Abstract
Microsporidia are a group of fungus-related intracellular parasites with severely reduced metabolic machinery. They lack canonical mitochondria, a Krebs cycle, and a respiratory chain but possess genes encoding glycolysis enzymes, a glycerol phosphate shuttle, and ATP/ADP carriers to import host ATP. The recent finding of alternative oxidase genes in two clades suggests that microsporidial mitosomes may retain an alternative respiratory pathway. We expressed the fragments of mitochondrial chaperone Hsp70 (mitHsp70), mitochondrial glycerol-3-phosphate dehydrogenase (mitG3PDH), and alternative oxidase (AOX) from the microsporidium Antonospora (Paranosema) locustae in Escherichia coli. Immunoblotting with antibodies against recombinant polypeptides demonstrated specific accumulation of both metabolic enzymes in A. locustae spores. At the same time comparable amounts of mitochondrial Hsp70 were found in spores and in stages of intracellular development as well. Immunoelectron microscopy of ultrathin cryosections of spores confirmed mitosomal localization of the studied proteins. Small amounts of enzymes of an alternative respiratory chain in merogonial and early sporogonial stages, alongside their accumulation in mature spores, suggest conspicuous changes in components and functions of mitosomes during the life cycle of microsporidia and the important role of these organelles in parasite energy metabolism, at least at the final stages of sporogenesis.
INTRODUCTION
The reductive evolution of mitochondria is a widely occurring feature of microaerophilic protists and obligate intracellular parasites from the phyla Heterokontophyta, Metamonada, Amoebozoa, Apicomplexa, and Microsporidia (10). Though mitochondrial remnants of some groups demonstrate extreme simplification, all “amitochondriate” eukaryotes studied to date have been shown to contain these organelles. Such widespread retention of mitochondrial homologues suggests their essential role for eukaryotic cells lacking canonical mitochondria. Biosynthesis of iron-sulfur clusters required for the maturation of diverse Fe/S proteins is one of the metabolic pathways found in mitochondrion-like organelles, hydrogenosomes, and mitosomes of amitochondriate protists (10). Hydrogenosomes of Trichomonas vaginalis (Metamonada) (24), mitochondrion-like organelles of human parasites Blastocystis spp. (Heterocontophyta) (23), and a single mitochondrion of Cryptosporidium (Apicomplexa) (1, 17, 37) may also be implicated in the core carbon and energy metabolism of parasites. The metabolic role of small, abundant (10 to 150 organelles per cell), double-membrane-bounded organelles called mitosomes that were found in Entamoeba histolytica (Amoebozoa) (26), Giardia lamblia (Metamonada) (18), microsporidium species Trachipleistophora hominis (36), and Encephalitozoon cuniculi (27, 34) is uncertain. Because these parasites lack any ATP-generating enzymes of mitochondrial origin, their mitosomes appeared not to be involved in energy metabolism (10). However, the genes encoding alternative oxidase (AOX) were recently discovered in two phylogenetic clades of microsporidia (35). Besides, all studied microsporidial species, with the exception of Enterocytozoon bieneusi (13), possess the genes encoding both cytosolic and mitochondrial components of the glycerol phosphate shuttle. This suggests that mitochondrial glycerol-3-phospate dehydrogenase (mitG3PDH) and AOX are coupled on the mitosomal inner membrane to provide an electron transfer from glycerol-3-phosphate to the oxygen through flavin adenine dinucleotide (FAD) and the ubiquinone pool. Thus, microsporidial mitosomes may be involved in parasite carbon and energy metabolism if they reoxidize reducing equivalents (NADH) produced by the Embden-Meyerhof pathway.
Surprisingly, the presence of an alternative respiratory pathway in parasite mitosomes could not be verified for most microsporidial species whose genomes have been studied thoroughly (E. cuniculi, Encephalitozoon intestinalis, E. bieneusi, Nosema bombycis, and Nosema ceranae). According to the phylogenetic system based on molecular analysis of the small subunit ribosomal DNA (rDNA) of 125 microsporidial species (32), these parasites belong to phylogenetic clade IV (Terresporidia) and are lacking AOX genes (35). To localize AOX and mitG3PDH in parasite mitosomes, we have chosen microsporidium Antonospora (Paranosema) locustae from phylogenetic clade II (Aquasporidia), infecting orthopteran insects. The genome of this parasite is partially sequenced and contains genes encoding both proteins (Antonospora locustae Genome Project; http://forest.mbl.edu/cgi-bin/site/antonospora01). Recognition of A. locustae mitG3PDH and AOX expressed in Saccharomyces cerevisiae by the protein import machinery of yeast mitochondria (5, 35) suggests indirectly their mitosomal localization. Besides, overexpression of A. locustae AOX in the bacterium Escherichia coli has demonstrated the ubiquinol-reducing activity of the parasite enzyme (35). Analysis of AOX found in distantly related microsporidium T. hominis (phylogenetic clade III [Marinosporidia]) has shown identical results (35).
