Abstract
The origin and early evolution of sex chromosomes are currently poorly understood. The Neurospora tetrasperma mating-type (mat) chromosomes have recently emerged as a model system for the study of early sex chromosome evolution, since they contain a young (<6 million years ago [Mya]), large (>6.6-Mb) region of suppressed recombination. Here we examined preferred-codon usage in 290 genes (121,831 codon positions) in order to test for early signs of genomic degeneration in N. tetrasperma mat chromosomes. We report several key findings about codon usage in the region of recombination suppression, including the following: (i) this region has been subjected to marked and largely independent degeneration among gene alleles; (ii) the level of degeneration is magnified over longer periods of recombination suppression; and (iii) both mat a and mat A chromosomes have been subjected to deterioration. The frequency of shifts from preferred codons to nonpreferred codons is greater for shorter genes than for longer genes, suggesting that short genes play an especially significant role in early sex chromosome evolution. Furthermore, we show that these degenerative changes in codon usage are best explained by altered selection efficiency in the recombinationally suppressed region. These findings demonstrate that the fungus N. tetrasperma provides an effective system for the study of degenerative genomic changes in young regions of recombination suppression in sex-regulating chromosomes.
INTRODUCTION
At present, little is known about the origin and early evolution of sex chromosomes. This is because most ancient sex chromosome systems are so highly deteriorated (e.g., Y in X/Y systems) that they retain few traces of the historical events driving their evolution (8, 9, 11, 12, 54). The limited data available to date about early stages of sex chromosome evolution have been derived from young sex chromosomes from certain plants (e.g., Silene [41]) and/or from neo-X/Y systems of Drosophila (4, 5). Thus, model systems for young sex chromosomes are needed (12). The Neurospora tetrasperma mating-type (mat) chromosomes have recently emerged as a model system for the study of early stages of sex chromosome evolution (42).
The filamentous ascomycete N. tetrasperma is a self-fertile (pseudohomothallic) organism presumed to have evolved from a self-incompatible (heterothallic) ancestor (15, 50). As with most filamentous ascomycetes, the N. tetrasperma mat chromosomes contain the mat locus, which comprises two dissimilar alleles (mat a and mat A idiomorphs) that regulate mating and sexual reproduction (10, 53). Pseudohomothallism in N. tetrasperma is associated with a specialized meiotic pathway. Specifically, the mat chromosomes contain a young (<6 million years ago [Mya]), large (>6.6-Mbp) segment of suppressed recombination, including the segment between the mat locus and the centromere, which ensures first division segregation of mating-type idiomorphs. Spindles align in parallel for the second meiotic cell division, ensuring that nuclei of opposite mating types are encased in each ascospore (50). Consequently, the two mat chromosomes coexist in separate haploid nuclei in both reproductive and vegetative tissue, consistent with a predominant heterokaryotic and self-fertile state (50). This life history trait differs from that of the closely related heterothallic haploid taxon Neurospora crassa, which undergoes normal recombination during meiosis, contains nuclei with a single mating type in each ascospore, and requires two partners containing nuclei of opposite types for mating and sexual reproduction (53).
The N. tetrasperma mat chromosomes share many of the key features associated with the dimorphic sex chromosomes of animals and plants. In particular, as in dimorphic sex chromosomes, the segment of recombination suppression is localized to the central region of the mat chromosomes (42, 44), is flanked by two normally recombining, pseudoautosomal (PA) regions (21, 22, 32, 42), and contains genes with markedly divergent alleles (32, 35, 42, 44). Furthermore, the region of suppressed recombination comprises at least two distinct evolutionary strata, i.e., regions of suppressed recombination that have arisen successively over time, including an older (stratum 1) and a younger (stratum 2) stratum (42). Such strata have been reported for a wide range of plant and animal sex chromosomes and for mating-type chromosomes from Microbotryum violaceum (11, 22, 27, 35, 52, 59). Altogether, it is evident that the N. tetrasperma mat chromosomes provide a framework with which to examine genomic alterations within the recombinationally suppressed regions of sex-regulating chromosomes.
Given that the recombinationally suppressed region in N. tetrasperma mat chromosomes is young, divergence levels among alleles are not saturated (42), and it has not undergone extensive gene loss or other typical signs of massive degeneration (9, 42, 54). Thus, degenerative genomic changes, if present at this early stage, are likely subtle. One genomic trait likely to serve as a signal of early-stage degeneration is synonymous codon usage. Synonymous codons are not used randomly, and in many organisms, natural selection favors the use of a subset of preferred codons (PR). Preferred codon levels (or bias in codon usage) within the genome have been associated with more efficient and accurate translation (16, 17, 57) and represent adaptation at the molecular level in protein-coding DNA (1, 2). In addition, it has been found that shorter genes have a greater bias in codon usage in certain animals and plants (for examples, see references 3 and 17), suggesting that these traits may have coevolved to further promote highly efficient/accurate translation. In total, one may conclude that marked shifts from preferred codons to nonpreferred codons (NPR) provide a means to test for early signs of genomic deterioration in the N. tetrasperma mat chromosomes (1, 5).
Given that each of the mat chromosomes is recombinationally suppressed in N. tetrasperma (42), degenerative codon changes may be expected to occur on both chromosomes. This feature contrasts with those of dimorphic sex chromosome systems, in which only one chromosome is nonrecombining (4, 12). Another factor that could influence the level and patterns of genomic deterioration among differentiating mat chromosomes in N. tetrasperma is exposure to haploid selection. In dimorphic sex chromosome systems of diploid plants and animals, the marked degeneration of the heterozygous nonrecombining chromosome may be facilitated by the sheltering of mutations by the homologous chromosome and by the restriction of haploidy to the gamete stage (12, 45). In contrast, sex chromosomes in diploid organisms with haploid sex determination systems (e.g., Bryophytes and certain Fungi) are expected to have similar, and low, degeneration levels in the sex chromosomes. This pattern is expected because sex chromosomes should be equally sheltered at the diploid stage, and only those genes not associated with haploid growth may accumulate deleterious recessive mutations (7, 30). The pseudohomothallic system of N. tetrasperma has features of both systems. In particular, the heterokaryotic nature of N. tetrasperma may allow the sheltering of recessive mutations on the mat chromosomes, paralleling that observed in diploid organisms (49). In turn, the fact that each nucleus is haploid in N. tetrasperma might also allow for haploid selection. Specifically, homokaryotic sectors may arise within the heterothallic mycelium, as has been observed in certain heterokaryotic basidiomycetes (56). In addition, findings that as many as 20% of the asexual and sexual spores produced in the laboratory for N. tetrasperma are homokaryotic (44, 48) suggest that a segment of the natural population is haploid. Thus, it is possible that the mat chromosomes may evolve in a manner similar to that of dimorphic sex chromosomes and/or similar to that of a haploid system. Further data are needed to ascertain whether the young recombinationally suppressed region in N. tetrasperma is subjected to degeneration and which of these factors drive such processes.
