Abstract
The yeast-hypha transition is an important virulence trait of Candida albicans. We report that the AGC kinase Sch9 prevents hypha formation specifically under hypoxia at high CO2 levels. sch9 mutants showed no major defects in growth and stress resistance but a striking hyperfilamentous phenotype under hypoxia (<10% O2), although only in the presence of elevated CO2 levels (>1%) and at temperatures of <37°C during surface growth. The sch9 hyperfilamentous phenotype was independent of Rim15 kinase and was recreated by inhibition of Tor1 kinase by rapamycin or caffeine in a wild-type strain, suggesting that Sch9 suppression requires Tor1. Caffeine inhibition also revealed that both protein kinase A isoforms, as well as transcription factors Czf1 and Ace2, are required to generate the sch9 mutant phenotype. Transcriptomal analyses showed that Sch9 regulates most genes solely under hypoxia and in the presence of elevated CO2. In this environment, Sch9 downregulates genes encoding cell wall proteins and nutrient transporters, while under normoxia Sch9 and Tor1 coregulate a minor fraction of Sch9-regulated genes, e.g., by inducing glycolytic genes. Other than in Saccharomyces cerevisiae, both sch9 and rim15 mutants showed decreased chronological aging under normoxia but not under hypoxia, indicating significant rewiring of the Tor1-Sch9-Rim15 pathway in C. albicans. The results stress the importance of environmental conditions on Sch9 function and establish a novel response circuitry to both hypoxia and CO2 in C. albicans, which suppresses hypha formation but also allows efficient nutrient uptake, metabolism, and virulence.
INTRODUCTION
Candida albicans is a normal inhabitant of human mucosal surfaces, but lowering of immune defenses can cause tenacious localized and severe systemic fungal disease. C. albicans parasitism depends on its ability to change cellular morphologies, especially to transform its spherical yeast to a filamentous true hyphal form (dimorphism). During recent years numerous regulators of the yeast-hypha transition have been described that are required to regulate dimorphism in general or only under specific environmental conditions (reviewed in reference 8). Under normoxia a main signaling pathway of hypha formation is triggerd by molecules present in serum, such as muramyl peptides, and by carbon dioxide (20, 43). This pathway involves an increased cyclic AMP (cAMP) level that relieves the repression of protein kinase A (PKA) catalytic subunits Tpk1 and Tpk2 (3, 35), which in turn activate downstream transcriptional regulators, including, as the global regulator, Efg1. Efg1 and Efg1-dependent transcription factors regulate expression of target genes required for hyphal morphogenesis, e.g., by inactivating negative regulators Tup1 and Nrg1 and by recruiting histone-modifying activities that close or open up chromatin structures (23, 39). Under hypoxia, the ability of C. albicans to form hyphae is effectively repressed (4, 35). Interestingly, some positive regulators of hypha formation under normoxia, including Efg1 and Flo8, are also needed as repressors of filamentation under hypoxia (6, 10, 33, 36). Past results (18) and data presented here indicate that relief of repression rather than induction is a crucial mechanism that allows expression of hypha-specific genes under several environmental conditions.
The PKA isoforms Tpk1 and Tpk2 are members of the so-called AGC group of kinases in eukaryotes, including PKA, PKB, PKC, and PKG, as well as phosphoinosite-dependent kinase (PDK) and ribosomal protein S6 kinase (reviewed in reference 17). AGC proteins have a common domain organization consisting of a central catalytic domain flanked by an N-terminal lipid-binding domain of pleckstrin homology and a C-terminal regulatory domain containing a conserved hydrophobic sequence that can be phosphorylated by an upstream kinase. Fungal AGC kinases respond primarily to the availability of sugars and nitrogen. An example is the PKB protein Sch9 in the yeast Saccharomyces cerevisiae (homolog of Akt in mammalian cells), which in rich media is phosphorylated and thereby activated by the target of rapamycin, TORC1 (40). Both Sch9 and TORC1 proteins colocalize to the vacuolar membrane, presumably to sense and/or to mobilize intracellular nutrients (32). In S. cerevisiae, sch9 mutants form smaller colonies and grow at a lower rate (because of a prolonged G1 phase), presumably because Sch9 is a positive regulator of transcription, especially of genes encoding ribosomal proteins and genes involved in nitrogen metabolism (9, 16, 42). Recent results indicate that Tor1 in C. albicans has a similar function but also regulates additional genes, including genes encoding adhesins (2). In S. cerevisiae Sch9 was also found to be required for hyperosmotic stress resistance by direct transcriptional activation of osmostress-responsive genes (27). On the other hand, under nutrient-rich conditions, Sch9 was found to collaborate with PKA to reduce stress sensitivity by blockage of the AGC kinase Rim15 that activates transcription factors Gis1 and Msn2/4, driving expression of stress protection genes (5, 28). In sch9 mutants or under nutrient-poor conditions, i.e., low Sch9 activity, these transcription factors are activated via Rim15 and increase stress protection and significantly prolong the chronological life span of yeast (12, 41).
In C. albicans, a homolog of S. cerevisiae Sch9 has been partially characterized, and it was shown that it is required for efficient growth, stress resistance, and filamentation under certain conditions (22, 25). Here we describe additional functions of the C. albicans Sch9 homolog and show that it is a component of a regulatory circuit sensing oxygen and CO2. Specifically, Sch9 is a positive regulator of yeast growth, in all environments, while under hypoxia and high carbon dioxide levels it strongly represses hypha formation. We demonstrate that transcriptomal effects of Sch9 are hypoxia and CO2 dependent and that other Sch9 functions, including its contribution to antifungal resistance and longevity, are dependent on this gaseous environment.
MATERIALS AND METHODS
Strains and growth conditions.
C. albicans strains are listed in Table 1. Strains were grown in yeast extract-peptone-dextrose (YPD) medium, YPS medium (1% yeast extract, 2% peptone, 2% sucrose), or in supplemented SD minimal medium at 30°C (33); solid media contained, in addition, 2% agar. Hypoxic growth under defined gaseous conditions was carried out using an Invivo200 hypoxia chamber (Ruskinn) with attached flasks of nitrogen, CO2, and compressed air (36). Liquid media were preequilibrated overnight under hypoxia/CO2 conditions before inoculation. Transformation of C. albicans strains was carried out using the spheroplast method as described previously (33). For hyphal induction in liquid, the strains were grown at 37°C in YP medium with 20% horse serum. To induce chlamydospore formation, cells were streaked out lightly on chlamydospore induction medium (cornmeal agar [Merck]–0.33% Tween 80), covered by a coverslip, and incubated for 5 days at 20°C (34).
Table 1.
