Abstract
We have previously shown that deletion of GOA1 (growth and oxidant adaptation) of Candida albicans results in a loss of mitochondrial membrane potential, ATP synthesis, increased sensitivity to oxidants and killing by human neutrophils, and avirulence in a systemic model of candidiasis. We established that translocation of Goa1p to mitochondria occurred during peroxide stress. In this report, we show that the goa1Δ (GOA31), compared to the wild type (WT) and a gene-reconstituted (GOA32) strain, exhibits sensitivity to inhibitors of the classical respiratory chain (CRC), including especially rotenone (complex I [CI]) and salicylhydroxamic acid (SHAM), an inhibitor of the alternative oxidase pathway (AOX), while potassium cyanide (KCN; CIV) causes a partial inhibition of respiration. In the presence of SHAM, however, GOA31 has an enhanced respiration, which we attribute to the parallel respiratory (PAR) pathway and alternative NADH dehydrogenases. Interestingly, deletion of GOA1 also results in a decrease in transcription of the alternative oxidase gene AOX1 in untreated cells as well as negligible AOX1 and AOX2 transcription in peroxide-treated cells. To explain the rotenone sensitivity, we measured enzyme activities of complexes I to IV (CI to CIV) and observed a major loss of CI activity in GOA31 but not in control strains. Enzymatic data of CI were supported by blue native polyacrylamide gel electrophoresis (BN-PAGE) experiments which demonstrated less CI protein and reduced enzyme activity. The consequence of a defective CI in GOA31 is an increase in reactive oxidant species (ROS), loss of chronological aging, and programmed cell death ([PCD] apoptosis) in vitro compared to control strains. The increase in PCD was indicated by an increase in caspase activity and DNA fragmentation in GOA31. Thus, GOA1 is required for a functional CI and partially for the AOX pathway; loss of GOA1 compromises cell survival. Further, the loss of chronological aging is new to studies of Candida species and may offer an insight into therapies to control these pathogens. Our observation of increased ROS production associated with a defective CI and PCD is reminiscent of mitochondrial studies of patients with some types of neurodegenerative diseases where CI and/or CIII dysfunctions lead to increased ROS and apoptosis.
INTRODUCTION
Candida albicans is a pathogen of skin and mucosa or causes blood-borne tissue invasion. To cause disease, the organism utilizes a variety of factors to adhere to, break down host substrates, and invade tissue. There is also evidence that both glycolytic and nonglycolytic metabolism is critical to survival of the organism in the host, a phenomenon termed host niche-specific metabolic adaptation (9, 14, 21, 41, 50). The paradigm is that when blood-borne glucose is plentiful, cells metabolize by glycolysis to produce energy. However, at host niches lacking sufficient glucose, such as within phagocytes, the organism adapts through other pathways, such as the glyoxylate cycle, to minimize carbon loss, while gluconeogenesis is used to increase cell glucose levels. Both cycles include participation of mitochondria. Carbon source preferences and respiratory circuits vary in usage among fungi and yeasts and reflect their life styles. For instance, model yeast depends almost exclusively upon fermentation as a source of energy. Saccharomyces cerevisiae is defined as a Crabtree-positive organism, which utilizes glucose repression of aerobic respiration, in contrast to others, like Candida albicans, which are Crabtree negative and rely upon the oxidation of substrates via the mitochondrial tricarboxylic acid (TCA) cycle to generate ATP even in the presence of glucose (7, 51). In fact, S. cerevisiae lacks complex I (CI) of the classical respiratory electron transport chain (ETC) as well as the alternative oxidase (AOX) and parallel respiratory chains, unlike Candida species (18, 55) (see also below).
There are three mitochondrial respiratory pathways in Candida species and other fungi: the classical respiratory chain (CRC), the alternative oxidase chain, and the parallel respiratory (PAR) chain (12, 18, 26–34, 43–46, 52, 58). Of these pathways, the CRC provides the largest amount of cellular ATP and is the major source of respiratory activity and O2 consumption. In Candida parapsilosis, the AOX and CRC pathways are constitutive while the PAR pathway is active only when the CRC and AOX pathways are blocked (46). This compensatory mechanism may allow the cells to adapt to stress conditions (33). The contributions of each of the three respiratory pathways can be distinguished by using specific inhibitors. Thus, rotenone, 2-thenoyltrifluoroacetone (TTFA), antimycin A (AA), potassium cyanide (KCN), and oligomycin inhibit CI, CII, CIII, CIV, and CV, respectively, of the CRC, while SHAM (salicylhydroxamic acid) is an inhibitor of the AOX pathway. KCN at higher concentrations (10 mM) is an inhibitor of both the PAR pathway and of CIV of the CRC pathway. In the CRC pathway, CI, CIII, and CIV oxidoreductases of the inner mitochondrial membrane (IM) produce H+ ions which are pumped to the intermembrane space (IMS). The generation of a membrane potential (ΔψM) is then used by the IM CV to synthesize ATP. CI functions through coenzyme Q (CoQ), which serves as an intermediate step for electron flow through CIII and CIV. CoQ also provides electrons for the AOX and PAR pathways. While the three respiratory pathways have been described in Candida species, less is known about complexes I to IV of the CRC and about the genes that are associated with the complex functions. A gene encoding the C. albicans NADH dehydrogenase CI protein (NDH51) when deleted resulted in a filamentation defect even at low concentrations of glucose (45). More recently, it was shown that in the same ndh51Δ mutant, the pyruvate dehydrogenase complex protein X (Pdx1) increased by 15-fold, but the filamentous defect remained (58). The Lpd1 of C. albicans, a dihydrolipomide dehydrogenase and part of the pyruvate dehydrogenase complex, also is required for filamentous growth (35). These data demonstrate a relationship between the CRC pathway and morphogenesis. Metabolic signal networks important to mitochondrial functions are also inadequately described although the Hog1 mitogen-activated protein kinase (MAPK) appears significant (5).
