Abstract
The polymorphic yeast Candida albicans exists in yeast and filamentous forms. Given that the morphogenetic switch coincides with the expression of many virulence factors, the yeast-to-hypha transition constitutes an attractive target for the development of new antifungal agents. Since an untapped therapeutic potential resides in small molecules that hinder C. albicans filamentation, we characterized the inhibitory effect of conjugated linoleic acid (CLA) on hyphal growth and addressed its mechanism of action. CLA inhibited hyphal growth in a dose-dependent fashion in both liquid and solid hypha-inducing media. The fatty acid blocked germ tube formation without affecting cellular growth rates. Global transcriptional profiling revealed that CLA downregulated the expression of hypha-specific genes and abrogated the induction of several regulators of hyphal growth, including TEC1, UME6, RFG1, and RAS1. However, neither UME6 nor RFG1 was necessary for CLA-mediated hyphal growth inhibition. Expression analysis showed that the downregulation of TEC1 expression levels by CLA depended on RAS1. In addition, while RAS1 transcript levels remained constant in CLA-treated cells, its protein levels declined with time. With the use of a strain expressing GFP-Ras1p, CLA treatment was also shown to affect Ras1p localization to the plasma membrane. These findings suggest that CLA inhibits hyphal growth by affecting the cellular localization of Ras1p and blocking the increase in RAS1 mRNA and protein levels. Combined, these effects should prevent the induction of the Ras1p signaling pathway. This study provides the biological and molecular explanations that underlie CLA's ability to inhibit hyphal growth in C. albicans.
INTRODUCTION
Over the past decades, opportunistic fungal infections have gained importance among hospital-acquired infections due to a growing community of individuals immunocompromised by HIV infection, cancer treatment, or organ transplantation (74). The opportunistic pathogen Candida albicans, a member of the normal human microbiota, inhabits the gastrointestinal and genitourinary tracts, mucous membranes, and skin. It is responsible for various forms of diseases, ranging from superficial infections of mucosal surfaces to severe, life-threatening systemic infections that largely depend on a host's physical and physiological conditions. C. albicans is the fourth leading cause of nosocomial infections and the most common fungal species causing bloodstream infections, with associated mortality rates of 38 to 49% (27, 70, 87). Treatment of such infections is complicated by a limited number of antifungal drugs, many of which have adverse side effects, and by emerging resistance to all clinically useful antifungals.
The success of C. albicans as a pathogen stems from its ability to deploy a full armada of activities that contribute to its virulence, including the production of proteases, adhesins, and phospholipases as well as its ability to switch morphologically and phenotypically (11, 48). The most studied morphological switch, the yeast-to-hypha transition, is induced by a variety of environmental cues, including elevated temperature, neutral or alkaline pH, nitrogen and/or carbon starvation, and growth in serum (30). A complex network of signaling pathways regulates hyphal growth (4, 12, 85), among which the Ras1p-cyclic AMP (cAMP)-protein kinase A (PKA) and the mitogen-activated protein (MAP) kinase pathways play major roles. Both cascades are controlled by the small GTPase Ras1p (53). Membrane-bound, Ras1p binds the Ras association domain of the adenylate cyclase Cyr1p, thereby stimulating the cyclization of ATP into cAMP (31). Upon binding cAMP, the regulatory subunit of PKA Bcy1p releases the catalytic subunits Tpk1p and Tpk2p, thereby activating PKA (16). The transcription factor Efg1p is a downstream target of PKA (5, 81). Ras1p also impinges on the MAP kinase pathway, downstream of which lies the transcription factor Cph1p (55). Other transcription factors involved in hyphal growth include Tec1p, Flo8p, Cph2p, Ume6p, Rim101p, and Czf1p (1, 13, 14, 22, 50, 51, 76, 88). Hyphal growth is negatively controlled by the general repressor Tup1p in association with the DNA-binding proteins Nrg1p and Rfg1p (8, 10, 42, 59). Activation of these signal transduction cascades modulates the expression of hypha-specific genes, many of which are involved in virulence (4, 12).
Although it has recently been demonstrated that the yeast-to-hypha transition is not always required for infectivity in systemic candidiasis (65), it is generally accepted that hyphal growth is critical for virulence in various types of C. albicans infections. Several lines of evidence link the yeast-to-hypha transition to pathogenicity, the first being that mutants locked in either yeast (cph1/cph1, efg1/efg1, and hgc1/hgc1) or filamentous (tup1/tup1 and nrg1/nrg1) forms are avirulent in systemic candidiasis (7, 56, 59, 89). Filamentation is required for C. albicans to evade phagocytes and escape from blood vessels (57, 71). Colonization of medical devices by biofilms depends on hyphal development and the expression of hypha-specific gene products (64). In parallel, using C. albicans strains in which hyphal growth can be manipulated externally, several groups have shown that inhibiting filamentation is a means by which virulence may be attenuated during systemic candidiasis (15, 75). Moreover, small molecules that block filamentation have been shown to exert a protective effect in mucosal candidiasis (36) and to reduce C. albicans-induced damage to endothelial cells (83). Not only do these findings demonstrate an association between filamentation and virulence in C. albicans, but they also suggest that the yeast-to-hypha transition may constitute a therapeutic target.
We recently isolated fatty acids from bovine whey that had the ability to inhibit germ tube formation in C. albicans (19). Given that the inability to switch to a hyphal form reduces the virulence potential of C. albicans, we characterized the inhibitory effect of conjugated linoleic acid (CLA) on hyphal growth and investigated its mechanism of action. We demonstrate that CLA inhibits hyphal development in a dose-dependent fashion in both liquid- and solid-inducing media. Global gene expression analysis reveals that CLA affects the expression of hypha-specific genes and of several morphogenesis regulators, including RAS1, TEC1, and UME6. We show that Ras1p is required for CLA to downregulate TEC1 expression levels. We demonstrate that in the presence of CLA, Ras1p cellular levels decrease. In addition, CLA causes the delocalization of Ras1p from the plasma membrane. These findings suggest that CLA inhibits hyphal growth in C. albicans by affecting Ras1p signaling and downregulating the expression of downstream targets, including TEC1 and UME6.
MATERIALS AND METHODS
Strains and growth conditions.
The yeast strains used in this study are listed in Table 1. Strains were streaked out onto YPD plates (1% yeast extract, 2% peptone, 2% dextrose, 2% Bacto agar) and grown at 30°C for 24 to 48 h. In all of the experiments performed, strains were propagated overnight in YPD at 30°C to an optical density at 600 nm (OD600) of ∼12 to 14. Hyphal inductions were performed at 37°C on solid and in liquid media, using Spider (55), Lee's (54), and buffered alkaline (pH 8.0) M199 (Wisent) media. Media were solidified with 2% Bacto agar (Oxoid). When necessary, media were supplemented with uridine (50 μg ml−1). Conjugated linoleic acid (CLA) (Cayman Chemicals) was diluted in ethanol as a 1,000× stock and added to media in concentrations ranging from 0 to 250 μM. Ethanol was used as drug vehicle (final concentration, ≤0.5%).
Table 1.
Candida albicans strains used in this study
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| SC5314 | Candida albicans wild-type clinical isolate | 33 | |
| CAI4 | ura3::λimm434/ura3::λimm434 | 32 | |
| ZK3379 | CAI4 | HWP1-lacZ-URA3 | 38 |
| MRC6 | CAI4 | fox2::hisG/fox2::hisG RPS10/rps10::URA3 | 72 |
| MRC10 | CAI4 | icl1::hisG/icl1::hisG RPS10/rps10::URA3 | 72 |
| HLC52 | CAI4 | efg1::hisG/efg1::hisG-URA3-hisG | 56 |
| CDH107 | CAI4 | ras1::hisG/ras1::hisG-URA3-hisG | 53 |
| BCa2-10 | CAI4 | tup1::hisG/tup1::hisG-URA3-hisG | 8 |
| BCa23-3 | CAI4 | nrg1::hisG/nrg1::hisG-URA3-hisG | 10 |
| DK129 | CAI4 | rfg1::hisG/rfg1::hisG-URA3-hisG | 42 |
| IIHB6 | CAI4 | tpk1::hisG/tpk1::hisG-URA3-hisG | 6 |
| TPO7.4 | CAI4 | tpk2::hisG/tpk2::hisG-URA3-hisG | 80 |
| WY-ZXD3 | CAI4 | RAS1/GFP-RAS1-hisG-URA3-hisG | 90 |
| RM1000 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | 62 | |
| MRC41 | RM1000 | ctf1::HIS1/ctf1::hisG RPS10/rps10::URA3 | 73 |
| BWP17 | ura3::λimm434/ura3::λimm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG | 86 | |
| CJN308 | BWP17 | tec1::Tn7-UAU1/tec1::Tn7-URA3 | 63 |
| SN95 | ura3::λimm434/URA3 iro1::λimm434/IRO1 arg4/arg4 his1/his1 | 66 | |
| SN152 | SN95 | ura3::λimm434/URA3 iro1::λimm434/IRO1 arg4/arg4 leu2/leu2 his1/his1 | 66 |
| DK318 | SN95 | arg4::ARG4/arg4 his1::HIS1/his1 | 1 |
| DK312 | SN152 | arg4::ARG4/arg4 leu2/leu2 his1/his1 ume6:: C.m.LEU2/ume6::C.d.HIS1 | 1 |
Hyphal growth assays in liquid and on solid media.
