Abstract
Human endocytic protein ITSN1 regulates actin reorganization by activating Rho family GTPases, such as Cdc42. The process is enhanced by ITSN binding of WASP, an effector of Cdc42 and a potent activator of actin polymerization. In the human pathogen Cryptococcus neoformans, endocytic protein Cin1 also interacts with Cdc42 and Wsp1, an uncharacterized WASP homolog, but the significance of these interactions remains unknown. Wsp1 contains several conserved domains, including a WASP homology 1 domain (WH1), a GTPase binding/Cdc42 and Rac interactive binding domain (GBD/CRIB), and a C-terminal domain composed of verprolin-like, central, and acidic motifs (VCA). Thus, Wsp1 exhibits domain compositions more similar to human WASP proteins than Saccharomyces cerevisiae Las17/Bee1, a WASP homolog lacking the GDB/CRIB domain. Wsp1 is not an essential protein; however, the wsp1 mutant exhibited defects in growth, cytokinesis, chitin distribution, and endocytosis and exocytosis. The wsp1 mutant was also unable to undergo genetic cross, produce the polysaccharide capsule, or secrete the enzyme urease. An in vitro phagocytosis assay showed a higher phagocytic index for the wsp1 mutant, whose ability to cause lethal infection in a murine model of cryptococcosis was also attenuated. Our studies reveal divergent evolution of WASP proteins in the fungal phylum and suggest that the conserved function of WASP proteins in the actin cytoskeleton may also impact fungal virulence.
INTRODUCTION
Cryptococcus neoformans is a basidiomycetous fungal pathogen that infects primarily immunocompromised individuals, causing meningoencephalitis, which is often fatal if left untreated (3, 19). This haploid organism has a bipolar mating system and undergoes genetic cross under laboratory conditions. In the environment, C. neoformans propagates primarily through budding, and yeast cells, probably in a desiccated form, and are thought to enter the host through the respiratory tract and cause infection (3, 19). Studies by Kopecka and colleagues indicated that C. neoformans has a fully developed actin cytoskeleton and it maintains actin dynamics similar to Saccharomyces cerevisiae during mitotic division and asexual reproduction (14).
In unicellular fungi, budding and the establishment of cell polarity are accomplished through dynamic changes of the actin cytoskeleton, which are regulated by an integrated signaling cascade of the Rho family of small GTPases, such as Rho, Rac, and Cdc42 (4, 25). Cdc42 is highly conserved among the eukaryotic organisms in both sequence and function (12). Cdc42 functions through binding of effector molecules that contain the GTPase-binding domain (GBD; also known as the Cdc42/Rac interactive binding [CRIB] domain). Among the effector proteins of Cdc42 that contain the CRIB/GBD domain, Wiskott-Aldrich syndrome (WAS) proteins (WASP) are most known for their ability to promote actin assembly (4).
WASP and similar but neuron-specific WASP (N-WASP) proteins are a family of human proteins (both are referred to as WASPs) whose mutations result in WAS, a rare X-chromosome-linked hereditary disease characterized by thrombocytopenia, eczema, and immunodeficiency (5). These disease symptoms are the result of abnormalities in cytoskeletal structures, polarization, and motility at the cellular level, and most studies of WASPs have focused on their roles in regulating branched filamentous actin through the Arp2/3 complex (see the reviews in references 27, 39, and 40). The WASPs are multimodular proteins that contain an N-terminal WASP homology 1 domain (WH1), a basic amino acid domain (B), a CRIB/GBD motif, a proline-rich domain (PR), and a C-terminal verprolin (V), central connecting (C), and acid amino acid (A) domain (collectively termed VCA) (27, 31). In its resting state, WASP is in an autoinhibitory conformation through the binding of the VCA domain to its B and CRIB/GBD domains (A to B and V to CRIB/GBD, respectively). Binding of GTP-Cdc42 to WASP through the CRIB/GBD domain, phosphatidylinositol bisphosphate (PIP2) to the B domain, or the SH3 domain to the PR domain all result in opening of the masked and autoinhibited VCA domain, leading to its activation of the Arp2/3 protein (15, 27, 31).
Despite the conserved function of WASPs, distinctions exist among various paralogs/homologs, resulting in a complexity in their filamentous actin activation mechanisms. In the plant model Arabidopsis thaliana, no WASP homolog was found, but instead a family of WAVE (verprolin-homologous) proteins that function to promote actin nucleation were found (15). The WAVE proteins also exist in mammals and, in contrast to WASPs that are activated by Cdc42, WAVE proteins are regulated by Rac (23). In fungi, S. cerevisiae Las17/Bee1, Schizosaccharomyces pombe Wsp1, and Candida albicans Wal1 are WASP homologs that lack the GBD/CRIB domain (9, 17, 18, 32). This indicates that a distinct activation mechanism is likely to be involved in the functions of these proteins (32). Consistent with this proposition, a study suggested that the activation of actin polymerization by Las17/Bee1 is indeed indirect (6). Not surprisingly, human WASP failed to complement an S. cerevisiae las17/bee1 mutant and restored functions (18), despite the finding that Las7 exhibited a role in the activation of actin filaments and some of the las17/bee1 yeast mutant phenotypes mimicked the cellular defects of Wiskott-Aldrich syndrome (18, 21).
C. albicans Wal1 exhibits a certain degree of domain similarities to S. cerevisiae Las17/Bee1 and S. pombe Wsp1, sharing up to 38% and 28%, respectively, in amino acid sequence identity (32). Similar to Las17/Bee1, Wal1 is required for polarity establishment, intracellular traffic, and vacuolar biogenesis in C. albicans and for the polarized growth of C. albicans hyphae (32).
Studies of C. neoformans have suggested that a number of proteins have a role in actin function. The Rho GTPases Cdc42 and Cdc420 are required for actin polarization under stress conditions and have also been mostly noted for functioning with Ras proteins to mediate thermal resistance, a role previously established for the Rac homolog of the fungus (1, 22, 30). GTPase Sav1, an S. cerevisiae Sec4/Rab8 homolog, is involved in post-Golgi complex secretion (38), a process that involves the rearrangement of the actin cytoskeleton (see the review in reference 35). Recently, we identified a novel cryptococcal intersectin protein, Cin1, and characterized its functions, including those in the intracellular transport and regulation of the actin cytoskeleton (28). As a part of the ongoing effort to elucidate the unique Cin1 pathway, we identified a WASP homolog, Wsp1, and showed that Wsp1 associated with Cin1 through an interaction between the Wsp1-PR and Cin1-SH3 domains (28). This implies that a functional mechanism similar to WASP of mammalian cells exists in C. neoformans.