The objective of the present study was to evaluate degenerative changes in preferred-codon usage within the young region of suppressed recombination in the N. tetrasperma mat chromosomes. For this purpose, we compared preferred-codon usage between mat a and mat A linked alleles among genes from various regions of the mat chromosomes (two pseudoautosomal regions and the younger [stratum 2] and older [stratum 1] regions of suppressed recombination), examined the role of gene length, and identified the possible mechanisms driving the degeneration.
MATERIALS AND METHODS
For our analysis, we generated DNA sequence data (coding sequences [CDS]) for mat a and mat A linked alleles for each of 290 genes located on the N. tetrasperma strain P4492 mat chromosomes by using Solexa technology. The mat a and mat A linked alleles were isolated and sequenced using haploid, single-mating-type component strains of the N. tetrasperma heterokaryotic strain P4492 (referred to here as P4492a and P4492A, respectively). The gene order within N. tetrasperma chromosomes was based on the order known for N. crassa, since these two genomes have been shown to be highly correlated (32). The gene order is available from the National Center for Biotechnology Information (NCBI) genome browser MapViewer (http://www.ncbi.nlm.nih.gov/mapview/). Following earlier conventions, we assumed that normally recombining gene alleles are identical or nearly identical in normally recombining genomic regions in a highly inbred taxon such as N. tetrasperma and that genes located in recombinationally suppressed regions show greater synonymous divergence (Ks) among their alleles over time (27, 35, 42, 43, 52).
Fungal strains.
We examined genomic sequence data derived from the two haploid, single-mating-type component strains (mat a and mat A) of the N. tetrasperma heterokaryotic strain P4492 (P4492a and P4492A, respectively) and from the N. crassa strain FGSC 2489. P4492 originates from the Perkins collection of Neurospora strains from nature, curated by the Fungal Genomics Stock Center (FGSC), University of Missouri. The FGSC identification numbers (IDs) for the mat a and mat A component strains are FGSC 9034 and FGSC 9033, respectively. N. tetrasperma strain P4492 has previously been shown to belong to phylogenetic lineage 1 of the N. tetrasperma species complex (43).
DNA sequencing.
The DNA sequences for the single-mating-type component strains P4492a and P4492A were generated by Geneservice, Source BioScience plc, using Solexa technology. The 55-bp paired-end DNA library was constructed with an insert size of ∼170 bp. The sequencing process followed the protocol for Genome Analyzer II (GA II), and the images from GA II were processed by the GA II software pipeline for the purpose of base calling. All 55-bp paired-end reads for P4492a and P4492A were mapped against the reference genome of N. tetrasperma FGSC 2508 (available from the DOE Joint Genome Institute [http://www.jgi.doe.gov]) using Maq (http://maq.sourceforge.net/) (36). The maximum number of mismatches allowed was set to 3 in the mapping process. The consensus sequences were filtered and called with a minimum read depth of 3, a minimum mapping quality of 40, and a minimum neighboring quality of 20. This mapping protocol is conservative, limiting false-positive mapping. Coding DNA, intergenic regions, and introns were identified from the consensus sequences of linkage group I (the mat chromosome) of P4492a and P4492A. For N. crassa, the DNA sequences and NCU IDs were obtained from the Neurospora crassa Database (annotation version 3; FGSC 2489) (http://www.broad.mit.edu/annotation/genome/neurospora/).
Identification of gene sequences.
A gene set of 290 genes was identified and utilized for analysis in our investigation (3 sequences per gene—1 each for P4492a, P4492A, and N. crassa—for a total of 870 sequences). For this purpose, we first identified a total of 2,309 N. crassa genes located on the mat chromosomes that have been defined by an NCU number at the Neurospora crassa Database (NCU IDs have been assigned to well-defined genes and to those encoding hypothetical and predicted proteins [http://broad.mit.edu/annotation/fungi/neurospora/]). The genes located on the mat chromosomes of P4492a and P4492A were subsequently identified by comparison to this N. crassa sequence database: the consensus sequences of linkage group I for P4492a and for P4492A were each defined as a database and compared to the N. crassa gene set (i.e., the CDS regions for each gene) using BLASTN (http://www.ncbi.nlm.nih.gov/). The N. crassa gene with the lowest E value (with a cutoff of ≤10−6) for a particular region of N. tetrasperma P4492a and P4492A linkage group I was considered a match. The genomic region contained within and surrounding each match for P4492a and for P4492A was isolated and aligned to the matching N. crassa CDS region using ClustalW (58). Introns and/or indel sequences identified within the P4492a and P4492A sequences from the alignment were removed. Genomic regions from P4492a and P4492A that matched an entire mRNA from N. crassa, including the start codon and the stop codon, and that did not contain unknown or ambiguous nucleotides were identified for our analysis. Each P4492a and P4492A gene was translated to ensure the accurate identification of reading frames. Synonymous substitution rates (Ks) between alleles of each of the 290 genes described above were determined using default settings in DnaSP (37). The final aligned gene sequences, with gaps removed, used for P4492a and for P4492A are provided in File S1 in the supplemental material (sequence designations ending with P4492_a and P4492A are from the mat a and mat A chromosomes, respectively).
Identification and analysis of preferred codons.