In animals, plants, and most fungi, mitochondria are indispensable for many cell activities (1, 2, 15, 16, 20, 32, 43, 50, 52, 59, 60). It is also evident that programmed cell death (PCD) can be initiated through a number of inducers including ROS, the origin of which can be defective mitochondria, as reported in both mammalian cells and fungi such as C. albicans (11, 13, 15, 16, 25, 26, 36, 48, 52, 54, 59). In contrast, cancer cells favor glycolysis, and therapies that focus upon metabolic reprogramming of mitochondria have been applied to induce chemosensitivity (19). Other inducers of PCD in fungi include the antifungal compounds amphotericin B and plagiochin E, farnesol, phytosphingosine, and the α-pheromone of C. albicans (3, 4, 17, 54, 56, 57, 59, 60). Farnesol (a quorum-sensing molecule) induced apoptosis through either metacaspase- or caspase-dependent pathways in C. albicans (60). The amphotericin B effect was observed in biofilms formed in vitro and could be reversed by the addition of an inhibitor of caspase production (4). Indications are that PCD occurs through apoptosis as deletion of the MCA1 of C. albicans, a homologue of the MCA1 metacaspase of S. cerevisiae, attenuates the oxidative stress cell death response as well as caspase activation (13). A recent observation on pheromone-induced death (PID) in C. albicans during mating indicates that the mechanism differs from that of S. cerevisiae, which requires calcineurin (3).
In our previous report on Goa1p of C. albicans, we showed its requirement for mitochondrial functions including the generation of membrane potential to provide ATP and respiration (8). Using a green fluorescent protein (GFP)-tagged Goa1p, we demonstrated its translocation to mitochondria, especially under stress conditions or when cells are grown on glycerol. GOA1 is also required for virulence in an invasive model of candidiasis (8). The objective of this study is to characterize the mitochondrial target of Goa1p of C. albicans and relate the loss of that target to cell functions. It is obvious that dysfunctional mitochondria result in profound effects on this fungus.
MATERIALS AND METHODS
Strains.
For all experiments, we used SC5314 (wild type [WT]) as well as the gene-reconstituted strain (GOA32) and the goa1 mutant (GOA31) (8).
Growth studies of strains with CRC and AOX inhibitors.
For drop plate assays, strains were grown overnight at 30°C in YPD (yeast extract, peptone, dextrose) broth; cells were washed and suspended in phosphate-buffered saline ([PBS] 0.1 M; pH 7.4) (8). The sensitivity of strains to inhibitors of complexes I to V and to AOX was evaluated as follows. Serial dilutions of cells (5 × 101 to 5 × 105 cells in 5 μl) were spotted onto YPD agar containing each inhibitor (1 μM rotenone, 6 μM TTFA, 8 μM antimycin A, 1 or 10 mM KCN, 20 μM oligomycin, and 2 mM SHAM [final concentrations]). Growth was evaluated after 48 h of incubation at 30°C.
Oxygen consumption.
Oxygen consumption was measured polarographically using a Clark-type electrode (model 5300; Yellow Springs Instruments, OH). Cells were grown overnight at 30°C in 10 ml of YPD broth and then diluted in fresh YPD broth for an additional 3 to 4 h until exponential growth was achieved (32). The cells were collected and washed with PBS, and an equal number of cells of each strain (5 × 108) was suspended in 2 ml of YPD broth. The cells were then loaded into a sealed 1.5-ml glass chamber. Changes in oxygen tension in the chamber were measured, and the respiratory rate was calculated as the consumption of oxygen over time. In order to quantify oxygen consumption by different respiratory pathways, strains were cultured in the presence of classical respiratory pathway inhibitors (6 μM TTFA, 10 μM rotenone, 10 μM antimycin A, or potassium cyanide [1 or 10 mM]) for 1 h while the AOX pathway was inhibited using 5 mM SHAM before cells were harvested, and O2 consumption was determined. Also, 200 μM flavone was used to measure the contribution of NADH dehydrogenases to respiration (57).
Transcription of AOX1, AOX2, and alternative NADH dehydrogenases, NDE1 and YMX6.