Here, we further characterized Wsp1 and examined its function in the growth, morphology, and virulence of the fungus. Through whole-gene and domain-specific gene disruption, we show that Wsp1 has multiple functions in maintaining cellular morphology, cytokinesis and chitin distribution, and endocytosis and exocytosis. We also show that Wsp1 is less resistant to phagocytosis in an in vitro model and avirulent in a murine model of cryptococcosis. Our study illustrates the important role of the first fungal WASP homolog containing the GBD/CRIB domain and provides evidence of divergence between different groups of fungi in the evolution of WASP/Las17 proteins.
MATERIALS AND METHODS
Strains, plasmids, and media.
C. neoformans var. neoformans (serotype D) strains were used in this study, with strains JEC21 and JEC20 as the standard MATα and MATa strains (16). All strains are listed in Table S1 of the supplemental material. Oligonucleotide primers for PCR amplification are listed in Table S2 of the supplemental material. The CDC42(Q61L) allele was obtained by overlapping PCR. The 5′ partial sequence was amplified by PCR with M13 reverse primer and primer PW1498 with pGS950 as the template, while the 3′ end was amplified with primer PW1497 and M13 forward primer, and also with pGS950 as the template. Overlap PCR was performed with primers PW1495 and PW1497, and the PCR product was then ligated into a TA vector and the sequence was verified. DNA plasmids used in this study are listed in Table S3 of the supplemental material. All culture media and reagents were prepared as described previously (28).
Mutant strain construction and complementation.
The wsp1::NEO and wsp1::NAT alleles were obtained by the split-marker approach, and the mutant strains were obtained by biolistic transformation, as described previously (28). A wsp1 mutant was complemented with the full-length WSP1 gene, including the promoter and terminator sequences, which were amplified through a two-step process. First, the 5′ sequence was amplified with primers PW1185 and PW1186 and the fragment ligated into plasmid pGS199, resulting in plasmid pGS764. Second, the 3′ sequence was amplified with primers PW1187 and PW1193 and inserted into pGS764, resulting in plasmid pGS781. The DNA sequence was verified by sequencing and the WSP1 gene linked to the NAT marker was reintroduced into the wsp1::NEO mutant by a second round of biolistic transformation.
To construct the WSP1-GBD allele, in which the sequence corresponding to amino acids 160 to 224 was deleted, overlap PCR was performed using two templates. One template was amplified with primer PW1188 and M13 forward primer from pGS462, and another with primer PW1189 and M13 reverse primer from pGS780. Overlap PCR was carried out with primers PW1187 and PW968, and the resulting PCR product was cloned (pGS783). The product was then cut with BamHI and XbaI and inserted into pGS324 to get the final construct pGS794. In this plasmid, the Wsp1-GBD allele is driven by a constitutively active glyceraldehyde phosphate dehydrogenase Gpd1 promoter (28). The plasmid was transformed into the wsp1 mutant strain, and transformants were verified by PCR. Expression levels were estimated by semiquantitative reverse transcription-PCR (see Fig. S1 in the supplemental material).
To generate the wsp1 mutant in DsRed-Sec4 (Sav1) strains, plasmid pGS1139 was first introduced into wild-type strain JEC21 by biolistic transformation. One of the transformants, GYS506, that exhibited robust red fluorescent signal under the microscope was transformed with the wsp1::NEO split-marker products to obtain GYS563. This transformant strain was used to examine the role of Wsp1 in Sec4/Sav1 localization.
Yeast two-hybrid assay.
Full-length WSP1 cDNA was released from plasmid pGS462 with EcoRI and BglII and inserted into pGBKT7, resulting in pGBKT7::WSP1 (pGS849). cDNA for CDC42 was synthesized with primer PW1495 and adaptor primer AUAP and cloned in the TA vector (Agilent), resulting in pGS950. Once verified by DNA sequencing, cDNA was cut with EcoRI and BglII and inserted into pGADT7, resulting in pGS1074. Plasmid DNA was transformed into the yeast AH109 strain, and interactions were assessed according to the method previously described (10, 24).
Fluorescence fusion protein construction and detection.
To construct the DsRed-Sec4 fusion protein, SEC4/SAV1 cDNA was amplified with PW1510 and AUAP and cloned (pGS1119). The SEC4/SAV1 fragment cut with BamHI and SpeI was inserted into pGS988 (28) to obtain the final construct, pGS1139. The DsRed-SEC4 fusion construct also has the GPD1 promoter.
Nuclear staining with 4′,6-diamino-2-phenylindole (DAPI) was performed as described previously using fresh cells grown overnight (28). Distributions of chitin and actin were observed by staining cells with calcofluor white (CFW) and rhodamine-conjugated phalloidin, respectively, as described previously (28). The vesicles and endosomes were observed by incubating appropriately prepared cells with FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide] (28). Cells were mounted on poly-l-lysine-coated glass slides and observed for fluorescence under a Zeiss Axio Imager 2 microscope (Carl Zeiss, Inc., Thornwood, NY).
Mutant phenotype characterization.
Assays for cell fusion, conjugation tube formation, and mating, as well as melanin and capsule production, were performed as described previously (28, 33). For growth, cells of an approximate optical density at (OD600) of 0.1 were serially diluted, and 5 μl each was spotted onto yeast extract-peptone-dextrose (YPD) and yeast nitrogen base plates for growth at 25, 30, and 37°C. To assay for urease, aliquots from saturated YPD broth cultures of wild-type JEC21, wsp1, and wsp1 WSP1 were grown in liquid YPD overnight with shaking at 30°C. Cells were collected by centrifugation and washed twice with sterile water. Cells were suspended in sterile water and counted by using a hemacytometer, followed by plating on YPD to ensure accuracy and viability of counts. A total of 6 × 105 cells were inoculated into 1.5 ml of Christensen's urea broth with or without urea. Cells were incubated up to 3 days at 30°C, and absorbance at 550 nm and CFU were estimated daily.
Phagocytosis assay.