The preferred codons for our analysis were determined using the complete genomewide codon usage data available for N. crassa at the Codon Usage Database (http://www.kazusa.or.jp/codon/). In particular, we identified the most frequent codons per amino acid; one preferred codon was identified for amino acids encoded by two, three, or four codons, and two preferred codons were identified for amino acids encoded by six codons. The final preferred-codon list contains 21 codons (see Table SA1 in the supplemental material), which are highly similar to those in the list of optimal codons (i.e., codons present in the most highly expressed genes) reported for other fungi; the latter was based on examination of only a subset of the genomic sequences (38). All codons not listed in Table SA1 in the supplemental material were defined as nonpreferred codons. In order to further verify the preferred-codon list, we determined the relative synonymous codon usage (RSCU) for each codon for the pseudoautosomal regions in N. tetrasperma by using CodonW (J. Peden) (http://codonw.sourceforge.net). RSCU values measure the usage of each codon relative to all synonymous codons for a particular amino acid; higher RSCU values denote greater usage of a specific codon relative to its synonymous codons. These RSCU values for the normally recombining region of N. tetrasperma (see Table SA2 in the supplemental material) are in agreement with the preferred-codon list estimated from N. crassa shown in Table SA1.
Using the preferred-codon list discussed above, we conducted a comparative analysis of preferred-codon frequency among genes, among alleles per gene, among genes of different lengths, and among the recombinationally suppressed and pseudoautosomal regions of N. tetrasperma. Codon usage counts were determined using CAIcal (47).
Assessment of the allele-specific switches from preferred to nonpreferred codons.
In order to identify specific switches from preferred codons (PR) to nonpreferred codons (NPR) within the recombinationally suppressed and pseudoautosomal regions of the N. tetrasperma mat chromosomes, we examined the codons found in N. tetrasperma P4492a, N. tetrasperma P4492A, and N. crassa for every codon position in our 290 genes. For this purpose, the DNA sequences for all 290 genes were concatenated in the sequential order in which they occur on the mat chromosomes of N. crassa. Using these data, we identified cases where N. crassa and one of the N. tetrasperma mat chromosomes contained the same preferred codon (i.e., the ancestral codon) while the other mat chromosome contained a different, nonpreferred codon. This is consistent with an allele-specific degenerative switch on one chromosome (from a preferred to a nonpreferred codon [PR → NPR]). Based on the same approach, we also identified allele-specific switches from a nonpreferred to a preferred codon (NPR → PR) associated with either the N. tetrasperma mat a or the N. tetrasperma mat A chromosome. Using these two variables, we measured the net excess of allele-specific switches to nonpreferred codons (PR → NPR) over switches to preferred codons (NPR → PR) (1) for each genomic region within each of the mat chromosomes. In addition, we standardized the number of excess switches from preferred to nonpreferred codons relative to the total number of codons examined per chromosomal segment (frequency [Fr]Excess NPR = excess PR → NPR switches per 1,000 codon positions) in order to directly compare values among genomic regions. A total of 25,502 codon positions from the pseudoautosomal regions, 18,419 codon positions for stratum 2, and 77,910 codon positions for stratum 1 were examined (a total of 121,831 codon positions).
Identification and analysis of introns.
In order to ascertain the role of mutational pressure in changes in preferred-codon usage in N. tetrasperma, we compared the GC contents of introns (GCI) to the GC contents of third codon positions (GC3) of the genes under study. For this purpose, we first identified introns in N. crassa based on the alignments between genomic DNA and the CDS regions (data are available from the Neurospora crassa Database). Alignments were conducted for each of the 290 genes included in this study by using ClustalW. Gaps identified within the genomic DNA that exceeded 30 nucleotides were identified as introns. For N. tetrasperma P4492a and P4492A, introns were identified by alignment of the consensus sequence (i.e., the genomic region previously defined for each gene) with the N. crassa CDS region; gaps in the genomic DNA were identified as introns. In some cases, the N. tetrasperma introns contained internal gaps relative to the N. crassa intron, which could result from small insertions (in N. crassa) or deletions (in N. tetrasperma) and/or from gaps in the Solexa sequence assembly. Introns from N. crassa, P4492a, and P4492A were aligned. and gaps were removed. For genes with more than one intron, the introns were concatenated prior to analysis.
RESULTS
Demarcations within the mat chromosomes.
As an initial step in our analysis, we identified the location of each of our 290 genes on the N. tetrasperma mat chromosomes. The various genomic regions on the mat chromosomes have previously been shown to occur in the following sequential order: pseudoautosomal region 1 (PA1), the younger region of suppressed recombination (stratum 2), the older region of suppressed recombination (stratum 1, containing the centromere), and pseudoautosomal region 2 (PA2). Based on the a priori assumption that normally recombining gene alleles are identical or nearly identical in N. tetrasperma (42, 43), as well as the demarcations on the mat chromosomes previously reported by Menkis et al. (42), we found that 228 of the 290 genes examined here are located in the region of suppressed recombination and 62 are located in the flanking pseudoautosomal regions. This region of suppressed recombination contains a high percentage of genes with a Ks of >0 in the comparisons of N. tetrasperma P4492a and P4492A alleles (>78% of genes in this region have diverged). We infer that 190 of the genes in the region of suppressed recombination are located within the older stratum 1, while 38 are in the younger, less divergent stratum 2 (Table 1; see also Table SA3 in the supplemental material). Of the 62 genes located in the PA regions, 32 are contained within PA1, the flanking region adjacent to stratum 2, and 30 are located in PA2, the flanking region adjacent to stratum 1 (Table 1; see also Table SA3). The two pseudoautosomal regions had a preponderance of genes with a Ks of zero (i.e., <3.3% of genes show a Ks of >0 [Table 1]).
Table 1.
Genes identified for the Neurospora tetrasperma single-mating-type component strains used in the present analysisa
| Genomic region | No. of genes identified | Alleles located on the mat a vs mat A chromosome |
|
|---|---|---|---|
| Mean Ks | Fraction (%) of genes with a Ks of >0 | ||
| Pseudoautosomal region 1 | 32 | 1.8 × 10−4 | 1/32 (3.1) |
| Region of suppressed recombination | |||
| Stratum 2 | 38 | 0.0038 | 13/38 (34.2) |
| Stratum 1 | 190 | 0.0180 | 149/190 (78.4) |
| Pseudoautosomal region 2 | 30 | 1.7 × 10−4 | 1/30 (3.3) |
Strains P4492a and P4492A.