C. albicans strains used in the study
| Strain | Genotype | Reference |
|---|---|---|
| CAF2-1 | URA3/ura3::imm434 | 13 |
| CAI4 | ura3::imm434/ura3::imm434 | 13 |
| CAS4 | As CAI4 but sch9::hisG/sch9::hisG | This work |
| CCS3 | As CAS4 but URA3/ura3::imm434 | This work |
| CCS4 | As CAS4 but LEU2/leu2::[ACT1p-SCH9 URA3] | This work |
| CCS5 | As CAS4 but leu2::[URA3] | This work |
| CAR23-7-5-1 | As CAI4 but rim15::hisG/rim15::hisG | This work |
| CCS6 | As CAR23-7-5-1 but URA3/ura3::imm434 | This work |
| AF1002 | As CAI4 but sch9::hisG/sch9::hisG-URA3-hisG rim15::hisG/rim15::hisG | This work |
| IIHB6 | As CAI4 but tpk1Δ::hisG-URA3-hisG//tpk1Δ::hisG | 3 |
| HPY300U (tpk1mut) | As CAI4 but tpk1Δ::hisG/tpk1Δ::hisG URA3/ura3::imm434 | 26 |
| TPO7.4 | As CAI4 but tpk2Δ::hisG/tpk2Δ::hisG-URA3-hisG | 35 |
| HPY400U (tpk2mut) | As CAI4 but tpk2Δ::hisG/tpk2Δ::hisG URA3/ura3::imm434 | 26 |
| JLC19 | As CAI4 but cph1Δ::hisG/cph1Δ::hisG-URA3-hisG | 21 |
| CKY230 | As CAI4 but czf1Δ::hisG/czf1Δ | 4 |
| MK106 | ace2Δ::FRT/ace2Δ::FRT | 19 |
Construction of sch9 and rim15 mutants.
Both alleles of SCH9 (ORF19.829/ORF19. 8449) were deleted in strain CAI4 by using the URA blaster method (13). A 1.0-kb sequence upstream of the open reading frame (ORF) was amplified by genomic PCR using primers CaSCH9-dis1/for and -rev; likewise, a 0.55-kb fragment of the 3′-unstranslated region (3′-UTR) was amplified using CaSCH9disII-for and -rev. Oligonucleotides used for PCR in this paper are listed in Table S1 of the supplemental material. Using BamHI/BglII sites at the fragment ends of both fragments were inserted into the BamHI/BglII sites flanking the hisG-URA3-hisG cassette in p5921. The BamHI/BglII 5.6-kb fragment of the resulting plasmid p5921-SCH9dis was used to sequentially disrupt both SCH9 alleles as described previously (13). Correct positioning of the SCH9 disruption cassette was verified by colony PCR and by genomic Southern blotting assays (data not shown).
In a similar approach, both alleles of RIM15 (ORF19.7044) were deleted in strain CAI4. A 1.1-kb fragment upstream of the ORF was amplified by genomic PCR using primers CaRIM15disI-for and -rev; likewise, a 1.1-kb fragment partially encompassing 500 bp of the ORF and 500 bp of the 3′-UTR was amplified using CaRIM15disII-for and -rev. Both flanking fragments were inserted into p5921 as described above. The BamHI/BglII 6.2-kb fragment of the resulting plasmid pCaRIM15/Blaster was then used to sequentially disrupt both RIM15 alleles, and correct integration of the disruption cassette was verified by genomic Southern blot assays (data not shown). The resultant strain was named CAR23-7-5-1. In addition, a sch9 rim15 double mutant strain (AF1002) was constructed by disrupting SCH9 in the background of strain CAR23-7-5-1, as described above.
Because the presence of URA3 at ectopic loci may influence C. albicans phenotypes, we reconstituted URA3 at its authentic locus in sch9 and rim15 mutants, as described previously (29). The resulting prototrophs were named CCS3 (sch9 mutant) and CCS6 (rim15 mutant).
Construction of SCH9 and RIM15 expression plasmids.
The SCH9 ORF was PCR amplified using genomic DNA of strain CAI4 using primers CaSCH9/BHI/for and -rev, which generate BamHI and BglII sites flanking the ORF. The 2.4-kb SCH9 ORF fragment was inserted into the BglII site downstream of the PCK1 promoter in pBI-1 (37) to generate pBI-CaSCH9 and downstream of the ACT1 promoter in pYW105 to construct pYW-CaSCH9. Both expression vectors also contain URA3 as a selection marker and the CaARS replicator. Likewise, the RIM15 ORF was amplified primers RIM15-Start and -Stop, which generate flanking BamHI sites. The 5.8-kb BamHI fragment was also cloned downstream of the ACT1 promoter in plasmid pDS1044-I to generate p2297R1.
Transcriptome of sch9 mutant.
Strains CAF2-1 (SCH9/SCH9) and CCS3 (sch9/sch9) were grown at 25°C in liquid YPS medium under an atmosphere of (i) air, (ii) 0.2% O2 (no CO2), or (iii) 0.2% O2 plus 6% CO2. Three independent cultures were used for each strain to isolate total RNA in the exponential growth phase and generate Cy3- or Cy5-labeled cDNAs as described previously (10). Labeled cDNAs representing wild-type and mutant transcripts were hybridized to genome-wide C. albicans microarrays (Eurogentec) representing all ORFs in duplicate, as described previously (10). Spot intensities were evaluated using program GenePix 6.0 for background substraction and GeneSpring 10.2 for normalization (LOWESS) and determination of the mean values (ratios of transcript level in the sch9 mutant relative to the SCH9 strain). Results are available at ArrayExpress under accession number E-MEXP-2407.
Virulence tests.
Female, 8- to 10-week-old, outbred CD1 mice were obtained from Harlan Nossan Laboratories (Milan, Italy) and housed at the Animal Facilities of the University of Perugia, Perugia, Italy. Procedures involving animals and their care were conducted in conformity with national and international laws and policies. For systemic infection, mice were inoculated intravenously on day zero with C. albicans CAF2-1, CCS3, or CCS4 strains (1 × 106/mouse in 0.5 ml saline). Infected animals were monitored daily for survival. The experiments were repeated 3 times by using 4 animals/experimental group. Differences between survival data for the groups were analyzed by using the Mann-Whitney U test.
RESULTS
sch9 and rim15 mutants.
In S. cerevisiae the Tor1-Sch9-Rim15 kinase cascade regulates various functions, including metabolism, stress response, and longevity (reviewed in reference 31). The B-type kinase Sch9 is known to repress the activity of Rim15, which positively regulates stress responses and chronological life span (38). The C. albicans genome contains genes (ORF19.829 and ORF19.7044) encoding close homologs of Sch9 and Rim15 (1). CaSch9 and ScSch9 have 66% overall identity, which rises to near-complete identity in the kinase catalytic domain; Rim15 homologs in both species share 35% overall identity, which increases to 60% between residues 1263 and 1408 in the kinase domain (see Fig. S1 in the supplemental material). Using the URA blaster protocol we disrupted both alleles of SCH9 and RIM15 in C. albicans CAI4. In the resulting homozygous mutants we reinserted the URA3 selection marker at its authentic locus to construct strains with an identical URA3 status as the control strain, CAF2-1 (7). Furthermore, mutant strains were reconstituted by integrating plasmids carrying the respective wild-type SCH9 or RIM15 ORFs.
Filamentation phenotypes of the sch9 mutant.