To quantify the inhibitory effect of CLA on C. albicans hyphal growth, β-galactosidase activity was measured using the C. albicans reporter strain ZK3379, in which lacZ is under the control of the hypha-specific HWP1 promoter (38). Briefly, cells from an overnight culture were washed in sterile distilled water and diluted to 1 × 106 cells ml−1 in prewarmed medium. Twenty-four-well polystyrene microplates (Costar 3526; Corning, NY) were seeded with 1 ml of inoculated medium per well. Working solutions of CLA were freshly prepared and added immediately to seeded wells in volumes of 5 μl, yielding final concentrations ranging from 0 to 50 μM. Microplates were incubated statically in a water bath set to 37°C for 4 h. β-Galactosidase activity was quantified as described previously (47). Data are represented as the means and standard deviations of results from replicate assays performed on three independent days. To assess the effect of CLA on colony morphology, C. albicans cells from an overnight culture were washed in sterile distilled water and spread (ca. 100 colonies per plate) on plates of solidified medium supplemented with ethanol or CLA (100 μM). Plates were incubated at 37°C for 3 to 4 days and photographed using a Leica MZ FLIII fluorescence stereomicroscope mounted with a Micropublisher camera. To examine the effect of CLA on hyphal growth of C. albicans wild-type and mutant strains, cells from an overnight culture were washed in sterile distilled water and diluted to a concentration of 1 × 106 cells ml−1 in prewarmed Spider medium supplemented with ethanol or CLA (25 μM). Flasks were shaken at 150 rpm at 37°C. Aliquots of cells were harvested at various time points, fixed with 3.7% formaldehyde for 30 min at room temperature, washed twice with 1× phosphate-buffered saline (PBS), and visualized by differential interference contrast (DIC) microscopy using an upright Nikon microscope with a 100× immersion oil objective and a 10× projection lens mounted with a Nikon DXM1200F digital camera. To distinguish hyphae from pseudohyphae, fixed cells were washed twice in water, stained for 5 min with 4′-6′diamidino-2-phenyl-indole (DAPI; 1 mg ml−1) diluted 1:1,000 and calcofluor white (1 mg ml−1) diluted 1:10, examined microscopically using epifluorescence, and classified according to the criteria of Sudbery et al. (82). To ensure that CLA treatment did not compromise cellular growth rates, growth curves were generated. SC5314 cells from an overnight culture were diluted to 1 × 106 cells ml−1 in Spider medium supplemented with ethanol or CLA (25 μM). Flasks were shaken at 150 rpm at 30°C. OD600 was measured every hour for 7 h. Data are means and standard deviations of results from duplicate biological samples. The effect of CLA (250 μM) on cellular growth rates in YPD at 30°C was assessed similarly.
Gene expression profiling.
SC5314 cells grown overnight in YPD at 30°C were washed in sterile distilled water and diluted to 5 × 106 cells ml−1 (OD600 of 0.1) in Spider medium supplemented with ethanol or CLA (100 μM). Cultures were shaken at 150 rpm at either 30°C or 37°C for 90 min. Cells were collected by vacuum filtration on 0.45-μm membrane filters (MF-Millipore membrane filters) and frozen in an ethanol bath at −80°C. Total RNA was isolated from quadruplicate independent biological samples using an RNeasy minikit (Qiagen). Briefly, frozen cells were thawed out in RNeasy buffer RLT at a ratio of 3:1 (vol/vol) buffer/pellet. Resuspended cells were divided into 1-ml aliquots in 2-ml screw-cap microcentrifuge tubes containing 0.6 ml of 0.5-mm-diameter acid-washed glass beads. Samples were homogenized 6 times, for 5 min each, in a BeadBeater set at maximum speed. Lysates were kept on ice between each cycle. Total RNA was extracted from homogenized samples according to the Qiagen RNeasy protocol. For the two-color microarray experiments, RNA from cells grown at 37°C or in the presence of CLA at 30°C or 37°C was compared to RNA from cells grown at 30°C. Four biological replicates were used in each experiment, which included two Cy3/Cy5 and two Cy5/Cy3 comparisons. cDNA labeling, microarray hybridization, washing, scanning, and statistical analysis methods were essentially performed as described previously (77). In each comparison, genes with statistically significant modulations were identified in volcano plots using a 2.0-fold cutoff point and a Welsh t test with a false-discovery rate of less than 5%.
Northern blot analysis.
Cells were grown as described above for gene expression profiling. RNA was prepared using the hot-phenol method. Fifteen micrograms of total RNA was separated on 1.2% agarose-7.5% formaldehyde denaturing gels and transferred by capillary action to Hybond-N+ nylon membranes (GE Healthcare Life Sciences). Probes were generated by PCR amplification of genomic DNA and purified using an Illustra GFX PCR DNA and gel band purification kit (GE Healthcare Life Sciences). The sequences of the primers used to generate all probes are listed in Table 2. Fifty nanograms of probe was labeled by random priming using Ready-To-Go DNA labeling beads (GE Healthcare Life Sciences) and [α-32P]dCTP. Unincorporated nucleotides were removed using Sephadex G-50 columns. Blots were hybridized overnight and washed at 65°C according to the method of Church and Gilbert (18), scanned using a Molecular Dynamics Typhoon phosphorimager, and quantified with ImageQuant software (version 5.0; Molecular Dynamics). Data are means and standard deviations of results from duplicate biological samples.
Table 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Product size (bp) |
|---|---|---|
| qPCR ACT1F | TCCAGAAGCTTTGTTCAGACCAGC | 170 |
| qPCR ACT1R | TGCATACGTTCAGCAATACCTGGG | |
| qPCR RAS1F | GTTGTTGTTGGAGGTGGTGGTGTT | 180 |
| qPCR RAS1R | GGCCAGATATTCTTCTTGTCCAGC | |
| qPCR TUP1F | CCAGCACCAACAACGTTTGACAGA | 176 |
| qPCR TUP1R | TGGGCCAACTCCAAGTCATACACT | |
| qPCR NRG1F | TGGTGATTTACTGGCCAACTCCCT | 180 |
| qPCR NRG1R | CATGTTGGCCATGGACATTGGTGT | |
| ACT1F probe | GTTGACCGAAGCTCCAATGAATCC | 629 |
| ACT1R probe | TGCATACGTTCAGCAATACCTGGG | |
| TEC1F probe | GTTACCACCACGAGCACTGGC | 486 |
| TEC1R probe | TGAAGGGTGTTGGCTATTATGCG |
qPCR analysis of C. albicans transcripts.
For quantitative PCR (qPCR) analysis, C. albicans cells from an overnight culture were diluted to 1 × 106 cells ml−1 in Spider medium supplemented with ethanol or CLA (25 μM) and grown at 37°C. Cells were collected at various time points (0, 30, 60, and 90 min) by vacuum filtration on 0.45-μm membrane filters (MF-Millipore membrane filters) and frozen in an ethanol bath at −80°C. Total RNA was isolated from duplicate independent biological samples for each condition and time point using the hot-phenol method. RNA was resuspended in 50 to 200 μl diethyl pyrocarbonate-treated water, quantified by a spectrophotometer (NanoDrop 2000; Thermo Scientific), and stored at −80°C. RNA samples were DNase digested (rDNase I; Ambion) and used as templates in qPCR amplification reactions to certify them as DNA free. The lack of a PCR product indicated that samples were not contaminated with genomic DNA. Five hundred nanograms of total RNA was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Two cDNAs were synthesized for each biological replicate. Quantitative PCR was carried out on a Rotor-Gene 6000 (Corbett Life Science). The primers were designed with PrimerQuest (Integrated DNA Technologies) and are listed in Table 2. The qPCR mixtures contained 12.5 μl FastStart SYBR green master mix (Roche Applied Science), 8.5 μl Milli-Q water, 200 nM each primer, and 1 μl cDNA product diluted 1:100, and the qPCRs were performed in duplicates. Generally, the difference between two threshold cycle (CT) values for the same sample was <0.5. Relative expression levels were calculated using the delta-delta CT method [2(CT for target condition − CT for ACT1 condition) − (CT for target at time zero − CT for ACT1 at time zero)], in which the condition was either ethanol or CLA treatment and ACT1 was the housekeeping gene.
Protein extraction and immunoblotting.
Wild-type and tagged strains were grown in Spider medium and harvested by following the same procedure as that described above for gene expression profiling. Total protein extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1% Na-deoxycholate [DOC], 0.1% SDS) containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitors (Complete EDTA-free tablets [Roche Applied Science]). Total extracts (50 μg) were resolved by SDS-PAGE (7.5%) and transferred to Hybond enhanced chemiluminescence (ECL) nitrocellulose membranes (GE Healthcare Life Sciences). To detect green fluorescent protein (GFP)-Ras1p, mouse anti-GFP antibodies (Roche Applied Science) (1:1,000 in Tris-buffered saline–Tween [TBS-T]–5% nonfat milk) were used. Gsp1p protein levels, shown as a loading control, were detected using rabbit anti-Gsp1p antibodies (3) (1:10,000 in TBS-T–5% nonfat milk) overnight at 4°C. Signals were detected using Lumi-Light Western blotting substrate (Roche Applied Science).
Microarray data accession number.
Microarray data sets can be found at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE25822.
RESULTS
CLA inhibits hyphal growth in C. albicans in response to various hypha-inducing conditions.