The divergence values among gene alleles suggest that recombination suppression might not be perfectly enforced within the central region of the mat chromosomes. For example, although a Ks of >0 was observed among alleles for many genes in stratum 1 and stratum 2 (Table 1), a substantial fraction of genes in these regions, particularly in the younger stratum 2, have a Ks of zero (like the pseudoautosomal regions) among the mat a and mat A linked alleles (stratum 2 has 65.8% of genes with a Ks of zero, while stratum 1 has 21.6% [Table 1]). It is possible that some level of gene conversion, the nonreciprocal transfer of DNA during meiosis, which homogenizes gene alleles (29), and/or crossover events occur during the early stages of recombination suppression in the mat chromosomes. It is also possible that recombination suppression has existed for too short a time to affect all genes in a similar manner, and that thus, there is inherent stochasticity in divergence among genes.
Preferred codons and gene length.
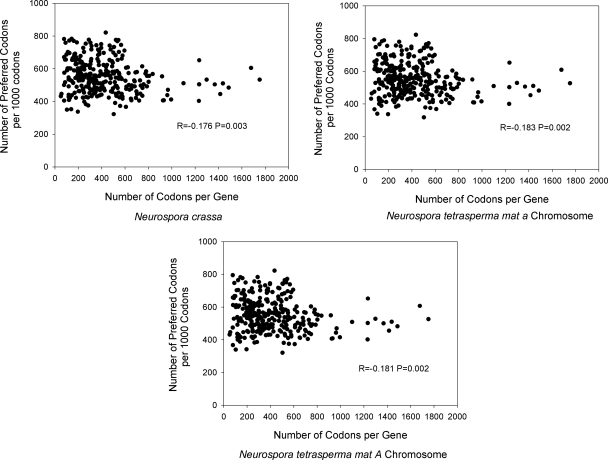
In order to assess the relationship between gene length and codon usage bias in Neurospora using the 290 genes examined here, we standardized the frequency of preferred codons for each gene under study as follows: number of preferred codons per 1,000 codons = number of preferred codons/total number of codons × 1,000. Our data show that the number of preferred codons per 1,000 codons is inversely correlated with gene length (i.e., the number of codons per gene) for N. crassa and for each of the N. tetrasperma strains (P4492a and P4492A) (P < 0.05) (Fig. 1; see also Table SA4 in the supplemental material). Nonetheless, we noted that the correlation coefficients are relatively low (R, ≤0.183 [Fig. 1]) and that the correlation is not universally detected across all gene length categories (see Table SA4). For example, the short (<300 codons) and medium-length (≥300 and <500 codons) genes have similar frequencies of preferred codons (mean, between 562.9 and 570.5 preferred codons per 1,000), while the longest genes (>500 codons) have statistically significantly (P < 0.05) lower numbers of preferred codons (between 526.2 and 529.2 preferred codons per 1,000 [see Table SA4]). Thus, the Neurospora genes show an inverse relationship between gene length and codon usage bias, similar to that reported for other organisms (e.g., 3, 17), with the caveat that short and medium-length genes (with as many as 500 codons) each have high levels of preferred-codon usage.
Fig. 1.
Relationship between gene length (number of codons) and the number of preferred codons per 1,000 codons for the 290 genes examined in this study. Pearson correlation coefficients and P values are shown.
Variation in preferred-codon levels among gene alleles.
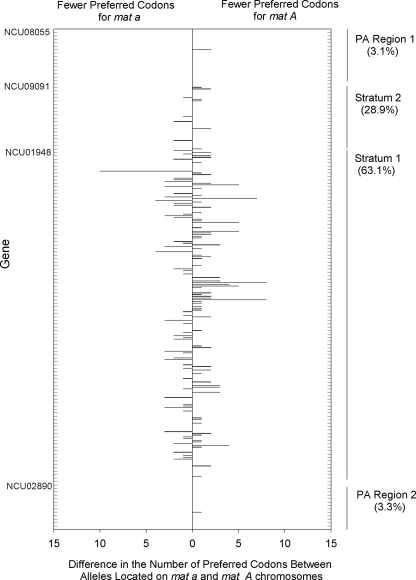
Marked divergence in the number of preferred codons was detected among the mat a and mat A linked alleles for genes located in the recombinationally suppressed region, but not in the pseudoautosomal regions, of the N. tetrasperma mat chromosomes. Specifically, the numbers of preferred codons were identical for mat a and mat A linked alleles in the vast majority of genes located in the pseudoautosomal regions, with less than 3.3% of genes showing differences (Fig. 2). In contrast, we found that 63.1% of genes in the older stratum (stratum 1) and 28.9% of genes in the younger stratum (stratum 2) of the recombinationally suppressed region had different levels of preferred codons among alleles. The magnitude of the differences among gene alleles differed markedly among the segments of the mat chromosomes, with much greater differentials observed for genes located in stratum 1 than in stratum 2, where one allele had an excess of as many as 10 preferred codons over the other. The data also reveal that the specific allele with more, or fewer, preferred codons could occur on either the mat a or the mat A chromosome and that this parameter was largely gene specific.
Fig. 2.
Differences in the number of preferred codons between alleles for the 290 genes located on the mat a and mat A chromosomes in Neurospora tetrasperma strain P4492. The NCU ID for the first gene in each chromosomal segment is shown. The percentage of genes that have differences in the number of preferred codons among alleles is shown for each chromosomal region. Two or more genes are represented within each grid point on the vertical scale. The genes are listed in the order in which they occur on the mat chromosomes.
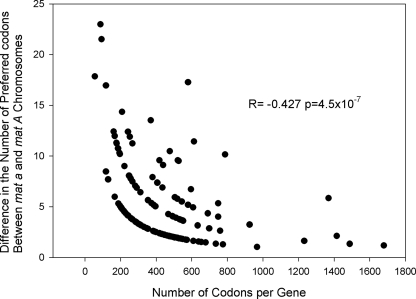
The data also show that the level of divergence in the frequency of preferred codons among homologous gene alleles in the recombinationally suppressed region is associated with gene length. For example, for each of the 228 genes in the centrally located region of suppressed recombination, we found that the absolute value of the difference in the number of preferred codons (per 1,000 codons) between the mat a and mat A linked alleles is statistically significantly inversely correlated with gene length (R = −0.427; P = 4.5 × 10−7 [Fig. 3 ]). Thus, these findings demonstrate that there is substantially greater divergence in the levels of preferred codons among homologous alleles for shorter genes than for longer genes; this is consistent with greater rates of genomic deterioration in shorter genes within the recombinationally suppressed region.
Fig. 3.
Absolute value of the difference in the number of preferred codons (per 1,000 codons) between the alleles of genes located in the region of suppressed recombination of the N. tetrasperma mat a versus mat A chromosomes, plotted against the number of codons per gene. Genes with no differences have been excluded.