In previous reports the sch9 mutant was shown to be completely defective in chlamydospore formation (25) and partially defective in growth and filamentation (22). We reassessed these phenotypes by using the URA3 reconstituted sch9 mutant CCS3 and the SCH9 URA3 reconstituted mutant CCS4. As described previously (22), colony sizes of the sch9 mutant on YPD agar were reduced and the doubling time of mutant CCS3 was increased to 1.65 h, compared to 1.2 h of the CAF2-1 control strain in YPD medium at 30°C. Standard plate tests using Lee's, Spider, or serum medium showed a slight delay, which presumably was related to reduced growth, but no major defects in hypha formation at 37°C (data not shown).
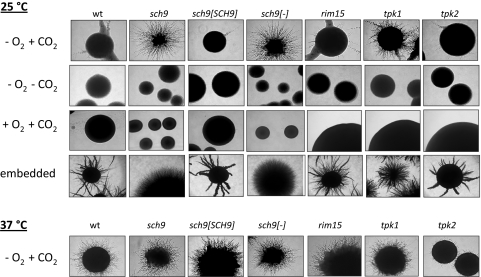
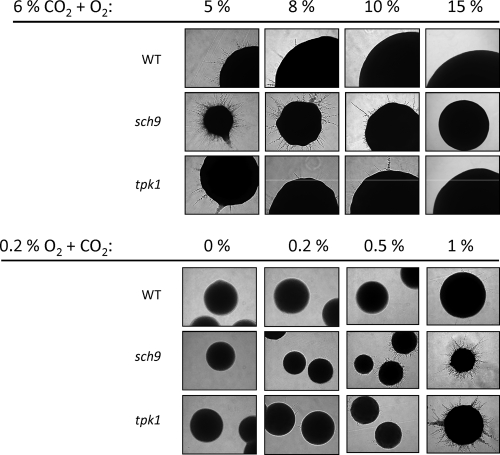
The sch9 mutant developed slighly smaller colonies than the control strain under hypoxia (0.2% O2) on YPS medium at 25°C and generated few or no hyphae emanating from colonies. However, strikingly, the presence of a high CO2 concentration under hypoxia (0.2% O2, 6% CO2) led to strongly increased filamentous growth of the sch9 mutant but not of the control strain or the SCH9 reconstituted mutant CCS4 (Fig. 1 A, top panel). Hyperfilamentation of the sch9 mutant was also observed during YPS-embedded growth, a condition presumably lowering oxygen and increasing CO2 levels (Fig. 1, top panel). To assess the threshold O2 and CO2 concentrations for this phenotype we carried out experiments at various concentrations of both gases. Under high CO2 (6%), hypha formation of the sch9 mutant declined gradually with increasing O2 concentrations and was almost completely absent at 10% O2 (Fig. 2, top). At low O2 levels (0.2% O2), hypha formation started at 0.5% CO2, and it was clearly apparent at 1% CO2 (Fig. 2, bottom). Unexpectedly, the ability of the sch9 mutant to hyperfilament under hypoxia at elevated CO2 levels was strongly influenced by temperature. At 37°C filamentation of the sch9 mutant was moderate and equaled the control and SCH9 reconstituted strains under all gaseous conditions (Fig. 1, bottom panel, shows results for the hypoxia–high-CO2 condition). We conclude that temperatures of <37°C, O2 concentrations of <10%, and CO2 concentrations of >1% trigger strong hypha formation of the sch9 mutant. Conversely, it follows that C. albicans wild-type cells use Sch9 to effectively suppress hypha formation under these growth conditions. Opposite to these results, filamentation of mutants lacking the regulator Efg1 was greatly induced under hypoxia, even in the absence of CO2 (33).
Fig. 1.
Colony phenotypes of C. albicans strains. Wild-type and mutant strains were grown on YPS agar for 3 days (25°C) or 2 days (37°C), and colonies were photographed. Strains were grown in an atmosphere of 0.2% O2 containing 6% CO2 (- O2 + CO2) or lacking CO2 (- O2 - CO2) or under 20% oxygen (normoxia) containing 6% CO2 (+ O2 + CO2). In addition, cells were grown embedded in YPS agar under air. Strains used were CAF2-1 (wild-type [wt]), CCS3 (sch9), CCS4 (sch9[SCH9]), CCS5 (sch9[-]), CCS6 (rim15), IIHB6 (tpk1), and TPO7.4 (tpk2).
Fig. 2.
Oxygen- and CO2-dependent filamentation. Strains CAF2-1 (wt), CCS3 (sch9), and IIHB6 (tpk1) were grown at 25°C on YPS agar either under 6% CO2 and the indicated concentrations of oxygen or under 0.2% oxygen and the indicated concentrations of CO2. Colony phenotypes were photographed after 3 days of growth.
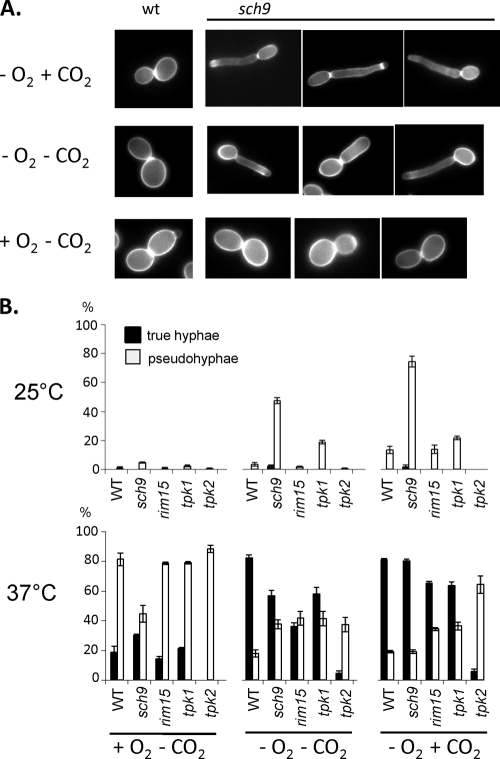
In liquid YPS medium, a particular sch9 phenotype was observed under hypoxia, since cells formed pseudohyphae while the control strain grew in yeast form (Fig. 3 A). This phenotype occurred irrespective of CO2 levels at 25°C under hypoxia but not under normoxia. At 37°C, under any gaseous condition, no major differences in filamentation were observed, since both the sch9 mutant and the control strain formed true hyphae, as well as pseudohyphae (Fig. 3B). Thus, in liquid media Sch9 suppresses pseudohypha formation under hypoxia. Comparison of sch9 mutant phenotypes during growth in liquid or on agar suggests that surface contact is yet another parameter influencing the activity of the Sch9 protein.
Fig. 3.
Dependence of cellular morphologies on hypoxia and CO2. (A) Cells of sch9 mutant CCS3 and control strain CAF2-1 (wild type [wt]) were grown in liquid YPS for 4 h at 25°C. Cells were grown for 4 h in low oxygen (0.2% O2) containing or not containing 6% CO2. Chitin was stained with calcofluor white (2 μg/ml), and cells were inspected by fluorescence microscopy. (B) Distribution of cell forms in various strains. Cells were grown as for panel A, and at least 100 cells were classified according to their cell form. Percentages of pseudohyphal and true hyphal forms are indicated by the white and black bars, respectively, while the remaining percentage is due to yeast-form cells. For strain designations, see Fig. 1.
