epigenetic memory is encoded in the well-characterized histone modifications that are specific to either active, silent, or “bookmarked” var genes (7, 10, 18, 29, 30). Perhaps DNA elements within var promoters and introns direct epigenetic memory by recruiting histone-modifying enzymes, such as the Sir2 complexes. The potential role that var introns play in the placement of appropriate epigenetic marks within the upstream regions of var genes is not clear. Recently, Epp and colleagues showed that the bidirectional promoters within var introns produce nuclear noncoding RNAs (ncRNAs) that associate closely with chromatin (15). The function, if any, of these ncRNAs is unknown. However, there are examples of RNA-based silencing mechanisms in other organisms, in which noncoding RNAs mediate the eventual recruitment of histone-modifying enzymes (33). A similar process may be at work in the case of var genes, and indeed, well-conserved orthologues of chromatin-modifying complexes are present in the P. falciparum genome (2).
A link between the var introns and epigenetic memory can be inferred from our work. The phenotypes of all the clones generated in this study were stable over many months in culture, regardless of drug selection. The four clones in which the concatameric arrangement was indistinguishable showed different, yet stable, luciferase phenotypes, suggesting that once silencing interactions between a var intron and promoter are established, they are maintained in subsequent generations. This conclusion is supported by work done on var gene switching rates. var genes are most likely to maintain their transcriptional status in the next generation, as switch rates are very low (16, 21). In the context of an infection, antigenic variation must be infrequent so that the entire repertoire of antigens is not exhausted and prematurely exposed to the host immune system.
In addition to DNA elements and local chromatin structure, other studies have indicated that large-scale chromosome organization may be important in var gene regulation. Our study raises questions about the existence of insulated var gene chromatin domains. In other organisms, the phenotypic effects of DNA control elements, such as insulators and boundaries, are often varied (4), as seen in our clones. Observations of the variability of silencing led us to design the dual-var transient-transfection experiment to explore the directionality of the intron's silencing effect. The results of the dual-var experiment can be interpreted in one of two ways: either the intron on the dual-var plasmid partially silences each reporter gene, or it silences one or the other at random. Considering previously published data on the exclusive nature of var promoter-intron pairing (17), the second interpretation is more likely. This would suggest that the intron has the intrinsic ability to interact with and silence promoters located either up- or downstream. This variability and directionality of silencing has implications for understanding var gene silencing in the context of the endogenous var gene clusters. More than half of the var gene repertoire is organized as closely spaced clusters of var genes, often arranged in tandem and occasionally head to head. There are a few introns within these clusters that are closer, in linear DNA distance, to the neighboring downstream var promoter than to their own upstream promoter. Whether this distance is an important factor in determining intron-promoter interactions is unknown. Frank and colleagues showed, and Dzikowski et al. confirmed, that pairing is required not only for silencing but also for inclusion in the mutually exclusive expression mechanism (14, 17). Given the results of our study, one can deduce that each endogenous var promoter-intron pair must be insulated from the influences of other nearby introns in order for strict monoallelic expression to work. Presumably, such insulating elements are not present in our transgenic constructs, and their absence might explain both the variegated phenotypes of our genotypically indistinguishable clones and the bidirectionality of intron-mediated silencing. Our observations may point to the existence of insulating, or boundary, elements within native var gene clusters that maintain chromatin domains. These domains could isolate promoter-intron pairs and might consist of loops, similar to what has been described for the insulators surrounding Drosophila heat shock genes or gypsy transposons (23). The loops may then be further organized into active and silent transcription hubs at the nuclear periphery. However, no boundary or insulating element or specialized chromatin structures have yet been identified in P. falciparum, although there are some data that strongly suggest the existence of loops tethered to the nuclear periphery (30). Further investigations should focus on the study of higher-order chromatin structures of native var genes.
A multilayered model best accounts for most of the research done on var gene regulation, but many large gaps exist. What is the nature of the interaction between var intron promoters and var upstream promoters? The sequence of var introns is highly conserved, and this element seems to be found in all var genes, regardless of whether they are expressed or silent. Further, its sequence does not change when a gene switches from the silent to the active state. Therefore, how is it that the one expressed var gene of the repertoire escapes the silencing effect of the intron? The unknown limiting factor that maintains strict monoallelic expression may be yet another layer of regulation. Effector molecules, such as noncoding RNAs and chromatin binding proteins, are just now being identified (15, 31, 37) but have not yet been cast as key players in the story of var gene regulation.
ACKNOWLEDGMENTS
We thank Adina Heinberg and Dacia Kwiatkowski for assistance with real-time PCR assays.
This work was supported by National Institutes of Health grant AI 52390. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation. K.W.D. is a Stavros S. Niarchos Scholar. L.S. is a student of the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional M.D.-Ph.D. Program and is supported by National Institutes of Health Medical Scientist Training Program Grant GM07739.
Footnotes
Published ahead of print on 11 February 2011.