In this study, we have overexpressed the fragments of A. locustae AOX, mitG3PDH, and mitochondrial Hsp70 (mitHsp70) genes in bacterium E. coli to raise polyclonal antibodies (Abs) and localize parasite proteins. Immunolocalization of mitHsp70, the specific marker of T. hominis (36) and E. cuniculi (27) mitosomes, was done because these organelles still have not been identified in A. locustae. Immunoblotting with Abs against recombinant proteins has demonstrated specific accumulation of AOX and mitG3PDH in mature A. locustae spores, whereas comparable amounts of mitHsp70 were found in intracellular development stages (meronts and sporonts) and spores. On ultrathin cryosections of mature spores, three studied proteins were localized in mitosomes found for the first time in microsporidium A. locustae. These data suggest that microsporidium mitochondrial remnants retain their role in energy metabolism, at least in the late stages of sporogenesis.
MATERIALS AND METHODS
A. locustae spores and prespore intracellular stages.
A. locustae spores and stages of intracellular development were isolated from the fat bodies of experimentally infected Locusta migratoria locusts containing sufficient numbers of spores and merogonial and sporogonial stages, as described previously (19).
PCR amplification of protein-encoding sequences.
The sequences encoding mitHsp70, AOX, and mitG3PDH were obtained from the A. locustae Genome Database (Marine Biological Laboratory at Woods Hole, funded by NSF award number 0135272; http://forest.mbl.edu/cgi-bin/site/antonospora01) and amplified with Pfu polymerase using a parasite genomic DNA template as previously described (6). The following specific primers were designed for PCR amplification of the full-size copy of the gene encoding mitG3PDH, and the 3′-end fragments of the mitHsp70 (1,747-bp) and AOX (621-bp) genes (the sites of restriction enzymes are noted by lowercase letters and the parts complementary to microsporidial gene sequences are underlined): mitG3PDH (forward), 5′-CGAAggatccGATGATAAACAAACGTACATACACC-3′; mitG3PDH (reverse), 5′-CTGAgaattcTTAAAGCATTCCAAGACCCAGGG-3′; mitHsp70 (forward), 5′- AGGAATagatctAGGAACAACAAACT-3′; mitHsp70 (reverse), 5′- TCTTggtaccTCAAAGCCGGGCCCTGACCTCCT-3′; AOX (forward), 5′-TTAAggatccGATCACACTCTTGGACCTGAGTAGA-3′; and AOX (reverse), 5′- AAGTaagcttAGTCTGCCATGCTGTGGTTTGTAT-3′.
DNA constructs for expression of A. locustae proteins.
The PCR products were gel purified, cleaved with the appropriate restriction enzymes, and inserted into the pRSET vector (Invitrogen, CA) digested with the same enzymes. The first 600 nucleotides downstream of the T7 promoter were sequenced to verify the correct amplification and insertion of fragments.
mitG3PDH.
The full-size mitG3PDH gene (1,773 kb) was cloned in plasmid pRSETb (Invitrogen, CA) at the BamHI/EcoRI sites. To eliminate the predicted N-terminal hydrophobic region, the construction was additionally digested with the HindIII enzyme, and the fragment flanked by the HindIII sites, present in the gene (position 385) and multiple cloning site (MCS) of vector, was gel purified and inserted in plasmid pRSETc cut with the same enzyme. Plasmids with the correct orientation of the gene fragment were selected using the inner SacII site (position 1436) and EcoRI site in MCS.
mitHsp70.
A PCR-amplified copy of the mitHsp70 gene fragment (1,747 bp) was digested at the BamHI inner site (position 1209 in the full-size gene) and the KpnI site introduced in the reverse primer. The gel-purified 3′-end fragment of 665 bp was cloned into pRSETb cut by BamHI and KpnI enzymes.
AOX.
A fragment of the AOX gene (621 bp) was amplified, digested, and cloned in the pRSETb plasmid at the BamHI/HindIII sites.
Heterologous expression and isolation of recombinant products.
Expression was done in BL21(DE3)-derived C41 and C43 (16) and BL21(DE3)pLys E. coli strains as previously described (6). After 18 h of cultivation at 37°C, bacterial cells were harvested by centrifugation at 3,000 × g for 10 min, washed with distilled water, and sonicated in 50 mM Tris-Cl buffer (pH 8.0).