Derepressed filamentation of the sch9 mutant does not require Rim15.
Because of the Tor1-Sch9-Rim15 cascade known in S. cerevisiae, we assumed that elevated activity of Rim15 was the cause for elevated filamentation of the sch9 mutant. To clarify this notion we constructed rim15 single and rim15 sch9 double mutants and examined their phenotypes.
In S. cerevisiae, Sch9 represses the function of Rim15 to activate genes involved in the stress response, sporulation, transition into stationary phase, and longevity (28). Unexpectedly, the C. albicans rim15 mutant CCS6 did not show any significant phenotypic differences compared to the control strain CAF2-1 or the reconstituted mutant CAR23-7-5-1[p2297R1]. No phenotypic abnormalities were observed with respect to growth rate, budding index in stationary phase, heat shock (53°C) sensitivity, osmotolerance, glycogen accumulation, or sensitivity to several antifungal agents; furthermore, hyphal development in liquid or on solid media with the inducers serum and GlcNAc was similar in rim15 and wild-type strains (data not shown). The only slight mutant phenotype arose during growth under hypoxia (with or without CO2), where filamentation was slightly increased in the rim15 mutant compared to control strains (Fig. 1, top panel). We conclude that Rim15 in C. albicans has a significantly different function than its homolog in S. cerevisiae and that it does not have a major role in growth or hypha formation under normoxia or hypoxia. Importantly, the sch9 rim15 mutant strain showed an identical phenotype under hypoxia and elevated CO2 as the sch9 single mutant, i.e., vigorous hypha formation (Fig. 4 A). Thus, released activity of Rim15 in cells lacking Sch9 is not the reason for the hyperfilamentous phenotype.
Fig. 4.
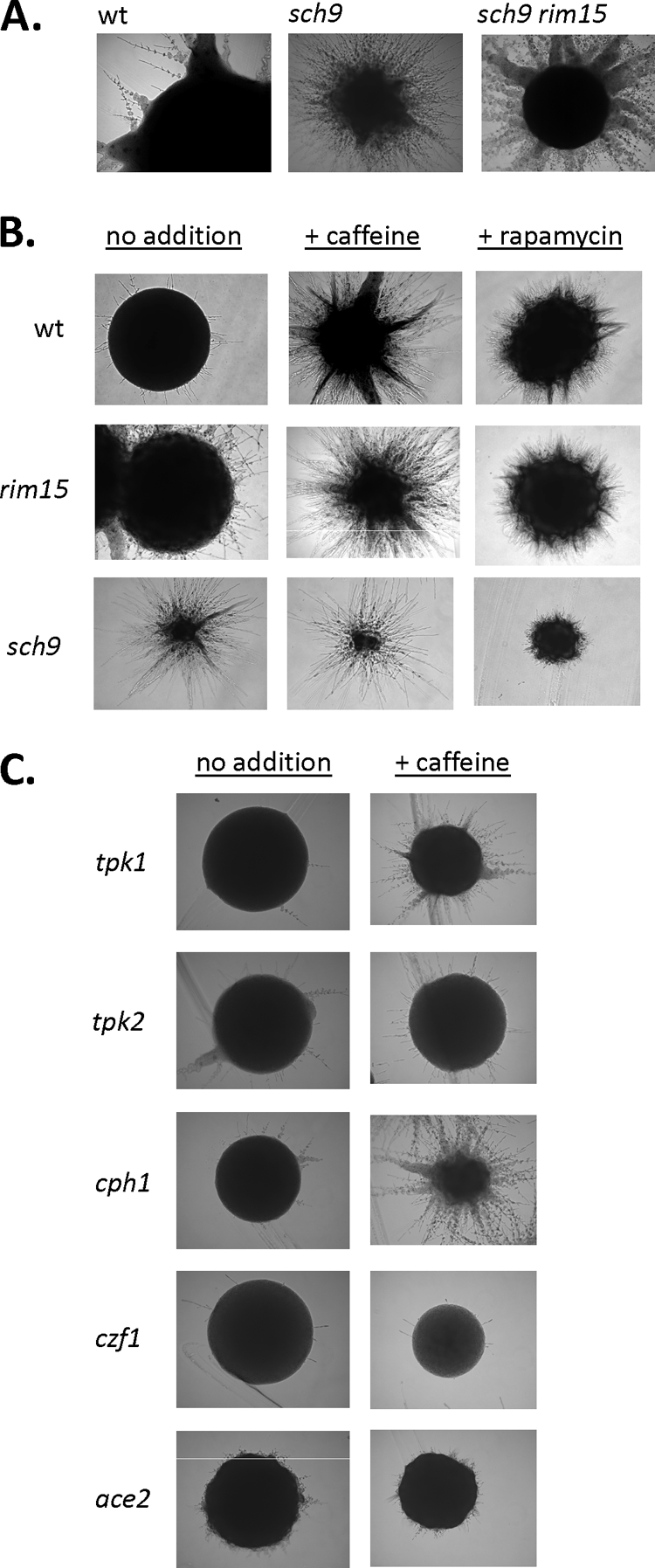
Hypoxic filamentation by C. albicans mutants. Single colonies of strains were grown on YPS agar at 25°C under an atmosphere of 0.2% O2 and 6% CO2; colonies were photographed after 3 days of growth. (A) Control strain CAF2-1, sch9 mutant CCS3, and sch9 rim15 double mutant AF1002 were compared. (B and C) Strains were grown on YPS medium without additions or in YPS medium containing 2 nM rapamycin or 1 mM caffeine. Strains included control strain CAF2-1, rim15 mutant CCS6, sch9 mutant CCS4, tpk1 mutant HPY300U, tpk2 mutant HPY400U, cph1 mutant JLC19, czf1 mutant CKY230, and ace2 mutant MK106.
Tor1 inhibition generates sch9 mutant-like hyperfilamentation.
Tor1 activity is inhibited by rapamycin and caffeine (2, 16, 41). Because Tor1 functions upstream of Sch9, we asked if Tor1 inhibition can generate a sch9 mutant phenotype in wild-type strains. This was indeed the case, since both rapamycin and caffeine induced strong hypha formation in the control strain and the rim15 mutant (Fig. 4B). Hyperfilamentation of the sch9 mutant was not stimulated further by either of these inhibitors, suggesting that Tor1 and Sch9 function in the same pathway. Importantly, rapamycin/caffeine-induced filamentation only occurred under conditions known to reveal the sch9 mutant hyperfilamentation phenotype, i.e., during hypoxic growth with elevated levels of CO2 (data not shown). Thus, these results indicate that the Tor1-Sch9 pathway strongly suppresses filamentation of C. albicans under specific environmental conditions.