REFERENCES
- 1. Aley S. B., Sherwood J. A., Howard R. J. 1984. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J. Exp. Med. 160:1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aravind L., Iyer L. M., Wellems T. E., Miller L. H. 2003. Plasmodium biology: genomic gleanings. Cell 115:771–785 [DOI] [PubMed] [Google Scholar]
- 3. Brolin K. J., et al. 2009. Simultaneous transcription of duplicated var2csa gene copies in individual Plasmodium falciparum parasites. Genome Biol. 10:R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bushey A. M., Dorman E. R., Corces V. G. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calderwood M. S., Gannoun-Zaki L., Wellems T. E., Deitsch K. W. 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278:34125–34132 [DOI] [PubMed] [Google Scholar]
- 6. Chen Q., et al. 1998. Developmental selection of var gene expression in Plasmodium falciparum. Nature 394:392–395 [DOI] [PubMed] [Google Scholar]
- 7. Chookajorn T., et al. 2007. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U. S. A. 104:899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deitsch K. W., Calderwood M. S., Wellems T. E. 2001. Malaria. Cooperative silencing elements in var genes. Nature 412:875–876 [DOI] [PubMed] [Google Scholar]
- 9. Deitsch K. W., Driskill C. L., Wellems T. E. 2001. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 29:850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duraisingh M. T., et al. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13–24 [DOI] [PubMed] [Google Scholar]
- 11. Dzikowski R., Deitsch K. W. 2008. Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 382:288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dzikowski R., Deitsch K. W. 2009. Genetics of antigenic variation in Plasmodium falciparum. Curr. Genet. 55:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dzikowski R., Frank M., Deitsch K. 2006. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS. Pathog. 2:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dzikowski R., et al. 2007. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 8:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Epp C., Li F., Howitt C. A., Chookajorn T., Deitsch K. W. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15:116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frank M., Dzikowski R., Amulic B., Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 64:1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frank M., et al. 2006. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum. J. Biol. Chem. 281:9942–9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freitas L. H., Jr., et al. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36 [DOI] [PubMed] [Google Scholar]
- 19. Gannoun-Zaki L., Jost A., Mu J. B., Deitsch K. W., Wellems T. E. 2005. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell 4:490–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner M. J., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horrocks P., Pinches R., Christodoulou Z., Kyes S. A., Newbold C. I. 2004. Variable var transition rates underlie antigenic variation in malaria. Proc. Natl. Acad. Sci. U. S. A. 101:11129–11134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joergensen L., et al. 2010. Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathog. 6:e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kadauke S., Blobel G. A. 2009. Chromatin loops in gene regulation. Biochim. Biophys. Acta 1789:17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirkman L. A., Su X. Z., Wellems T. E. 1996. Plasmodium falciparum: isolation of large numbers of parasite clones from infected blood samples. Exp. Parasitol. 83:147–149 [DOI] [PubMed] [Google Scholar]
- 25. Kraemer S. M., Smith J. D. 2003. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 50:1527–1538 [DOI] [PubMed] [Google Scholar]
- 26. Kyes S., Pinches R., Newbold C. 2000. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 105:311–315 [DOI] [PubMed] [Google Scholar]
- 27. Kyes S. A., et al. 2003. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 48:1339–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lavstsen T., Salanti A., Jensen A. T. R., Arnot D. E., Theander T. G. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Rubio J. J., et al. 2007. 5′ Flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lopez-Rubio J. J., Mancio-Silva L., Scherf A. 2009. Genome wide localization of heterochromatin couples clonally variant gene regulation to perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190 [DOI] [PubMed] [Google Scholar]
- 31. Mancio-Silva L., Rojas-Meza A. P., Vargas M., Scherf A., Hernandez-Rivas R. 2008. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 121:2046–2053 [DOI] [PubMed] [Google Scholar]
- 32. Milner D. A., Jr., Montgomery J., Seydel K. B., Rogerson S. J. 2008. Severe malaria in children and pregnancy: an update and perspective. Trends Parasitol. 24:590–595 [DOI] [PubMed] [Google Scholar]
- 33. Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457:413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mok B. W., et al. 2008. Default pathway of var2csa switching and translational repression in Plasmodium falciparum. PLoS. ONE 3:e1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Montgomery J., et al. 2007. Differential var gene expression in the organs of patients dying of falciparum malaria. Mol. Microbiol. 65:959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Donnell R. A., et al. 2001. An alteration in concatameric structure is associated with efficient segregation of plasmids in transfected Plasmodium falciparum parasites. Nucleic Acids Res. 29:716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pérez-Toledo K., et al. 2009. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 37:2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ralph S. A., Scheidig-Benatar C., Scherf A. 2005. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. U. S. A. 102:5414–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Scherf A., et al. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17:5418–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scherf A., Lopez-Rubio J. J., Riviere L. 2008. Antigenic variation in Plasmodium falciparum. Annu. Rev. Microbiol. 62:445–470 [DOI] [PubMed] [Google Scholar]
- 42. Tonkin C. J., et al. 2009. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 7:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trager W., Jensen J. B. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 44. Voss T. S., et al. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439:1004–1008 [DOI] [PubMed] [Google Scholar]
- 45. Voss T. S., et al. 2007. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol. Microbiol. 66:139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winter G., et al. 2003. The 3D7var5.2 (var(COMMON)) type var gene family is commonly expressed in non-placental Plasmodium falciparum malaria. Mol. Biochem. Parasitol. 127:179–191 [DOI] [PubMed] [Google Scholar]