Yeast cells were collected from 10-ml cultures of the WT, GOA31, and GOA32 strains grown at 30°C for 16 h in YPD broth. Cells were washed twice with PBS and divided into three equal aliquots, two of which were treated with 5 mM H2O2 or 5 mM SHAM for 30 min while the third was left untreated for the same time. Total RNA was extracted using Trizol following a PBS wash. For H2O2- and SHAM-treated samples, cells were washed two additional times with PBS. The quality and concentration of RNAs were measured by a nano-spectrophotometer, and approximately 1 μg of the total RNA was subjected to first-strand cDNA synthesis (QuantiTect Reverse Transcription; Qiagen). Real-time PCR assays were performed with 20-μl reaction volumes that contained 1× iQ SyBR green Supermix, including a 0.2 μM concentration of each primer and 8 μl of a 1:8 dilution of each cDNA from each strain. A standard curve for each gene was established from WT cells grown at 30°C. WT cDNA was prepared in a series of dilutions (1:4 to 1:256) for each experiment. The primers used for real-time PCR expression analysis were the following: for 18S rRNA, CGCAAGGCTGAAACTTAAAGG (forward) and AGCAGACAAATCACTCCACC (reverse); for AOX1, GCAACTCCAATCCCAAATCAC (forward) and ACACGATAAACTCCTGCTTCAG (reverse); for AOX2, TCTCCAGCTTTCCATCAACC (forward) and TGGGTAACTGTCACATTCTCAC (reverse) (30, 31); for NDE1, TGCTAACCCAACTCCAAAGG (forward) and CCAGACCAAATCAGCAACAG (reverse); for YMX6, CATCCAACGACCCTAAGATCC (forward) and AATGTTCAGCCACCCCAG (reverse).
Quantitative reverse transcription-PCR (qRT-PCR) for each experiment was performed in triplicate (Bio-Rad iQ5), and the transcription of AOX1, AOX2, NDE1, and YMX6 genes was normalized to 18S RNA levels. Data are presented as the means ± standard deviations (SD). The 2−ΔΔCT (where CT is threshold cycle) method of analysis was used to determine the fold change in gene transcription (31).
Spheroplast and mitochondrial preparations.
Cells were grown in 250 ml of YPD broth overnight at 30°C, harvested by centrifugation (5,000 rpm for 10 min), washed once with 50 ml of cold water and once with buffer A (1 M sorbitol, 10 mM MgCl2, 50 mM Tris-HCl [pH 7.8]), and then centrifuged at 5,000 rpm for 10 min.
Cells were then suspended in buffer A (50 ml) supplemented with 30 mM dithiothreitol (DTT) for 15 min at room temperature with shaking (100 rpm) and then collected and suspended in 15 ml of buffer A with 1 mM DTT containing 100 mg of Zymolyase 20T (Seikagaku Biobusiness, Inc.) per 15 g of pelleted cells. Shake cultures (100 rpm) were incubated at 30°C for 60 min or until 90% of cells were converted into spheroplasts (as determined by light microscopy). Digestion was stopped by adding 15 ml of ice-cold buffer A. Then spheroplasts were washed twice with buffer A. Crude preparations of mitochondria were isolated as follows: spheroplasts were suspended in 10 ml of cold buffer B (0.6 M mannitol, 1 mM EDTA, 0.5% bovine serum albumin [BSA], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM Tris-HCl [pH, 7.4]) and then broken mechanically using a Dounce homogenizer on an ice bath (18). Cell debris was removed by low-speed centrifugation (1,000 × g for 10 min). The supernatants containing mitochondria were centrifuged at 10,500 × g for 10 min, and the pellet was washed twice with 20 ml of ice-cold buffer C (0.6 M mannitol, 1 mM EDTA, 1% BSA, 10 mM Tris-HCl [pH 7.0]). Mitochondria were suspended in 1 ml of buffer D (0.6 M mannitol, 10 mM Tris-HCl, [pH 7.0]), and the protein content was determined by the Biuret method.
Purification of mitochondria.
Mitochondrion purification was performed using a two-step gradient according to standard procedures (10, 53). Two milliliters of PB1 buffer (0.3 M mannitol, 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], 0.1% [wt/vol] BSA, pH 7.5) containing 30% (vol/vol) Percoll was placed at the bottom of a centrifugation tube (Beckman, Inc.). Then, 2 ml of PB2 buffer (0.3 M sucrose, 10 mM TES, 0.1% [wt/vol] BSA, pH 7.5) containing 20% Percoll (vol/vol) was layered on top of PB1 buffer. The crude mitochondrial preparation was layered on top of the gradient, which was then centrifuged at 40,000 × g for 45 min at 4°C. A whitish band (purified mitochondria) was collected from the interface of the Percoll gradients. Purified mitochondria were washed twice by centrifugation in 3 ml of PB1 buffer for 10 min at 12,600 × g.
Mitochondrial pellets were suspended in 1 ml of cold S1 buffer (272 mM sucrose, 40 mM HEPES, 150 mM KCl, pH 7.5), incubated on ice for 15 min, diluted with 5 ml of S2 buffer (40 mM HEPES, 150 mM KCl, pH 7.5), and centrifuged at 100,000 × g for 30 min. The pellets were suspended in cold S3 buffer (solubilization medium; 750 mM 6-aminohexanoic acid [ε-amino-n-caproic acid], 50 mM bis-Tris-HCl, 0.5 mM EDTA, pH 7.0) at a concentration of 2 mg of protein/ml. Freshly prepared 10% (wt/vol) n-dodecyl-β-d-maltoside (DMM) in water (200 μl) was added at a ratio of 1 g of detergent to 1 g of protein. The pellets were homogenized by repeated pipetting on ice for 5 min and centrifuged at 108,000 × g for 10 min at 4°C. Supernatants were collected, and protein determinations were done by the Biuret method and blue native polyacrylamide gel electrophoresis (BN-PAGE) (see below). Coomassie staining was also used to visualize proteins.
Enzymatic assays of CRC CI to CIV.
