Abstract
Cryptococcus neoformans is an AIDS-associated human fungal pathogen and the most common cause of fungal meningitis, with a mortality rate over 40% in AIDS patients. Significant advances have been achieved in understanding its disease mechanisms. Yet the underlying mechanism of a high frequency of cryptococcal meningitis remains unclear. The existence of high inositol concentrations in brain and our earlier discovery of a large inositol transporter (ITR) gene family in C. neoformans led us to investigate the potential role of inositol in Cryptococcus-host interactions. In this study, we focus on functional analyses of two major ITR genes to understand their role in virulence of C. neoformans. Our results show that ITR1A and ITR3C are the only two ITR genes among 10 candidates that can complement the growth defect of a Saccharomyces cerevisiae strain lacking inositol transporters. Both S. cerevisiae strains heterologously expressing ITR1A or ITR3C showed high inositol uptake activity, an indication that they are major inositol transporters. Significantly, itr1a itr3c double mutants showed significant virulence attenuation in murine infection models. Mutating both ITR1A and ITR3C in an ino1 mutant background activates the expression of several remaining ITR candidates and does not show more severe virulence attenuation, suggesting that both inositol uptake and biosynthetic pathways are important for inositol acquisition. Overall, our study provides evidence that host inositol and fungal inositol transporters are important for Cryptococcus pathogenicity.
INTRODUCTION
Cryptococcus neoformans is a major human fungal pathogen and the causative agent of fatal cryptococcal meningoencephalitis. It is the leading cause of fungal meningitis, with an estimated ∼1 million cases worldwide in patients with AIDS and organ transplants and in those who are immunocompromised by cancer chemotherapy or by other treatments (1, 9, 11, 47, 49). Due to the medical significance and genetic tractability of C. neoformans, extensive studies have been conducted on the mechanism of its virulence. Several signal pathways important for Cryptococcus virulence have been identified (1, 5, 27, 33, 34, 63). However, fungal virulence is a complex trait, and new virulence-determining mechanisms remain to be explored.
Inositol is essential for cellular structure and intracellular signaling regulation of all eukaryotes. Inositol-derived products have been reported to be important for pathogenicity in fungi and parasites. Enzymes involved in sphingolipid biosynthesis and degradation pathways, such as inositol-phosphoryl ceramide synthase 1 (Ipc1) (35) and inositol phosphosphingolipid-phospholipase C1 (Isc1) (53), have been found to promote pathogenicity in C. neoformans. The importance of sphingolipids for pathogenicity has also been recognized in some parasitic pathogens, including Leishmania and Trypanosoma species, and related enzymes have been proposed as potential drug targets (36, 37, 45, 60, 67). The diacyglycerol (DAG)-protein kinase C1 (Pkc1) signaling pathway is critical for virulence factor production and pathogenicity in C. neoformans (18, 19, 24, 25). The glycophosphatidylinositol (GPI)-anchored glycolipid is derived from inositol and is necessary for pathogenicity in Candida species (61).
There are nine inositol stereoisomers, and myo-inositol is the common one (here, referred to as inositol unless otherwise specified). There are two main sources by which fungal cells can acquire inositol. One is the internal synthesis of inositol. Intracellular glucose can be converted into inositol in a multiple-step biosynthetic pathway (14, 50). Inositol can also be imported from the extracellular environment via inositol transporters. The inositol transporter (ITR) gene family is part of the sugar transporter superfamily and plays an important role in inositol sensing in fungi, including Saccharomyces cerevisiae (30, 31, 42–44, 51), Candida albicans (12, 28, 50), C. neoformans (64), and Schizosaccharomyces pombe (40). Studying ITR genes in fungi has clinical significance as ITR genes identified in lower eukaryotes, such as fungi (28) and protozoa (15, 39, 52), are proton coupled and differ both kinetically and pharmacologically from the sodium-dependent inositol transporter system (SMIT) in humans. Therefore, they have the potential to be developed as potential targets for drug treatment.
Inositol plays a unique role in the development of cryptococci. It is known that C. neoformans is one of a few yeast organisms that can utilize inositol as a sole carbon source (6, 23). Recently, we identified inositol as one important compound for stimulating the sexual reproduction of this yeast (64, 65). Furthermore, an unusually large ITR gene family has been identified in C. neoformans based on the sequence homology with known ITR genes in other organisms. Functional characterization revealed that this ITR gene family is important for both sexual reproduction and fungal virulence (64, 65). Unlike most fungi that contain one or two inositol transporters, the H99 strain carries 10 genes that are considered ITR candidates, and most of them are located in the telomere regions of the chromosomes (64). The ITR homologues can be divided into two distinct groups (I and II). The seven members of group I (Itr1, Itr1a, Itr2, Itr3, Itr3a, Itr3b, and Itr3c) share strong sequence homology with well-characterized ITR genes in other fungi, such as S. cerevisiae and C. albicans. Group II has three members (Itr4, Itr5, and Itr6) that are more distantly related ITR genes that have been characterized in other yeasts but are closely related to HGT19, potential a second ITR in C. albicans (12). However, the role of HGT19 in inositol function has not yet been confirmed.
Cryptococcal meningitis is the predominant form of cryptococcal infection and causes high mortality, but its disease mechanism is still poorly understood. It has been suggested that the high rate of cryptococcal meningitis may be related to the high inositol levels found in mammalian brains (38). The inositol concentration in human cerebrospinal fluid (CSF) is around 22 mg/liter, compared to an average 2.8 mg/liter in plasma, which makes the human brain the location with the most abundant free inositol (17, 55–58). High inositol levels are also found in animal brains (7, 17). Gene expression profiling during experimental cryptococcal meningitis in rabbits showed that the inositol 3-phosphate synthase gene (INO1) and inositol monophosphatase were highly expressed during brain infection, further suggesting a potential role of inositol in the development of cryptococcal meningitis (59). Because Cryptococcus contains an expanded ITR gene family and can utilize inositol as a sole carbon source, we hypothesize that sensing and utilizing host inositol play important roles during pathogen-host interactions. The high levels of inositol in the human brain could, then, be one reason why this pathogen so frequently causes meningitis. Our previous functional studies on the seven group I ITR genes revealed that individual single mutations had no clear impact on fungal virulence and that this may be due to functional redundancy, but the ino1 itr1a double mutant did show virulence attenuation. This result suggests that inositol acquisition is important for the pathogenicity of C. neoformans (64).
In this study, we continue a functional analysis of ITR homologues in C. neoformans to understand their role in virulence of C. neoformans. We expressed ITR4, ITR5, and ITR6 in an S. cerevisiae heterologous expression system that had previously been used to express the other seven ITR genes in C. neoformans (64). Inositol uptake assays for all 10 Saccharomyces strains expressing Cryptococcus ITR genes revealed that Itr1a and Itr3c are two major inositol transporters. Significantly, itr1a itr3c double mutants and ino1 itr1a itr3c triple mutants showed significant virulence attenuation in murine infection models. Additional ITR genes were activated in the triple mutant background to partially compensate for the inositol defect. These results support the hypothesis that inositol acquisition in C. neoformans is important for the Cryptococcus-host interaction.
MATERIALS AND METHODS
Strains, media, and growth conditions.
C. neoformans strains used in this study are listed in Table 1. Strains were grown at 30°C on yeast extract-peptone-dextrose (YPD) agar medium and synthetic (SD) medium. V8 medium (pH 5.0) was used for mating assays. Modified MS medium (Murashige and Skoog medium) was used for mating and sporulation assays and was prepared as previously described (65). Yeast nitrogen base (YNB) medium without inositol was purchased from Sigma-Aldrich. Niger seed medium was used to test for melanin production. Dulbecco's modified Eagle's (DME) medium for assessing capsule production was prepared as previously described (3). All other media were prepared as described previously (1, 64).