The gene fragment encoding the C-terminal part of chaperone mitHsp70 was expressed in C41 cells. The same yield of recombinant product was observed with or without IPTG (isopropyl-β-d-thiogalactopyranoside) induction of T7 RNA polymerase expression. In contrast to most foreign proteins forming insoluble inclusion bodies (IBs) in E. coli, the C-terminal polypeptide of mitHsp70 was accumulated in soluble form and successfully purified by one-step immobilized metal ion affinity chromatography (IMAC). Sonicated bacterial cells were centrifuged at 18,000 × g for 10 min; 10 mM imidazole, 0.3 M NaCl (final concentrations), and a 1/10 volume of Ni-CAM HC resin (Sigma-Aldrich, MO) (equilibrated with 50 mM Tris-Cl buffer [pH 8.0] containing 0.3 M NaCl and 10 mM imidazole) were added to the soluble fraction. After gentle stirring overnight at 4°C, the resin was carefully washed, and recombinant protein was eluted with equilibration solution containing 0.25 M imidazole.
Expression of the full-size mitG3PDH gene in E. coli was not effective, though we used C41, C43, and BL21(DE3)pLys strains specially designed for production of heterologous toxic proteins. To excise the N-terminal hydrophobic region, predicted by TMHMM version 2.0 and HMMTOP servers, we have expressed the lengthy 3′-end part of the gene, starting with the inner HindIII site at position 385. The most effective expression of this fragment was observed with IPTG-induced C41 cells, whereas IPTG-induced C43 cells were more suitable for production of recombinant AOX. Recombinant mitG3PDH and AOX were accumulated as insoluble IBs in E. coli. They were spun down at 1,500 × g for 10 min and carefully washed with the same buffer solution containing 0.05% Triton X-100. The recombinant proteins were dissolved by the resuspending of IBs in a solution of 50 mM Tris-Cl and 8 M urea followed by the removal of insoluble debris at 18,000 × g for 5 min.
Production, purification, and depletion of polyclonal Abs.
Recombinant mitG3PDH and AOX polypeptides solubilized in 8 M urea were diluted 10-fold with distilled water, mixed with an equal volume of Freund's adjuvant (Sigma, MO) (complete for the first injection and incomplete for the next ones), and used for immunization. Purified by IMAC, mitHsp70 was dialyzed against TBS (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) before mixing it with adjuvant. Rabbits were immunized by four intramuscular injections (300 μg protein per injection) at 10-day intervals. Ten days after the last immunization, 15 ml of blood was collected. Immune sera were analyzed by immunoblotting, and specific Abs were purified against recombinant proteins transferred to a nitrocellulose membrane as described previously (6). The preparation of Abs against the α subunit of pyruvate dehydrogenase (PDH) and the Sec13 subunit of the COPII complex was described in recent publications (6, 8).
To deplete anti-mitHsp70 Abs, soluble proteins of P. locustae spores were separated by SDS-PAGE in 12% gel, transferred to nitrocellulose membrane, and stained with Ponceau S. The major band of 75 kDa was carefully cut out, blocked in TBS with 2% bovine serum albumin (BSA) for 1 h, and washed with TBS. Affinity-purified anti-mitHsp70 Abs stored at −20°C in the presence of 50% glycerol and BSA (1 mg/ml) were diluted 1:7 with TBS and incubated in the presence of membrane for 3 h at room temperature. Incubation was followed by aspiration of Abs into another tube and washing of the membrane with 0.2 M glycine-Cl (pH 2.5) and TBS. After three rounds of depletion, Abs were concentrated with Centricon centrifugal filter units (Millipore, MA) and stored at −20°C in the presence of BSA (1 mg/ml) plus 50% glycerol.
Antibodies produced in this study are available to the research community. Please contact the corresponding author by e-mail.
Preparation of samples for immunoblotting.
A. locustae spores were broken in TS solution (25 mM Tris-Cl [pH 8.0], 0.3 M sucrose) by vortexing with 2.5-mm glass balls (BDH, United Kingdom) for 30 min, and homogenate was cleared by centrifugation at 100 × g for 10 min. The stages of intracellular development (meronts and sporonts) were purified by centrifugation in Percoll density gradient (19) and ruptured by sonication. For comparative analysis of spores and intracellular stages, both samples were equilibrated in protein concentration by Bradford's method (4). To analyze the intrasporal localization of proteins, cleared spore homogenate was centrifuged in a series at 20,000 × g for 20 min and at 200,000 × g for 1 h. The pellets were resuspended with the volume of TS brought up to that of the final supernatant and assayed by immunoblotting. All manipulations with microsporidial native proteins were performed at 4°C.
SDS-PAGE and immunoblotting were carried out as previously described (6).
Ultrathin cryosectioning and immunogold labeling.