Excess of allele-specific switches from preferred to nonpreferred codons.
We determined the net excess of synonymous allele-specific switches to nonpreferred codons based on comparisons of PR → NPR and NPR → PR switches (Table 2) at codon positions where amino acids were conserved among the mat chromosomes. Marked differences were found in the level of synonymous allele-specific codon switches among the various segments of the N. tetrasperma mat chromosomes (Table 2). For the pseudoautosomal regions, we found exceptionally low numbers of allele-specific switches within the N. tetrasperma mat a and mat A chromosomes. In these regions, we observed only two allele-specific switches from a preferred to a nonpreferred codon (PR → NPR; on the N. tetrasperma mat A chromosome) and one switch from a nonpreferred to a preferred codon (NPR → PR; on the mat a chromosome) (Table 2). Thus, these data are consistent with normal recombination in the pseudoautosomal regions. In the recombinationally suppressed stratum 2, however, we found elevated levels of allele-specific changes in preferred-codon usage. Specifically, we detected nine allele-specific switches from a preferred to a nonpreferred codon on the mat a chromosome. Only four cases were found where a nonpreferred codon had switched to a preferred codon; thus, we conclude that there is a net excess of five nonpreferred codons in stratum 2 on the mat a chromosome. Similar observations were made for alleles located on the mat A chromosome, where we found a net gain of four allele-specific nonpreferred codons in stratum 2 (Table 2). The greatest level of allele-specific switches was detected in stratum 1. Specifically, we found that a total of 232 allele-specific synonymous switches had occurred from a preferred to a nonpreferred codon on the mat a chromosome, while only134 were detected in the opposite direction (nonpreferred to preferred) (this difference is statistically significant [P, <0.05] by Fisher's exact test) (Table 2), resulting in an excess of 98 nonpreferred codons among the gene alleles examined on this chromosome. Similarly, 263 allele-specific switches from preferred to nonpreferred codons were detected for the mat A chromosome, while only 128 switches from nonpreferred to preferred codons were found (P, <0.05 by Fisher's exact test), yielding an excess of 135 nonpreferred codons. Standardization of the number of excess switches from preferred to nonpreferred codons relative to the total number of codons examined per chromosomal segment (FrExcess NPR) further verified the differences between the various genomic regions. In particular, the frequency of excess switches from preferred to nonpreferred codons was found to be 4.7-fold higher in stratum 1 than in stratum 2 for the mat a chromosome (calculated as FrExcess NPR(stratum1)/FrExcess NPR(stratum2), or 1.26/0.27) and 7.9-fold higher for the mat A chromosome (1.73/0.22) (Table 2), consistent with greater degeneration with extended periods of recombination suppression. Taken together, these findings show that there is a net excess of allele-specific switches from preferred to nonpreferred codons within the recombinationally suppressed regions in N. tetrasperma mat chromosomes (but not in the pseudoautosomal regions) and that the level of degeneration increases with longer periods of recombination suppression. Furthermore, these data also reveal that there is a marked excess of preferred-to-nonpreferred codon switches on both the mat a and the mat A chromosomes and thus that there is not a single “degenerative” chromosome in N. tetrasperma.
Table 2.
Numbers of synonymous allele-specific switches from preferred to nonpreferred codons and from nonpreferred to preferred codons that have occurred on the Neurospora tetrasperma mat a and mat A chromosomes (relative to Neurospora crassa)
| N tetrasperma chromosomea | Type of allele-specific switchb | Type of codon located in: |
No. of switchesc | Excess of switches from PR to NPRd | FrExcess NPRe | ||
| N. crassa | N. tetraspermamat a | N. tetraspermamat A | |||||
| Pseudoautosomal regions | |||||||
| mat a | PR to NPR | PR | NPR | PR | 0 | −1 | −0.04 |
| NPR to PR | NPR | PR | NPR | 1 | |||
| mat A | PR to NPR | PR | PR | NPR | 2 | +2 | 0.08 |
| NPR to PR | NPR | NPR | PR | 0 | |||
| Stratum 2 | |||||||
| mat a | PR to NPR | PR | NPR | PR | 9 | +5 | 0.27 |
| NPR to PR | NPR | PR | NPR | 4 | |||
| mat A | PR to NPR | PR | PR | NPR | 7 | +4 | 0.22 |
| NPR to PR | NPR | NPR | PR | 3 | |||
| Stratum 1f | |||||||
| mat a | PR to NPR | PR | NPR | PR | 232 A | +98 | 1.26 |
| NPR to PR | NPR | PR | NPR | 134 B | |||
| mat A | PR to NPR | PR | PR | NPR | 263 A | +135 | 1.73 |
| NPR to PR | NPR | NPR | PR | 128 B | |||
The total numbers of codon positions examined for the pseudoautosomal region, stratum 2, and stratum 1 are 25,502, 18,419, and 77,910, respectively.
PR, preferred codons; NPR, nonpreferred codons.
Within each stratum, values followed by different letters indicate a statistically significant difference (P < 0.05) between the number of switches from PR to NPR and the number of switches from NPR to PR.
Data shown are based on total numbers of switches from PR to NPR and from NPR to PR where the amino acid is conserved. When all possible switches, regardless of amino acid status, are included, the net excess remains the same for the pseudoautosomal region; the values for stratum 2 are +5 for both the mat a and mat A chromosomes; and the values for stratum 1 are +100 for mat a and +138 for mat A.
Frequency of excess PR-to-NPR switches per 1,000 codon positions, calculated as (number of PR-to-NPR switches − number of NPR-to-PR switches)/total codon positions examined × 1,000.
Three codon positions in stratum 1 had a codon switch on both the mat a and mat A chromosomes and were excluded from these results.
The net excess of synonymous allele-specific switches to nonpreferred codons described above was determined using PR → NPR and NPR → PR switches for synonymous codon positions where the amino acid was conserved (Table 2). Examination of the total excess of allele-specific switches among preferred and nonpreferred codons, i.e., independent of amino acid status and including both synonymous and nonsynonymous changes, yielded highly similar results. We found a net excess of 100 allele-specific switches from preferred to nonpreferred codons on the mat a chromosome and 138 on the mat A chromosome for stratum 1 (Table 2, footnote d; see also Table SA5 in the supplemental material). The similarity of the findings is consistent with the fact that the vast majority of allele-specific codon switches were synonymous (95.8% [mat a] and 93.5% [mat A] of allele-specific codon switches in stratum 1 were synonymous).