From overnight cultures grown at 30°C, enzyme activities of complexes I to IV (CI to CIV) were measured spectrophotometrically by standard methods (10, 38). Crude mitochondrial preparations were first treated with two cycles of freeze-thawing in a hypotonic solution (25 mM K2HPO4 [pH 7.2], 5 mM MgCl2), followed by a hypotonic shock in H2O. A total of 20 μg of mitochondrial protein from each strain was used in each enzyme assay. Substrates (electron donor and electron acceptor) varied depending upon which complex was to be assayed. The reaction for each enzymatic assay was terminated by adding a complex-specific inhibitor. The activities for each complex were indicated by the rate of substrate oxidation or reduction as nM/min/mg of protein. Each complex was assayed in the presence of inhibitors of other complexes to ensure that the activity reflected only the enzyme complex of interest.
Complex I (NADH:ubiquinone oxidoreductase).
Mitochondrial protein was dissolved in 0.8 ml of H2O and incubated for 2 min at 37°C and then mixed with 0.2 ml of a solution containing 50 mM Tris, pH 8.0, 5 mg/ml BSA, 0.24 mM KCN, 4 μM antimycin A, and 0.8 mM NADH, the substrate for CI. The reaction was initiated by introducing an electron acceptor, 50 μM DB (2,3-dimethoxy-5-methyl-6-n-decyl-1,4 benzoquinone). Enzyme activity was followed by a decrease in absorbance of NADH at 340 nm minus that at 380 nm using an extinction coefficient of 5.5 mM−1 cm−1 (38). Additionally, rotenone (4 μM) was introduced into the reaction mixture to quantify the rotenone-sensitive complex activity.
Complex II (succinate:ubiquinone oxidoreductase).
Mitochondrial protein was added to a solution of 10 mM KH2PO4 (pH 7.8), 2 mM EDTA, 1 mg/ml BSA, 80 μM DCPIP (acceptor; 2,6-dichlorophenolindophenol), 4 μM rotenone, 0.24 mM KCN, 4 μM antimycin A, and 0.2 mM ATP (total of 1 ml). As the donor substrate, 10 mM succinate was added, and the reaction mixture was incubated for 10 min at 30°C. A decrease in absorbance due to the reduction of DCPIP was measured at 600 nm for 5 min using an extinction coefficient of 13.0 mM−1 cm−1. To measure CII-specific activity, 1 mM TTFA (thenoyltrifluoroacetone) was added.
Complexes II and III (succinate:cytochrome c reductase).
The assay for CII plus CIII was done under conditions similar to those for CII described above using 10 mM succinate as the donor substrate. The reaction was initiated following the addition of 40 μM oxidized cytochrome c (incubation, 10 min at 30°C), followed by the addition of 1 mM TTFA for an additional 5 min. The extinction coefficient for cytochrome c reduction at 550 nm is 18.5 mM−1 cm−1 (9).
Complex IV.
The CIV assay was performed at 550 nm, and the decrease in absorbance resulting from the oxidation of reduced cytochrome c was measured. To reduce cytochrome c, 5 mg of l-ascorbic acid was mixed with 100 mg of cytochrome c in 8 ml of 10 mM potassium phosphate buffer, pH 7.0. The reduced cytochrome c solution was then dialyzed against 10 mM potassium phosphate buffer, pH 7.0, for 20 h at 4°C with three changes of buffer. Reduced cytochrome c was then brought to a final volume of 10 ml with 0.1 M phosphate buffer (pH 6.8). In a 1-ml reaction medium, 20 μg of mitochondrial protein was added to a buffer containing 10 mM KH2PO4 (pH 6.5), 0.25 M sucrose, 1 mg/ml BSA, and 10 μM reduced cytochrome c. Changes in absorbance were measured following the oxidation of cytochrome c using 2.5 mM lauryl maltoside to permeabilize mitochondria. Complex specificity was determined in the presence of 0.24 mM KCN (3 min for each part of the assay).
Blue native polyacrylamide gel electrophoresis.
Mitochondrial protein was concentrated by vacuum centrifugation (53). Ten microliters of BN sample buffer (2×) was mixed with 20 μl of each sample (∼60 to 80 μg of protein) and loaded onto a BN-PAGE gradient gel (4 to 16%) (Invitrogen, Inc.). One ml of 2× BN sample buffer consisted of 1.5 M 6-aminohexanoic acid, 0.05 M bis-Tris (pH 7.0), 65 μl of 10% DMM, 20 μl of proteinase inhibitor mixture, and 100 μl of glycerol. Electrophoresis was performed in an X-Cell SureLock mini-cell system (Invitrogen) with 200 ml of cathode buffer in the upper (inner) buffer chamber and 150 ml of anode buffer in the lower (outer) buffer chamber. The cathode buffer was prepared from a 10× stock of 500 mM Tricine, 150 mM bis-Tris (pH 7.0; or 75 mM imidazole), and 0.02% Serva Blue G-250, supplemented with 0.02% DDM. The 1× anode buffer consisted of 50 mM bis-Tris, pH 7.0. Electrophoresis was carried out at 4°C and 65 V for 1 h and then raised to 120 V overnight. An in-gel enzyme assay for CRC CI was accomplished as follows: gels were rinsed briefly twice with MilliQ water and equilibrated in 0.1 M Tris-HCl, pH 7.4 (reaction buffer), for 20 min. The gels were then incubated in fresh reaction buffer with 0.2 mM NADH–0.2% nitroblue tetrazolium (NBT) for 1 h. Reactions were stopped by fixing the gels in 45% methanol–10% (vol/vol) acetic acid, and then gels were destained overnight in the same solution. Image processing of gels was done using ImageJ software (23).