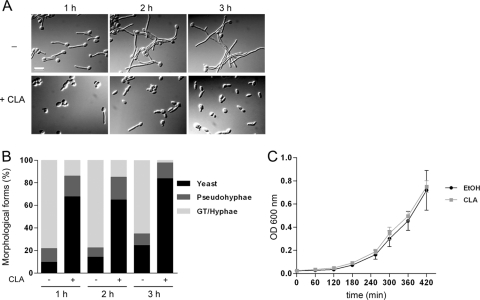
Several fatty acids, including conjugated linoleic acid (CLA), were recently shown to inhibit Candida albicans germ tube formation in various hypha-inducing liquid media (19). We first assessed the inhibitory effect of CLA on hypha formation induced in different conditions using the C. albicans reporter strain HWP1p-lacZ, in which β-galactosidase activity reflects the amount of hyphal growth in cultures (38). In Spider, Lee's, and M199 (pH 8.0) media, CLA inhibited hyphal growth in a dose-dependent fashion, albeit to various extents (Fig. 1A). In all media, 50 μM of the fatty acid reduced β-galactosidase activity by more than 80% compared to the level for ethanol-containing cultures, confirming that the presence of CLA impeded filamentation. In Spider medium supplemented with 25 μM CLA, cells grew as yeasts and short pseudohyphae, reflecting reduced β-galactosidase activity levels (Fig. 1A and B). CLA also disrupted filamentation on solid media, being more effective in Spider medium (Fig. 1C). However, hyphal growth inhibition by CLA was medium dependent. For instance, in RPMI 1640 and YPD-10% fetal bovine serum (FBS) liquid media, CLA inhibited filamentation, but effective concentrations tended to be higher (19; data not shown). In addition, CLA had no effect on hyphal growth induced on solid YPD-10% FBS, synthetic low-ammonia dextrase (SLAD), or yeast nitrogen base (YNB) supplemented with N-acetylglucosamine or upon embedding of cells in yeast extract-peptone (YP) medium (data not shown).
Fig. 1.
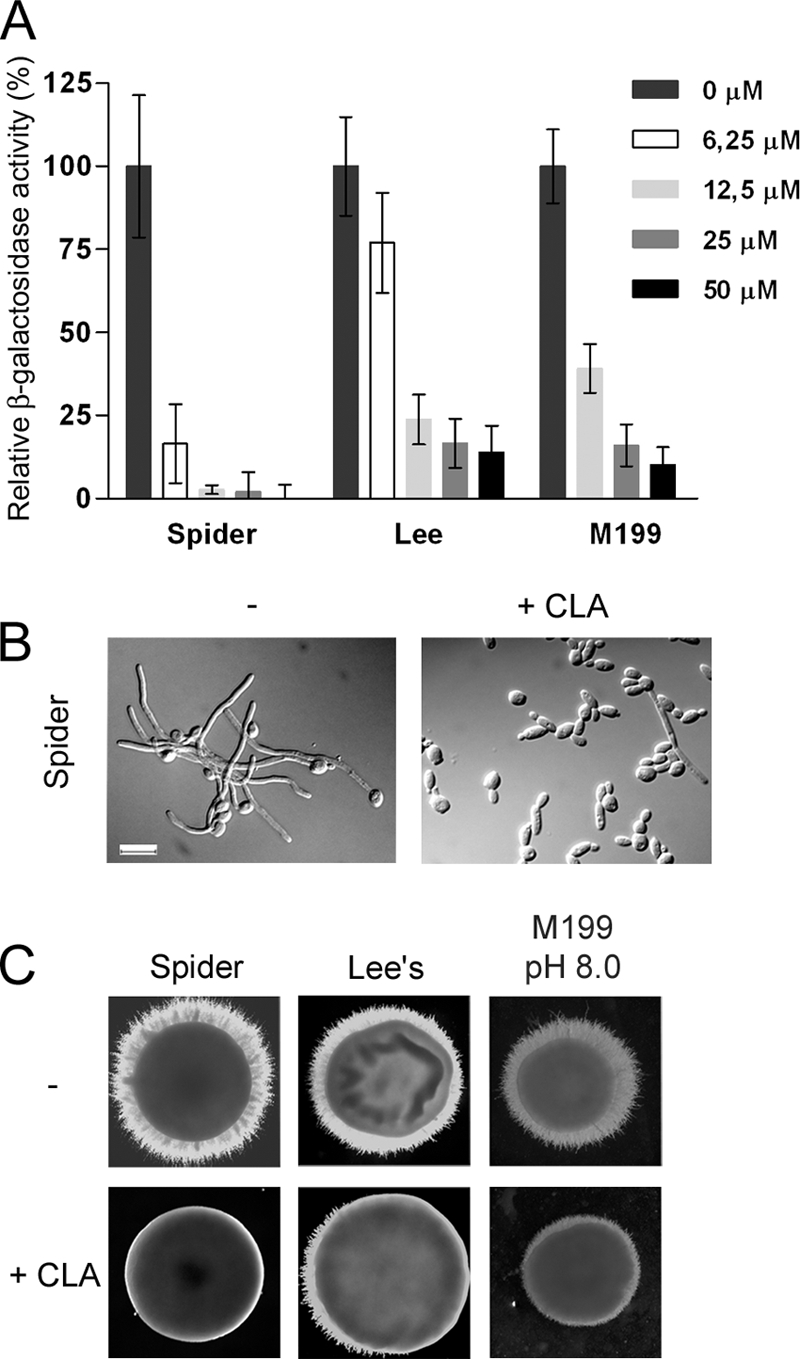
Conjugated linoleic acid (CLA) inhibits hyphal growth in Candida albicans. (A) The effect of CLA on C. albicans hyphal growth was measured by using the C. albicans reporter strain ZK3379. Cells were induced to filament in Spider, Lee's, and M199 (pH 8.0) media supplemented with CLA (0 to 50 μM) for 4 h at 37°C. β-Galactosidase activity in CLA-treated cultures was measured and normalized to that of untreated cultures, in which β-galactosidase activity was set as 100%. Data are means and standard deviations of results from duplicate assays performed on three independent days. (B) Aliquots of cells grown in Spider medium in the absence or presence of 25 μM CLA for 4 h at 37°C were harvested and visualized at ×100 magnification using DIC optics. Bar = 10 μm. (C) Filamentous growth of C. albicans SC5314 was induced on solid media supplemented with ethanol or 100 μM CLA. Plates were incubated at 37°C for 3 to 4 days.
Other fatty acids, such as oleic, linoleic, and α- and γ-linolenic acids also modulated hyphal growth in C. albicans. Levels of β-galactosidase activity, as well as cellular and colony morphology, indicated that all fatty acids, like CLA, interfered to different extents with C. albicans hyphal growth induced under various conditions (data not shown). In addition, fatty acids, including CLA, inhibited filamentation in Candida tropicalis and C. dubliniensis (data not shown), in Aspergillus fumigatus, and in Fusarium graminearum (20; data not shown), suggesting the response to fatty acids and to CLA may be conserved among other fungi.
CLA impedes germ tube formation without affecting cellular growth.
With the use of the HWP1p-lacZ strain and β-galactosidase quantification, CLA and other fatty acids were shown to abrogate, delay, or decrease the induction of HWP1 promoter activity, indirectly suggesting that hyphal growth was impaired (Fig. 1A) (19). To assess which step of the yeast-to-hypha transition was affected by CLA, we examined the morphology of cells induced to filament in Spider medium in the absence or presence of CLA. Untreated cells showed long germ tubes and/or very short filaments and long hyphae at the 1- and 3-h time points, respectively. At the same time points, most CLA-treated cells appeared as elongated yeasts or showed elongating buds that resembled germ tubes (Fig. 2A). With the use of the criteria established by Sudbery et al. (82), cells were stained with DAPI and calcofluor white and classified as yeast, pseudohyphae, or germ tubes/hyphae. As seen in Fig. 2B, 80 to 90% of untreated cells formed hyphae and pseudohyphae over the 3-h time course. In contrast, the majority (70 to 80%) of cells grown in the presence of CLA remained in the yeast form. While some CLA-treated cells did initiate germ tube formation, elongating filaments were not detected by the 3-h time point, indicating that these cells had resumed pseudohyphal and yeast growth modes. These findings suggest that CLA prevents germ tube formation and hyphal elongation from occurring. CLA also blocked hyphal elongation in cells that were already engaged in the hyphal growth program (data not shown).
Fig. 2.
CLA impedes germ tube formation of Candida albicans without affecting cellular growth. (A) C. albicans SC5314 cells were induced to filament in Spider medium at 37°C in the absence or presence of 25 μM CLA. Aliquots of cells were visualized as described for Fig. 1B. Bar = 10 μm. (B) Quantification of yeast, pseudohyphae, germ tubes (GT) and hyphae in C. albicans cultures shown in panel A. n was >150 for each condition and time point. (C) Yeast cells were grown as described for panel A at 30°C. OD600 was measured at various time points. Data are means and standard deviations of results from duplicate biological samples.
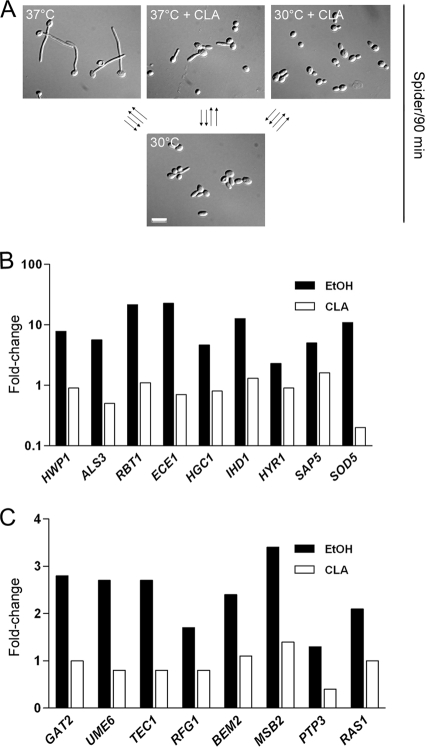
C. albicans filamentation can be inhibited by cytotoxic or cytostatic molecules (83). Since CLA interfered with hyphal growth, we assessed whether cellular growth rates at 30°C in Spider medium were affected by ethanol or CLA. Under these conditions, germ tube formation is slightly induced at early time points (Fig. 3A), but hyphal growth is not maintained and cells eventually resume budding growth (data not shown). The growth rates of untreated and CLA-treated cells were identical, suggesting that CLA does not inhibit hyphal growth by exerting cytotoxic or cytostatic effects (Fig. 2C). Similar results were obtained for yeast cells grown in the absence or presence of CLA in YPD medium (data not shown).