Since Tor1 inhibition was able to generate a sch9 mutant-like phenotype, we were able to conveniently test if other suspected regulators of hypoxic filamentation are required for this phenotype. In both tpk1 and tpk2 mutants, hyperfilamentation under hypoxia and high CO2 was greatly reduced compared to the control strain or the cph1 mutant known to be filamentation defective on agar surfaces (21) (Fig. 4C). Likewise, in the czf1 and ace2 mutants, known to be defective in hypoxic filamentation (4, 14, 19, 24), caffeine was unable to trigger hyperfilamentation. These results indicate that Tpk1, Tpk2, Czf1, and Ace2 allow hypha formation under hypoxia and elevated CO2, if repression by the Tor1-Sch9 pathway does not occur.
Antifungal resistance and longevity of sch9 and rim15 mutants.
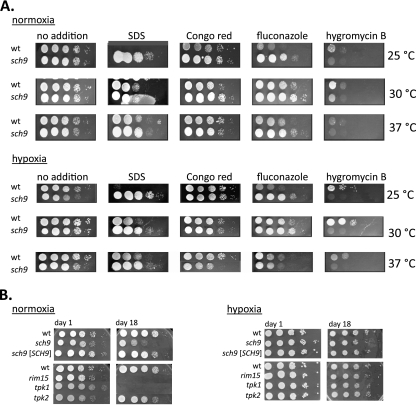
Standard drop dilution tests were performed to evaluate the sensitivity of the sch9 mutant CAS4 and the control strain CAI4 in different oxygen and temperature environments (Fig. 5 A). The sch9 mutant was significantly more resistant to SDS and fluconazole than the control strain, especially at 25°C. In turn, the sch9 mutant was significantly more sensitive than the control strain to hygromycin B, especially under hypoxia. We conclude that lack of Sch9 leads to increased or reduced sensitivity, depending on the type of antifungal agent. Furthermore, we note that the oxygen level and temperature have significant effects on antifungal susceptibilities.
Fig. 5.
Environment-dependent sensitivities of the sch9 mutant. (A) Sensitivities to antifungals. Strains were grown in air (normoxia) or under hypoxia (99.9% nitrogen) at the indicated temperatures. Five-microliter drops of 10-fold dilutions containing 105 to 101 cells of strain CAI4 (wild type [wt]) and sch9 mutant CAS4 were spotted on YPD agar containing 0.06% SDS, 200 μg/ml Congo red, 5 μg/ml fluconazole, or 200 μg/ml hygromycin B. (B) Longevity experiment. Strains were grown in liquid YPS medium at 25°C under normoxia or under hypoxia (0.2% O2, 6% CO2) for 1 day or 18 days. Viability of cells after this time was assayed by a drop dilution test as for panel A. Strains tested were control strain CAF2-1, sch9 mutant CCS5, reconstituted sch9 mutant CCS4, rim15 mutant CCS6, tpk1 mutant IIHB6, and tpk2 mutant TPO7.4.
The yeast S. cerevisiae has been used as a model system to study chronological cellular aging, and nutrient-rich environments were shown to reduce longevity by activating Sch9 via Tor1, which inactivates Rim15 and prevents its role in stress responses (12, 41). In spite of these antagonistic roles of Sch9 and Rim15, both C. albicans sch9 and rim15 were unable to survive for long in stationary phase in cultures grown under normoxia, compared to the control strain (Fig. 5B). Unexpectedly, however, following hypoxic growth in stationary phase, both mutants survived, similar to the control strain (Fig. 5B). This protective effect of hypoxia also occurred in the absence of elevated CO2 levels (data not shown). The results suggest that both Sch9 and Rim15 are required for longevity specifically under normoxia, while they are dispensable for survival under hypoxia. Furthermore, these results again indicate different regulatory wiring of Sch9 proteins in C. albicans and S. cerevisiae.
Transcriptome of sch9 mutant.
In S. cerevisiae Sch9 is a target of the Tor kinase (40), and inhibition of Tor1 in C. albicans by rapamycin indicated that Tor1 function is largely conserved between both fungal species (2). To explore if the functions of Tor1 are mediated via Sch9 in C. albicans, we performed transcriptomal comparisons of the sch9 mutant CCS3 and the control strain CAF2-1. These comparisons were carried out for cells growing under normoxia but also under hypoxia (with or without CO2), because the above phenotypes had suggested specific roles for Sch9 under this condition.
In all environments, levels of 310 transcripts were regulated at least 2-fold in the sch9 mutant relative to the control strain. Venn diagrams and functional categories are shown in Fig. 6. Regulated genes are listed in Table S2 of the supplemental material. Surprisingly, the majority of Sch9-dependent up- or downregulation genes were detected solely under hypoxia. For example, 33 transcripts were downregulated in the sch9 mutant only under normoxia, 104 transcripts under hypoxia, and only 7 transcripts were downregulated under both conditions (Fig. 6). Interestingly, about one-third of genes down- or upregulated by Sch9 under hypoxia depended on the presence of CO2. This finding demonstrates the function of Sch9 under hypoxia and shows its importance for CO2-dependent induction and repression of gene expression. To confirm some of the transcriptomal data, we compared levels of specific transcripts in the sch9 mutant CCS3 to the reconstituted mutant CCS4 by quantitative. The results demonstrated a largely similar Sch9-mediated environmental regulation compared to the array experiments for 3 key transcripts, including ERG11, ECE1, and RNR22, while the CZF1 and ORF19.5831 transcripts showed a different regulation (see Table S3 in the supplemental material). The aberrant result of the latter transcripts is probably due to the fact that in CCS4 a single SCH9 allele driven by the ACT1 promoter was used for complementation; thus, the different dosage and regulation of Sch9 may partially alter its transcriptomal effects.
Fig. 6.
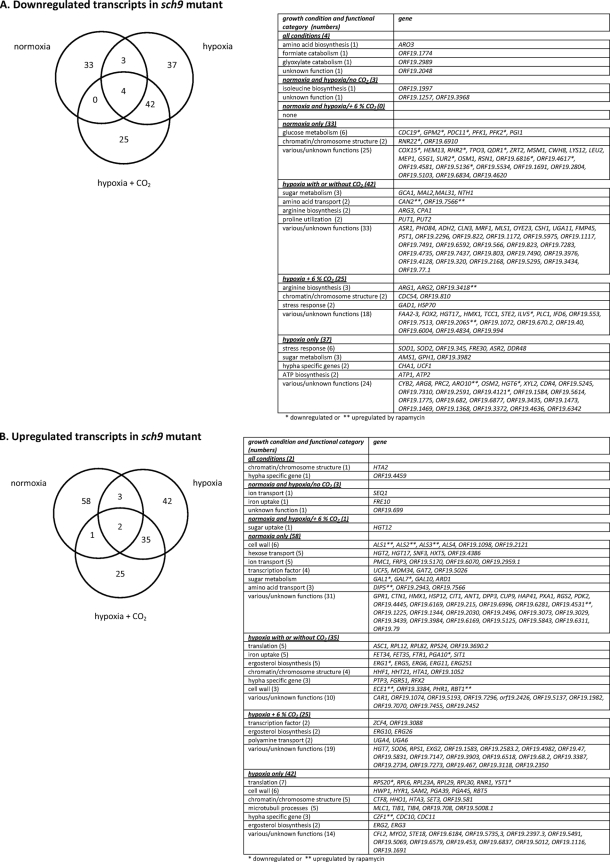
Down- and upregulated transcription in the sch9 mutant CCS3 relative to wild-type strain CAF2-1. The Venn diagrams depict numbers of genes regulated at least 2-fold under the 3 conditions (air, 0.2% O2, or 0.2% O2 plus 6% CO2); affected genes are listed below. A total of 144 and 166 transcripts were down- and upregulated, respectively, in the sch9 mutant. (A) Downregulated genes; (B) upregulated genes. Genes also regulated by rapamycin (2) are marked with asterisks (*, downregulated; **, upregulated genes).