Flow cytometry assays of ROS.
Intracellular ROS production was detected by staining cells with the ROS-sensitive fluorescent dye DCFDA (2′,7′-dichlorofluorescein diacetate; Sigma) using a FACScan flow cytometer (488 nm; Becton Dickinson) (36). Cells from 25-ml cultures grown at 30°C overnight in YPD medium were collected and washed twice with PBS. The pellets were suspended to 1 × 106 cells in 1 ml of PBS and treated with or without 50 μM DCFDA for 30 min at 30°C in the dark. Cell fluorescence in the absence of DCFDA was used to verify that background fluorescence was similar per strain. Cells from each DCFDA-treated sample were collected and washed twice with PBS after staining. Then, propidium iodide (PI) was added to each sample to measure the dead cells prior to the DCFDA assays. The mean fluorescence for ROS was quantified only in live and standard-sized cells.
Chronological life span.
Strains were inoculated in 5 ml of YPD broth and incubated overnight at 30°C. For each strain, 106 cells were inoculated into 50 ml of synthetic glucose (SC) medium without or with 5 mM H2O2 (final concentration) (2). For treated cultures, H2O2 was added daily to maintain concentration. Every 24 h for 21 days, 0.5 ml from each culture was diluted to 2.0 × 103 cells/ml, and 100-μl and 250-μl aliquots were added to YPD agar plates. The plates were incubated for 48 h at 30°C, and the numbers of CFU were determined. For each strain and time point, the percent survival was determined by dividing the number of CFU by the cell number obtained by microscopic counts. The percentage of viable cells reflecting the chronological life span was determined for each time point.
Determinations of apoptosis in strains by flow cytometry. (i) Caspase assay.
Caspase activity was measured using a FLICA (for fluorochrome-labeled inhibitor of caspases) Poly-Caspase Apoptosis Detection Kit (Immunochemistry Technologies, LLC) based on FAM-VAD-FMK (where FAM is carboxyfluorescein and FMK is fluoromethyl ketone), which binds covalently to active caspases once it enters cells. After unbound FAM-VAD-FMK or FLICA reagent is washed away, the remaining green fluorescent signal is a direct measure of the quantity of caspase-positive cells. The optimal excitation range for FLICA is 488 to 492 nm, and the emission range is 515 to 535 nm.
Activity was measured at zero time (overnight cultures; control), and cells were grown or incubated for a period equivalent in time to 10 or 20 generation times (GT) in YPD medium or for 2 and 10 generations in cells treated with 10 mM H2O2. Cells were washed with PBS twice prior to the caspase assays. For optimal FLICA labeling in C. albicans, cells were adjusted to 2 × 106 in 300 μl of PBS buffer, and 10 μl of 30× FLICA solution was added. Cells were incubated for 1 h at 37°C in the dark and then washed twice with 1 ml of the wash buffer recommended by the manufacturer. Stained cells were suspended in 400 μl of the same wash buffer and treated with 2 μl of propidium iodide (200 μg/ml) for 20 min at room temperature. A second aliquot of FLICA-treated cells that was not stained with PI was set aside. The percentage of cells with positive PI staining and negative caspase staining by flow cytometry was excluded from caspase measurements. An equal volume of nonstained cells for each time point was set up as a negative control for each experiment.
(ii) TUNEL assays.
In addition to caspase assays, we also measured the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) in the same set of strains. The TUNEL assay includes a terminal deoxynucleotidyltransferase (TdT) that adds Br-dUTP (bromolated dUTP nucleotides) to DNA breaks. The incorporation of Br-dUTP results in a green fluorescent signal (fluorescein isothiocyanate [FITC]) once anti-bromodeoxyuridine (BrdU) antibody is added. An Apo-BrdU in situ DNA fragmentation assay kit (BioVision) was used to identify DNA strand breaks.
Measurements of TUNEL utilized cells of each strain for similar numbers of generation times as used in the caspase assays. Equal numbers of cells for each strain were used in the assays. Cells were grown as indicated above for caspase assays, again with or without peroxide. Cells were washed with PBS twice, and one half of the total was treated with 10 mM H2O2. Cell numbers were adjusted to 5 × 106 in 0.5 ml of PBS buffer and fixed by adding 5 ml of 1% (wt/vol) p-formaldehyde in PBS on ice for 15 min. After cells were washed twice in PBS, the pellets were suspended in 0.5 ml of PBS and 5 ml of ice-cold 70% ethanol and incubated for 30 min on ice. The residual ethanol was then removed by centrifugation, and cells were washed twice with buffer. Pelleted cells (∼5 × 106) were suspended in 50 μl of DNA labeling solution, incubated for 1 h at 37°C in the dark, and then washed twice with 1 ml of rinse buffer provided in the kit. Treated cells were suspended again in 0.1 ml of anti-BrdU antibody and incubated for 30 min at room temperature in the dark. Immediately, the positively stained cells were measured by flow cytometry (excitation wavelength/emission wavelength, 488/520 nm). An equal volume of nonstained cells was used as a negative control. PI staining was used to determine the percentage of dead cells.
RESULTS
Drop plate assays: GOA31 is sensitive to rotenone and SHAM inhibitors of mitochondrial respiratory pathways.