J774A.1 macrophages of four to five passages were collected and counted, and viability was assessed with trypan blue stain (28). Macrophages were seeded on coverglasses in 12-well plates and stimulated with 50 units/ml mouse recombinant gamma interferon (R&D Systems), 0.3 μg/ml Escherichia coli 0111:B4 lipopolysaccharide (Sigma), and 25 μl/ml pooled mouse serum (JEC21 infected) before exposure to yeast cells at a 2:1 multiplicity of infection. Mixed cells were incubated at 5% CO2, 37°C for 4 h and observed using an inverted microscope (Olympus CV41). Noningested yeast cells were removed by washing three times in 37°C sterile phosphate-buffered saline (Fisher Scientific). For phagocytic index determination, coverglasses were fixed in methanol and stained with Giemsa stain (Sigma), and three to five fields per coverglass were photographed under a microscope (Zeiss Axio Imager 2) outfitted with a Zeiss digital camera. At least 120 macrophages were counted for ingested yeast cells per infecting strain, and the phagocytic index is reported as the number of ingested yeast cells per 100 macrophages.
Virulence assessment.
BALB/c mice were inoculated via the lateral tail vein with 2.5 × 105 fresh-grown cells as determined by counting with a hemacytometer and viability plating on YPD medium (>95% viability). For wsp1 mutants that exhibited morphological defects, individual chains or clusters of cells were considered a single cell. Mice were monitored twice daily and sacrificed when moribund or at 90 days postinfection. Brains, kidneys, and lungs were dissected for CFU determinations. Organs were weighed and homogenized in 1 ml of sterile water containing 50 μg/ml chloramphenicol and plated on YPD following serial dilution. CFU were determined after 3 to 5 days of growth at 30°C. Homogenates were also stained with India ink and observed in a blinded manner.
For histological studies, brains were preserved in 4% buffered formalin, processed, embedded, and sectioned at 5-μm thickness. Sections were allowed to adhere to glass slides, stained with periodic acid-Schiff stain (PAS; Sigma), and observed in a blinded manner. Representative areas were photographed under a microscope (Olympus BX51) equipped with a digital camera (Olympus).
RESULTS
Wsp1 contains a GBD/CRIB domain unique among known fungal WASPs.
In the process of characterization of Cin1, a cryptococcal intersectin homolog that plays multiple functions in C. neoformans, we identified the WSP1 gene (CNBE5050 [www-sequence.stanford.edu/cgi-bin/cneoformans]) encoding a WASP homolog, Wsp1 (GenBank accession number HQ399545), and we found that Cin1 interacts with the proline-rich domain of Wsp1 in a yeast two-hybrid assay (28). This finding suggested that Cin1 might regulate, analogous to human intersectin ITSN1, the actin cytoskeleton through interactions with Wsp1 and Rho family GTPase Cdc42 (GenBank accession number HQ645962) (28). To identify the function of Wsp1, we characterized the full-length WSP1 gene, generated wsp1 mutants, and complemented the wsp1 mutants with the wild-type WSP1 gene. We also generated the GBD/CRIB domain-specific mutant strains by transformation of the wsp1 mutant strain with the WSP1-GBD mutant allele driven by a constitutively active Gpd1 promoter.
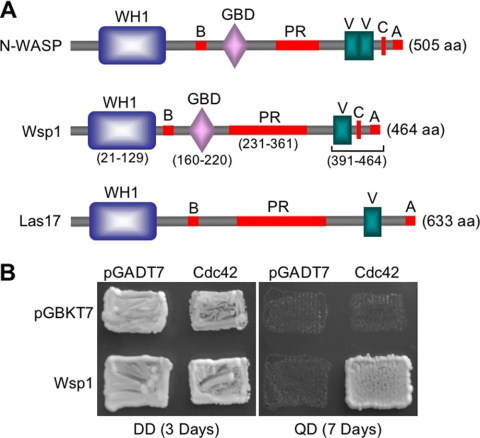
Amino acid sequence analysis indicated that Wsp1 shares limited homology with several fungal WASP-like proteins, such as S. cerevisiae Las17/Bee1 (overall amino acid identity, 32%) and Candida albicans Wal1 (29%) and to human WASPs (29% amino acid identity). Interestingly, in addition to the N-terminal WH1 domain, the PR domain, and the C-terminal VCA domain, Wsp1 contains a Rho GBD/CRIB domain (Fig. 1 A). The GBD/CRIB domain, consisting of approximately 17 amino acids, commonly occurs in mammalian WASPs (27a). It also exists in Ustilago maydis (UM03687; overall amino acid identity, 62%) but is not present in S. cerevisiae Las17/Bee1 or C. albicans Wal1 (Fig. 1A). Wsp1 was shown previously to exhibit a physical interaction with Cin1 through the PR domain (28) and, consistent with the GBD/CRIB domain containing WASPs, Wsp1 was shown to interact with cryptococcal Rho GTPase Cdc42 in a yeast two-hybrid screening (Fig. 1B). Since the GBD/CRIB domain plays a key role in the autoinhibition/activation of WASP function, its presence in Wsp1 and mediation of an interaction with Cdc42 suggest that a regulatory mechanism similar to WASP but distinct from Las17/Bee1/Wal1 is present in C. neoformans.
Fig. 1.
C. neoformans Wsp1 is homologous to WASP/Las17 proteins. (A) Comparison of domain organization between Wsp1 (CNBE5050), human n-WASP (GenBank accession number NP_003932), and S. cerevisiae Las17/Bee1 (GenBank accession number CAA99390). WH1, WASP homology 1 domain; B, basic domain; GBD, GTPase binding domain; PR, proline-rich domain; V: verprolin domain; C, central or cofilin homology domain; A, acidic domain. (B) Wsp1 interacts with Cdc42 in a yeast two-hybrid assay. DD, double dropout medium (SD -Leu -Trp); QD, quadruple dropout medium (SD -Leu -Trp -His -Ade).
Wsp1 is required for growth at 37°C, cytokinesis, and chitin distribution.