Table 1.
Strains used in this study
| C. neoformans var. grubii strain | Description | Reference or source |
|---|---|---|
| H99 | MATα wild type | 48 |
| KN99a | MATa wild type | 41 |
| CDX99 | MATα itr1a::NAT | 64 |
| CDX100 | MATaitr1a::NEO | 64 |
| CDX166 | MATα itr3c::NAT | 64 |
| CDX167 | MATaitr3c::NAT | 64 |
| UBCINO1 | MATα ino1::NEO | 64 |
| CUX8 | MATaino1::NEO | 64 |
| UBCINO11 | MATα ino1::NEO INO1 | 64 |
| CUX46 | MATα itr1a::NEO itr3c::NAT | This study |
| CUX47 | MATaitr1a::NEO itr3c::NAT | This study |
| CUX57 | MATα ino1::NEO itr1a::NEO itr3c::NAT | This study |
| CUX58 | MATaino1::NEO itr1a::NEO itr3c::NAT | This study |
| CUX80 | MATα itr1a::NEO itr3c::NAT ura5 | This study |
| CUX74 | MATα itr1a::NEO itr3c::NAT ura5 ITR1A-URA5 | This study |
| CUX75 | MATα ino1::NEO itr1a::NEO itr3c::NAT ura5 | This study |
| CUX76 | MATaino1::NEO itr1a::NEO itr3c::NAT ura5 | This study |
| CUX86 | MATα ino1::NEO itr1a::NEO itr3c::NAT ura5 INO1 ITR1A-URA5 | This study |
Database and sequence information.
All fungal ITR genes and other sequences were obtained from a variety of databases via the web link (http://fungal.genome.duke.edu/). All sequences for C. neoformans var. grubii were from the Broad Institute (http://www.broadinstitute.org/). Related ITR sequences in S. cerevisiae and C. albicans were obtained from the Saccharomyces genome database and Candida genome database. Phylogeny trees were generated using ClustalX, version 2.0 (32), and viewed via Treeview X (46).
Detection of ITR gene expression using quantitative reverse transcription-PCR (qRT-PCR).
To test how the ITR genes respond to the presence of environmental myo-inositol, both in vitro and in vivo, we measured the mRNA levels for all 10 ITR genes under different conditions via quantitative real-time PCR (qPCR). Cultures of C. neoformans var. grubii wild-type strain H99 and its mutant strains were grown on YPD medium for 24 h at 30°C. Collected cells were washed with distilled H2O (dH2O), and pellets were used for total RNA extraction. Tissues infected by H99 or its mutants were also collected from infected animals right after they were terminated, and tissues were homogenized using a homogenizer and spun down. Pellets were washed with phosphate-buffered saline (PBS) and used for RNA preparation. Total RNAs were extracted using Trizol reagents (Invitrogen) and purified with a Qiagen RNeasy Cleanup Kit (Qiagen) following the manufacturer's instructions. Purified RNAs were quantified using a Nanodrop instrument (Thermo Scientific). The same approach was used to prepare RNAs from itr1 mutant strains.
First-strand cDNAs were synthesized using a Superscript III cDNA synthesis kit (Invitrogen) following the instructions provided by the manufacturer. Expression of ITR genes and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was analyzed with the comparative threshold cycle (CT) method using Brilliant SYBR green qPCR reagents (Stratagene) as described previously (64).
Inositol uptake assay.
Full-length cDNAs of the ITR4, ITR5, and ITR6 genes were amplified from C. neoformans H99 total cDNA and were cloned into the yeast expression vector pTH19 (22), under the control of the ADH1 promoter. Each ITR expression plasmid was introduced into an S. cerevisiae strain lacking both inositol transporters, Itr1 and Itr2 (Scitr1 Scitr2) for heterologous expression as previously described (64). The expression of each Cryptococcus ITR in this yeast heterologous system was verified by RT-PCR using gene-specific primers (see Table S1 in the supplemental material). Yeast strains were tested for growth on YPD medium at 30°C and 37°C.
The inositol uptake assay protocol was adapted in part from protocols of Jin and Seyfang (28) and Chen and Reynolds (12). In brief, the S. cerevisiae control strain BY4742, the S. cerevisiae itr1 itr2 mutant strain, and mutant strains expressing ITR genes from C. neoformans were grown in YPD liquid cultures overnight at 30°C. Cells were diluted in YPD medium to an optical density at 600 nm (OD600) of 1.0, grown at 30°C, and collected at an OD600 of 5.0 by centrifugation at 2,600 × g for 5 min. Cells were then washed twice with PBS at 4°C and resuspended in 2% glucose to a final concentration of 2 × 108 cells/ml as determined by a hemacytometer. For the uptake assay, the reaction mixture (200 μl) contained 2% glucose, 40 mM citric acid-KH2PO4 (pH 5.5), 0.15 μM myo-[2-3H]inositol (1 μCi/μl; MP BioMedicals). Additional 200 μM unlabeled inositol (Sigma-Aldrich) was added to the reaction mixtures for competition assays. Equal volumes of the reaction and cell mixtures (60 μl each) were warmed to 30°C and mixed for the uptake assay, which was performed for 10 min at 30°C. As negative controls, mixtures were kept at 0°C (on ice) during the 10-min incubation. Aliquots of 100 μl were removed and transferred onto prewetted Metricel filters (1.2 μm pore size) on a vacuum manifold. The filters were washed four times each with 2 ml of ice-cold water. The washed filters were removed and added to liquid scintillation vials for measurements on a PerkinElmer Tri-Carb 2900TR scintillation counter.
Generation of itr1a itr3c mutants and ino1 itr1a itr3c mutants.
In mating assays, C. neoformans cells of opposite mating types were mixed and cocultured on V8 or MS agar medium at 25°C in the dark for 10 days, and filamentation was examined by light microscopy. Spore production was visualized by microscopy and photographed. Basidiospores were dissected from matings performed on MS medium.
To generate itr1a itr3c double mutants, a mating between α itr1a::NEO and a itr3c::NAT mutants was conducted, and spores were visualized and isolated with an MSM microscope system (Singer Instrument, England). Genomic DNA was isolated from all progeny that grew on YPD medium with both nourseothricin and G418. PCR was used to screen for ITR1A gene deletion with primers JH16703/JH8994 and JH16705/JH16706 and for ITR3C gene deletion with primers JH19471/JH8994 and JH19469/JH19470 (see Table S1 in the supplemental material). To generate ino1 itr1a itr3c triple mutants, a mating between α itr1a::NEO itr3c::NAT double mutant and a ino1::NEO mutant was conducted, and spores were isolated. Triple mutants were screened first by PCR on cultures initiated from single spores. All mutants confirmed by PCR were further confirmed by Southern blot analyses. The mating type of each confirmed mutant strain was determined by PCR using mating type-specific primers and by genetic crossing.
To generate complemented strains of the itr1a itr3c double mutant, ura5 mutant strains were generated by selecting colonies grown on agar plates containing 0.1% 5-fluoroorotic acid (5-FOA). A genomic DNA fragment that contained a 1.5-kb upstream region of the ITR1A open reading frame (ORF), the ITR1A gene ORF, and its 500-bp downstream region was amplified by PCR. The ITR1A PCR fragment was fused with the URA5 selective marker gene at its C terminus in an overlap PCR. The ITR1A-URA5 overlap PCR product was biolistically transformed in both itr1a itr3c mutant strains.
Assays for melanin and capsule production.