Almost none of the genes regulated in the sch9 mutant were found to be downregulated in the presence of rapamycin, i.e., by Tor1 inactivation (2). While Tor1 activity was required to induce a large set of genes encoding components of the translational machinery, none of these genes was Sch9 dependent for expression. Twelve genes involved in translation are even repressed under hypoxia, since their levels increased in the sch9 mutant under this condition (e.g., RPS20/24, RPL6/12/23/24/30/82). The only similarity of rapamycin treatment with sch9 mutation was downregulation of genes involved in glucose metabolism (e.g., of the glycolytic genes CDC19, GPM2, PDC11, and PFK2). Sch9 deficiency also downregulated additional genes involved in sugar metabolism but only under hypoxia, e.g., the GRA1 gene encoding glucoamylase and strongly the MAL2 and MAL31 genes involved in maltose metabolism.
A total of 166 genes were upregulated in the sch9 mutant, thus reflecting a repressive function of Sch9 on these genes. The only similarities of these genes to genes regulated in rapamycin-treated cells were genes involved in cell wall structure and morphogenesis, including ALS1, ALS2, ALS3, ECE2, RBT1, and CZF1 (marked in Fig. 6). Upregulation of the latter 3 genes was observed only in hypoxic cells supporting increased pseudohyphal growth of the sch9 mutant under hypoxia. Surprisingly, Sch9 represses a large number of genes involved in metabolite transport, since genes encoding transporters for hexoses, ions, iron, amino acids, and polyamine, which were upregulated in the sch9 mutant. The suppression of transport genes may be related to the assumption that increased activity of Sch9 reflects high nutrient levels, and high-affinity uptake systems may not be needed under this condition. Furthermore, several ERG genes were upregulated in the sch9 mutant under hypoxia (ERG10 and ERG26, only in the presence of CO2). Because these genes are known to be upregulated under hypoxia (33), it appears that Sch9 dampens this response, presumably to prevent membranes from losing fluidity, which is already compromised by lowered biosynthesis of unsaturated fatty acids.
Finally, we also note that some genes were regulated oppositely in the sch9 mutant and by rapamycin treatment of the control strain, including the CAN2 permease gene and GAL1/GAL7 genes, which were down- and upregulated, respectively. We conclude that only a minor subset of genes regulated by Sch9 is also regulated via Tor1.
Virulence of the sch9 mutant.
To confirm the contribution of Sch9 to C. albicans virulence, which was suggested previously (22), we used strains containing URA3 at its native locus to exclude any effects due to extragenic URA3 localization (7). Mice were infected intravenously with control strain CAF2-1, sch9 mutant strain CCS3, or the SCH9 reconstituted strain CCS4. Median survival times of mice infected with the control CAF2-1 strain and mutant CCS3 were 5.5 days and 27 days, respectively (Table 2). Forty percent of mice infected with the mutant survived until the end of the observation period (60 days). In contrast, after inoculation of the reconstituted mutant CCS4, survival of mice was as short as that of animals infected with the control strain. Thus, we confirmed that Sch9 is essential for virulence of C. albicans. Reduced virulence appears not to be caused by impaired biofilm formation, since in a static model of biofilm formation in normoxia or hypoxia with and without CO2 (36) the sch9 mutant did not show significant defects (data not shown).
Table 2.
Virulence of C. albicans strains in a mouse model of hematogenously disseminated infection
| C. albicans straina | Relevant genotype | MSTb | D/Tc | Survival range (days) |
|---|---|---|---|---|
| CAF2-1 | SCH9/SCH9 ura3/URA3 | 5.5 | 12/12 | 2–8 |
| CCS3 | sch9/sch9 ura3/URA3 | 27* | 7/12 | 7 to >60 |
| CCS4 | sch9/sch9 ura3/ura3 LEU2/ leu2::[ACT1p-SCH9 URA3] | 7 | 12/12 | 2–10 |
CD1 mice were infected intravenously with 1 × 106 C. albicans cells.
Median survival time (in days). *, P < 0.01 (mutant versus wild-type strain CAF2-1).
Number of dead mice (D) at 60 days over the total number of animals infected (T).
DISCUSSION
The human body contains numerous niches that are low in oxygen but increased in CO2 levels that may become colonized by C. albicans and other pathogens (reviewed in reference 11). We report here that C. albicans has a high inherent propensity to filament under this condition but that this property is repressed by the Sch9 kinase. At oxygen levels of <10% O2 and >1% CO2, the action of Sch9 is needed to downregulate hypha formation, possibly to evade immune responses directed against the hyphal growth form. Recently, glucan unmasked in hyphae and other hypha-associated components has been shown to directly interact with host components (30, 38). Thus, it appears that Sch9 activity could be an important requirement for commensal yeast growth of C. albicans, e.g., within phagocytotic vacuoles or in the gut, which contains low oxygen and high CO2 levels. Hyperfilamentation of the sch9 mutant was detected experimentally during growth on agar and at temperatures of <37°C. Agar is a polysaccharide that in an actual infection event may mimic glycostructures on the surface of host cells or in the extracellular matrix. The fact that hyperfilamentation of the sch9 mutant was observed at lower temperatures may reflect our present ignorance of additional factors regulating hypha formation under hypoxic conditions and high CO2 levels. For example, switching to and stabilization of the opaque growth form was found to occur only at lower temperatures and only recently was shown to depend on elevated CO2 levels even at 37°C (15). Some C. albicans components, including Efg1 and Flo8 transcription factors, have already been described that repress hypha formation during embedded growth or under hypoxia at lower temperatures (6, 33), but repression by these proteins did not require CO2, as Sch9 does. We conclude that Sch9 has a novel regulatory function, since it integrates sensing of hypoxia, CO2, and surface contact to supppress filamentation of C. albicans.