When selecting transformants from G418-containing selection medium, we found generally two types of colonies at 30°C, namely, a faster-growing type and a type that grew slowly. The latter contained the wsp1::NEO allele, indicating that Wsp1 might be required for normal growth. On YPD without the drug, the requirement of Wsp1 for normal growth was subtle at 25 and 30°C but most apparent at 37°C (Fig. 2). Growth was mostly restored in the complemented wsp1 WSP1 (Fig. 2). However, the WSP1-GBD mutant, which contains a partial WSP1 allele without the GBD domain driven by the Gpd1 promoter (28) had partially restored growth at 37°C (Fig. 2). As indicated by time-lapse microscopic photography, the wsp1 mutant showed delayed cell division (see Fig. S2 in the supplemental material), which is consistent with the growth assay results at 25°C (Fig. 2).
Fig. 2.
Wsp1 is required for normal growth. The wild type and wsp1, wsp1 WSP1, and WSP1-GBD mutant strains were serially diluted, and 5 μl of each was spotted on YPD medium. Growth was allowed for 48 h at the indicated temperatures. Two independent isolates of each strain were used to compare results with the wild-type strain (JEC21).
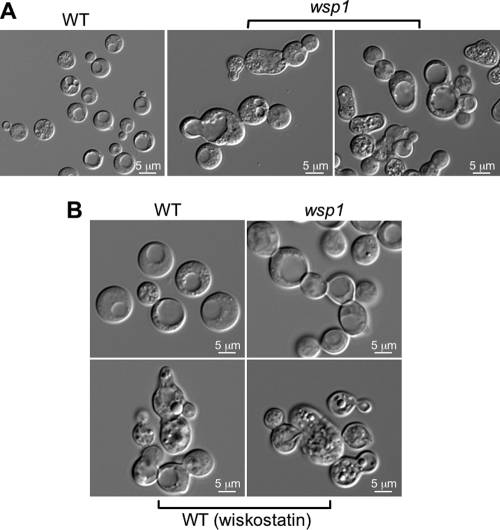
Microscopy examination of wsp1 mutant cells showed a gross defect in morphology. In contrast to unicellular budding of the wild-type strain, the wsp1 mutant cells were abnormal, with cells jointed and clustered, indicating a defect in cytokinesis (Fig. 3 A). The severity of cell clustering, which could not be separated by mechanical forces such as vortex or sonication, varied depending on incubation time, with the most severe clustering seen in cultures that were 3 days or older. In a quantitative measurement following 24 h of growth in YPD, unicellular cells (8.3 μm, average diameter) accounted for nearly half of the cells observed (196/399), in contrast to 90% (157/175) seen in the wild-type strain (6.38 μm, average diameter). The clustering of the wsp1 mutant was similar to cells grown in the presence of 10 μM wiskostatin, an inhibitor of mammalian WASPs (Fig. 3B).
Fig. 3.
Wsp1 is required for normal morphology. (A) The wsp1 mutant exhibits various degree of abnormalities in morphology. The wild-type strain and wsp1 mutants were grown at 30°C in liquid YPD overnight (24 h). The severity of abnormality positively correlated with the culture incubation time. (B) The mammalian WASP-inhibiting agent wiskostatin (10 μM) induces a morphological defect in C. neoformans that is similar to disruption of the WSP1 gene. Cells were grown in YPD medium with or without wiskostatin at 30°C for 3 days.
When stained with the DNA dye DAPI, single nucleated cells were found in the wild-type, complemented, and WSP1-GBD strains in almost all of the cells observed, and most nuclei appeared as distinctively round and solid DAPI-stained masses (Fig. 4 A). In the wsp1 mutant cells, a nuclear staining pattern was found, but DAPI-stained materials were rather diffuse, suggesting that wsp1 may also have abnormal nuclei (Fig. 4A).
Fig. 4.
Wsp1 is required for proper chitin distribution. (A) The nuclear morphology is altered as the result of the WSP1 gene disruption. Nuclear staining with DAPI was apparent in the wsp1 and wild-type strains, but the edges of the nuclei in the wsp1 mutant were rugged, in contrast to the wild-type strains, in which nuclei appeared as solid individual masses stained by DAPI. (B) The wild-type strain, and wsp1 WSP1 and WSP1-GBD strains showed an even distribution of CFW-stained material around the cell periphery, indicating normal chitin localization, in contrast to the wsp1 mutant, which showed aggregated chitin staining inside the cells. Cells were stained as described previously (28) and observed under a Zeiss Axio Imager 2 microscope.
Since chitin is important for cell separation, we examined the distribution of chitin by staining cells with CFW. An even distribution of chitin was observed near the cell wall and along the cortex in the wild-type and control strains (Fig. 4B), but not in the wsp1 mutant. In this mutant, chitin appeared abnormal, either as uneven layers near the cell periphery or randomly distributed dense patches (Fig. 4B). The complemented and the WSP1-GBD allele-specific mutant strains exhibited the chitin distribution of the wild type (Fig. 4B).
Wsp1 is required for actin cytoskeleton, endocytosis, and exosomal localization of Sec4/Sav1 GTPase.
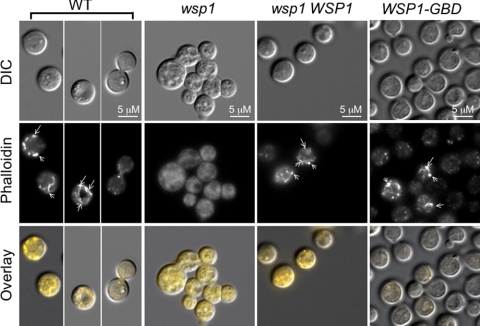
Actin and the actin cytoskeleton also participate in cell separation, and so we performed rhodamine-conjugated phalloidin staining to observe actin cables and patches. Actin cables are bundles of unbranched filaments involved in long distance vesicle trafficking, whereas actin patches are composed of branched actin filaments and proteins necessary for intracellular trafficking (26, 29, 37). In C. neoformans, the intersectin Cin1 protein also mediates endocytosis of FM4-64 and the exocytotic transport of a fluorescent-labeled laccase fusion protein, Lac1-DsRed (28). The actin patch had a cortical distribution pattern near the plasma membrane, whereas the long actin cable was present but did not appear in all cell stages (Fig. 5, wild-type [WT], wsp1 WSP1, and WSP1-GBD strains). Consistently, disruption of the WSP1 gene resulted in disappearance of the actin cable and significantly reduced actin patches (Fig. 5). The occurrence of the cable and patch in the wsp1 mutant was 1 and 4.6%, respectively, whereas the wild-type and complemented strains all showed actin patches and cables (Fig. 5).
Fig. 5.