Melanin production was assayed by inoculating C. neoformans strains into 2 ml of YPD liquid medium, incubating the culture overnight at 30°C, and spotting 5 μl of each culture with 10× series dilutions on Niger seed agar medium and L-3,4-dihydroxyphenylalanine (l-DOPA) medium. The agar plates were incubated at 30°C or 37°C for 2 days, and pigmentation of fungal colonies was assessed and photographed. To examine capsule production, 5 μl of overnight cultures was inoculated on DME agar medium and incubated at 30°C for 3 days. Capsule was visualized with india ink negative staining and observed with an Olympus CX41 microscope equipped with an Infinity digital camera (Olympus).
To quantify the production of melanin by different strains, the activity of laccase enzyme was measured as previously described (26). Single colonies of each strain were inoculated in 5 ml of YPD liquid medium and incubated at 30°C overnight. A total of 108 cells from each overnight culture were inoculated in to 25 ml of l-DOPA medium in 125-ml flasks and incubated at 30°C for 16 h with shaking at 250 rpm. The cultures were further incubated at 25°C for 24 h with shaking at 250 rpm. One milliliter of each culture was centrifuged, and the supernatants were spectrophotometrically read for optical density at a wavelength of 475 nm.
Murine infection.
Cryptococcus strains were grown at 30°C overnight, and cultures were washed twice with 1× phosphate-buffered saline (PBS) by centrifugation and resuspended at a final concentration of 2 × 106 CFU/ml. Groups of 10 female A/Jcr mice (NCI-Frederick, MD) were used for each infection. For the intranasal inhalation model, mice were intranasally infected with 105 yeast cells of each strain in 50 μl of PBS as previously described (13). For the intravenous injection model, 5 × 104 yeast cells in a 100-μl volume for each strain were inoculated via tail vein injection. For the intracerebral injection model, mice were sedated with a xylazine-ketamine combination, and the top of the head was sterilized using antiseptic until the hair was thoroughly wet. A total of 500 yeast cells in 50 μl of PBS were directly injected into the cerebrum. Animals that appeared moribund or in pain were sacrificed by CO2 inhalation. Survival data from the murine experiments were statistically analyzed between paired groups using a log rank test and the PRISM program, version 4.0 (GraphPad Software) (P values of <0.01 were considered significant).
Histopathology and organ fungal burden.
Infected animals were sacrificed at 3 and 7 days postinfection. Lungs and brains were isolated and fixed in 10% formalin solution and sent to the University of Medicine and Dentistry of New Jersey (UMDNJ) core facility for section preparation. Tissue slides were stained with hematoxylin and eosin (H&E) and examined by light microscopy. At each time point of the infection, infected lungs and brains were also isolated and homogenized using a homogenizer (Ultra-Trra T8; IKA) in 1× PBS buffer. Resuspensions were diluted, and 50 μl of each dilution was spread on YPD medium with antibiotics, and colonies were counted after 3 days of incubation at 30°C.
RESULTS
Cryptococcus Itr1a and Itr3c are two major inositol transporters with high inositol uptake activity.
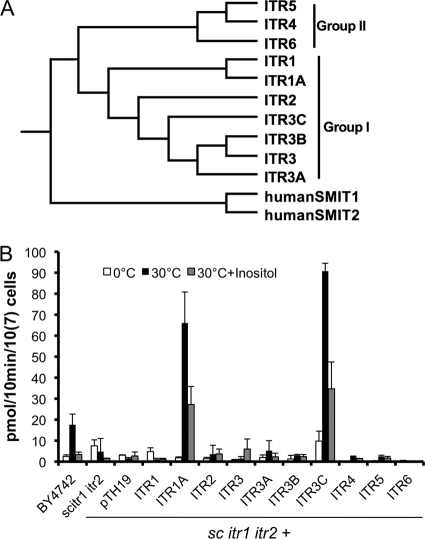
Two groups (group I and group II) of potential inositol transporters with a total of 10 members were identified in the H99 genome (Fig. 1 A). In our previous studies, the group I seven ITR genes of C. neoformans were expressed in an S. cerevisiae mutant strain with deletions of both ITR1 and ITR2 (Scitr1 Scitr2), and we determined that ITR1A and ITR3C could rescue the growth defect of the Scitr1 Scitr2 mutant (64). In this study, we also expressed Cryptococcus group II ITR genes (ITR4, ITR5, and ITR6) in the Saccharomyces tester mutant to further understand the role of each ITR in inositol sensing and uptake. Full-length cDNAs of ITR4, ITR5, and ITR6 were cloned into the yeast expression vector pTH19 under the control of the ADH1 promoter. The expression of these ITR genes was confirmed by RT-PCR (see Fig. S1 in the supplemental material). Strains expressing ITR4, ITR5, and ITR6 could not complement the growth defect of Scitr1 Scitr2 (see Fig. S1). The failure of expression of ITR genes other than ITR1A or ITR3C to complement the defect could reflect either the instability of the overexpressed ITR genes or their lack of functional inositol transporter activity.
Fig. 1.
Saccharomyces strains expressing ITR1A or ITR3C have high inositol uptake activity. (A) Phylogenetic tree of ITR gene family in C. neoformans (Itr1 to Itr6). The full-length protein sequences were used for alignment. The phylogram was generated using ClustalX, version 2.1, viewed by TreeView. (B) Inositol uptake analysis of Cryptococcus ITR genes expressed in a yeast heterologous system. Yeast cells were mixed with 3H-labeled inositol and incubated at 0°C or 30°C for 10 min. Additional 200 μM unlabeled inositol was added to the reaction mixture for a competition assay as a control. Error bars indicate standard deviations of data from at least three independent experiments. sc, S. cerevisiae.
To better understand the role of each ITR in inositol function, inositol uptake activity was measured for all 10 Saccharomyces strains expressing C. neoformans ITR genes by using 3H isotope-labeled myo-inositol (myo-[2-3H]inositol). Uptake assays were performed at 30°C by incubating yeast cells with myo-[2-3H]inositol for 10 min. The results showed that yeast strains expressing ITR1A and ITR3C had very high uptake activity, which is consistent with their ability to complement the growth defect. None of the others showed significant inositol uptake activity (Fig. 1B). Our results indicated that Itr1a and Itr3c are major inositol transporters.
Expression of INO1 is not altered by inositol level or mutation of ITR1A and ITR3C.
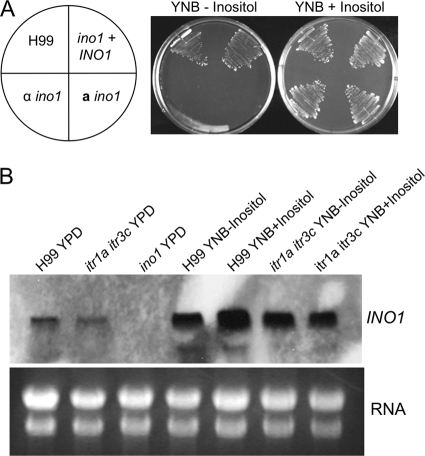
There are two active pathways that can supply inositol for fungal cellular function: importing inositol using ITRs or producing inositol via the Ino1-controlled biosynthetic pathway. A C. neoformans ino1 mutant is inositol auxotrophic and cannot survive when inoculated on a medium without inositol (Fig. 2 A). To further understand the role of the biosynthetic pathway, we measured the expression of the INO1 gene under different inositol conditions. Overnight cultures of H99 were grown in YPD liquid medium, YNB medium lacking inositol, or YNB with 1% inositol. Total RNAs were prepared, and INO1 expression was detected in a Northern blot assay. Our results showed that INO1 is expressed under all tested conditions (Fig. 2B). The expression of INO1 was much higher when Cryptococcus cultures were incubated on minimal medium than when cultured on rich medium (YPD), suggesting that INO1 expression is regulated by nutrient availability.