In addition to allele-specific switches from preferred to nonpreferred codons (and vice versa), we also determined the frequency of other types of preferred codon changes for each of the 121,831 codon positions examined in this study (see Table SA5 in the supplemental material). For example, we found a relatively high number of codon positions in which N. crassa contained a preferred codon and N. tetrasperma contained nonpreferred codons that were identical for P4492a and P4492A (and vice versa) in stratum 1 and stratum 2. This is indicative of a codon switch that occurred either in the N. crassa or the N. tetrasperma lineages (prior to recombination suppression). Table SA5 in the supplemental material provides a summary of the frequencies of all types of preferred codon changes among the mat chromosomes.
Mutation versus selection.
In order to assess whether mutational or selective forces drive the evolution of preferred Neurospora codons, we compared the GC contents of third codon positions (GC3) and the GC contents of introns (GCI) (55, 60) among the 290 genes examined here. If mutational pressures drive preferred-codon usage in Neurospora, a direct association between GC3 and GCI would be expected. The results show no evidence of a correlation between GC3 and GCI for N. crassa genes or for genes located on the N. tetrasperma P4492 mat a and mat A chromosomes (se Fig. SA1 in the supplemental material). Given that preferred Neurospora codons end in G or C nucleotides (see Table SA1 in the supplemental material), this is consistent with a lack of a role for mutational bias. Thus, the data suggest that selective pressure variation is driving changes in preferred-codon usage in the mat chromosomes.
The finding that codon usage in N. tetrasperma mat chromosomes is driven largely/primarily by selection (and not mutational bias) suggests that the divergence in preferred-codon usage in the segment of suppressed recombination in the mat chromosomes is attributable to altered selective constraints. Nonetheless, it is conceivable that small-scale/localized mutational biases could also play a role (39). In particular, gene-specific mutational biases associated with the region of suppressed recombination could drive the observed divergence (e.g., allele-specific mutational biases, specific to each gene). In order to assess this possibility, we compared the difference in the frequency of preferred codons to the difference in the frequency of GC nucleotides in introns for mat a versus mat A linked alleles from among genes located in the region of suppressed recombination (using the genes showing differences in preferred codons and in GCI per allele). The data show that the direction of the differences in preferred-codon frequency among alleles per gene has no association with the difference in GCI frequency (see Fig. SA2 in the supplemental material). For example, among the genes where the mat a linked allele has a higher number of preferred codons than the mat A linked allele, fewer than half (44.1%) have a parallel trend in GC frequency (i.e., higher GCI frequency in the mat a than in the mat A linked allele). In fact, 55.9% of genes have the opposite GCI trend (lower GCI frequency in the mat a linked allele). Genes with a greater frequency of preferred codons in mat A than in mat a linked alleles also show a lack of association between preferred-codon frequency and GCI frequency (see Fig. SA2 in the supplemental material). Thus, these data indicate that mutational bias has no substantive relationship at the intragene level with the divergence of preferred-codon usage in the region of suppressed recombination within the mat chromosomes.
DISCUSSION
The net excess of allele-specific codon switches from preferred to nonpreferred codons within the recombinationally suppressed regions of the N. tetrasperma mat chromosomes (Table 2; Fig. 2) is consistent with genomic degeneration. This effect is time dependent, as indicated by higher levels of allele-specific changes in the older stratum 1 than in stratum 2 (Table 2). The accumulation of nonpreferred codons is likely the result of reduced selective efficiency in regions of suppressed recombination. The nonindependence of gene alleles in chromosomal regions with suppressed recombination is believed to reduce the effective population size, giving rise to decreased selective efficiency and genetic hitchhiking (26, 28). For example, the fixation of accumulated deleterious mutations, such as switches from preferred to nonpreferred codons, in nonrecombining regions may be promoted by processes such as background selection (which promotes the fixation of mildly deleterious mutations [11]) and/or by Muller's ratchet (with the stochastic loss of chromosome classes with the lowest levels of mutations [46]). Positive selection might also contribute to degeneration in regions with suppressed recombination; for instance, positive-selection events may drag deleterious mutations at linked sites to fixation via selective sweeps (6, 11, 51). In this regard, the net decline in preferred-codon usage in the recombinationally suppressed region of the N. tetrasperma mat chromosomes is likely driven by relaxed purifying selection and/or by positive-selection events.
The lack of an association between preferred-codon usage and GCI suggests that mutational pressure does not play a significant role in the degeneration of the N. tetrasperma mat chromosomes (see Fig. SA1 and SA2 in the supplemental material). Biased gene conversion toward GC-ending codons has been proposed to be a potential force driving GC content in coding regions and, thus, preferred-codon evolution (19, 23, 24, 25, 33). This is an unlikely factor in our findings, because such events have been associated primarily with regions of very high recombination in outbreeding species (19), whereas we found preferred-codon degeneration within a region of suppressed recombination within the inbreeding species N. tetrasperma. In addition, biased gene conversions have been predicted to have similar impacts on coding and intron regions (20) and to preferentially convert AT to GC (40), neither of which was found in our data (see Fig. SA2 in the supplemental material; we found elevated numbers of switches from GC-ending to AT-ending codons). Accordingly, the totality of our data point toward the conclusion that degeneration in preferred-codon usage in the mat chromosomes results from altered selective pressure in the young regions of suppressed recombination.
Codon changes and gene length.
The elevated bias in codon usage in shorter genes (CDS regions) found here (Fig. 1) has been reported for certain animals and plants (e.g., Drosophila, Arabidopsis. Caenorhabditis, and Populus [3, 14, 17]). This might result from enhanced gene expression (3, 18) and/or from higher recombination rates in shorter genes (13), each of which likely leads to enhanced selection pressure for preferred-codon usage (3, 13). The findings of markedly higher levels of degenerative changes in preferred-codon usage for shorter genes (Fig. 3) suggest that they are highly susceptible to genomic alterations at the onset of recombination suppression. It may be speculated that a history of highly efficient selection for preferred-codon usage in shorter genes, possibly resulting from elevated gene expression and/or recombination rates (3, 13), makes these genes most vulnerable to reduced/altered selection pressures after the onset of recombination suppression (11). Given that the accumulation of deleterious mutations is a key factor shaping the degeneration of Y chromosomes (11, 34), our data suggest that short genes might play a relatively greater role than longer genes in the earliest stages of degeneration. Further studies will be needed to ascertain the precise reasons why elevated levels of degenerative codon changes are inherent to shorter genes.