A. locustae mature spores were fixed with 4% formaldehyde and 1% glutaraldehyde in 0.2 M PHEM buffer (60 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 25 mM HEPES, 2 mM MgCl2, 10 mM EDTA, pH 7.4) and washed in the same buffer. The samples were embedded in 10% gelatin, cooled in ice, and cut into 1-mm3 blocks in the cold room. The blocks were infused with 2.3 M sucrose at 4°C for at least 2 h and frozen in liquid nitrogen. Sections, 45 to 60 nm thick, were cut at −120°C or −110°C using Ultracut R/FCS (Leica, Italy) equipped with an antistatic device (Diatome, PA) and diamond knife. Ultrathin sections were picked up in a mix of 1.8% methylcellulose and 2.3 M sucrose (1:1) as described previously (15). Cryosections were collected on Formvar/carbon-coated copper slot grids and then incubated with Abs diluted 1:50 (depleted anti-mitHsp70 Abs) or 1:100 (anti-AOX and anti-G3PDH Abs) in PBS with 0.1% BSA-c (Aurion, Netherlands) followed by protein A-conjugated gold particles of different sizes (20). Double immunolabeling was performed as described previously (20, 33), with optimal combinations of gold particle sizes. To reduce specificity problems with double labeling (when two rabbit antibodies are used) following treatment with the first Abs and protein A, sections were additionally fixed with 1% glutaraldehyde for 5 min, washed with 0.12% glycine, and blocked in PBS containing 0.01% BSA-c, as previously recommended (33). Additionally, both sequences of the introduction of Abs and combinations of gold particles of different size were tested. After labeling, sections were treated with 1% glutaraldehyde, counterstained with uranil acetate, and embedded in methylcellulose uranyl acetate as described previously (21). Automated datum acquisition was carried out on a Tecnai12 electron microscope at 120 kV (FEI/Philips Electron Optics) equipped with a slow-scan charge-coupled-device (CCD) camera.
RESULTS
Specific accumulation of metabolic enzymes in microsporidial spores.
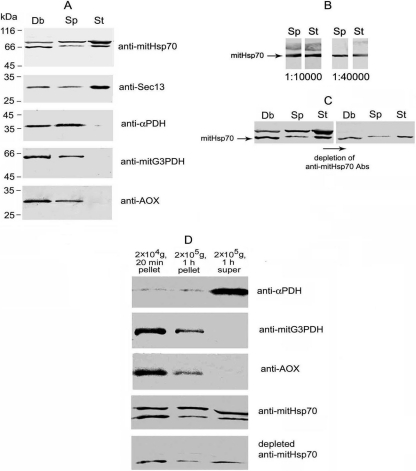
A Western blot assay with Abs against recombinant polypeptides has shown the presence of the proteins in A. locustae mature spores (Fig. 1 A). Anti-mitHsp70 Abs recognized two bands of 66 and about 75 kDa. Abs against mitG3PDH and AOX reacted with single bands of about 64 and 30 kDa, respectively. Like the α subunit of PDH (another enzyme of carbon metabolism), mitG3PDH and AOX were undetectable in stages of intracellular development (meronts and sporonts) isolated by Percoll density gradient centrifugation. Importantly, it cannot be interpreted as a result of protein leakage or degradation during the isolation procedure because comparable amounts of molecular chaperon mitHsp70 and the Sec13 subunit of COPII (coat complex involved in intracellular traffic) were found in the same samples of spores and stages of intracellular development.
Fig. 1.
Western blot assay of A. locustae proteins. (A) Samples of spore homogenate cleared by centrifugation at 100 × g for 10 min (Sp) and stages of intracellular development (meronts and sporonts) ruptured by sonication (St) were equalized in protein concentration, and 20 μg was analyzed by immunoblotting. This experiment has clearly demonstrated specific accumulation of metabolic enzymes in microsporidial spores. To estimate protein diffusion from destroyed spores, an aliquot of crude debris (Db) resuspended in TS equal to the volume of homogenate was analyzed. (B) Double band recognized by anti-Hsp70 Abs represents two different proteins because serial dilution of anti-mitHsp70 immune serum up to 1:40,000 has resulted in drastic weakening of upper major band, whereas 66-kDa protein has remained specifically stained. (C) Specific decoration of 66-kDa protein by anti-mitHsp70 Abs depleted against major parasite protein of 75 kDa has confirmed the existence of unique epitopes in mitHsp70 polypeptide chain. Protein samples of Sp, St, and Db were prepared as described in the legend to panel A. (D) To analyze intrasporal localization of studied proteins, cleared spore homogenate was centrifuged in a series at 20,000 × g for 20 min and at 200,000 × g for 1 h, and the pellets were resuspended, with the volume of TS brought up to that of the final supernatant prior to Western blot assay. The pelleting of mitG3PDH, AOX, and most of mitHsp70 by high-speed centrifugation has suggested their localization in some membrane compartment.
Identification of A. locustae mitHsp70.