Fig. 2.
INO1 expression is not regulated by inositol. (A) ino1 mutants are inositol auxotrophic. Yeast cells of H99, the α ino1 mutant, the a ino1 mutant, and its complemented strains were streaked on YNB medium with or without 1% inositol. Cultures were incubated at 30°C for 48 h. (B) Overnight cultures of H99, the itr1a itr3c mutant (CUX46), and the ino1 itr1a itr3c mutant (CUX57) grown on YPD or YNB medium without inositol or with 1% inositol were prepared and their total RNAs were purified. Total RNAs were separated on a denatured gel and transferred to a nylon membrane. The INO1 gene fragment was used as the probe for detecting the expression of INO1 transcripts. Total RNAs stained with ethidium bromide were used as loading controls.
To elucidate the potential effect of the inositol uptake on inositol biosynthesis, we generated the itr1a itr3c double mutants because of their importance in inositol uptake. The expression of the INO1 gene in this double mutant background was analyzed by Northern blotting. Interestingly, the level of INO1 expression observed in the itr1a itr3c double mutant background was similar to that of the wild type under all three culture conditions, suggesting that INO1 expression was not altered by different expression of ITR genes. This result also indicated that the expression of INO1 is not regulated by the availability of inositol in the medium since similar mRNA levels were detected in both wild-type and mutant cultures prepared from medium with or without supplemental inositol (Fig. 2B).
Itr1a and Itr3c are required for sexual reproduction.
Although we previously found that inositol is important for stimulating fungal mating, each inositol transporter single mutant still produced normal dikaryotic mating hyphae and normal sporulation except the itr1 mutant, which showed a mating defect in bilateral mating assays (64). Because of the importance of Itr1a and Itr3c in inositol uptake when they are expressed in S. cerevisiae, mating assays were performed in itr1a itr3c double mutants. Although no obvious mating defect was observed in unilateral mating assays between itr1a itr3c double mutants and the wild type, bilateral mating results (itr1a itr3c × itr1a itr3c) showed that such double mutants had a clear defect in the production of mating hyphae on both MS and V8 mating media, further suggestive of their functional importance (Fig. 3).
Fig. 3.
itr1a itr3c mutants showed a mating defect. Unilateral and bilateral mating assays were performed in both MS and V8 media with itr1a itr3c double mutants and ino1 itr1a itr3c triple mutants. Mating cultures were incubated at room temperature in the dark for 5 days before photography. Significant reduction of mating hyphal production was observed in the bilateral mating assays of the double mutants and triple mutants.
Itr1a and Itr3c are important for fungal virulence.
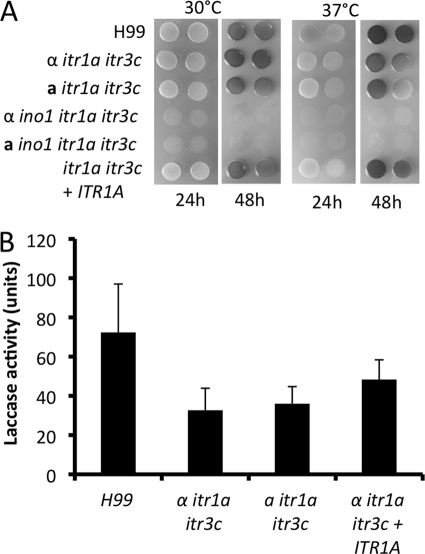
In vitro analyses showed that the itr1a itr3c double mutant produced normal capsules and had normal growth at 37°C. Mutants produced normal melanin when incubated on Niger seed agar medium (data not shown). However, a modest melanin defect was observed when the mutants were incubated on l-DOPA agar medium (Fig. 4 A). The difference in melanin production between these two melanin detection media (Niger seed medium and l-DOPA medium) may be due to different substrate concentrations. Laccase activity analyses also revealed that the double mutants showed lower laccase activity (Fig. 4B). Such results indicate that Itr1a and Itr3c may also play a role in virulence factor development.
Fig. 4.
itr1a itr3c mutants showed a melanin defect and reduced laccase activity. (A) itr1a itr3c double mutants showed modest melanin defect on l-DOPA medium. Overnight cultures of H99, itr1a itr3c mutants (CUX46 and CUX47), and the complemented strains (CUX74) as well as ino1 itr1a itr3c triple mutants (CUX57 and CUX58) were washed with dH2O and spotted on l-DOPA medium. Melanin production was photographed after plates were incubated at either 30°C or 37°C for 24 h. Triple mutants could not grow on l-DOPA medium due to lack of inositol in the medium. (B) Laccase activity assays were performed as described in Materials and Methods. Results presented were analyzed based on three independent replicates.
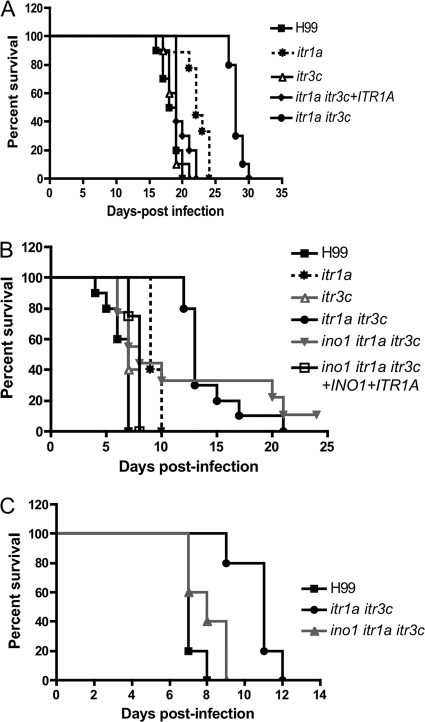
We examined the virulence of itr1a itr3c mutants in a murine inhalation model. Our results showed that mice infected by wild-type H99 or the itr3c mutant survived for less than 22 days postinoculation while mice infected by the itr1a itr3c double mutant had a prolonged survival rate between 27 and 30 days. Because itr3c single mutants did not show any visible defect in our studies, we generated functionally complemented strains of itr1a itr3c double mutants by reintroducing a copy of only the ITR1A gene. Mice infected by the complemented strain had a survival rate similar to that of the wild type, an indication of functional complementation. The itr1a mutant strain also showed modest virulence attenuation but was not statistically significant (P > 0.1). These results indicate that the itr1a itr3c double mutant had significant virulence attenuation (Fig. 5 A) and that both Itr1a and Itr3c are required for full virulence.
Fig. 5.
itr1a itr3c mutants showed significant virulence attenuation in murine infection models. (A) Female A/Jcr mice were inoculated intranasally with the following strains: H99, itr1a mutant (CDX99), itr3c mutant (CUX42), and itr1a itr3c double mutant (CUX46) and its complement strain (CUX74). Groups of 10 mice were infected with 1 × 105 yeast cells each strain. (B) Survival curves of infected mice in a murine intravenous injection model. Female A/Jcr mice were inoculated via tail veins with the following strains: H99, the itr1a itr3c mutant (CUX46), the ino1 itr1a itr3c mutant (CUX57), and the complemented strain (CUX74). Groups of 10 mice were infected with 5 × 104 yeast cells each strain. (C) Groups of 10 mice were infected by H99, CUX46, or CUX57 via intracerebral injection with 500 yeast cells per mouse. Animals were monitored for clinical signs of cryptococcal infection and sacrificed at predetermined clinical endpoints that predict imminent mortality.