Patterns of degeneration.
Our present data reveal that the degeneration in preferred-codon usage occurs in the recombinationally suppressed region for each of the two mat chromosomes of N. tetrasperma (Table 2). This finding corresponds with the fact that each of the mat chromosomes is recombinationally suppressed, and it is consistent with trends expected for systems with haploid sex determination, i.e., degeneration on both chromosomes (7). Although both of the mat chromosomes of N. tetrasperma show degeneration, their degeneration levels differ; for instance, in stratum 1, the excess of allele-specific switches from preferred to nonpreferred codons in mat A versus mat a is 37 (calculated as 135 − 98), which is >27% more in mat A (Table 2). This suggests a deviation from the expectation of similar levels of degeneration for organisms with extensive haploid selection. Furthermore, in contrast to theoretical predictions for organisms with extended haploid stages of development, our data do not suggest that there has been a low level of degeneration in the region of suppressed recombination. For example, in stratum 1, the excesses of allele-specific switches from preferred to nonpreferred codons were 1.26 and 1.73 per 1,000 codons for mat a and mat A, respectively, values more than 21-fold higher than those for the pseudoautosomal regions (≤0.08 per 1,000 codons) (Table 2). Moreover, we found that the net differences in the preferred-codon levels among mat-linked alleles are also substantial (the preferred codon differential for some genes is >10 [Fig. 2]). Thus, it is possible that the recombinationally suppressed region in N. tetrasperma mat chromosomes evolves in a hybrid manner, with degeneration on both chromosomes as expected under haploid systems (Table 2) and with substantial degeneration and unequal levels of degeneration along the mat linked alleles, as predicted for dimorphic sex chromosomes (Table 2; Fig. 2).
Haploid selection is believed to be a possible contributing factor in the lack of degeneration reported for recombinationally suppressed segments of the large mating-type locus (>100 kb) of Cryptococcus neoformans. In this system, no losses in gene functionality or changes in chromosome size have been found (23). However, it is not known whether C. neoformans shows more-subtle degenerative genomic changes, such as a decline in preferred-codon usage, as reported here. Notably, C. neoformans can exist as a haploid or a diploid and often occurs as haploid cells in the environment (23); thus, there may be marked opportunity for haploid selection to limit degeneration in this taxon. The opportunity for haploid selection may be lower in N. tetrasperma. Haploid selection in N. tetrasperma might arise due to homokaryotic sectors in the heterokaryon or due to the presence of occasional haploid individuals within the population (48, 56). However, the predominance of a heterokaryotic state in N. tetrasperma may permit the sheltering of mutations among haploid nuclei during the life cycle (49), a feature that might facilitate greater degeneration in this species than in organisms that exist primarily or solely as haploid individuals.
The complex pattern of degeneration in the region of suppressed recombination in N. tetrasperma could result from the combined effects of a predominant heterokaryotic state (31) that allows both haploid selection and mutational sheltering. It is also possible that other factors, such as chromosome-specific structural rearrangements on the mat A chromosome (32) and/or rare outcrossing or interspecific hybridization events (31, 43), may contribute to the pattern of degeneration.
Conclusions.
Based on the present data, several key conclusions can be drawn regarding genomic changes associated with the young regions of recombination suppression in N. tetrasperma mat chromosomes. First, the early stages of recombination suppression are characterized by marked and largely independent degeneration in preferred-codon usage among gene alleles. Second, the level of degeneration is magnified over longer periods of recombination suppression, as evidenced by substantially higher levels of degeneration in stratum 1 than in stratum 2. Third, both mat a and mat A chromosomes are subjected to degeneration, i.e., there is no single “degenerative” chromosome, in contrast to the pattern in ancient dimorphic (X/Y) systems, although a greater level of degeneration was found on the mat A than on the mat a chromosome. Further studies from other recently emerging model systems of young sex chromosomes (e.g., Silene [41]) will be needed to ascertain whether this pattern of degeneration is a universal feature of early stages of recombination suppression or is specific to the N. tetrasperma mat chromosomes. Fourth, our data indicate that the rate of divergence in preferred-codon usage is greater for short genes, which have evolved for highly efficient translation. In the future, as more genomic data for N. tetrasperma become available, studies should focus on the transposition frequency and the onset of gene-inactivating mutations within the region of restricted recombination in order to reveal whether these other degenerative traits are also inherent to early sex chromosome evolution.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge research funding from the Royal Swedish Academy of Sciences (Hierta-Retzius research grant) (C.A.W.), The Royal Physiographic Society in Lund (C.A.W.), The Lars Hierta Minne Foundation (C.A.W.), the Wenner-Gren Foundation (C.A.W.), and The Swedish Research Council (H.J.).