Fig. 3.
Expression levels of selected differentially expressed genes. (A) Transcriptional profiles of untreated cells at 37°C and CLA-treated cells at 37°C and 30°C were obtained by independently comparing the levels for each experimental condition, i.e., 37°C, 37°C with CLA, and 30°C with CLA, to those for the control condition, growth at 30°C. Four biological replicates were used in each experiment, which included two Cy3/Cy5 and two Cy5/Cy3 comparisons. Representative micrographs of cells used in the microarray experiments are shown. Bar = 10 μm. Fold change values are shown for hypha-specific genes (B) and genes involved in signal transduction (C) in untreated and CLA-treated cells at 37°C. The significantly differentially expressed genes were obtained by comparing the transcriptional profile of cells at 37°C with that of CLA-treated cells at 37°C and are listed in Table S4 in the supplemental material. Data are fold change values. EtOH, ethanol.
Gene expression analysis.
To gain further insight into the inhibitory effect of CLA on hyphal growth, we performed global gene expression profiling of cells grown in Spider medium in the absence or presence of CLA for 90 min. Transcriptional profiles of untreated cells at 37°C and CLA-treated cells at 37°C and 30°C were obtained by independently comparing the levels for each experimental condition, i.e., 37°C, 37°C with CLA, and 30°C with CLA, to those for the control condition, growth at 30°C. Each experiment was performed in quadruplicate: RNA was prepared from four independent biological replicates and used to perform four independent hybridizations (Fig. 3A). Only genes that were modulated 2-fold up or down with a P value of <0.05 were deemed significantly differentially expressed. Data presented in Tables S1, S2, and S3 in the supplemental material show the transcriptional profiles of each experimental condition compared to the control condition. The transcriptional profiles of the experimental conditions were then compared to one another, i.e., 37°C versus 37°C with CLA and 30°C with CLA versus 37°C with CLA, resulting in two other transcriptional profiles (Tables S4 and S5) and the scatter plots in Fig. S1A. All significantly differentially expressed genes originating from the five transcriptional profiles generated were organized by hierarchical clustering, yielding a global transcriptional profile of 714 modulated transcripts (Table S6 and Fig. S1B). The hierarchical clustering revealed that 61 genes were downregulated under all three experimental conditions (Fig. S1B). Gene Ontology (GO) term analysis revealed these genes were involved in RNA metabolic processes, ribosome biogenesis, translation, and transcription, reflecting a repression of the translational machinery. These findings suggest that experimental growth conditions, i.e., 37°C and CLA treatment, are less favorable than control growth conditions, i.e., 30°C, and result in a metabolic decrease (61).
We first chose to investigate the transcriptional profile of cells grown in Spider medium at 37°C. Gene expression analysis showed that 520 genes were modulated upon growth at 37°C (see Table S1 in the supplemental material). Of those genes, 198 were upregulated while 322 were downregulated. Interestingly, ∼30% of the upregulated genes have been described as being induced during the yeast-to-hypha transition in Lee's medium (34) (Table S1). Hypha-specific genes such as ECE1, RBT1, IHD1, SOD5, HWP1, ALS3, and HGC1 were highly induced (34, 61, 89). Genes involved in signal transduction, including the GTPase gene RAS1, the adenylate cyclase gene CYR1, the mucin-like signaling protein gene MSB2, and the Rho1p GTPase-activating protein (GAP) gene BEM2, were also upregulated (Tables 3 and S1). In addition, several transcription factors known to be involved in hyphal growth were among the 198 upregulated genes and included CPH1, CPH2, TEC1, BCR1, and UME6 (Table 3). These transcription factors are components of the MAP kinase, the Ras1p-cAMP-PKA, and the Tup1p-Nrg1p signaling pathways (1, 43, 50, 51, 55, 63, 88), except CPH2, which appears to function independently of known signaling cascades (51). Other upregulated genes encoded transcription factors Gat2p and Cas4p (34), while orf19.6705 encodes a nucleotide exchange factor. Taken together, the transcriptional profile of cells grown at 37°C suggests that several signaling pathways participate in orchestrating the hyphal growth program in Spider medium.
Table 3.
Selected genes upregulated during the yeast-to-hypha transition in Spider medium
| Systematic name | Gene name | Function | Fold change |
|---|---|---|---|
| orf19.1187 | CPH2 | Transcriptional activator of hyphal growth | 3.0 |
| orf19.6705 | Putative guanyl nucleotide exchange factor with Sec7p domain | 2.9 | |
| orf19.4056 | GAT2 | Putative DNA-binding transcription factor | 2.8 |
| orf19.5908 | TEC1 | TEA/ATTS transcription factor involved in regulation of hypha-specific genes | 2.7 |
| orf19.1822 | UME6 | Transcription factor; required for wild-type hyphal extension | 2.7 |
| orf19.1693 | CAS4 | Protein of RAM cell wall integrity signaling network; role in cell separation; required for hyphal growth | 2.5 |
| orf19.723 | BCR1 | Transcription factor required for wild-type biofilm formation | 2.3 |
| orf19.4433 | CPH1 | Transcription factor required for mating and hyphal growth on solid media | 2.1 |
| orf19.5148 | CYR1 | Adenylate cyclase | 2.0 |
| orf19.1760 | RAS1 | RAS signal transduction GTPase; regulates cAMP and MAP kinase pathways | 2.1 |
| orf19.1490 | MSB2 | Mucin family member, possible sensor of cell wall damage | 3.4 |
| orf19.6573 | BEM2 | Putative Rho1p GTPase-activating protein (GAP) | 2.4 |
Next, we examined the responses of cells to CLA at 30°C and 37°C. We had designed the microarray experiments to be able to distinguish between the transcriptional changes associated with morphological differences induced by growth at 37°C and those elicited specifically by the fatty acid. However, cultures of C. albicans grown in Spider medium at 30°C still contained a significant number of pseudohyphal cells (Fig. 3A), making it impossible to fully eliminate morphogenesis as a variable. Thus, transcriptional profiles of CLA's effects at both temperatures turned out to be highly similar, as seen in the scatter plot (see Fig. S1A in the supplemental material). Indeed, when the transcriptional profiles of CLA-treated cells at 37°C and those at 30°C were directly compared, only 33 genes were found to be differentially modulated (see Table S5 in the supplemental material), and many of these have previously been shown to be temperature regulated (28). We thus focused our functional analysis on the transcriptional profile of cells exposed to CLA at 37°C compared to that of cells grown at 30°C. CLA treatment resulted in the modulation of 296 transcripts (Table S2). One hundred fifty-five genes were upregulated while 141 were downregulated. GO-term analysis showed that upregulated genes belong to the categories “unknown biological function” (31%), “response to chemical stimulus” (17.4%), “carbohydrate metabolic processes” (16.8%), and “lipid metabolic processes” (16.1%). Fatty acids are nonfermentable carbon sources which are converted to acetyl-coenzyme A (acetyl-CoA) by β-oxidation. Acetyl-CoA drives the glyoxylate cycle, yielding oxaloacetate, which is converted to glucose via gluconeogenesis. Transcript levels of hallmark genes of the β-oxidation (FAA21, POX1, PXP2, POX1-3, ECI1, POT1, FOX2, FOX3, PEX5, CAT2, and ANT1), glyoxylate (ICL1, MLS1, and MDH1-3), and gluconeogenesis (FBP1) pathways increased in the presence of CLA, which may reflect a flow of carbon from fatty acids to acetyl-CoA to glucose. Most of these genes have been described as being induced upon internalization of C. albicans by macrophages (57) or by oleic acid (73). Genes encoding glycolytic enzymes, including PGK1, PGI1, and FBA1, were also upregulated in the presence of CLA, which may suggest that both glycolysis and gluconeogenesis are occurring simultaneously. In addition, a group of genes involved in transport (15.5%), including TPO4, PDR16, CDR11, CDR4, RTA3, and FLU1, were upregulated in the presence of CLA. These genes encode transmembrane transporters which play roles in phospholipid, fatty acid, or drug transmembrane transport.
To address the mechanism of action of CLA, we compared the transcriptional profiles of cells grown in the absence or presence of CLA at 37°C. Gene expression analysis revealed that 150 genes were significantly differentially expressed (see Table S4 in the supplemental material). To facilitate data mining, fold change ratios were generated by normalizing data obtained for CLA-treated cells to that obtained for untreated cells, resulting in 72 and 78 genes with upregulated (>2-fold) and downregulated (<0.5-fold) fold change ratios, respectively (Table S4). GO-term analysis revealed that the subset of 72 CLA-upregulated genes was enriched in genes implicated in “lipid metabolic processes” (27.4%) and “transport” (15.1%). Most of the genes involved in transmembrane transport, fatty acid β-oxidation and peroxisome biogenesis, the glyoxylate cycle, and gluconeogenesis were among the genes induced in CLA-treated cells (Table S2) discussed in the previous section. Because cells may be metabolizing CLA and converting it to glucose, we examined whether the CLA-mediated inhibition of hyphal growth was linked to fatty acid metabolism. To do so, we analyzed the effect of CLA on filamentation of the fox2/fox2, icl1/icl1, and ctf1/ctf1 mutant strains, which filament normally but cannot assimilate fatty acids (72, 73). CLA inhibited hyphal growth in all three strains, suggesting that fatty acid metabolism is not involved in the inhibition of filamentation (data not shown).