Wsp1 is required for maintenance of proper actin dynamics. Rhodamine-conjugated phalloidin staining showed that actin cables and filaments were present in the wild-type, wsp1 WSP1 complemented, and WSP1-GBD domain-specific allele strains but disappeared from the wsp1 mutant. The cortical distribution of actin patches and filaments (indicated by arrows) are seen in the wild-type, wsp1 WSP1-complemented, and WSP1-GBD domain-specific allele strains. The wsp1 mutant showed faint background staining instead. The method of staining was as described previously (28).
Internalization of FM4-64 was not found in the wsp1 cells at 15 or 30 min or longer after the exposure, which was similar to results for the cin1 mutant (28), in contrast to the wild-type strain, where endosomes appearing as ring-like structures were apparent and abundant (Fig. 6). Internalization of FM4-64 was normal in the complemented strain (wsp1 WSP1) and the domain-specific allele transformant (WSP1-GBD) (Fig. 6).
Fig. 6.
The endocytosis of FM4-64 requires the function of Wsp1. The wild-type, wsp1, wsp1 WSP1, and WSP1-GBD strains exhibited the attachment of FM4-64 to the plasma membrane when the dye was first introduced. However, at 15 and 30 min (data for 30 min are shown), FM4-64 is internalized by all strains except wsp1, shown as intracellular bright ring-like structures, which are early endosomes and/or vacuoles. Cells were prepared and stained with FM4-64 as described previously (28). Note that the strain containing the Wsp1-GBD allele showed normal dye uptake.
To further examine whether Wsp1 is required for exocytosis, we examined the localization of a fluorescently labeled Sec4/Sav1 protein. In S. cerevisiae, Sec4 is essential for vesicle-mediated exocytic secretion and associated with exosomes to regulate the polarized delivery of transport vesicles to the exosomes at the plasma membrane (7). In C. neoformans, the Sec4 homolog Sav1 regulates the secretory process of the polysaccharide component glucuronoxylomannan (GXM) (38). Consistent with the location of Sec4 in S. cerevisiae, DsRed-Sec4/Sav1 was found as solid dots (presumably exosomes) near the plasma membrane of the wild-type cells, but the signal was mostly diffuse, and no solid dots were found near the plasma membrane in the wsp1 mutant strain (Fig. 7), suggesting a defect in exosomal distribution.
Fig. 7.
Wsp1 is required for exosomal localization of a DsRed-Sec4/Sav1 fusion protein. The fusion gene was introduced into a sec4/sav1 mutant strain, and fluorescence was observed under a Zeiss Axio Imager 2 microscope. Images were photographed with an attached Zeiss camera and processed using Adobe Photoshop CS4. Sec4/Sav1 functions in the secretory pathway, and the DsRed-Sec4 protein is mostly associated with exosomes (exocysts) attached to the plasma membrane in wild-type cells (top center). In the wsp1 strain, however, the fusion protein appears to have a diffuse pattern and only a small amount of fluorescence appears in a dotted pattern, suggesting a reduced exosomal localization and defect in plasma membrane attachment (bottom center).
Wsp1 is important for pheromone-responsive mating and virulence characteristics.
The actin cytoskeleton plays a central role in the maintenance of cell morphology and in the intracellular transport of vesicular bodies containing pheromone receptors and enzymes such as laccases (28). Since the wsp1 mutant was defective in displaying actin patches and cables, we examined additional characteristics of the mutants, including pheromone responses and mating, and other virulence factors, such as the production of melanin pigment and polysaccharide capsules. The early pheromone response can be observed through a confrontation assay in which cells are monitored for the production of hypha-like filaments in response to the presence of the opposite mating partner (34). The wsp1 mutant of either mating type had a defect in conjugation tube formation in comparison to the wild-type, complemented (wsp1 WSP1), and the WSP1 allele without the GBD domain (WSP1-GBD) (Fig. 8 A). Genetic cross between two wsp1 mutants resulted in sterility, while cross between a wsp1 mutant and a normal mating partner generated fewer mating-specific filaments than the wild-type and complemented strains (Fig. 8B). The defect in mating was not as apparent in the WSP1-GBD strain in the unilateral cross than bilateral cross, where both partners contain the WSP1-GBD allele (Fig. 8B). This suggests that Wsp1 is required for mating and that some of this function might be attributed to the GBD domain. Moreover, when the MATα wsp1 mutant linked to the NEO marker was mixed, respectively, with the MATa wsp1 mutant with the NAT marker (bilateral) or wild-type (unilateral) strain of the opposite mating type, spotted on V8 agar, collected after 24 h, and plated on YPD medium containing both G418 and NAT (28), no colonies were found for the bilateral cross, in contrast to a large amount of cells seen in the cross between two wild-type strains and visible cells from the unilateral cross (Fig. 8C). This indicates that Wsp1 is required for genetic cross, and Wsp1 from one of the strains partially suppresses the defect of the wsp1 mutant.
Fig. 8.
Wsp1 is required for pheromone-responsive mating. (A) Formation of conjugation tubes, which are filaments produced in response to the presence of pheromones by the opposite mating type strain, can be seen in the wild-type, wsp1 WSP1, and Wsp1-GBD strains (after 2 days at 2°C). The MATα cells normally produce more abundant filaments than MATa cells. The wsp1 strains of either mating type failed to produce conjugation tubes (after 3 days at 25°C). (B) Wsp1 and the GBD domain are required for mating. In the unilateral cross, Wsp1 protein by one of the strains was at least partially sufficient for function. Mating of the two strains without GBD domains resulted in a significant reduction in mating-specific filament formation. (C) Fusion assay results indicate that the wsp1 mutants failed to undergo nuclear fusion, and Wsp1 from one of the parents can provide partial function (7 days at 25°C).
In capsule-inducing Dulbecco's modified Eagle's medium (DMEM), the wsp1 mutant cells showed no capsule formation by 2 days at 30°C, in comparison to the wild-type and complemented strains (Fig. 9 A). On dopamine medium, which induces melanin pigment at 30°C, the wsp1 mutant exhibited reduced melanin production that was restored by reintroducing the WSP1 gene into the mutant (Fig. 9A). Additionally, the WSP1-GBD allele only partially complemented the defect of wsp1 in melanin production (Fig. 9A). Moreover, at 37°C, the wsp1 mutant exhibited less pigmentation in medium, indicating decreased urease secretion (Fig. 9A). In a quantitative urease assay in which 6 × 105 cells were initially inoculated, the absorbance (at 550 nm) measurement was consistent with the urease secretion reduction in the wsp1 mutant (Fig. 9B, left chart). The decrease was partially restored in the complemented strain (Fig. 9B, left chart). Cells in Christensen's urea broth were also evaluated for CFU plating to ensure equivalent cell counts were used (Fig. 9B, right graph). Collectively, these assay results suggested that the defect of the wsp1 strain might render it less effective in infection of the host.