Because of the high inositol concentrations in human and animal brains, ITRs may be important during brain infection by affecting either fungal cell dissemination and blood brain barrier (BBB) crossing or fungal growth in the brain. Because we used an inhalation model, death of the mice in the experiments could have been caused by multiple organ failures even before the full progression of brain infection. To address this concern, we performed additional virulence assays by using other animal models. An intravenous injection model was used to specifically check virulence during dissemination and during brain infection. In this tail vain injection model, yeast cells are able to bypass the establishment of local infection in the lung and disseminate into the brain directly via the bloodstream. Ten female A/Jcr mice per group were used in our studies, and each mouse was injected with 5 × 104 yeast cells; infected mice were carefully monitored, and their survival rates were recorded. Our results found that itr1a itr3c double mutants had significant virulence attenuation compared to wild-type H99 and the complemented strain (Fig. 5B).
We also performed the virulence assay using a murine intracerebral injection model to specifically investigate the role of Itr1a and Itr3c in brain infection. Five hundred yeast cells were injected into the cerebrum of each mouse directly. Mice infected by H99 died rapidly by 7 days postinjection, while animals infected by the itr1a itr3c mutant survived up to 12 days, indicating significant virulence attenuation of the mutant strain during brain infection (Fig. 5C).
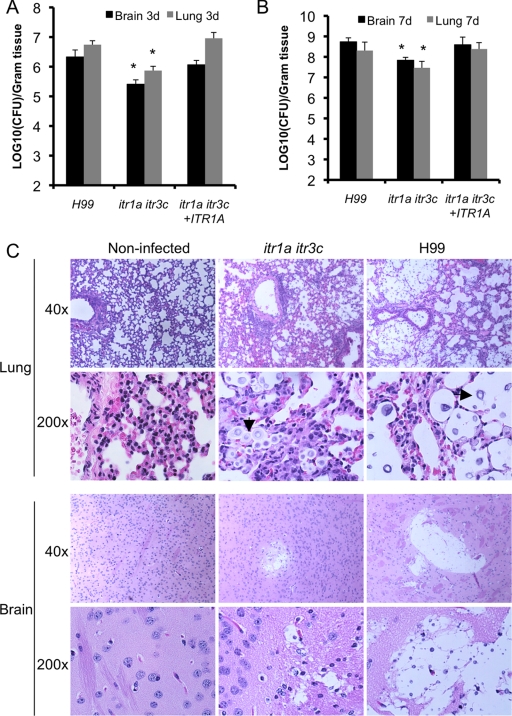
To understand why itr1a itr3c double mutants have a virulence defect, the fungal burdens in infected lungs and brains at different time points of intravenous infection were measured by yeast CFU count. At 3 or 7 days postinfection, compared with mice infected with wild-type strain H99, mouse brains and lungs infected by the double mutant showed significant CFU reductions (Fig. 6 A and B). Histopathology results of infected lungs also demonstrated less severe lesion development in lungs and brains infected by the double mutant (Fig. 6C). These observations suggest that the itr1a itr3c mutant showed attenuated virulence during both lung and brain infection.
Fig. 6.
H&E staining of lung and brain tissues infected by H99 or the itr1a itr3c mutant. (A) A/Jcr mice were infected with 5 × 104 cells per mouse via tail vein injection, and infected lungs and brains were harvested at 3 days and 7 days postinoculation. Fungal burdens in organs infected by the itr1a itr3c mutant (CUX46) were compared with organs infected by H99. The number of yeast CFU per gram organ were measured in brain and lung homogenates. Each error bar indicates the standard error of the mean for values from three animals. The asterisk indicates statistical significance. (B) H&E-stained slides prepared as described in Materials and Methods. Arrows denote yeast cells in the infected tissue.
The ino1 itr1a itr3c mutant strain showed attenuated virulence.
To evaluate the potential functional redundancy between the inositol uptake/sensing pathway and the inositol biosynthetic pathway, we generated the ino1 itr1a itr3c triple mutants in both mating types and analyzed their role in fungal virulence using murine models. The ino1 itr1a itr3c mutant showed normal growth both at 30°C and 37°C (data not shown) but showed a clear defect on mating filament production in bilateral mating assays (Fig. 3). In our virulence assay using the intravenous infection model with 5 × 104 cells as an initial inoculum, it was observed that 50% of mice infected by the triple mutant showed an increased survival rate, an indication that this mutant strain also showed attenuated virulence at a similar level as the itr1a itr3c double mutant (Fig. 5B). But the virulence attenuation phenotype was not as strong as that of the itr1a itr3c double mutants in the intracerebral infection model (Fig. 5C).
The itr1a itr3c mutants showed defects in inositol uptake activity.
To understand whether the virulence attenuation and mating defect of the itr1a itr3c mutants were due to the defect in inositol uptake activity, we performed inositol uptake assays for H99, the itr1a itr3c double mutant, and the ino1 itr1a itr3c triple mutant following the method described in Materials and Methods. Our results showed that all three strains had similar uptake activities when a small amount of inositol was available. However, when the inositol concentrations in the reaction mixture were higher than 0.5 μM, mutant cells could import much less free inositol in 10 min than the wild-type strain H99 (Fig. 7). Interestingly, the triple mutant showed a higher inositol uptake rate than the double mutant under the same conditions, suggesting that remaining ITR genes could be activated in the triple mutant to compensate for the mutation of INO1 in the itr1a itr3c double mutant background.
Fig. 7.
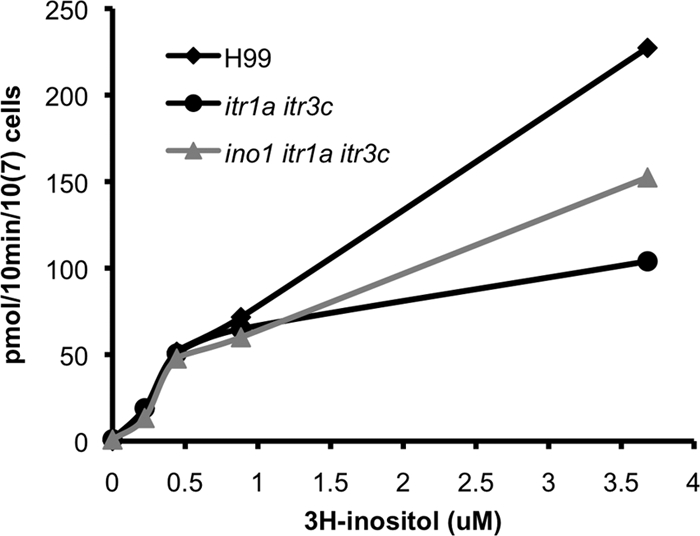
The itr1a itr3c mutant had a lower inositol uptake rate than the wild-type H99. Inositol uptake assays were performed for C. neoformans strains (H99, CUX46, and CUX57) following the protocol described in Materials and Methods with several different concentrations of radioactively labeled inositol (0, 0.22 μM, 0.44 μM, 0.88 μM, and 3.68 μM). Representative results from three repeats were presented.
Additional ITR genes were induced in the double mutant and the triple mutant.