Given that lipid metabolism was not impeding hyphal growth, we focused on the subset of 78 CLA-downregulated genes (see Table S4 in the supplemental material). GO-term analysis showed that genes involved in “filamentous growth” (20.8%) and “pathogenesis” (16.9%) were among the subset of CLA-downregulated genes. Indeed, the expression levels of hypha-specific genes, including HWP1, ALS3, RBT1, ECE1, HGC1, IHD1, HYR1, SAP5, and SOD5, were greatly reduced in CLA-treated cells compared to the levels for untreated cells (Fig. 3B and Table S4). CLA also blocked the induction of transcription factors involved in hyphal growth, such as GAT2, UME6, TEC1, and RFG1 (Fig. 3C). A smaller (5.2%) yet more interesting category of CLA-downregulated transcripts was enriched in genes involved in signal transduction, such as RAS1, BEM2, MSB2, and PTP3 (Fig. 3C). As seen in Table 3, transcript levels of these genes increased during the yeast-to-hypha transition, except for PTP3 (Table S4). CLA either prevented the induction or lowered the induction levels of RAS1, BEM2, and MSB2. As for PTP3, which encodes a protein tyrosine phosphatase, its transcript levels were repressed in CLA-treated cells (Fig. 3C and Table S4). Taken together, the transcriptional data suggest that CLA negatively affects the expression of hypha-specific genes as well as genes known to regulate the yeast-to-hypha transition.
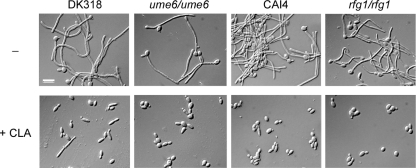
UME6 and RFG1 are not required for CLA-mediated inhibition of hyphal growth.
Gene expression analysis revealed that CLA reduced the expression levels of genes encoding regulators of hyphal growth, including TEC1, UME6, RFG1, and RAS1 (Fig. 3C). To determine whether these regulators were required for CLA's effect on hypha formation, we examined the responses of the ume6/ume6 and rfg1/rfg1 mutant strains to CLA when these strains were induced to filament in Spider medium at 37°C. As seen in Fig. 4, CLA inhibited hyphal growth in all parental strains. Interestingly, CLA inhibited filamentation independently of UME6 and RFG1, indicating that neither gene is required for CLA's inhibitory effect on hyphal growth (Fig. 4). The ras1/ras1 and tec1/tec1 mutants could not be analyzed in a similar fashion, since these strains failed to filament in Spider medium as previously reported (our observations and references 39, 53, and 90).
Fig. 4.
UME6 and RFG1 are not required for CLA-mediated hyphal growth inhibition. The DK318, CAI4, ume6/ume6 (DK312), and rfg1/rfg1 (DK129) strains were grown in Spider medium at 37°C in the absence or presence of 25 μM CLA for 4 h. Cells were visualized as described for Fig. 1B. Bar = 10 μm.
CLA downregulates TEC1 expression in a Ras1p-dependent manner.
Based on the previous screen, morphogenesis regulators potentially mediating CLA's effect on hyphal growth were narrowed down to Ras1p and Tec1p, since both were required for filamentation in Spider medium (data not shown). In its active GTP-bound form, Ras1p activates the adenylate cyclase Cyr1p, stimulating the cyclization of ATP to cAMP (31). Upon binding cAMP, Bcy1p, the regulatory subunit of PKA, releases the functionally redundant catalytic subunits Tpk1p and Tpk2p, thereby enabling their activity (16). The transcription factor Efg1p, an important regulator of hyphal growth, is a downstream target of PKA (5). In addition, Efg1p also regulates TEC1 expression (50). Thus, Tec1p constitutes a downstream target of the Ras1p-cAMP-PKA pathway.
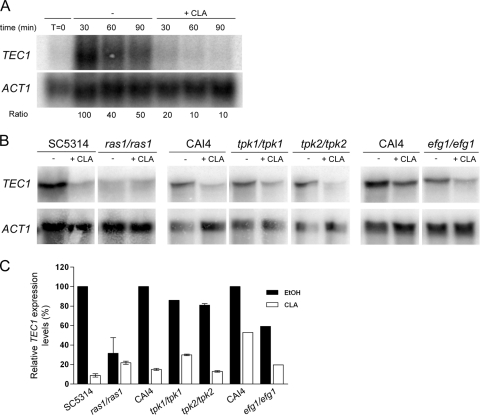
Gene expression analysis showed that CLA reduced TEC1 expression levels (Fig. 3C). Given that TEC1 is induced upon the yeast-to-hypha transition, we examined how CLA affected the kinetics of expression of TEC1. We performed a time course analysis of TEC1 transcript levels in cells grown in Spider medium in the absence or presence of CLA. As expected, TEC1 expression was induced in untreated cells during the yeast-to-hypha transition (Fig. 5A). In contrast, TEC1 mRNA levels were downregulated by an average of 5-fold in CLA-treated cells compared to the level for untreated cells, thus confirming transcriptional profiling results. Similar trends were also observed with the use of quantitative PCR analysis (data not shown). Furthermore, Tec1p protein levels were reduced in the presence of CLA, thus following the same expression pattern as the TEC1 transcript (data not shown).
Fig. 5.
TEC1 downregulation by CLA is Ras1p dependent. (A) Quantitative Northern blot analysis was used to examine the kinetics of expression of TEC1 in SC5314 cells grown in Spider medium at 37°C in the absence or presence of 100 μM CLA. TEC1 transcript levels were quantified and normalized to those for the ACT1 loading control. Ratios were obtained by normalizing TEC1 transcript levels to those for untreated cells obtained at the 30-min time point, which were set as 100%. (B) TEC1 expression levels in the parental, ras1/ras1 (CDH107), tpk1/tpk1 (IIHB6), tpk2/tpk2 (TPO7.4), and efg1/efg1 (HLC52) strains grown as described for panel A for 90 min. TEC1 transcript levels were quantified and normalized to those for the ACT1 loading control. (C) Data presented are the relative TEC1 expression levels obtained by normalizing TEC1 transcript levels to those for parental untreated cells, which were set as 100%. Data are means and standard deviations of results from two independent hybridizations performed on duplicate biological samples. A single hybridization was performed using RNA isolated from the CAI4 and efg1/efg1 strains.
A possible role for the Ras1p-cAMP-PKA signaling pathway in mediating CLA's effect on TEC1 expression was then investigated. Northern blot analysis revealed that in Spider medium, TEC1 induction was mostly Ras1p dependent (Fig. 5B and C). Interestingly, the effect of CLA on TEC1 expression was reduced in the absence of RAS1. In the parental strain, TEC1 transcript levels decreased ∼10-fold in CLA-treated cells compared to the level for untreated cells, while in the ras1/ras1 mutant strain, the downregulation reached only 1.5-fold (Fig. 5C). On the other hand, TEC1 induction in Spider medium and its downregulation by CLA did not depend on the presence of either TPK1 or TPK2 (Fig. 5B and C). These results may be explained by the functional redundancy of the two PKA isoforms (6). Since Efg1p is a regulator of TEC1 expression, we examined its role in mediating CLA's effect on TEC1 mRNA levels. Northern analysis revealed that TEC1 induction in Spider medium depended partially on EFG1, as TEC1 transcript levels were 1.7-fold lower in the efg1/efg1 mutant strain than in the parental strain (Fig. 5B and C). However, EFG1 was not required for CLA's repressive effect on TEC1, as the magnitudes of TEC1 downregulation by CLA (∼2-fold) were similar in parental and mutant strains (Fig. 5C). Additionally, it should be noted that CLA did not modulate EFG1 mRNA and protein levels (data not shown). Taken together, these results implicate RAS1, but not TPK1, TPK2, or EFG1, in mediating CLA's repressive effect on TEC1 expression.
CLA reduces GFP-Ras1p protein levels and affects its localization.