We thank John Taylor (UC Berkeley), Louise Glass (UC Berkeley), Don Natvig (University of New Mexico), David Jacobson (UC Berkeley), and the JGI (DOE Joint Genome Institute) for access to their FGSC 2508 data, used as a reference in the assembly of our N. tetrasperma Solexa data, prior to its publication. We also thank the two anonymous reviewers for valuable comments on our manuscript.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Akashi H. 1994. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics 136:927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akashi H. 1995. Inferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akashi H. 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11:660–666 [DOI] [PubMed] [Google Scholar]
- 4. Bachtrog D. 2003. Adaptation shapes patterns of genome evolution on sexual and asexual chromosomes in Drosophila. Nat. Genet. 34:215–219 [DOI] [PubMed] [Google Scholar]
- 5. Bachtrog D. 2003. Protein evolution and codon usage bias on the neo-sex chromosomes of Drosophila miranda. Genetics 165:1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bachtrog D. 2004. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36:518–522 [DOI] [PubMed] [Google Scholar]
- 7. Bull J. J. 1978. Sex chromosomes in haploid dioecy: a unique contrast to Muller's theory for diploid dioecy. Am. Nat. 112:245–250 [Google Scholar]
- 8. Bull J. J. 1983. Evolution of sex determination mechanisms. Benjamin Cummings, Menlo Park, CA [Google Scholar]
- 9. Carvalho A. B. 2002. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 12:664–668 [DOI] [PubMed] [Google Scholar]
- 10. Casselton L. A. 2008. Fungal sex genes—searching for the ancestors. Bioessays 30:711–714 [DOI] [PubMed] [Google Scholar]
- 11. Charlesworth B., Charlesworth D. 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charlesworth D., Charlesworth B., Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128 [DOI] [PubMed] [Google Scholar]
- 13. Comeron J. M., Kreitman M., Aguadéb M. 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151:239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cutter A. D., Wasmuth J. D., Washington N. L. 2008. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 178:2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dettman J. R., Jacobon D. J., Taylor J. W. 2003. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57:2703–2720 [DOI] [PubMed] [Google Scholar]
- 16. Duret L. 2000. tRNA gene number and codon usage in the C. elegans genome are coadapted for optimal translation of highly expressed genes. Trends Genet. 16:287–289 [DOI] [PubMed] [Google Scholar]
- 17. Duret L., Mouchiroud D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 96:4482–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eisenberg E., Levanon E. Y. 2003. Human housekeeping genes are compact. Trends Genet. 19:362–365 [DOI] [PubMed] [Google Scholar]
- 19. Eyre-Walker A. 1993. Recombination and mammalian genome evolution. Proc. Biol. Sci. 252:237–243 [DOI] [PubMed] [Google Scholar]
- 20. Eyre-Walker A. 1999. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics 152:675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fraser J. A., Heitman J. 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51:299–306 [DOI] [PubMed] [Google Scholar]
- 22. Fraser J. A., Heitman J. 2005. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 15:645–651 [DOI] [PubMed] [Google Scholar]
- 23. Fraser J. A., et al. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:2243–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galtier N., Duret L. 2007. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 23:273–277 [DOI] [PubMed] [Google Scholar]
- 25. Galtier N., Piganeau G., Mouchiroud D., Duret L. 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159:907–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gordo I., Charlesworth B. 2001. Genetic linkage and molecular evolution. Curr. Biol. 11:R684–R686 [DOI] [PubMed] [Google Scholar]
- 27. Handley L. L., Ceplitis H., Ellegren H. 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167:367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill W. G., Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8:269–294 [PubMed] [Google Scholar]
- 29. Holliday R. 1974. Molecular aspects of genetic exchange and gene conversion. Genetics 78:273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hood M. E. 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160:457–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacobson D. 1995. Sexual dysfunction associated with outcrossing in Neurospora tetrasperma, a pseudohomothallic ascomycete. Mycologia 87:604–617 [Google Scholar]
- 32. Jacobson D. J. 2005. Blocked recombination along the mating-type chromosomes of Neurospora tetrasperma involves both structural heterozygosity and autosomal genes. Genetics 171:839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khakhlova O., Bock R. 2006. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46:85–94 [DOI] [PubMed] [Google Scholar]
- 34. Kondrashov A. S. 1998. Measuring spontaneous deleterious mutation process. Genetica 102–103:183–197 [PubMed] [Google Scholar]
- 35. Lahn B. T., Page D. 1999. Four evolutionary strata on the human X chromosome. Science 286:964–967 [DOI] [PubMed] [Google Scholar]
- 36. Li H., Ruan J., Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Librado P., Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 38. Lloyd A. T., Sharp P. M. 1993. Synonymous codon usage in Kluyveromyces lactis. Yeast 9:1219–1228 [DOI] [PubMed] [Google Scholar]
- 39. Marais G., Mouchiroud D., Duret L. 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc. Natl. Acad. Sci. U. S. A. 98:5688–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marais G., Charlesworth B., Wright S. I. 2004. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marais G. A., et al. 2008. Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr. Biol. 18:545–549 [DOI] [PubMed] [Google Scholar]
- 42. Menkis A., Jacobson D. J., Gustafsoon T., Johannesson H. 2008. The mating-type chromosome in the filamentous asycomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4:e1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Menkis A., Bastiaans E., Jacobson D. J., Johannesson H. 2009. Phylogenetic and biological species diversity within the Neurospora tetrasperma complex. J. Evol. Biol. 22:1923–1936 [DOI] [PubMed] [Google Scholar]
- 44. Merino S. T., Nelson M. A., Jacobson D. J., Natvig D. O. 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143:789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muller H. J. 1914. A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 17:325–336 [Google Scholar]
- 46. Muller H. J. 1964. The relation of recombination to mutational advance. Mutat. Res. 106:2–9 [DOI] [PubMed] [Google Scholar]
- 47. Puigbò P., Bravo I. G., Garcia-Vallve S. 2008. CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct 3:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raju N. B. 1992. Functional heterothallism resulting from homokaryotic conidia and ascospores in Neurospora tetrasperma. Mycol. Res. 96:103–116 [Google Scholar]
- 49. Raju N. B. 2009. Neurospora as a model fungus for studies in cytogenetics and sexual biology at Stanford. J. Biosci. 34:139–159 [DOI] [PubMed] [Google Scholar]
- 50. Raju N. B., Perkins D. D. 1994. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora and Podospora. Dev. Genet. 15:104–118 [DOI] [PubMed] [Google Scholar]
- 51. Rice W. R. 1987. Genetic hitch-hiking and the evolution of reduced genetic activity of Y sex chromosome. Genetics 116:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sandstedt S. A., Tucker P. K. 2004. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 14:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiu P. K. T., Glass N. L. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3:183–188 [DOI] [PubMed] [Google Scholar]
- 54. Skaletsky H., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837 [DOI] [PubMed] [Google Scholar]
- 55. Smith N., Eyre-Walker A. 2001. Synonymous codon bias is not caused by mutation bias in G+C-rich genes in humans. Mol. Biol. Evol. 18:982–998 [DOI] [PubMed] [Google Scholar]
- 56. Stenlid J. 2008. Population biology of forest decomposer basidiomycetes. Br. Mycol. Soc. Symp. Ser. 28:105–122 [Google Scholar]
- 57. Stoletzki N., Eyre-Walker A. 2007. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 24:374–381 [DOI] [PubMed] [Google Scholar]
- 58. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Votintseva A. A., Filatov D. A. 2009. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics 182:1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolfe K., Sharp H. P. M., Li W.-H. 1989. Mutation rates differ among regions of the mammalian genome. Nature 337:283–285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.