Fig. 9.
Formation of polysaccharide capsule and melanin pigment and secretion of urease enzyme are attenuated in the wsp1 mutant. (A) The wild-type, wsp1, wsp1 WSP1, and WSP1-GBD strains were incubated in DMEM for 2 days at 30°C. Cells were stained with India ink, and the capsule was observed under a Zeiss Axio Imager 2 microscope (top panel). Bar, 5 μm. Melanin was induced on dopamine medium, and cells were allowed to grow at 30°C for 2 days (middle panel). Urease secretion was observed after incubation of all strains in Christensen's medium for 2 days at 37°C. The wsp1 strain secreted no ureases, even when the incubation time was increased to 4 days, while no qualitative difference in phenol red color shift was seen between the wild-type, wsp1 WSP1, and WSP1-GBD strains. (B) Cells were adjusted to approximately equal density (absorbance at 550 nm), and the liquid urease assay results suggested that urease secretion is indeed decreased in the wsp1 mutant. The decrease was partially restored in the complemented strain (left graph). *, statistically significant difference (P < 0.05). Viability of cells in Christensen's urea broth was confirmed by CFU plating to ensure equivalent cell counts (right graph).
Wsp1 is required for resistance to phagocytosis and for survival in a murine model of cryptococcosis.
To examine the likely role of Wsp1 in the interaction between the fungus and its host, we employed an in vitro macrophage model and a murine animal virulence model. Following 4 h of interaction between cryptococcal cells and J774A.1 macrophages, the phagocytic index (ingested yeast cells per 100 macrophages) for the wsp1 mutant was 20.7 ± 5.9 (mean ± standard deviation), whereas the index for the wild type was 2.6 ± 2.2, indicating that the mutant cells were taken up at a significantly greater degree than the wild type. Unexpectedly, the phagocytic index for the complemented wsp1 WSP1 mutant strain was near the level of the wsp1 mutant (21.5 ± 15.9) in two independent experiments.
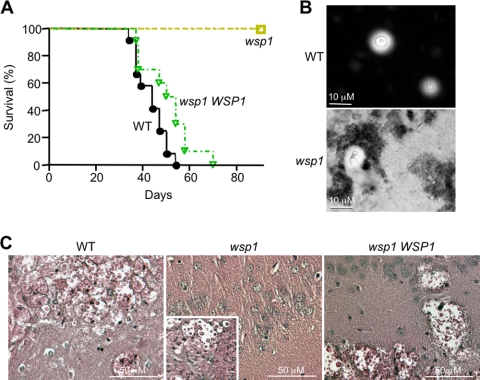
Virulence testing showed that mice infected with the wild type or the complemented mutant exhibited median survival of 44 and 52 days, respectively (Fig. 10 A). These survival curves were not significantly different, as determined by the Gehan-Breslow-Wilcoxon test (P = 0.0684). In contrast, the wsp1 mutant-infected mice showed no sign of disease at 90 days postinfection (Fig. 10A). Surprisingly, despite showing no sign of disease, wsp1-infected animals had detectable CFU in seven of eight tested brains, three of eight lungs, and one kidney sample. All CFU were confirmed to express resistance to G418, consistent with the observation that the wsp1 cells were isolated from these mice. When CFU were determined for the mouse brains infected with the wsp1 mutant, they were 2 to 3 logs lower than those infected by the wild type and the complemented strains.
Fig. 10.
Wsp1 is required for virulence in a murine animal model. (A) Disruption of WSP1 results in a complete loss of infectivity in BALB/c mice. The wild-type, wsp1, and wsp1 WSP1 strains were inoculated into mice via lateral tail veins, and mice were observed twice daily until 90 days postinfection. The wsp1-infected mouse remained symptom free, whereas those infected with control strains exhibited progressive symptoms and morbidity. (B) Brain smears stained with India ink show cryptococcal cells. They were abundant in those infected with the wild type (top panel), but their occurrence was rare in wsp1-infected mice (bottom). (C) PAS stain of brains collected from mice infected with the wild-type, wsp1, and wsp1 WSP1 strains. The insert in the middle panel (wsp1) details the only brain of four collected that had cryptococcal cells (see text for descriptions).
Microscopic examination of brain homogenates stained with India ink showed that the wsp1 cells from infected, although apparently healthy, animals had aberrant morphologies and inconsistent capsule formation, and some were difficult to visualize compared to the wild type or the complemented strains (Fig. 10B). Upon histological examination, PAS staining of the formalin-fixed brain tissues in WT-infected animals showed focal and foamy lesions in the brain in both observed specimens, with no tropism for a specific area of the brain (Fig. 10C). The wild-type cell diameter varied, and most cells were spherical (Fig. 10C). The complemented wsp1 WSP1 strain-infected mice showed more extensive, diffuse lesions in the brain than those infected with the wild-type strain, and sections had an overall foamy appearance (Fig. 10C). In contrast, the wsp1 mutant led to cryptococcal cells in only one of four observed specimens. Of the four brain specimens observed histologically by PAS staining, three exhibited normal brain architecture and neither aberrant morphology nor cryptococcal cells were observed (Fig. 10C). The wsp1-infected mouse brain with an observable lesion had only one focal lesion within the brain tissue, and this was not localized to the outer edge or in the ventricles of the brain (Fig. 10C, insert). Additionally, the lesion contained cryptococcal cells of various but overall larger sizes, with a greater number of ovoid and pointed cells, which were verified to contain the wsp1 mutant allele by PCR. While this remains an exception, the overall observations are consistent with survival data and CFU estimation and support a necessary role for Wsp1 in cryptococcus-host interactions and wild-type virulence.
DISCUSSION
Wsp1 indicates divergence of WASPs in fungi.