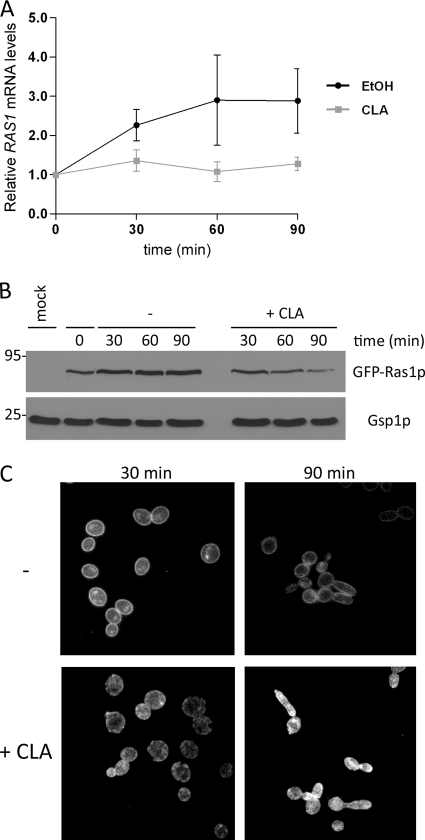
Transcriptional profiling showed that RAS1 expression was upregulated 2.1-fold during the yeast-to-hypha transition but that it was not induced in CLA-treated cells (Fig. 3C). We examined CLA's effect on RAS1 transcription at earlier time points by performing a time course analysis of RAS1 mRNA levels in cells grown in Spider medium in the absence or presence of CLA. Quantitative PCR analysis revealed that RAS1 transcript levels increased in untreated cells by an average of 2.7-fold, confirming results obtained by gene expression analysis (Fig. 6A). On the other hand, CLA completely abrogated RAS1 induction, as its mRNA levels remained relatively unchanged compared to levels at the zero time point (Fig. 6A). We next investigated CLA's effect on Ras1p protein levels and localization using the WY-ZXD3 strain expressing a GFP-Ras1 fusion protein previously developed for similar analyses (90). We monitored GFP-Ras1p cellular levels in cells induced to filament in Spider medium in the absence or presence of CLA. In untreated cells, GFP-Ras1p levels increased gradually, as did its mRNA levels (Fig. 6B). Unexpectedly, in CLA-treated cells, GFP-Ras1p levels declined with time, as seen at the 90-min time point (Fig. 6B). Thus, GFP-Ras1p did not follow the same pattern as its transcript, which remained constant (Fig. 6A). Taken together, these results suggest that CLA treatment reduces the steady-state levels of GFP-Ras1p.
Fig. 6.
CLA reduces GFP-Ras1p protein levels and affects its localization. (A) SC5314 cells were grown in Spider medium at 37°C in the absence or presence of 25 μM CLA and harvested at the indicated time points. Transcript levels of RAS1 were measured by quantitative PCR and normalized to those for ACT1. Relative expression levels were obtained by normalizing data for each time point to data obtained at time zero. Data are means and standard deviations for duplicate biological samples. (B) Ras1p protein levels were analyzed using a strain expressing GFP-Ras1p. Total protein extracts were prepared from SC5314 (mock) and GFP-Ras1p (WY-ZXD3) strains grown as described for panel A in the absence or presence of 100 μM CLA. Western blotting was performed using anti-GFP antibodies. Gsp1p, shown as a loading control, was detected using antibodies raised against S. cerevisiae Gsp1p. Molecular masses (kDa) are indicated on the left. (C) GFP-Ras1p-expressing cells were grown in Spider medium at 30°C in the absence or presence of 100 μM CLA. Aliquots of cells were removed and examined directly at ×100 magnification using epifluorescence.
We then examined GFP-Ras1p localization in untreated and CLA-treated cells. To facilitate comparisons, we examined GFP-Ras1p in yeast cells grown in Spider medium in the absence or presence of CLA at 30°C. Under such conditions, the GFP-Ras1p expression patterns were identical to those at 37°C, arguing that CLA's effect on GFP-Ras1p levels was not temperature dependent (data not shown). As seen in Fig. 6C (top panel), untreated cells demonstrated a strong fluorescent signal at the plasma membrane, confirming previous observations (90). Additionally, GFP-Ras1p could be seen in punctate patches within cells, especially at the 30-min time point. In contrast, GFP-Ras1p could barely be detected at the plasma membrane in CLA-treated cells. Instead, the fluorescent signal appeared diffuse throughout the cytoplasm or concentrated in patches within cells (Fig. 6C). Moreover, the “patch” phenotype was maintained, becoming more obvious at later time points (data not shown). These findings indicate that CLA causes the delocalization of GFP-Ras1p from the plasma membrane.
CLA affects the Tup1p-Nrg1p signaling pathway.
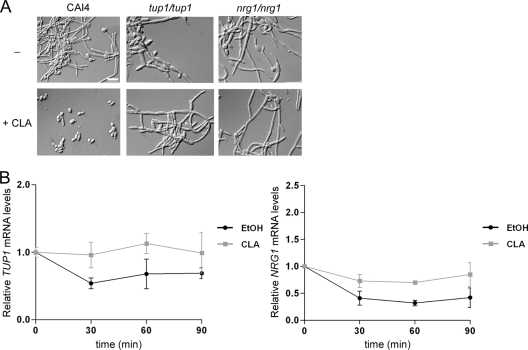
It has been established that the MAP kinase, Ras1p-cAMP-PKA, and Tup1p-Nrg1p signaling pathways make independent contributions to filamentation (9). The hyphal growth repressor Tup1p functions with the DNA-binding proteins Nrg1p and Rfg1p to negatively regulate hyphal growth and hyphal gene expression (8, 10, 42, 46, 59). Given that Tup1p and Nrg1p are involved in mediating the inhibitory effect of farnesol on filamentation (45), their role in the CLA-mediated hyphal growth inhibition was then investigated. We examined the responses of the tup1/tup1 and nrg1/nrg1 mutant strains to CLA when these strains were induced to filament in Spider medium at 37°C. While CLA inhibited hyphal growth in the parental strain CAI4, the tup1/tup1 and nrg1/nrg1 mutant strains remained filamentous, even when higher CLA concentrations were used (Fig. 7A). This indicates that CLA, like farnesol, inhibits filamentation by affecting a pathway requiring Tup1p and Nrg1p.
Fig. 7.
CLA affects the Tup1p-Nrg1p signaling pathway. (A) The CAI4, tup1/tup1 (BCa2-10), and nrg1/nrg1 (BCa23-3) strains were grown in Spider medium at 37°C in the absence or presence of 25 μM CLA for 4 h. Aliquots of cells were harvested and visualized at ×100 magnification by DIC optics. Bar = 10 μm. (B) C. albicans SC5314 cells were grown as described for panel A and harvested at the indicated time points. Transcript levels of TUP1 and NRG1 were measured as described for Fig. 6A.
TUP1 and NRG1 are modulated at the transcriptional level upon the yeast-to-hypha transition (10, 45). According to transcriptional profiling, TUP1 and NRG1 were not significantly differentially expressed in response to CLA, but this could be due to their low expression levels. Thus, we assessed how CLA affected the kinetics of expression of TUP1 and NRG1. Quantitative PCR revealed that TUP1 and NRG1 transcript levels decreased ∼2-fold during the 90-min time course in untreated cells (Fig. 7B). In contrast, CLA prevented the downregulation of both repressors, as TUP1 and NRG1 mRNA levels remained unchanged compared to initial levels. These findings suggest that CLA may inhibit hyphal growth by preventing the relief of repression exerted by the Tup1p-Nrg1p pathway.
DISCUSSION
While efforts have been put forth to elucidate the molecular mechanisms underlying the yeast-to-hypha transition in Candida albicans, small molecules affecting the morphogenetic switch have been identified in concurrent studies. Our findings have enabled us to add CLA to the growing list of molecules that modulate hyphal growth, which includes farnesol, dodecanol, fatty acids and lipid metabolites, rapamycin, and geldanamycin as well as histone deacetylase inhibitors (2, 19, 23, 37, 40, 45, 60, 68, 79). CLA was effective at inhibiting hyphal growth in most hypha-inducing media (Fig. 1). However, its inhibitory activity was significantly reduced in media containing FBS or N-acetylglucosamine, in RPMI 1640, or upon embedding of cells in YP medium (data not shown). The medium-dependent inhibitory effect of CLA on hyphal growth may be due to the nature or robustness of hypha-inducing signals, to the enhanced growth capacity of nutritionally rich media (61, 83), to the nonspecific lipid-binding capacity of serum albumin in FBS (52), or to the poor solubility of CLA in aqueous media (73).
Is CLA directly inhibiting filamentation or is this effect mediated by one of its metabolites? Fatty acids are nonfermentable carbon sources metabolized by C. albicans to acetyl-CoA and to glucose via β-oxidation, the glyoxylate cycle, and gluconeogenesis (57). Hallmark genes of these three biochemical pathways were upregulated in CLA-treated cells (see Tables S2 to S4 in the supplemental material), suggesting that CLA was possibly being converted to glucose. We ruled out that fatty acid metabolism was involved in the CLA-mediated hyphal growth inhibition by confirming that CLA blocked filamentation of the fox2/fox2, icl1/icl1, and ctf1/ctf1 mutant strains, which are unable to metabolize fatty acids (72, 73; data not shown). However, fatty acids are not only metabolized via β-oxidation, they can also be derived into oxygenated lipid metabolites. In C. albicans, arachidonic acid was shown to be a precursor for the production of 3,18-dihydroxy-5,8,11,14-eicosatetraenoic acid (3,18-di-HETE), an eicosanoid found exclusively in hyphae which was suggested to play a role in morphogenesis (25). Fatty acids such as linoleic and arachidonic acids are also derived into prostaglandins and leukotrienes, which reportedly affect hyphal development (29, 67). Indeed, prostaglandin E2 and thromboxane B2 were shown to enhance the yeast-to-hypha transition (44, 68, 69). Thus, while our results demonstrate that molecules derived from CLA's metabolism played no role in the CLA-mediated hyphal growth inhibition, we cannot exclude the possibility that oxygenated CLA derivatives may be involved in the effect.