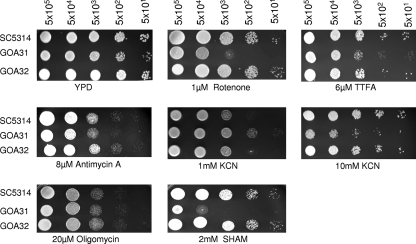
We tested each strain for sensitivity to inhibitors of the CRC and AOX respiratory pathways using drop plate assays with and without inhibitors. As shown in Fig. 1, GOA31 was more sensitive to rotenone (CI) than WT and GOA32 (GOA1-reconstituted) cells but about equally as sensitive as control strains to antimycin A (CIII), TTFA (CII inhibitor), and oligomycin (CV inhibitor). Interestingly, GOA31 grew somewhat better in the presence of 1 mM KCN (CIV inhibitor) but was growth inhibited at 10 mM KCN. At this concentration, both the CIV and PAR pathways are inhibited. GOA31 was also more sensitive to SHAM, an inhibitor of the AOX pathway, than the control strains (Fig. 1). These inhibitor sensitivity data suggest that the mutant has a dysfunctional CI and CIV and AOX pathway.
Fig. 1.
Drop plate assays. All strains were grown in YPD agar with or without inhibitors of complexes I to V (CI to CV) and the AOX pathway at the cell concentrations indicated. The concentration of each inhibitor is also shown. Cultures were incubated for 48 h and photographed. GOA31 (goa1Δ/goa1Δ) is more sensitive than control strains to the CI CRC and the AOX pathway inhibitors.
Oxygen consumption is reduced in GOA31 by CRC CI and CIV inhibitors.
Ruy et al. have shown that the CRC and AOX pathway inhibitors that reduce respiration also inhibit growth in C. albicans (52). Therefore, our objective in these experiments was to correlate the growth data from drop plate assays (Fig. 1) with oxygen consumption experiments. To do this, we measured oxygen consumption (nM O2 consumption/min/ml/Klett unit) in all strains using inhibitors of the CRC and AOX respiratory pathways, as well as the NADH dehydrogenases Nde1p and Ymx6p. For the wild type and GOA32 strains, it is apparent that when cells are treated with each inhibitor of the CRC pathway, oxygen consumption drops significantly compared to untreated conditions (Table 1). For all three strains, rotenone (CI inhibitor) caused a greater reduction in respiration than all other inhibitors. The sensitivity to rotenone is approximately two times greater than that of KCN compared to untreated GOA31 cells. Strain GOA31 respires at about 18 to 20% of the levels of the WT and the GOA32 strains in untreated cultures (Table 1). Likewise, rotenone caused the most inhibition in O2 consumption in GOA31 (∼100-fold reduction compared to untreated cells) while 1 mM KCN reduced respiration by about 50%, suggesting deficiencies in CRC CI and CIV. In comparison, when cells were treated with the AOX inhibitor SHAM, respiration in WT and GOA32 decreased by about 30 to 50% compared to untreated cells, but from GOA31 cells, in the presence of SHAM, oxygen consumption doubled compared to untreated GOA31 cells. This observation suggests a compensatory mechanism under these conditions that could be due to the PAR pathway since both the CRC and AOX pathways in GOA31 cells appear compromised in cultures, as indicated by the introduction of the AOX inhibitor SHAM. The PAR pathway is believed to be engaged when the AOX and CRC pathways are not, as described in C. parapsilosis (46). In addition, inhibition of the compensatory increase in respiration in GOA31 by flavone in SHAM-treated cells was observed (Table 1), suggesting that the NADH dehydrogenases may be associated with the compensatory effect. Further, KCN does abrogate the compensatory reaction, suggesting that CIV is contributory to the compensatory reaction. Also, the combination of TTFA and flavone reduced respiration by 50% in GOA31 compared to levels in control strains, indicating that a CI defect is responsible for the reduced respiration in the absence of inhibitors.
Table 1.
Consumption of oxygen for C. albicans WT, GOA31, and GOA32 strains in the presence of ETC inhibitors
| ETC inhibitor | Oxygen consumption in the indicated strain (nmol/min/ml/Klett unit) |
||
|---|---|---|---|
| WT | GOA31 | GOA32 | |
| No inhibitor | 309.6 ± 43.0 | 61.1 ± 10.8 | 320.8 ± 37.0 |
| 10 μM Rotenone | 17.6 ± 1.05 | 0.6 ± 0.3 | 10.8 ± 3.08 |
| 10 μM AA | 58.6 ± 10.5 | 46.0 ± 12.0 | 55.2 ± 13.5 |
| 1 mM KCN | 59.58 ± 3.0 | 32.43 ± 4.12 | 58.16 ± 10.01 |
| 10 mM KCN | 41.66 ± 3.4 | 25.66 ± 3.56 | 45.37 ± 3.86 |
| 2 mM Na3N | 62.6 ± 13.0 | 47.1 ± 10.0 | 54.1 ± 8.01 |
| 5 mM SHAM | 165.7 ± 28.3 | 148.0 ± 25.6 | 204.5 ± 36.6 |
| 5 mM SHAM + 1 mM KCN | 65.64 ± 4.63 | 46.97 ± 3.56 | 67.26 ± 8.69 |
| 6 μM TTFA | 83.04 ± 10.63 | 52.88 ± 4.53 | 90.41 ± 15.33 |
| 200 μM Flavone | 89.49 ± 5.24 | 63.96 ± 8.67 | 99.09 ± 18.86 |
| 6 μM TTFA + 200μM flavone | 82.70 ± 3.86 | 42.75 ± 3.78 | 80.60 ± 2.63 |
| 5 mM SHAM + 200μM flavone | 60.23 ± 7.53 | 39.90 ± 5.21 | 67.18 ± 8.51 |
Thus, in GOA31, the PAR pathway may take on increased importance in maintaining respiratory capacity. Still, growth of GOA31 is considerably reduced by SHAM in drop plates, so our data with GOA31 do not always support the paradigm that respiration inhibitors also affect growth. But there is evidence that in the case of rotenone, growth and respiration are considerably affected. We address the rotenone data in experiments with CI below.