The fact that the repressive function of Sch9 is apparent only at high CO2 levels under hypoxia raises a question about the Sch9 signaling pathway. Interestingly, we also obtained a sch9 mutant-like hyperfilamentation phenotype by repressing Tor1 activity by rapamycin or caffeine (2, 41). Because Tor1 is a known upstream activator of Sch9 in S. cerevisiae (40), it can be assumed that a Tor1-Sch9 pathway acts as a repressor of filamentation. On the other hand, Rim15, the AGC kinase downstream of Sch9 in S. cerevisiae (28, 41), was not required for the sch9 mutant phenotype, as shown by sch9 rim15 double mutants and by rapamycin/caffeine-induced hyperfilamentation of a sch9 mutant. Deletion of Rim15 in C. albicans also did not generate growth or stress defects known for S. cerevisiae rim15 mutants (28), indicating that the roles of Rim15 have diverged significantly in both fungal species. While Rim15 was dispensable for hypoxia/CO2-triggered filamentation, the transcription factors Czf1 and Ace2, which had previously been shown to be involved in filamentation under hypoxia or agar embedding (14, 24), appeared to be required for hyperfilamention in the case of the nonfunctional Tor1-Sch9 suppression pathway. Czf1 has been suggested to downregulate Efg1, which under hypoxia is a strong repressor of hypha formation, independent of elevated CO2 levels (14, 33, 34). The model in Fig. 7 involves the Tor1-Sch9 pathway in the Efg1 circuitry by proposing that in wild-type cells Sch9 inactivates/downregulates Czf1, thereby increasing the repressor activity of Efg1. This process appears necessary to counteract the bicarbonate-stimulated increase in adenylate cyclase activity, cAMP levels, and PKA activity, which stimulate filamentation (20). Although other mechanisms are possible, the proposed pathway can explain that C. albicans, in specific hypoxic/CO2-containing environments, prevents filamentation and favors growth in the yeast form.
Fig. 7.
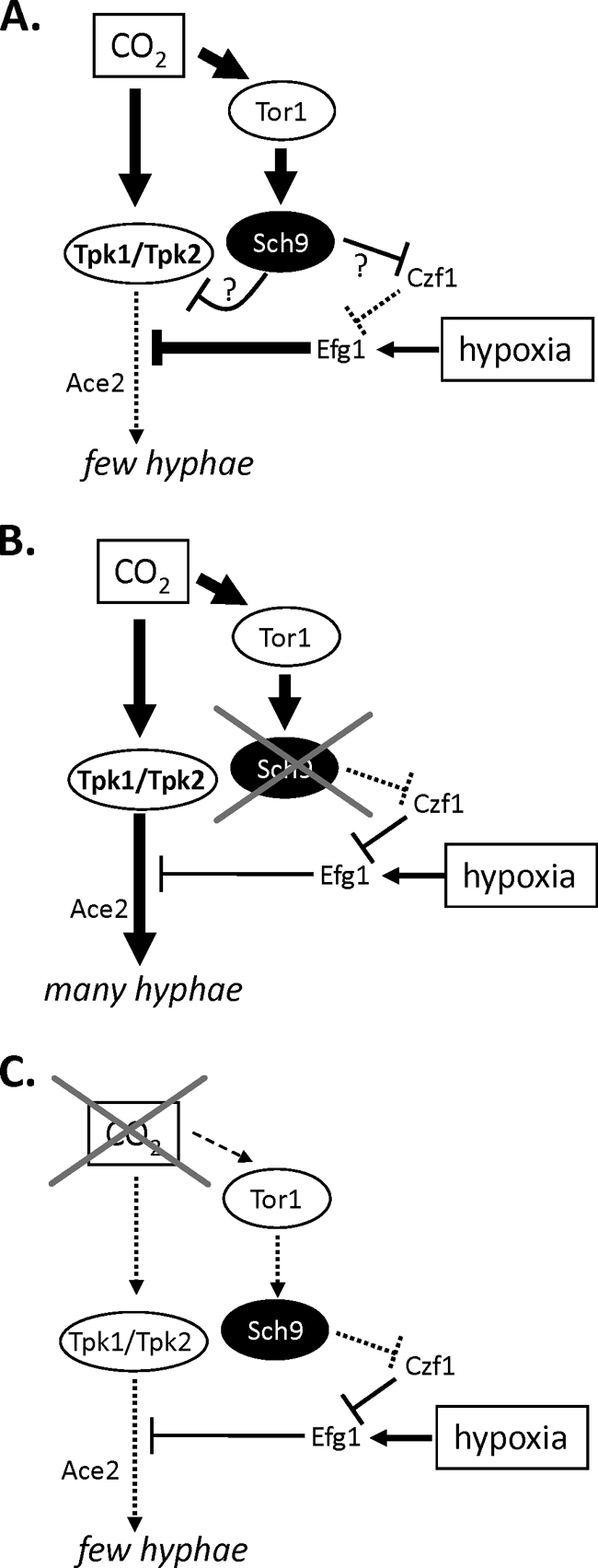
Model for the regulation of filamentation by Sch9. (A) Wild-type cells grown under hypoxia at high CO2 levels. Under hypoxia, Efg1 acts as a repressor of filamentous growth (14, 33, 34), but Czf1 limits its repressor activity (4, 14). Under high CO2 levels, the PKA isoforms Tpk1 and Tpk2 are activated, presumably by bicarbonate-mediated activation of adenylate cyclase (20). It is proposed that PKA is downregulated in certain environments (surface growth, temperatures of <37°C) by the Tor1-Sch9 pathway to prevent hypha formation. It is yet unclear if Sch9 increases Efg1 repressor activity by inactivation of Czf1, as shown, or by another mechansm, e.g., by blocking PKA activity. Ace2 is required for hypoxic filamentation (24) by a yet-unknown mechanism. (B) Lack of Sch9 or Tor1 activity during hypoxic growth under high CO2 fails to repress the CO2-triggered increase in PKA activity, leading to hyperfilamentation. (C) A moderate level of Efg1 repressor activity under hypoxic growth and low CO2 levels suffices to block hypha formation at low PKA activity.
Sch9 appears to be a general regulator of growth and morphogenesis of C. albicans because of additional phenotypes and aberrant gene regulation in the sch9 mutant. Mutants showed increased pseudohyphal growth in liquid medium that occurred only under hypoxia but was independent of CO2 levels. Furthermore, in the sch9 mutant, sensitivity to hygromycin B but also resistance to fluconazole, SDS, and Congo red were increased. The latter two resistance properties we observed are at variance with results of Liu et al. (22), who used different drug concentrations and strain backgrounds. Interestingly, resistance properties of the C. albicans control strain were significantly influenced by oxygen levels and temperature, e.g., because hygromycin, SDS, and fluconazole resistance increased under hypoxia and higher temperatures. Increased fluconazole resistance may be due to elevated ergosterol levels in the sch9 mutant under hypoxia, in which transcripts of several ERG genes are increased. A surprising phenotype of the sch9 mutant was its short chronological life span in the stationary growth phase. This finding is opposite to results for S. cerevisiae, for which Rim15 has been established as the key kinase to allow long-term survival by inducing stress response genes; the upstream Tor1-Sch9 kinases inactivate Rim15 during nutrient-rich growth, but they release Rim15 in stationary phase because they are downregulated (12, 41). Thus, it appears that Sch9 is not involved in regulating chronological life span in C. albicans, whereas the decreased life span of a rim15 mutant is consistent with the S. cerevisiae results and identifies the only significant phenotype of Rim15 depletion in C. albicans. Unexpectedly, the dependence of C. albicans longevity on Sch9 and Rim15 under normoxia was not paralleled under hypoxia, where both proteins were not needed for long-term survival. This result again stresses the different regulatory circuitries operating under hypoxia and normoxia, which in the past were reported apparent with regard to filamentation and biofilm formation (10, 33, 36).