We used global gene expression profiling as a means to investigate the transcriptional profile of cells induced to filament in the presence of CLA. Temperature shift in Spider medium promotes a hyphal growth program similar to the one induced upon filamentation in Lee's medium (see Table S1 in the supplemental material) (34). Spider and Lee's media have similar compositions, containing a source of fermentable carbon (mannitol or glucose), various amino acids, and salts, which may explain why the two transcriptional profiles were similar. Transcript levels of hypha-specific genes, such as ECE1, ALS3, and HYR1, and of several key regulators of hyphal growth, including CPH1, CPH2, TEC1, BCR1, UME6, GAT2, RAS1, and CYR1, increased during filamentation in Spider medium (Tables 3 and S1). However, the induction levels of several of these genes were lower than in other studies (34, 61). This discrepancy may have been due to hypha-inducing signals being weaker in Spider medium or to filamentation being somewhat induced under our control condition (Fig. 3A). Nonetheless, the transcriptional data indicated roles for the Ras1p-cAMP-PKA, the MAP kinase, the CPH2-TEC1, and the Tup1p-Nrg1p signaling pathways. These results confirmed that hyphal growth in Spider medium is regulated by a network of known signaling pathways which are activated simultaneously and converge (or do not converge) onto many of the same target transcription factors (4, 9, 12).
Comparing the transcriptional profiles of untreated and CLA-treated cells revealed that CLA affected the expression of genes encoding signal transducers and transcription factors, including RAS1, TEC1, UME6, and RFG1 (Fig. 3C). We examined CLA's effect on hyphal growth in strains deleted for each of these genes and showed that UME6 and RFG1 were dispensable for CLA-mediated inhibition of filamentation (Fig. 4). Similar conclusions for RAS1 and TEC1 could not be drawn, as both genes are required for hyphal growth in Spider medium (our observations and references 39, 53, and 90). Given that TEC1 lies downstream of the Ras1p-cAMP-PKA signaling pathway, we showed that CLA decreased TEC1 expression levels in a mostly Ras1p-dependent manner (Fig. 5B and C). Unexpectedly, while TEC1 induction was partially Efg1p dependent, CLA downregulated TEC1 mRNA levels independently of EFG1 (Fig. 5B). These results demonstrated that CLA inhibited filamentation by affecting the Efg1p-independent branch of the Ras1p signaling pathway. Likewise, the Hsp90p inhibitor geldanamycin was shown to modulate hyphal growth by affecting the Ras1p-cAMP-PKA signaling pathway independently of Efg1p (79).
Transcriptional profiling further revealed that CLA affected RAS1 expression (Fig. 3C). Indeed, CLA blocked the increase in RAS1 transcript levels, which occurred upon the yeast-to-hypha transition (Fig. 6A). Quite unexpectedly, while RAS1 mRNA levels remained constant in CLA-treated cells, GFP-Ras1p levels declined gradually (Fig. 6B). CLA also affected localization of GFP-Ras1p to the plasma membrane (Fig. 6C). Decreased GFP-Ras1p levels could be due to a decrease in its mRNA translation or to its degradation. However, several lines of evidence indicate that Ras delocalization results in its degradation and ultimately reduces its cellular levels. For instance, in Saccharomyces cerevisiae, a mutant Ras2p protein that could not be targeted to the membrane had lower cellular levels than the wild-type protein (24). Furthermore, farnesylthiosalicylic acid treatment was shown to dislodge Ras from the membrane and induce its degradation, resulting in lower Ras levels (35). In light of our findings, we can only speculate that the reduced GFP-Ras1p steady-state levels seen in CLA-treated cells stem from the protein being delocalized from the plasma membrane.
Several reasons may account for CLA's effect on GFP-Ras1p localization. First, the fatty acid may interfere with the posttranslational modifications of GFP-Ras1p. Ras proteins are modified by the addition of C-terminal lipids, such as farnesyl and palmitoyl moieties (84). Such modifications are involved in membrane association and subcellular localization, which are critical for Ras biological activities. Posttranslational modifications of Ras also play a role in filamentation in C. albicans, as compounds that prevent Ras prenylation were shown to inhibit hyphal growth (58). Second, CLA may also affect GFP-Ras1p localization by modifying the lipid composition of membranes. By being incorporated into membrane phospholipids, unsaturated fatty acids such as CLA may alter membrane structure and function, influencing the interaction of resident proteins with the plasma membrane (17). For instance, the polyunsaturated fatty acid docosahexaenoic acid (DHA) was shown to decrease membrane association of Ras by weakening its interactions with phospholipid acyl chains (21, 78). Interestingly, DHA also affected GFP-Ras1p localization to the plasma membrane in C. albicans (data not shown). The upregulation of RTA3 transcript levels in CLA-treated cells may be genetic evidence that the fatty acid is modifying membrane lipid composition (see Tables S2 to S4 in the supplemental material). Indeed, the S. cerevisiae RTA3 homologue (RSB1) was shown to be induced when plasma membrane glycerophospholipid asymmetry was altered (41). Nonetheless, further studies are warranted to address the underlying mechanism by which CLA exerts its effect on GFP-Ras1p membrane localization.
The biological activity of Ras is dictated by its subcellular localization. In S. cerevisiae, the farnesylated, membrane-bound form of Ras2p is approximately 100 times more effective in activating adenylate cyclase than its cytoplasmic form (49). Thus, it is tempting to suggest that CLA modulates the Ras1p signaling pathway by affecting the localization of Ras1p to the plasma membrane (Fig. 6C) and by abrogating the increases in RAS1 mRNA and protein levels (Fig. 6A and B). CLA would prevent the activation of the Ras1p pathway, resulting in the downregulation of TEC1 expression levels and inhibiting hyphal growth in C. albicans (Fig. 8). Additionally, CLA may also inhibit hyphal growth by preventing the relief of repression exerted by the Tup1p-Nrg1p pathway (Fig. 7B). Because the hyphal growth program is controlled by a network of signaling pathways, we expect the effect of CLA, like that of farnesol, to be multifactorial, in that the fatty acid could affect more than one signaling pathway (52). Nonetheless, our findings that CLA modulates RAS1 mRNA and protein levels in C. albicans demonstrate the broad-spectrum therapeutic properties of CLA, as the Ras pathway regulates filamentous growth and virulence in other fungi (26).
Fig. 8.
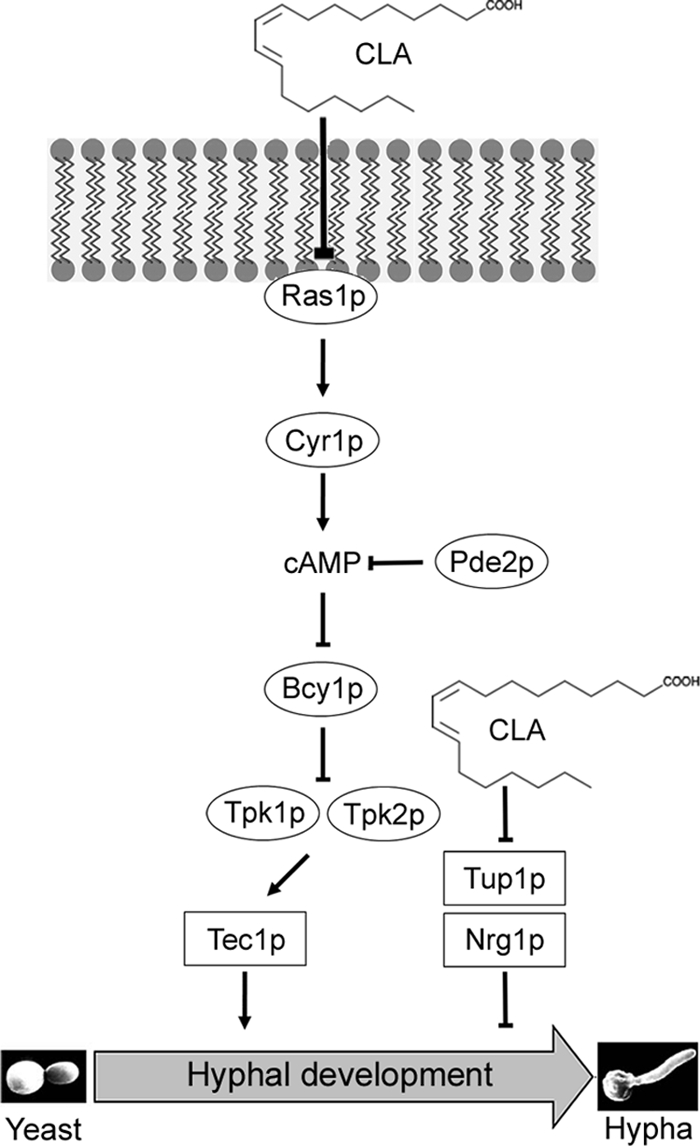
Proposed model underlying the mechanism by which CLA inhibits hyphal growth in Candida albicans. CLA inhibits the increase in RAS1 mRNA and protein levels and affects Ras1p membrane localization. Combined, these CLA-mediated effects impede the activation of the Ras1 signaling pathway and ultimately downregulate TEC1 expression and inhibit hyphal growth. In addition, CLA inhibits hyphal growth by preventing the relief of repression exerted by the Tup1p-Nrg1p complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Kolter (Harvard Medical School), M. Whiteway and D. Harcus (Biotechnology Research Institute, National Research Council of Canada), A. P. Mitchell (Carnegie Mellon University), D. Kadosh (University of Texas, San Antonio), H. Liu (University of California, Irvine), J. Ernst (Düsseldorf Universität, Germany), M. C. Lorenz (University of Texas Medical School, Houston), and M. Raymond and L. de Repentigny (Université de Montréal) for strains and plasmids, C. Beaurepaire (Biotechnology Research Institute, National Research Council of Canada) for RNA labeling and microarray hybridizations, M. Clément for insightful discussions, and P. C. Hallenbeck for critical reading of the manuscript.
J.S. is supported by studentships from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Fonds Québécois de la Recherche Sur la Nature et les Technologies (FQRNT), and the Faculty of Graduate Studies of the Université de Montréal. P.B. is the recipient of the Saputo Research Chair in Biomedical Dairy Products Optimization of the Université de Montréal.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 25 February 2011.