Since anti-mitHsp70 Abs recognized two proteins of similar molecular weights, it was necessary to find out which band (or if both of them) corresponds to the mitochondrial chaperon. Serial dilution of immune serum up to 1:40,000 resulted in a weakening of the upper major band (Fig. 1B), whereas the 66-kDa protein was still specifically recognized. To get stronger evidence that the double band represents two different proteins, of which the lower one is mitHsp70, A. locustae spore proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane, and the major band of 75 kDa was carefully cut out to deplete anti-mitHsp70 Abs. Specific staining of the 66-kDa band by depleted Abs (Fig. 1C) has confirmed the existence of unique epitopes in the mitHsp70 polypeptide chain.
Alongside the gene with the sequence encoding mitHsp70, at least three other genes of the conservative Hsp70 family are present in the A. locustae genome. Relatively high similarity (35 to 40% identity) between the amino acid sequences of mitHsp70 and another nonmitochondrial chaperone of 74 kDa suggests such cross-reactivity of Abs raised against the mitochondrial protein.
Association of mitHsp70, AOX, and mitG3PDH with spore membranes.
As shown in Fig. 1A, more than half of the α PDH subunit and 75-kDa protein, recognized by anti-mitHsp70 Abs, were extracted from A. locustae spores crushed in the presence of 0.3 M sucrose. In contrast, the major parts of mitHsp70, mitG3PDH, and AOX were found in crude debris pelleted by centrifugation of the spore homogenate at 100 × g for 10 min. The hindered diffusion of proteins from destroyed spores suggested their association with some membrane compartments. Further centrifugation of cleared spore homogenate confirmed the membrane localization of AOX and mitG3PDH because most enzymes were pelleted by high-speed centrifugation at 20,000 × g for 20 min and they were completely removed from the soluble fraction by ultracentrifugation (Fig. 1D). Since both enzymes are the components of a mitochondrial electron transport chain in other eukaryotic groups (11, 22), their membrane localization was expected. Most of mitHsp70 was also pelleted by high-speed centrifugation, but the final supernatant still contained a considerable amount of the chaperone. This result may be explained by the outflow of soluble matrix proteins from partially destroyed organelles.
Immunocytochemical detection of A. locustae mitosomes.
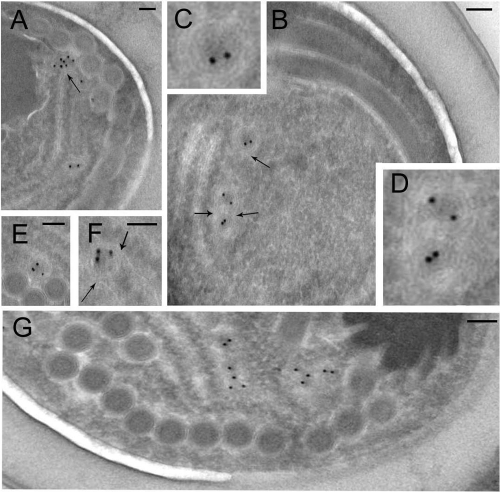
To visualize mitosomes in A. locustae mature spores, we used immunogold staining of ultrathin cryosections with affinity-purified anti-mitHsp70 Abs additionally depleted against an unspecific 75-kDa band. Abs gave highly specific labeling over small (50- to 200-nm) structures of round profiles bounded by double membranes resembling mitosomes of other microsporidial species (Fig. 2). About 3 or 4 such mitosomes were found in each spore section, and some of them appeared to be aggregated in pairs (Fig. 2B and G). As expected, most gold particles localized over the inner area of structures representing the mitosomal matrix. A. locustae mitosomes appeared to be in close contact with endoplasmic reticulum (ER) cisternae (Fig. 2A, B, and F), but the cryosection technique did not provide enough contrast to make a final conclusion about such links.
Fig. 2.
Identification of mitosomes in A. locustae spores. Ultrathin cryosections of mature spores were stained with affinity-purified and depleted anti-mitHsp70 Abs. Immunoelectron microscopy shows specific labeling of mitosomes, small (50- to 200-nm) structures of round profiles bounded by a double membrane (A to G). About 3 or 4 mitosomes were found per spore section, and some of them were aggregated in pairs (B, G). Mitosomes from panel B are shown enlarged (C, D). As expected, most of the labeling was present over the inner area of the organelles (mitosomal matrix). Microsporidial mitosomes appeared to be in close contact with ER cisternae (arrows). Gold grain size, 10 nm. Scale bar, 0.1 μm.
Immunolocalization of AOX and mitG3PDH in A. locustae mitosomes.