Transcription of AOX1, AOX2, and the NDE1 and YMX6 NADH dehydrogenases.
To explain the increased respiration in GOA31 and poor growth in drop plate assays (Fig. 1) in the presence of SHAM, we also measured AOX1 and AOX2 transcription by quantitative PCR (qPCR) (30, 31). However, in C. albicans there are also two alternative NADH dehydrogenase orthologs (NDE1 and YMX6) of the S. cerevisiae NDE1 and NDI1 that could account for the compensatory increase in respiration (42). Thus, transcription of each gene was followed in untreated strains as well as in strains treated with 5 mM H2O2 and SHAM (Fig. 2 and 3).
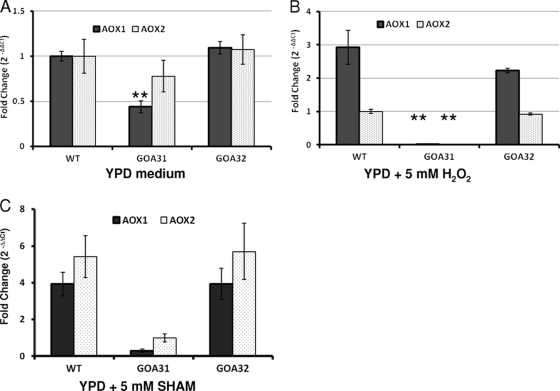
Fig. 2.
Transcription of the AOX genes of C. albicans (AOX1 and AOX2) in WT, GOA31 (goa1Δ/goa1Δ), and GOA32 (GOA1/goa1Δ) strains. Data are presented as the mean fold change (2−ΔΔCT) ± SD. All experiments were performed in triplicate. **, P < 0.001, GOA31 versus WT or GOA32. Overnight cultures were used for RNA extraction. Growth media are indicated. Peroxide or SHAM was added for an additional 30 min prior to extraction.
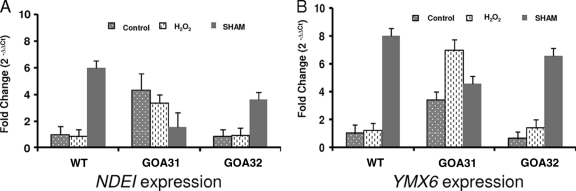
Fig. 3.
Transcription of the alternative NADH dehydrogenases NDE1 and YMX6 in strains of C. albicans. Cells were either not treated (control) or treated with 5 mM H2O2 for 30 min; RNA was extracted, and qPCR was performed using primers described in Materials and Methods. GOA31 (goa1Δ/goa1Δ) cells show higher expression in untreated and peroxide-treated cells than control cells but smaller increases following SHAM treatment.
For cells grown for 30 min in the absence of H2O2, transcription of AOX1 and AOX2 is similar in the WT and GOA32 strains, but in GOA31 AOX1 but not AOX2 is significantly reduced (Fig. 2A). Interestingly, in peroxide-treated cells, AOX1 but not AOX2 increases 2- to 3-fold in WT and GOA32 cells while transcription of both genes in GOA31 could not be detected (Fig. 2B). These data also indicate an important role for Aox1p and Aox2p in adaptation of peroxide-stressed cells. In SHAM-treated WT and GOA32 cells, both AOX1 and AOX2 expression slightly increased but not in GOA31 cells (Fig. 2C). We believe that the reduction of AOX1 in untreated cells and of AOX1 and AOX2 in peroxide-stressed cells in GOA31 is a direct consequence of the loss of GOA1.
Transcription levels of NDE1 and YMX6 were similar in both parental and GOA32 strains with or without peroxide or SHAM treatments (Fig. 3A and B). However, in GOA31, transcription of both genes was greater than in control strains in untreated or peroxide-treated cells, but it was reduced in SHAM-treated cells. Nevertheless, transcription of both genes occurred in GOA31. This observation together with the reduction in the respiration-compensatory increase in GOA31 by flavone (Table 1) suggests that Nde1p and Ymx6p may participate in the compensatory augmentation of respiration in SHAM-treated cells.
Complex I activity is decreased in the goa1Δ mutant (GOA31).
To understand which of the complexes of the CRC pathway is (are) defective enzymatically and to validate the inhibitor data described above, we assayed each complex (CI to CIV) enzymatically. Previously, we had shown that GOA31 compared to control strains had a negligible mitochondrial membrane potential and reduced ATP synthesis, which could point to reduced activity of CI, CIII, and/or CIV during respiration (8). Based upon our observations of inhibitor studies, we pursued experiments to determine the enzymatic activity of complexes I to IV (Table 2). CV was not assayed since growth inhibition of strain GOA31 was not observed with oligomycin (Fig. 1). Our data indicate that the specific activity of CI (nM/min/mg of protein) of GOA31 was decreased by about 80% compared activity in WT cells, while CII, CIII, and CIV activities ranged from 0 to 20% less in GOA31 than in WT cells (P < 0.001, GOA31 versus wild-type cells for CI). These assays were done with total mitochondrial protein from each strain in contrast to experiments described below, which were performed with CI protein only. We believe that the marginal effect of GOA1 deletion on CII, CIII, and CIV activity is related to the severe loss of CI activity that may affect downstream complexes. Strain GOA32 had specific activities for all complexes that ranged from 90 to 100% of wild-type cells (Table 2).