REFERENCES
- 1. Banerjee M., et al. 2008. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19:1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bastidas R. J., Heitman J., Cardenas M. E. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog. 5:e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belhumeur P., et al. 1993. GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in Saccharomyces cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol. Cell. Biol. 13:2152–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bockmuhl D. P., Ernst J. F. 2001. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 157:1523–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bockmuhl D. P., Krishnamurthy S., Gerads M., Sonneborn A., Ernst J. F. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243–1257 [DOI] [PubMed] [Google Scholar]
- 7. Braun B. R., Head W. S., Wang M. X., Johnson A. D. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun B. R., Johnson A. D. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105–109 [DOI] [PubMed] [Google Scholar]
- 9. Braun B. R., Johnson A. D. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in candida albicans. Genetics 155:57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Braun B. R., Kadosh D., Johnson A. D. 2001. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J. 20:4753–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown A. J., Odds F. C., Gow N. A. 2007. Infection-related gene expression in Candida albicans. Curr. Opin. Microbiol. 10:307–313 [DOI] [PubMed] [Google Scholar]
- 12. Brown A. J. P., Argimon S., Gow N. A. R. 2007. Signal transduction and morphogenesis in Candida albicans, p. 167–194 In Howard R. J., Gow N. A. R. (ed.), Biology of the fungal cell, 2nd ed. The Mycota VIII, 2nd ed. Springer-Verlag, Berlin, Germany [Google Scholar]
- 13. Brown D. H., Jr., Giusani A. D., Chen X., Kumamoto C. A. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651–662 [DOI] [PubMed] [Google Scholar]
- 14. Cao F., et al. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carlisle P. L., et al. 2009. Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U. S. A. 106:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cassola A., et al. 2004. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3:190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chapkin R. S., et al. 2008. Bioactive dietary long-chain fatty acids: emerging mechanisms of action. Br. J. Nutr. 100:1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Church G. M., Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clement M., Tremblay J., Lange M., Thibodeau J., Belhumeur P. 2007. Whey-derived free fatty acids suppress the germination of Candida albicans in vitro. FEMS Yeast Res. 7:276–285 [DOI] [PubMed] [Google Scholar]
- 20. Clement M., Tremblay J., Lange M., Thibodeau J., Belhumeur P. 2008. Purification and identification of bovine cheese whey fatty acids exhibiting in vitro antifungal activity. J. Dairy Sci. 91:2535–2544 [DOI] [PubMed] [Google Scholar]
- 21. Collett E. D., Davidson L. A., Fan Y. Y., Lupton J. R., Chapkin R. S. 2001. n-6 and n-3 polyunsaturated fatty acids differentially modulate oncogenic Ras activation in colonocytes. Am. J. Physiol. Cell Physiol. 280:C1066–C1075 [DOI] [PubMed] [Google Scholar]
- 22. Davis D., Wilson R. B., Mitchell A. P. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davis-Hanna A., Piispanen A. E., Stateva L. I., Hogan D. A. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 67:47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deschenes R. J., Broach J. R. 1987. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol. Cell. Biol. 7:2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deva R., Ciccoli R., Schewe T., Kock J. L., Nigam S. 2000. Arachidonic acid stimulates cell growth and forms a novel oxygenated metabolite in Candida albicans. Biochim. Biophys. Acta 1486:299–311 [DOI] [PubMed] [Google Scholar]
- 26. D'Souza C. A., Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349–364 [DOI] [PubMed] [Google Scholar]
- 27. Edmond M. B., et al. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239–244 [DOI] [PubMed] [Google Scholar]
- 28. Enjalbert B., Nantel A., Whiteway M. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erb-Downward J. R., Noverr M. C. 2007. Characterization of prostaglandin E2 production by Candida albicans. Infect. Immun. 75:3498–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ernst J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146(8):1763–1774 [DOI] [PubMed] [Google Scholar]
- 31. Fang H. M., Wang Y. 2006. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 61:484–496 [DOI] [PubMed] [Google Scholar]
- 32. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 34. Goyard S., et al. 2008. The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol. Biol. Cell 19:2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haklai R., et al. 1998. Dislodgment and accelerated degradation of Ras. Biochemistry 37:1306–1314 [DOI] [PubMed] [Google Scholar]
- 36. Hisajima T., et al. 2008. Protective effects of farnesol against oral candidiasis in mice. Microbiol. Immunol. 52:327–333 [DOI] [PubMed] [Google Scholar]
- 37. Hnisz D., Majer O., Frohner I. E., Komnenovic V., Kuchler K. 2010. The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6:e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hogan D. A., Vik A., Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212–1223 [DOI] [PubMed] [Google Scholar]
- 39. Homann O. R., Dea J., Noble S. M., Johnson A. D. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5:e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hornby J. M., et al. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ikeda M., Kihara A., Denpoh A., Igarashi Y. 2008. The Rim101 pathway is involved in Rsb1 expression induced by altered lipid asymmetry. Mol. Biol. Cell 19:1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kadosh D., Johnson A. D. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kadosh D., Johnson A. D. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalo-Klein A., Witkin S. S. 1990. Prostaglandin E2 enhances and gamma interferon inhibits germ tube formation in Candida albicans. Infect. Immun. 58:260–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kebaara B. W., et al. 2008. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot. Cell 7:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khalaf R. A., Zitomer R. S. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kippert F. 1995. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol. Lett. 128:201–206 [DOI] [PubMed] [Google Scholar]
- 48. Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 49. Kuroda Y., Suzuki N., Kataoka T. 1993. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science 259:683–686 [DOI] [PubMed] [Google Scholar]
- 50. Lane S., Birse C., Zhou S., Matson R., Liu H. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988–48996 [DOI] [PubMed] [Google Scholar]
- 51. Lane S., Zhou S., Pan T., Dai Q., Liu H. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1. Mol. Cell. Biol. 21:6418–6428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Langford M. L., Atkin A. L., Nickerson K. W. 2009. Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans. Future Microbiol. 4:1353–1362 [DOI] [PubMed] [Google Scholar]
- 53. Leberer E., et al. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 54. Lee K. L., Buckley H. R., Campbell C. C. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148–153 [DOI] [PubMed] [Google Scholar]
- 55. Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 56. Lo H. J., et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 57. Lorenz M. C., Bender J. A., Fink G. R. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McGeady P., Logan D. A., Wansley D. L. 2002. A protein-farnesyl transferase inhibitor interferes with the serum-induced conversion of Candida albicans from a cellular yeast form to a filamentous form. FEMS Microbiol. Lett. 213:41–44 [DOI] [PubMed] [Google Scholar]
- 59. Murad A. M., et al. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742–4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Murzyn A., Krasowska A., Stefanowicz P., Dziadkowiec D., Lukaszewicz M. 2010. Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS One 5:e12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nantel A., et al. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Negredo A., Monteoliva L., Gil C., Pla J., Nombela C. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143(2):297–302 [DOI] [PubMed] [Google Scholar]
- 63. Nobile C. J., Mitchell A. P. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 15:1150–1155 [DOI] [PubMed] [Google Scholar]
- 64. Nobile C. J., Nett J. E., Andes D. R., Mitchell A. P. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Noble S. M., Johnson A. D. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Noverr M. C., Erb-Downward J. R., Huffnagle G. B. 2003. Production of eicosanoids and other oxylipins by pathogenic eukaryotic microbes. Clin. Microbiol. Rev. 16:517–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Noverr M. C., Huffnagle G. B. 2004. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72:6206–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Noverr M. C., Phare S. M., Toews G. B., Coffey M. J., Huffnagle G. B. 2001. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect. Immun. 69:2957–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Phan Q. T., Belanger P. H., Filler S. G. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ramirez M. A., Lorenz M. C. 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 6:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ramirez M. A., Lorenz M. C. 2009. The transcription factor homolog CTF1 regulates β-oxidation in Candida albicans. Eukaryot. Cell 8:1604–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Richardson M., Lass-Florl C. 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14(Suppl. 4):5–24 [DOI] [PubMed] [Google Scholar]
- 75. Saville S. P., et al. 2006. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob. Agents Chemother. 50:3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schweizer A., Rupp S., Taylor B. N., Rollinghoff M., Schroppel K. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435–445 [DOI] [PubMed] [Google Scholar]
- 77. Sellam A., Tebbji F., Nantel A. 2009. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot. Cell 8:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seo J., Barhoumi R., Johnson A. E., Lupton J. R., Chapkin R. S. 2006. Docosahexaenoic acid selectively inhibits plasma membrane targeting of lipidated proteins. FASEB J. 20:770–772 [DOI] [PubMed] [Google Scholar]
- 79. Shapiro R. S., et al. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sonneborn A., et al. 2000. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 35:386–396 [DOI] [PubMed] [Google Scholar]
- 81. Stoldt V. R., Sonneborn A., Leuker C. E., Ernst J. F. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 83. Toenjes K. A., et al. 2005. Small-molecule inhibitors of the budded-to-hyphal-form transition in the pathogenic yeast Candida albicans. Antimicrob. Agents Chemother. 49:963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wennerberg K., Rossman K. L., Der C. J. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843–846 [DOI] [PubMed] [Google Scholar]
- 85. Whiteway M., Bachewich C. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70 [DOI] [PubMed] [Google Scholar]
- 87. Wisplinghoff H., et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 88. Zeidler U., et al. 2009. UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res. 9:126–142 [DOI] [PubMed] [Google Scholar]
- 89. Zheng X., Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhu Y., et al. 2009. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol. Microbiol. 74:862–875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.