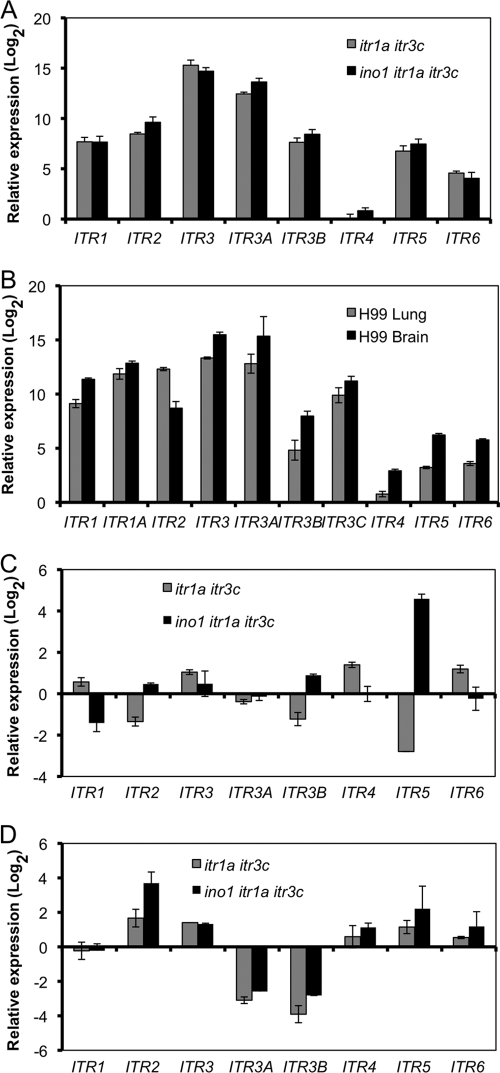
Because ino1 mutants could not survive on medium without inositol (Fig. 3A), blocking both inositol internal biosynthetic pathway and inositol uptake activity would be lethal, a phenomenon called “inositol-less death.” The possible reason that the ino1 itr1a itr3c triple mutant did not show more severe virulence attenuation than the itr1a itr3c double mutant could be either that additional ITR genes play an increased role in inositol uptake or that the remaining ITR genes become active in the triple mutant background. Such a hypothesis is consistent with our inositol uptake results showing that the triple mutant had a higher uptake rate than the double mutant (Fig. 7). To test this hypothesis, we first measured the expression of ITR genes in the double mutant and the triple mutant and compared their expression with levels in the wild type when all strains were cultured in YPD rich medium. Our qRT-PCR results showed that the expression levels of most ITR genes were significantly increased in both the double mutant and the triple mutant (Fig. 8 A). We also performed qRT-PCR to analyze the expression of all 10 ITR genes in lung and brain tissues that were infected by either H99, the double mutant, or the triple mutant strain in a intravenous injection model. Our qRT-PCR results showed that the expression of most ITR genes was highly induced in both lung and brain tissues infected by H99 compared to expression in H99 culture prepared on YPD rich medium (Fig. 8B). Compared to ITR expression in tissues infected by H99, ITR5 expression was highly induced in tissues infected by the triple mutant in both lung and brain. Meanwhile, the expression levels of ITR2, ITR3, ITR4, and ITR6 were induced in brain tissues infected by either the double mutant or the triple mutant (Fig. 8C and D). These results confirm that additional ITR genes are activated during infection with mutants lacking the two major ITR genes. The group II ITRs (Itr4, Itr5, and Itr6) may play an important role during Cryptococcus-host interactions, especially in the itr1a itr3c mutant backgrounds.
Fig. 8.
Comparison of the expression of Cryptococcus ITR genes in in vitro cultures or in infected lungs and brains using qRT-PCR. (A) RNAs were purified from cultures of H99 or the itr1a itr3c (CUX46) or the ino1 itr1a itr3c (CUX57) mutant that were grown on YPD medium overnight. (B to D) RNAs were isolated from either infected mouse lungs (B and C) or infected mouse brains (B and D) at the endpoint of the infection experiment. Gene expression was measured by qRT-PCR in triplicate, and the comparative CT method was used for the relative quantification. Values are expressed as relative expression (log2) (means ± SD) of ITR genes, normalized to the GAPDH gene endogenous reference and relative to either H99 overnight liquid culture (A and B), lungs infected by H99 (C), or brains infected by H99 (D). cDNAs from noninfected lung was used as a negative control to make sure that only the fungal GAPDH gene was amplified. Error bars indicate the standard deviations from three independent PCR results.
DISCUSSION
Our previous studies revealed that inositol is important for the sexual reproduction of C. neoformans and that this fungus may utilize inositol from plants to complete its sexual cycle in nature (65). We have also showed that C. neoformans contains a large inositol transporter (ITR) gene family, which is very unusual in fungi (64). In accord with the biological significance of inositol and inositol transporters (ITRs), it has been well documented that human and animal brains contain high inositol levels and that abnormal brain inositol levels are related to a variety of important human diseases such as bipolar disorder (7, 54–56, 58). C. neoformans is the most common cause of fungal meningitis in AIDS patients. Hence, it is important to investigate whether high inositol levels in brains play a role in the development of cryptococcal meningitis. However, the functional redundancy of the inositol acquisition system in C. neoformans complicates the study of these transporters.
Our previous studies revealed that most single transporter deletion mutants do not have any obvious defective phenotype (64). Although the ino1 itr1 and ino1 itr1a double mutants showed more clear defects in mating and virulence, it was still difficult to assess the function of each ITR, including its inositol uptake activity. In our previous study, we have utilized a yeast heterologous expression system that is based on a Saccharomyces mutant strain lacking inositol uptake activity caused by deletion of both inositol transporters ITR1 and ITR2 to assess the function of each ITR of C. neoformans (64). Here, we utilized these heterologous expression strains to assess inositol uptake activity of each Cryptococcus ITR and identified that Itr1a and Itr3c are major inositol transporters with high inositol uptake activity. Inositol uptake assays in C. neoformans strains also revealed that Itr1a and Itr3c are important for inositol uptake (Fig. 7). Because the itr1a itr3c double mutant had a much lower uptake rate than the wild-type strain only when a large amount of free inositol was available, Itr1a and/or Itr3c likely functions as a low-affinity transporter. Such an outcome is consistent with our early finding that both ITR1A and ITR3C are highly expressed during inositol induction based on our previous real-time PCR (qPCR) analyses (64). Among the rest of the ITR genes, high-affinity inositol transporters could exist. It is unclear why expressing the other eight ITR genes in this yeast heterologous system failed to complement the growth defect of the Saccharomyces mutant and did not show clear inositol uptake activity. Although all of them are expressed, based on RT-PCR analysis, it is possible that either overexpressed proteins are not stable or are not expressed properly in S. cerevisiae. Western blot analysis will be required to address this possibility, which is beyond the scope of this study. It is also possible that some of them are not bona fide inositol transporters and, instead, function as either transporters for other sugars or as inositol sensors. Our previous study has demonstrated that Itr1 may be a sensor based on its expression pattern and mutant phenotype (64). More study will be required to determine whether Itr1 functions as a sensor.
In addition to acquiring inositol through uptake by ITR genes, fungal cells can also synthesize inositol by utilizing other sugars. The INO1 gene encodes a rate-determining enzyme in the inositol biosynthetic pathway. Our results showed that INO1 expression was not regulated by the availability of extracellular inositol; instead, it was upregulated by the nutrient limitation condition. Such an outcome is consistent with earlier studies in C. neoformans, which suggest that expression of Cryptococcus Ino1 and cellular phosphatidylinositol (PI) levels are not regulated by inositol (38, 62), but these results differ from what has been observed in S. cerevisiae (10) and Candida glabrata (8). In S. cerevisiae, Ino1 expression is highly regulated by inositol concentrations. A key inositol regulatory system has been identified that involves two positive regulators, Ino2 and Ino4, which form a heterodimer via their basic helix-loop-helix (bHLH) domains and bind to the promoter of INO1 (2). The negative regulator Opi1 represses the expression and binding of the Ino2-Ino4 heterodimer (29). These inositol acquisition regulators in S. cerevisiae regulate not only phospholipid biosynthesis but also inositol uptake through ITR1 expression (2, 21, 31). Interestingly, we could not find homologs of these three proteins in C. neoformans by BLAST search with H99 genome database. Thus, it is possible that the regulatory system in C. neoformans is very different from that in S. cerevisiae, and the role of Ino1 and its regulatory system in C. neoformans remains to be determined.