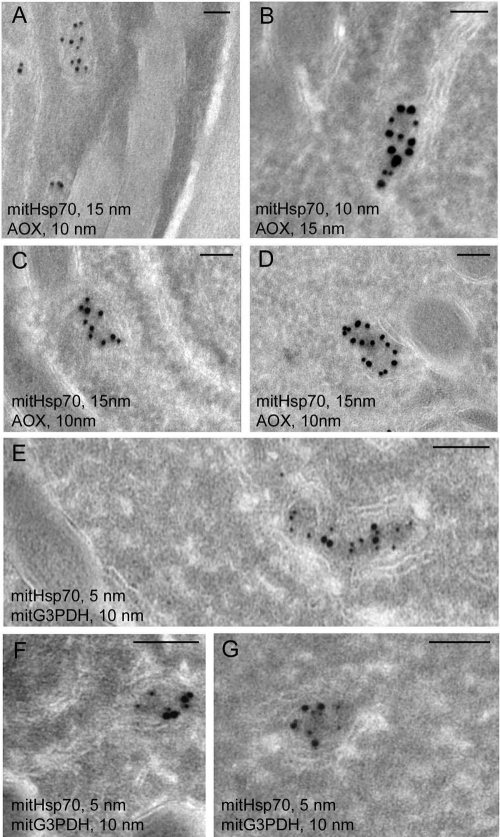
Double labeling of spore cryosections for affinity-purified anti-AOX, anti-mitG3PDH, and depleted anti-mitHsp70 Abs demonstrated the colocalization of both enzymes of the alternative respiratory chain with the mitosomal marker (Fig. 3). As expected, AOX and G3PDH were found mostly on the inner mitosomal membrane around the slightly more electron-dense matrix. Though the inner membrane may form structures resembling folds in microsporidial mitosomes (31), staining with anti-AOX and anti-mitG3PDH Abs depicted exactly the contours of some organelles (Fig. 3C and G). This result demonstrated that in spite of drastic reduction of the mitochondrial remnants in microsporidia, mitosomes of A. locustae spores contain the protein components of an alternative respiratory chain.
Fig. 3.
Immunolocalization of A. locustae AOX and mitG3PDH. Double labeling of spore cryosections with anti-AOX and anti-mitHsp70 Abs shows specific decoration of mitosomes and colocalization of both proteins in these organelles (A to D). Immunoelectron microscopy of spore cryosections with anti-mitG3PDH and anti-mitHsp70 Abs has shown similar colocalization of both proteins in A. locustae mitosomes (E to G). AOX and mitG3PDH labeling was seen mostly over the mitosomal membranes around the more electron-dense matrix. Scale bar, 0.1 μm.
DISCUSSION
Western blot analyses have shown specific accumulation of PDH subunits (6), AOX, mitG3PDH (present study), and phosphofructokinase (our unpublished data) in A. locustae mature spores and their low content in the stages of intracellular development (meronts and sporonts) isolated by Percoll density gradient centrifugation. The small amount of metabolic enzymes in the intracellular stages was not due to their leakage, degradation, or poor extraction. In the same samples of spores and merogonial plus early sporogonial stages, we have detected commensurable amounts of the Sec13 subunit of the COPII coat (3, 8), two bands recognized by anti-Hsp70 Abs (present study), and SNARE protein syntaxin (our unpublished data). Thus, expression patterns of the enzymes of core carbon metabolism and other housekeeping proteins appear to be different in the life cycle of microsporidia. During their intracellular development, parasites completely rely on the metabolic system of the infected host cells, using numerous transporters, like unique ATP/ADP translocases (12, 27). After formation of thick, complex walls, microsporidial spores should exploit their own ATP-generating system to provide long-term survival (up to several years) in the environment and invasion of new host cells. Besides, in the absence of a citric acid cycle and ATP citrate lyase, microsporidial glycolytic enzymes, PDH, and acetyl-coenzyme A (CoA) synthetase may be responsible for the synthesis of the nucleocytosolic pool of acetyl-CoA, an important compound for many metabolic reactions and protein acetylation. For example, acetylation of histones may activate chromatin transcription (25) in germinating spores, and the acetylation of structural proteins may be important for their assemblage (9).
The finding of two components of an alternative respiratory pathway and the mitosomal marker mitHsp70 in A. locustae spores allowed us to describe mitosomes in a microsporidial species from phylogenetic clade II (Aquasporidia) (32) and to demonstrate mitosomal localization of AOX and mitG3PDH. Immunolocalization of two components of alternative respiratory chain “mitG3PDH-ubiquinone Q pool—AOX” in the mitosomal membrane has confirmed that mitochondrial remnants maintain their role in the energy metabolism of microsporidia. However, in contrast to mitHsp70, both enzymes were not found in merogonial and early sporogonial stages of intracellular development. It suggests that the composition and functions of microsporidial mitosomes undergo conspicuous changes throughout the life cycle. Cyclic changes in the metabolism and structure of mitochondria take place in life cycles of many parasites. Since such unique properties of parasite mitochondria provide promising targets for chemotherapy (14), further investigation of microsporidial mitosomes could be prospective.