The transcriptomal analyses revealed that 310 genes were up- or downregulated more than 2-fold in the sch9 mutant. Sch9 regulates only a fraction of genes that are dependent on the Tor1 kinase. Such genes include genes involved in the metabolism of sugars, which are both Sch9 and Tor1 dependent, and genes encoding cell wall components, which are repressed by both proteins (2). Sch9 also represses numerous transport activities of C. albicans under various conditions, reflecting the role of Sch9 in an abundance metabolism, which is similar to the functions of PKA isoforms in the presence of nutrients. Importantly, we discovered that under hypoxia Sch9 regulates many genes that are unaffected under normoxia and that the presence of CO2 elicits yet another pattern of Sch9-dependent gene activities. Genes that are up- or downregulated by Sch9 only under hypoxia in the presence of 6% CO2 may be responsible for the hyperfilamentation phenotype of the sch9 mutant. The functions of such genes are largely unknown or represent various functional classes. Therefore, it remains to be determined how many of the respective 25 up- or 25 downregulated genes are involved in a CO2-sensing pathway that comprises Sch9. Future experiments will clarify whether, for example, transcription factors encoded by the ZCF4 and ORF19.3088 genes regulate hypoxia- and CO2-dependent genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Brunke (Jena) for help with the evaluation of microarray data.
This project was funded by the Deutsche Forschungsgemeinschaft (SFB590) and by EU project “Galar Fungail II” (MRTN-CT-2003-504148). Funding for A.F. was provided by the Jürgen Manchot Stiftung Düsseldorf.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Arnaud M. B., et al. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33:D358–D363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastidas R. J., Heitman J., Cardenas M. E. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bockmühl D. P., Krishnamurthy S., Gerads M., Sonneborn A., Ernst J. F. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243–1257 [DOI] [PubMed] [Google Scholar]
- 4. Brown D. H., Jr., Giusani A. D., Chen X., Kumamoto C. A. 1999. Filamentous growth of Candida albicans in response to physical environmental cues, and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651–662 [DOI] [PubMed] [Google Scholar]
- 5. Cameroni E., Hulo N., Roosen J., Winderickx J., De Virgilio C. 2004. The novel yeast PAS kinase Rim 15 orchestrates G0-associated antioxidant defense mechanisms. Cell Cycle 3:462–468 [PubMed] [Google Scholar]
- 6. Cao F., et al. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng S., et al. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71:6101–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cottier F., Mühlschlegel F. A. 2009. Sensing the environment: response of Candida albicans to the X factor. FEMS Microbiol. Lett. 295:1–9 [DOI] [PubMed] [Google Scholar]
- 9. Crauwels M., et al. 1997. The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology 143:2627–2637 [DOI] [PubMed] [Google Scholar]
- 10. Doedt T., et al. 2004. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15:3167–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ernst J. F., Tielker D. 2009. Responses to hypoxia in fungal pathogens. Cell. Microbiol. 11:183–190 [DOI] [PubMed] [Google Scholar]
- 12. Fabrizio P., Pozza F., Pietcher S. D., Gendron C. M., Longo V. D. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288–290 [DOI] [PubMed] [Google Scholar]
- 13. Fonzi W. A., Irwin M.-Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giusani A. D., Vinces M., Kumamoto C. A. 2002. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics 160:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang G., Srikantha T., Sahni N., Yi S., Soll D. R. 2009. CO(2) regulates white-to-opaque switching in Candida albicans. Curr. Biol. 19:330–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huber A., et al. 2009. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23:1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacinto E., Lorberg A. 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410:19–37 [DOI] [PubMed] [Google Scholar]
- 18. Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelly M. T., et al. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol. Microbiol. 53:969–983 [DOI] [PubMed] [Google Scholar]
- 20. Klengel T., et al. 2005. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15:2021–2026 (Erratum, 15:2177.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H., Köhler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 22. Liu W., Zhao J., Li X., Li Y., Jiang L. 2010. The protein kinase CaSch9p is required for cell growth, filamentation and virulence in the human fungal pathogen Candida albicans. FEMS Yeast Res. 10:462–470 [DOI] [PubMed] [Google Scholar]
- 23. Lu Y., et al. 2008. Efg1-mediated recruitment of NuA4 to promoters is required for hypha-specific Swi/Snf binding and activation in Candida albicans. Mol. Biol. Cell 19:4260–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mulhern S. M., Logue M. E., Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nobile C. J., Bruno V. M., Richard M. L., Davis D. A., Mitchell A. P. 2003. Genetic control of chlamydospore formation in Candida albicans. Microbiology 149:3629–3637 [DOI] [PubMed] [Google Scholar]
- 26. Park H., et al. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 27. Pascual-Ahuir A., Proft M. 2007. The Sch9 kinase is a chromatin-associated transcriptional activator of osmostress-responsive genes. EMBO J. 26:3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pedruzzi I., et al. 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12:1607–1613 [DOI] [PubMed] [Google Scholar]
- 29. Prill S. K.-H., et al. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546–560 [DOI] [PubMed] [Google Scholar]
- 30. Robinson M. J., et al. 2009. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J. Exp. Med. 206:2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rohde J. R., Bastidas R., Puria R., Cardenas M. E. 2008. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr. Opin. Microbiol. 11:153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roosen J., et al. 2005. PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol. Microbiol. 55:862–880 [DOI] [PubMed] [Google Scholar]
- 33. Setiadi E. R., Doedt T., Cottier F., Noffz C., Ernst J. F. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen-sensing- and Efg1p-regulatory networks. J. Mol. Biol. 361:399–411 [DOI] [PubMed] [Google Scholar]
- 34. Sonneborn A., Bockmühl D. P., Ernst J. F. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514–5517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sonneborn A., et al. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of the human pathogen Candida albicans. Mol. Microbiol. 35:386–396 [DOI] [PubMed] [Google Scholar]
- 36. Stichternoth C., Ernst J. F. 2009. Hypoxic adaptation by Efg1 regulates biofilm formation of Candida albicans. Appl. Environ. Microbiol. 75:3663–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stoldt V. R., Sonneborn A., Leuker C., Ernst J. F. 1997. Efg1, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor P. R., et al. 2007. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 8:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tebarth B., et al. 2003. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 329:949–962 [DOI] [PubMed] [Google Scholar]
- 40. Urban J., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26:663–674 [DOI] [PubMed] [Google Scholar]
- 41. Wanke V., et al. 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69:277–285 [DOI] [PubMed] [Google Scholar]
- 42. Wei Y., Zhang X. F. 2009. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle 8:4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu X. L., et al. 2008. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4:28–39 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.