Identification of Itr1a and Itr3c as major transporters allowed us to focus on the functional significance of these two ITR genes. Mutagenesis analyses revealed that the itr1a itr3c double mutants showed significant defects in both sexual reproduction and virulence, further demonstrating the importance of inositol transporters on the fungal life cycle and disease development. Similar to finding in our earlier studies, itr1a itr3c double mutants demonstrated no obvious defects pertaining to two major virulence factors (production of capsule and growth at 37°C). However, the double mutant did exhibit reduced melanin production and laccase activity when assayed on l-DOPA medium, suggesting that ITR genes are involved in melanin production. It is unclear whether the modest melanin defect is part of the cause of virulence attenuation. Because no significant virulence defect was observed in either itr1a or itr3c single mutants, the virulence defect of itr1a itr3c double mutants suggested that Itr1a and Itr3c are both required for full virulence, and they likely play a redundant role in inositol function. It remains unclear how inositol and fungal inositol transporters affect fungal virulence and whether this fungus utilizes inositol as a carbon source, a signaling molecule, or both (see Fig. S2 in the supplemental material). Our findings of reduced yeast CFU in lungs and brains infected by the itr1a itr3c mutant at all time points postinfection suggest that this mutant may have decreased fitness in vivo, which could be due to a defect in host inositol utilization.
Although a significant virulence defect was observed, the mutant still caused infection resulting in morbidity and mortality in the rodent models. While Itr1a and Itr3c appear to be the only major ITR genes, based on uptake assays in the yeast heterologous system, it is possible that other ITR genes also play a role in inositol sensing or uptake or that additional ITR candidates are activated in the double mutant background to partially compensate the defect of inositol acquisition. Also because the inositol internal biosynthetic pathway is still active, the potential functional redundancy between inositol biosynthesis and inositol uptake could compensate the defect of itr1a itr3c double mutants. We thus generated the ino1 itr1a itr3c triple mutants and evaluated the infectivity of such mutants. Interestingly, the triple mutant also showed virulence attenuation both in intravenous and intracerebral infection models but to a lesser extent than the double mutant in the intracerebral injection model. Because of the essentiality of Ino1 in inositol biosynthesis and the importance of Itr1a and Itr3c in inositol uptake, it is likely that other remaining ITR genes are activated for function in the triple mutant since completely blocking both pathways would trigger the so call inositol-less death and become lethal. Other remaining ITR genes must play a role in inositol acquisition in the triple mutant since this mutant is viable and has normal growth. Our inositol uptake results also agreed with this notion since the triple mutant still could acquire inositol at a rate even better than the itr1a itr3c double mutant.
Our qRT-PCR results on the expression of ITR genes shown in Fig. 8 revealed several interesting findings. First of all, compared to the expression of ITR genes in H99, the expression of the remaining ITR genes except ITR4 was highly induced in either the double mutant or the triple mutant background when these strains were cultured on YPD rich medium. Such an outcome suggests that there is a functional redundancy within these ITR genes and that Itr1a and Itr3c are important for function, which is consistent with our previous study (64). Because the double mutant still showed inositol uptake activity but at a much lower uptake rate when abundant free inositol was available, the increased expression of remaining ITR genes did not fully rescue the defect of mutant uptake activity. Second, most ITR genes, including ITR1A and ITR3C, were highly induced in infected lung and brain during infection compared with the in vitro culture condition. Compared to the H99 infection, the expression of ITR5 was highly induced in lung tissues infected by the triple mutant. The expression of ITR2 and the group II ITR genes (ITR4, ITR5, and ITR6) was also induced in brain tissues infected by the triple mutant compared to H99 infection. These outcomes confirmed our hypothesis that remaining ITR genes are activated to overcome the defect of inositol acquisition in the triple mutants, which could potentially explain why the triple mutant did not show more severe virulence attenuation in our murine models. The role of group II ITR genes remains unknown both in C. neoformans and in other fungi, including Candida species. Our RT-PCR results suggest that they may have an important function during infection, and this lends itself to future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Issar Smith and Carol Newlon for critical reading of the manuscript and valuable discussion and James Kronstad for providing the ino1 mutants. We thank John Perfect and Arturo Casadevall for valuable discussion and support on this project and Jason Stajich for valuable suggestions and discussions on bioinformatics analyses. We also acknowledge use of the C. neoformans genome sequences at Duke University, the Broad Institute, and the TIGR database.
This work was supported by NIH grant AI069397 to D.S.P. and UMDNJ institutional start-up funds to C.X.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Alspaugh J. A., Perfect J. R., Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambroziak J., Henry S. A. 1994. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 269:15344–15349 [PubMed] [Google Scholar]
- 3. Bahn Y. S., Hicks J. K., Giles S. S., Cox G. M., Heitman J. 2004. Adenylyl cyclase-associated protein AcaI regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3:1476–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Bahn Y. S., et al. 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5:57–69 [DOI] [PubMed] [Google Scholar]
- 6. Barnett J. A. 1976. The utilization of sugars by yeasts. Adv. Carbohydr. Chem. Biochem. 32:125–234 [DOI] [PubMed] [Google Scholar]
- 7. Battaglia F. C., Meschia G., Blechner J. N., Barron D. H. 1961. The free myo-inositol concentration of adult and fetal tissues of several species. Q. J. Exp. Physiol. Cogn. Med. Sci. 46:188–193 [DOI] [PubMed] [Google Scholar]
- 8. Bethea E. K., Carver B. J., Montedonico A. E., Reynolds T. B. 2010. The inositol regulon controls viability in Candida glabrata. Microbiology 156:452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Casadevall A., Perfect J. R. 1998. Cryptococcus neoformans. ASM press, Washington, DC [Google Scholar]
- 10. Chang H. J., Jesch S. A., Gaspar M. L., Henry S. A. 2004. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168:1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chayakulkeeree M., Perfect J. R. 2006. Cryptococcosis. Infect. Dis. Clin. North Am. 20:507–544,v–vi [DOI] [PubMed] [Google Scholar]
- 12. Chen Y. L., Kauffman S., Reynolds T. B. 2008. Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 76:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox G. M., Mukherjee J., Cole G. T., Casadevall A., Perfect J. R. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68:443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donahue T. F., Henry S. A. 1981. Inositol mutants of Saccharomyces cerevisiae: mapping the ino1 locus and characterizing alleles of the ino1, ino2 and ino4 loci. Genetics 98:491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drew M. E., et al. 1995. Functional expression of a myo-inositol/H+ symporter from Leishmania donovani. Mol. Cell. Biol. 15:5508–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reference deleted.
- 17. Fisher S. K., Novak J. E., Agranoff B. W. 2002. Inositol and higher inositol phosphates in neural tissues: homeostasis, metabolism and functional significance. J. Neurochem. 82:736–754 [DOI] [PubMed] [Google Scholar]
- 18. Gerik K. J., Bhimireddy S. R., Ryerse J. S., Specht C. A., Lodge J. K. 2008. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 7:1685–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerik K. J., et al. 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58:393–408 [DOI] [PubMed] [Google Scholar]
- 20. Reference deleted.