Finally, we should stress that physiological features are variable inside the microsporidial group and mentioned considerations do not concern species from clade IV (Terresporidia). First, evidence of a peculiar metabolism in terrestrial microsporidia was obtained by Undeen and Vander Meer (29). Those authors demonstrated that spore germination in aquatic microsporidia Edhazardia aedis (clade I, Aquasporidia), Vavraia culicis (clade III, Marinosporidia), and Anncaliia (Brachiola and Nosema) algerae (clade V, Aquasporidia) was accompanied by the loss of trehalose with a concomitant increase in glucose/fructose concentration. Meanwhile, no changes in sugar content were observed with germinating spores of six terrestrial microsporidia from clade IV, including species Vairimorpha necatrix, Vairimorpha lymantriae, Nosema disstriae, and Nosema apis. The finding of alternative oxidase genes in genera Trachipleistophora, Glugea, Spraguea (clade III, Marinosporidia), and Antonospora (clade II, Aquasporidia), alongside their absence in all sequenced genomes of parasites from clade IV (genera Nosema, Encephalitozoon, and Enterocytozoon), confirmed this originality (35). Though the genomes of Encephalitozoon and Nosema species contain both components of the glycerol phosphate shuttle, E. cuniculi mitG3PDH was not recognized by the import machinery of yeast mitochondria (5) (in contrast to the A. locustae enzyme) and its mitosomal localization was not confirmed by immunofluorescent assay (34). Moreover, most genes for core carbon metabolism were not found in the genome of human parasite E. bieneusi (2, 13).
The rationale for these discrepancies as well as their role in carbon metabolism in microsporidia physiology are still not clear. The presence of an alternative respiratory chain in spores of aquatic species suggests their oxygen requirement and some exchange with the water environment. The transition of the parasite to terrestrial life (the infecting of terrestrial hosts) and the long adaptation of spores to dry-air conditions might result in the loss of alternative respiration, slowing down spore metabolism (trehalase activity) and the enlargement of the ubiquinone pool by its redistribution from mitosomes to other spore membranes. On the one hand, this speculation conforms to low-trehalase activity in terrestrial microsporidia (29) and the relocalization of E. cuniculi mitG3PDH outside the mitosomes (34). On the other hand, AOX-containing spores of A. locustae retain infectivity in spite of long storage in a dry wheat bran formulation (grasshopper suppression agent Nolo Bait). These findings suggest that adaptations to terrestrial hosts could occur independently in different microsporidial groups and sometimes be convergent. We did not find any reduction of trehalose content or increase of glucose concentration in spores of the cricket Gryllus bimaculatus parasite Paranosema grylli (clade II, Aquasporidia) during discharge of their polar tubes (7). In this respect, P. grylli seems to be close to terrestrial microsporidia from clade IV. Interestingly, the activity of the P. grylli trehalose-hydrolyzing enzyme (α,α-trehalase) has an acid pH optimum (7), as was found for aquatic microsporidium A. algerae from clade V (28). At the same time, the pH optimum of homologous enzyme from terrestrial parasite N. apis (clade IV) is neutral (30).
ACKNOWLEDGMENTS
We thank Y. Y. Sokolova and Y. S. Tokarev for critical reading of the manuscript.
We acknowledge financial support from the Russian Foundation of Basic Research (RFBR N 08-04-01358).
Footnotes
Published ahead of print on 4 February 2011.
REFERENCES
- 1. Abrahamsen M. S., et al. 2004. Complete genome sequence of the apicomplexan Cryptosporidium parvum. Science 304:441–445 [DOI] [PubMed] [Google Scholar]
- 2. Akiyoshi D. E., et al. 2009. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 5:e1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beznoussenko G. V., et al. 2007. Analogs of the Golgi complex in microsporidia: structure and avesicular mechanisms of function. J. Cell Sci. 120:1288–1298 [DOI] [PubMed] [Google Scholar]
- 4. Bradford M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 5. Burri L., Williams B. A. P., Bursac D., Lithgow T., Keeling P. J. 2006. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc. Natl. Acad. Sci. U. S. A. 103:15916–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dolgikh V. V., et al. 2009. Heterologous expression of pyruvate dehydrogenase E1 subunits of the microsporidium Paranosema (Antonospora) locustae and immunolocalization of the mitochondrial protein in amitochondrial cells. FEMS Microbiol. Lett. 293:285–291 [DOI] [PubMed] [Google Scholar]
- 7. Dolgikh V. V., Semenov P. B. 2003. Trehalose catabolism in microsporidia Nosema grylli spores (rus.). Parazitologiia 37:333–342 [PubMed] [Google Scholar]