The actin cytoskeleton underlies essential cellular activities in higher eukaryotic cells by regulating processes, including cell division, motility, polarized growth, endocytosis, and exocytosis. WASP has a role in actin dynamics by activating the Arp2/3 protein complex through its C-terminal domain to nucleate actin. WASP also interacts with other proteins through additional domains, such as WH1, B, and GBD/CRIB, to integrate signals for the regulation of actin dynamics. Despite divergence in evolution between higher and lower eukaryotes resulting in differences in details, this mechanism of actin regulation appears to be largely conserved in fungi. In S. cerevisiae, Las17/Bee1 is required for the assembly of cortical actin patches, budding, and cytokinesis, and disruption of the WASP homolog LAS17/BEE1 resulted in a striking change in actin filament organization. Intriguingly, Las17/Bee1 does not contain the GBD/CRIB binding site, which is a conserved feature involved in the autoinhibition/activation of mammalian WASP functions. Since the regulation of mammalian WASP proteins can involve autoinhibition or/and trans-inhibition, it is generally suggested that trans-inhibition may be applied to Las17/Bee1 function. While WASP can also be activated by binding of SH3 domain-containing proteins to its PR-rich domain, the activity of Las17/Bee1 can be inhibited by SH3 proteins, such as SlaI and Bbc1 in S. cerevisiae (for a review see reference 20). Studies of S. pombe Wsp1 and C. albicans Wal1, which are Las17/WASP homologs, demonstrated that these WASP homologs are conserved in function and indicated that fungal WASPs may not contain the GBD/CRIB binding site (17, 32).
We previously identified a multifunctional endocytic protein, Cin1, from the basidiomycetous fungus Cryptococcus neoformans and established a role of Cin1 in the regulation of intracellular transport, actin dynamics, growth, and virulence of the fungus (28). Cin1 shares domain architecture and similar functions with human intersectin ITSN1, and homologs of Cin1 cannot be identified in S. cerevisiae or C. albicans, which has Pan1, an archetypal endocytic protein involved in endocytosis, actin cytoskeleton, and signaling (8, 36). Wsp1 was shown to interact with Cin1 and Cdc42, indicating these three proteins likely participate in a complex pathway integrating various signals to regulate multiple processes, including endocytosis and exocytosis, actin dynamics, and signaling (35). While a detailed Cin1-mediated endocytic pathway(s) remains to be illustrated, the unique compositions of Cin1 and Wsp1 proteins, as well as the recent identification of SH3 domains specific to basidiomycetous Ste50 (13), all suggest divergent evolution of multidomain adaptor proteins within the fungal phylum.
Wsp1 has conserved functions in intracellular transport, actin dynamics, and signaling of C. neoformans.
The presence of a GBD/CRIB domain in Wsp1 suggests that Wsp1 might be subjected to regulation by autoinhibition/activation. A Wsp1-GBD allele-specific mutant was thus included in the general characterization. In short, with the exception of growth assay and bilateral cross results, where reduction of growth (at 37°C) and reduction of mating-specific filaments were apparent, the role of the GBD/CRIB domain appeared to be subtle. This was rather unexpected, in comparison to mammalian WASPs, where an increase in actin polymerization has been reported (2). We reasoned that there are likely two explanations for this. First, GBD/CRIB is not the only region responsible for autoinhibition/activation, and the B domain preceding the GBD/CRIB domain may also be required. The B domain interacts with phosphatidylinositol 4,5-bisphosphate to potentiate the effect of the binding between Cdc42 and the GBD/CRIB domain (for a review, see reference 39). A more noticeable effect of Wsp1 on actin might be more apparent if both B and GBD/CRIB domains were deleted. Alternately, since Wsp1 is a conserved effector of Cdc42 in actin regulation, as demonstrated in many other systems (4), the effect of a Wsp1-GBD/CRIB domain-specific mutant allele may be more apparent if Cdc42 is rendered in a constitutively inactivated state. Nevertheless, further characterization of functions by Wsp1 and Cdc42 and their interactions are warranted.
We have identified Wsp1, based on the assumption that Cin1, like ITSN1, functions by interacting with Wsp1/WASP and Rho GTPase Cdc42 to regulate the actin cytoskeleton. Wsp1 was found to interact with Cin1 in our previous study (28) and to bind with Cdc42 in a yeast two-hybrid assay (Fig. 1B). We reasoned that Wsp1 likely functions in a conserved fashion analogous to WASP, and we proceeded to examine its cellular function. As expected, the wsp1 mutant displayed defects in growth, cytokinesis, chitin distribution, and endocytosis. These functions are in line with those of WASP and Las17/Bee1 protein. Moreover, since the actin cytoskeleton and intracellular transport determine the display of factors contributing to fungal interactions with macrophages and survival in hosts, the wsp1 mutant was unable to cause infection in a murine model. Many of these phenotypes of the wsp1 mutant are quite similar to those of the cin1 mutant, albeit less severe. Further genetic epistasis and biochemical analyses are warranted to explore the relationships between the two in the regulation of actin cytoskeleton and other functions.
Wsp1 underscores the importance of actin cytoskeleton in growth, differentiation, and virulence.
In humans, WAS is a primary immunodeficiency disease involving both T and B lymphocytes, which are cells responsible for providing protection against certain viral and fungal infections (T cells) and precursors to antibody-producing cells (B cells) in normal individuals. T cells from WAS patients fail to proliferate and to secrete interleukin-2 after anti-CD3 stimulation, and lymphocytes of WAS patients also show cytoskeletal abnormalities (27). The disease is caused by missense mutations in the WASP genes, especially in the binding sites for the WASP-interacting proteins (WIP) that regulate WASP function (11).
Our examination of a C. neoformans mutant strain in which the entire WSP1 gene encoding Wsp1 was disrupted provided a direct demonstration for the importance of such proteins. Our phenotypic characterization implicates a role for Wsp1 in growth, cytokinesis, and actin distribution of the fungus. The loss of Wsp1 function resulted in the reduced ability for the fungus to infiltrate macrophages and infect a murine model animal. Clearly, while the human WAS diseases were the results of altered or attenuated protein functions, phenotypes exhibited by the wsp1 mutant of C. neoformans provided a direct assessment on the effect of the loss of protein function. It is therefore feasible to propose that study of Wsp1 function can provide an alternate but direct approach to explore human WASP function. True to the “simpler” model of C. neoformans versus much more complex mammalian systems, no WIP proteins have been identified from the C. neoformans genome, suggesting that Wsp1 may be subject to a level of regulation far less complicated than that of the mammalian systems. Thus, regulation by Cdc42, a putative upstream activator, and Cin1, whose SH3 and RhoGEF domains bind and potentially activate Wsp1, all point out the importance of exploring the genetic relationship among these three proteins. Such explorations are no doubt imperative in the study of C. neoformans and in identifying novel anticryptococcal agents.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. E. Cutler for comments and Sarah Martin for technical assistance.