- 21. Graves J. A., Henry S. A. 2000. Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics 154:1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harashima T., Heitman J. 2005. Gα subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol. Biol. Cell 16:4557–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Healy M. E., Dillavou C. L., Taylor G. E. 1977. Diagnostic medium containing inositol, urea, and caffeic acid for selective growth of Cryptococcus neoformans. J. Clin. Microbiol. 6:387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heung L. J., Kaiser A. E., Luberto C., Del Poeta M. 2005. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J. Biol. Chem. 280:28547–28555 [DOI] [PubMed] [Google Scholar]
- 25. Heung L. J., Luberto C., Plowden A., Hannun Y. A., Del Poeta M. 2004. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J. Biol. Chem. 279:21144–21153 [DOI] [PubMed] [Google Scholar]
- 26. Hicks J. K., Bahn Y. S., Heitman J. 2005. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 4:1971–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Idnurm A., et al. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753–764 [DOI] [PubMed] [Google Scholar]
- 28. Jin J. H., Seyfang A. 2003. High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149:3371–3381 [DOI] [PubMed] [Google Scholar]
- 29. Kumme J., Dietz M., Wagner C., Schuller H. J. 2008. Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr. Genet. 54:35–45 [DOI] [PubMed] [Google Scholar]
- 30. Lai K., Bolognese C. P., Swift S., McGraw P. 1995. Regulation of inositol transport in Saccharomyces cerevisiae involves inositol-induced changes in permease stability and endocytic degradation in the vacuole. J. Biol. Chem. 270:2525–2534 [DOI] [PubMed] [Google Scholar]
- 31. Lai K., McGraw P. 1994. Dual control of inositol transport in Saccharomyces cerevisiae by irreversible inactivation of permease and regulation of permease synthesis by INO2, INO4, and OPI1. J. Biol. Chem. 269:2245–2251 [PubMed] [Google Scholar]
- 32. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 33. Lengeler K. B., et al. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loftus B. J., et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luberto C., et al. 2001. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 15:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin K. L., Smith T. K. 2006. Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem. J. 396:287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mina J. G., et al. 2009. The Trypanosoma brucei sphingolipid synthase, an essential enzyme and drug target. Mol. Biochem. Parasitol. 168:16–23 [DOI] [PubMed] [Google Scholar]
- 38. Molina Y., Ramos S. E., Douglass T., Klig L. S. 1999. Inositol synthesis and catabolism in Cryptococcus neoformans. Yeast 15:1657–1667 [DOI] [PubMed] [Google Scholar]
- 39. Mongan T. P., Ganapasam S., Hobbs S. B., Seyfang A. 2004. Substrate specificity of the Leishmania donovani myo-inositol transporter: critical role of inositol C-2, C-3 and C-5 hydroxyl groups. Mol. Biochem. Parasitol. 135:133–141 [DOI] [PubMed] [Google Scholar]
- 40. Niederberger C., et al. 1998. Exogenous inositol and genes responsible for inositol transport are required for mating and sporulation in Schizosaccharomyces pombe. Curr. Genet. 33:255–261 [DOI] [PubMed] [Google Scholar]
- 41. Nielsen K., et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nikawa J., Hosaka K. 1995. Isolation and characterization of genes that promote the expression of inositol transporter gene ITR1 in Saccharomyces cerevisiae. Mol. Microbiol. 16:301–308 [DOI] [PubMed] [Google Scholar]
- 43. Nikawa J., Hosaka K., Yamashita S. 1993. Differential regulation of two myo-inositol transporter genes of Saccharomyces cerevisiae. Mol. Microbiol. 10:955–961 [DOI] [PubMed] [Google Scholar]
- 44. Nikawa J., Tsukagoshi Y., Yamashita S. 1991. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266:11184–11191 [PubMed] [Google Scholar]
- 45. Oliveira M. M., Einicker-Lamas M. 2000. Inositol metabolism in Trypanosoma cruzi: potential target for chemotherapy against Chagas' disease. An. Acad. Bras Cienc. 72:413–419 [DOI] [PubMed] [Google Scholar]
- 46. Page R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 47. Park B. J., et al. 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23:525–530 [DOI] [PubMed] [Google Scholar]
- 48. Perfect J., Ketabchi N., Cox G. M., Ingram C. W., Beiser C. L. 1993. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J. Clin. Microbiol. 31:3305–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pukkila-Worley R., Mylonakis E. 2008. Epidemiology and management of cryptococcal meningitis: developments and challenges. Expert Opin. Pharmacother. 9:551–560 [DOI] [PubMed] [Google Scholar]
- 50. Reynolds T. B. 2009. Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: making it and taking it. Microbiology 155:1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson K. S., Lai K., Cannon T. A., McGraw P. 1996. Inositol transport in Saccharomyces cerevisiae is regulated by transcriptional and degradative endocytic mechanisms during the growth cycle that are distinct from inositol-induced regulation. Mol. Biol. Cell 7:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seyfang A., Landfear S. M. 2000. Four conserved cytoplasmic sequence motifs are important for transport function of the Leishmania inositol/H(+) symporter. J. Biol. Chem. 275:5687–5693 [DOI] [PubMed] [Google Scholar]
- 53. Shea J. M., Kechichian T. B., Luberto C., Del Poeta M. 2006. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect. Immun. 74:5977–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shetty H. U., Holloway H. W., Acevedo L. D., Galdzicki Z. 1996. Brain accumulation of myo-inositol in the trisomy 16 mouse, an animal model of Down's syndrome. Biochem. J. 313:31–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shetty H. U., Holloway H. W., Schapiro M. B. 1996. Cerebrospinal fluid and plasma distribution of myo-inositol and other polyols in Alzheimer disease. Clin. Chem. 42:298–302 [PubMed] [Google Scholar]
- 56. Spector R. 1988. Myo-inositol transport through the blood-brain barrier. Neurochem. Res. 13:785–787 [DOI] [PubMed] [Google Scholar]
- 57. Spector R., Lorenzo A. V. 1975. Myo-inositol transport in the central nervous system. Am. J. Physiol. 228:1510–1518 [DOI] [PubMed] [Google Scholar]
- 58. Spector R., Lorenzo A. V. 1975. The origin of myo-inositol in brain, cerebrospinal fluid and choroid plexus. J. Neurochem. 25:353–354 [DOI] [PubMed] [Google Scholar]
- 59. Steen B. R., et al. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Suzuki E., et al. 2008. Trypanosomatid and fungal glycolipids and sphingolipids as infectivity factors and potential targets for development of new therapeutic strategies. Biochim. Biophys. Acta 1780:362–369 [DOI] [PubMed] [Google Scholar]
- 61. Trinel P. A., et al. 1999. The Candida albicans phospholipomannan is a family of glycolipids presenting phosphoinositolmannosides with long linear chains of β-1,2-linked mannose residues. J. Biol. Chem. 274:30520–30526 [DOI] [PubMed] [Google Scholar]
- 62. Vincent V. L., Klig L. S. 1995. Unusual effect of myo-inositol on phospholipid biosynthesis in Cryptococcus neoformans. Microbiology 141:1829–1837 [DOI] [PubMed] [Google Scholar]
- 63. Wang P., Heitman J. 1999. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr. Opin. Microbiol. 2:358–362 [DOI] [PubMed] [Google Scholar]
- 64. Xue C., et al. 2010. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio 1:e00084–e00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xue C., Tada Y., Dong X., Heitman J. 2007. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe 1:263–273 [DOI] [PubMed] [Google Scholar]
- 66. Reference deleted.
- 67. Zhang O., et al. 2009. Degradation of host sphingomyelin is essential for Leishmania virulence. PLoS Pathog. 5:e1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.