- 8. Dolgikh V. V., Senderski I. V., Pavlova O. A., Beznoussenko G. V. 2010. Expression of vesicular transport genes in avisicular cells of microsporidia Paranosema (Antonospora) locustae. Cell Tissue Biol. 4:136–142 [Google Scholar]
- 9. Hammond J. W., Cai D., Verhey K. J. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20:71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hjort K., Goldberg A. V., Tsaousis A. D., Hirt R. P., Embley T. M. 2010. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janssen M. J., et al. 2002. Photolabeling identifies an interaction between phosphatidylcholine and glycerol-3-phosphate dehydrogenase (Gut2p) in yeast mitochondria. Biochemistry 41:5702–5711 [DOI] [PubMed] [Google Scholar]
- 12. Katinka M. D., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:450–453 [DOI] [PubMed] [Google Scholar]
- 13. Keeling P. J., et al. 2010. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol. Evol. 2:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kita K., Miyadera H., Saruta F., Miyoshi H. 2001. Parasite mitochondria as a target for chemotherapy. J. Health Sci. 47:219–239 [Google Scholar]
- 15. Liou W., Geuze H. J., Slot J. W. 1996. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106:41–58 [DOI] [PubMed] [Google Scholar]
- 16. Miroux B., Walker J. E. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 17. Mogi T., Kita K. 2010. Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol. Int. 59:305–312 [DOI] [PubMed] [Google Scholar]
- 18. Regoes A., et al. 2005. Protein import, replication, and inheritance of a vestigial mitochondrion. J. Biol. Chem. 280:30557–30563 [DOI] [PubMed] [Google Scholar]
- 19. Seleznev K. V., et al. 1995. Fractionation of different life cycle stages of microsporidia Nosema grylli from crickets Gryllus bimaculatus by centrifugation in Percoll density gradient for biochemical research. J. Eukaryot. Microbiol. 42:288–292 [Google Scholar]
- 20. Slot J. W., Geuze H. J. 1985. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur. J. Cell Biol. 38:87–93 [PubMed] [Google Scholar]
- 21. Slot J. W., Geuze H. J., Gigengack S., Lienhard G. E., James D. E. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sluse F. E., Jarmuszkiewicz W. 1998. Alternative oxidase in the branched mitochondrial respiratory network: an overview on structure, function, regulation, and role. Braz. J. Med. Biol. Res. 31:733–747 [DOI] [PubMed] [Google Scholar]
- 23. Stechmann A., et al. 2008. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr. Biol. 18:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Steinbüchel A., Müller M. 1986. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 20:57–65 [DOI] [PubMed] [Google Scholar]
- 25. Sterner D. E., Berger S. L. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tovar J., Fischer A., Clark C. G. 1999. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 32:1013–1021 [DOI] [PubMed] [Google Scholar]
- 27. Tsaousis A. D., et al. 2008. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453:553–556 [DOI] [PubMed] [Google Scholar]
- 28. Undeen A. H., ElGazzar L. M., Vander Meer R. K., Narang S. 1987. Trehalose levels and trehalase activity in germinated and ungerminated spores of Nosema algerae (Microspora: Nosematidae). J. Invertebr. Pathol. 50:230–237 [Google Scholar]
- 29. Undeen A. H., Vander Meer R. K. 1999. Microsporidian intrasporal sugars and their role in germination. J. Invertebr. Pathol. 73:294–302 [DOI] [PubMed] [Google Scholar]
- 30. Vandermeer J. W., Gochnauer T. A. 1971. Trehalase activity associated with spores of Nosema apis. J. Invertebr. Pathol. 17:38–41 [DOI] [PubMed] [Google Scholar]
- 31. Vávra J. 2005. “Polar vesicles” of microsporidia are mitochondrial remnants (“mitosomes”)? Folia Parasitol. 52:193–195 [PubMed] [Google Scholar]
- 32. Vossbrinck C. R., Debrunner-Vossbrinck B. A. 2005. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol. 52:131–142 [DOI] [PubMed] [Google Scholar]
- 33. Webster P. 1999. The production of cryosection through fixed and cryoprotected biological material and their use in immunocytochemistry, p. 49–75In Hajibagheri N. (ed.), Methods in molecular biology: electron microscopy method and protocol, vol. 117 Numana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 34. Williams B. A., Cali A., Takvorian P. M., Keeling P. J. 2008. Distinct localization patterns of two putative mitochondrial proteins in the microsporidian Encephalitozoon cuniculi. J. Eukaryot. Microbiol. 55:131–133 [DOI] [PubMed] [Google Scholar]
- 35. Williams B. A., et al. 2010. A broad distribution of the alternative oxidase in microsporidian parasites. PLoS Pathog. 6:e1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams B. A. P., Hirt R. P., Lucocq J. M., Embley T. M. 2002. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418:865–869 [DOI] [PubMed] [Google Scholar]
- 37. Xu P., et al. 2004. The genome of Cryptosporidium hominis. Nature 431:1107–1112 [DOI] [PubMed] [Google Scholar]