This study was supported in part by NIH grants (AI054958 and AI074001) and funding from the Research Institute for Children, New Orleans, LA.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Ballou E. R., Nichols C. B., Miglia K. J., Kozubowski L., Alspaugh J. A. 2010. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol. Microbiol. 75:763–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cai L., Bear J. E. 2008. Peering deeply inside the branch. J. Cell Biol. 180:853–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casadevall A., Perfect J. R. 1998. Cryptococcus neoformans. ASM Press, Washington, DC [Google Scholar]
- 4. Cotteret S., Chernoff J. 2002. The evolutionary history of effectors downstream of Cdc42 and Rac. Genome Biol. 3:REVIEWS0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Derry J. M., Ochs H. D., Francke U. 1994. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78:635–644 [DOI] [PubMed] [Google Scholar]
- 6. Evangelista M., et al. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118–122 [DOI] [PubMed] [Google Scholar]
- 7. Guo W., Roth D., Walch-Solimena C., Novick P. 1999. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18:1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang B., Cai M. 2007. Pan1p: an actin director of endocytosis in yeast. Int. J. Biochem. Cell Biol. 39:1760–1764 [DOI] [PubMed] [Google Scholar]
- 9. Huang T. Y., Renaud-Young M., Young D. 2005. Nak1 interacts with Hob1 and Wsp1 to regulate cell growth and polarity in Schizosaccharomyces pombe. J. Cell Sci. 118:199–210 [DOI] [PubMed] [Google Scholar]
- 10. James P., Halladay J., Craig E. A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jin F. J., Maruyama J., Juvvadi P. R., Arioka M., Kitamoto K. 2004. Adenine auxotrophic mutants of Aspergillus oryzae: development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 68:656–662 [DOI] [PubMed] [Google Scholar]
- 12. Johnson D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klosterman S. J., Martinez-Espinoza A. D., Andrews D. L., Seay J. R., Gold S. E. 2008. Ubc2, an ortholog of the yeast Ste50p adaptor, possesses a basidiomycete-specific carboxy terminal extension essential for pathogenicity independent of pheromone response. Mol. Plant Microbe Interact. 21:110–121 [DOI] [PubMed] [Google Scholar]
- 14. Kopecka M., et al. 2001. Microtubules and actin cytoskeleton in Cryptococcus neoformans compared with ascomycetous budding and fission yeasts. Eur. J. Cell Biol. 80:303–311 [DOI] [PubMed] [Google Scholar]
- 15. Kurisu S., Takenawa T. 2009. The WASP and WAVE family proteins. Genome Biol. 10:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon-Chung K. J., Edman J. C., Wickes B. L. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee W. L., Bezanilla M., Pollard T. D. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151:789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li R. 1997. Bee1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J. Cell Biol. 136:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitchell T. G., Perfect J. R. 1995. Cryptococcosis in the era of AIDS: 100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moseley J. B., Goode B. L. 2006. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70:605–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naqvi S. N., Zahn R., Mitchell D. A., Stevenson B. J., Munn A. L. 1998. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 8:959–962 [DOI] [PubMed] [Google Scholar]
- 22. Nichols C. B., Perfect Z. H., Alspaugh J. A. 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63:1118–1130 [DOI] [PubMed] [Google Scholar]
- 23. Okuda M., Ikeda K., Namiki F., Nishi K., Tsuge T. 1998. Tfo1: an Ac-like transposon from the plant pathogenic fungus Fusarium oxysporum. Mol. Gen. Genet. 258:599–607 [DOI] [PubMed] [Google Scholar]
- 24. Palmer D. A., Thompson J. K., Li L., Prat A., Wang P. 2006. Gib2, a novel Gβ-like/RACK1 homolog, functions as a Gβ subunit in cAMP signaling and is essential in Cryptococcus neoformans. J. Biol. Chem. 281:32596–32605 [DOI] [PubMed] [Google Scholar]
- 25. Park H. O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qualmann B., Kessels M. M., Kelly R. B. 2000. Molecular links between endocytosis and the actin cytoskeleton. J. Cell Biol. 150:111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramesh N., Geha R. 2009. Recent advances in the biology of WASP and WIP. Immunol. Res. 44:99–111 [DOI] [PubMed] [Google Scholar]
- 27a. Rohatgi R., Ho H. Y., Kirschner M. W. 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150:1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen G., Whittington A., Song K., Wang P. 2010. Pleiotropic function of intersectin homologue Cin1 in Cryptococcus neoformans. Mol. Microbiol. 76:662–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith M. G., Swamy S. R., Pon L. A. 2001. The life cycle of actin patches in mating yeast. J. Cell Sci. 114:1505–1513 [DOI] [PubMed] [Google Scholar]
- 30. Vallim M. A., Nichols C. B., Fernandes L., Cramer K. L., Alspaugh J. A. 2005. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 4:1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veltman D. M., Insall R. H. 2010. WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell 21:2880–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walther A., Wendland J. 2004. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich syndrome protein homolog Wal1p. Eukaryot. Cell 3:471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang P., Cutler J. E., King J., Palmer D. 2004. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 3:1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang P., Perfect J. R., Heitman J. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang P., Shen G. 24 January 2011. The endocytic adaptor proteins of pathogenic fungi: charting new and familiar pathways. Med. Mycol. doi:10.3109/13693786.2011.553246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wendland B., Emr S. D. 1998. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141:71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang H. C., Pon L. A. 2002. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 99:751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoneda A., Doering T. L. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 17:5131–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X., Machesky L. M. 22 April 2010. N-WASP. UCSD Nat. Mol. Pages. doi:10.1038/mp.a000091.01 [Google Scholar]
- 40. Zigmond S. H. 2000. How WASP regulates actin polymerization. J. Cell Biol. 150:F117–F120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.