Table 2.
GOA31 of C. albicans has dysfunctional CI activity
| CRC complex | Specific activity in the indicated strain (nM/min/mg of protein)a |
||
|---|---|---|---|
| WT | GOA31 | GOA32 | |
| I | 1,470 ± 80 | 320 ± 55* | 1,380 ± 88 |
| II | 1,640 ± 23 | 1,340 ± 100 | 1,410 ± 105 |
| II + III | 1,480 ± 66 | 1,180 ± 68 | 1,480 ± 71 |
| IV | 170 ± 34 | 150 ± 12 | 170 ± 20 |
*, P < 0.001, GOA31 versus the WT and GOA32 strains.
BN-PAGE.
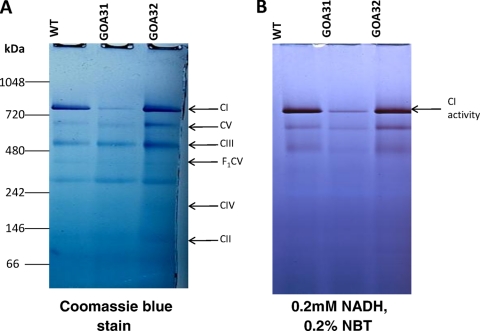
CI was also evaluated in GOA31 and control strains for both quantity (Coomassie staining) and enzyme activity by blue native polyacrylamide gel electrophoresis (BN-PAGE) (Fig. 4). We determined that CI is reduced in amount by ∼80 to 85% in GOA31 cells compared to the gel density ratios of CI/CIII and CI/CV to WT or GOA32 cells by ImageJ (Fig. 4A). Also, the in situ assay of CI enzyme activity demonstrated that strain GOA31 was reduced by approximately 85% compared to control strains (Fig. 4B). Of importance, the specific activity of CI is the same in WT cells as in GOA32 cells (Fig. 4A and B, compare the protein content and enzyme activity). This observation could imply several roles for Goa1p including a regulatory activity. Thus, by cell-based assays (Fig. 1 and Table 1), protein content, or enzymatic assays, our data support the hypothesis that GOA31 has an apparent defect in CI.
Fig. 4.
BN-PAGE electrophoresis of C. albicans CRC complex proteins in WT, GOA31 (goa1Δ/goa1Δ), and GOA32 (GOA1/goa1Δ) cells. (A) Coomassie stain of purified proteins of CI to CIV. Equal concentrations of total mitochondrial proteins from all strains were separated by BN-PAGE and stained accordingly. CI is significantly reduced in GOA31 compared to levels in the matched set of control strains, WT and GOA32. The molecular markers indicated on the left side are NativeMark (unstained protein standard; Invitrogen). (B) The in-gel enzyme activity of CI in BN-PAGE was assayed within 60 min after incubating the gel in reaction medium (0.1 M Tris-HCl, pH 7.4, 0.2 mM NADH as a substrate, and 0.2% NBT). Less CI enzyme activity in the GOA31 (goa1Δ/goa1Δ) strain was observed with NBT than in the WT and GOA32 (GOA1/goa1Δ) strains.
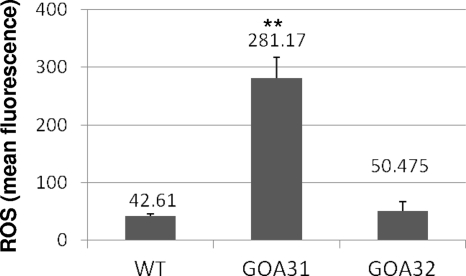
ROS production is increased in GOA31.
Dysfunction of CRC activity, especially CI and CIII in human neurodegenerative diseases and CI in fungi, is associated with an increase in cell ROS (2, 11, 15, 17, 40, 49). Therefore, to determine if a similar effect occurs in GOA31, a quantitative flow cytometry assay to measure total cell ROS of all strains was employed. The fluorescent dye 2′,7′-dichlorofluorescein diacetate (DCFDA) is widely used in both mammalian and fungal cells to measure intracellular ROS. Once the compound diffuses into cells, it is hydrolyzed into 2′,7′-dichlorofluorescein (DCFH) by cell esterases and accumulates in viable cells. As shown in Fig. 5, the level of ROS was dramatically increased by ∼7-fold in strain GOA31 compared to WT and GOA32 levels. The small amount of ROS in stationary-phase cells of WT and GOA32 strains may reflect an adaptation by regulatory mechanisms that include Goa1p either directly or indirectly.
Fig. 5.
ROS production in strains. Cells were obtained from overnight-grown cultures, washed twice with PBS, and incubated with DCFDA, which specifically reacts with ROS in viable cells. We show that ROS in GOA31 (goa1Δ/goa1Δ) was increased but not in the WT or GOA32 (GOA1/goa1Δ) strain (**, P < 0.001).
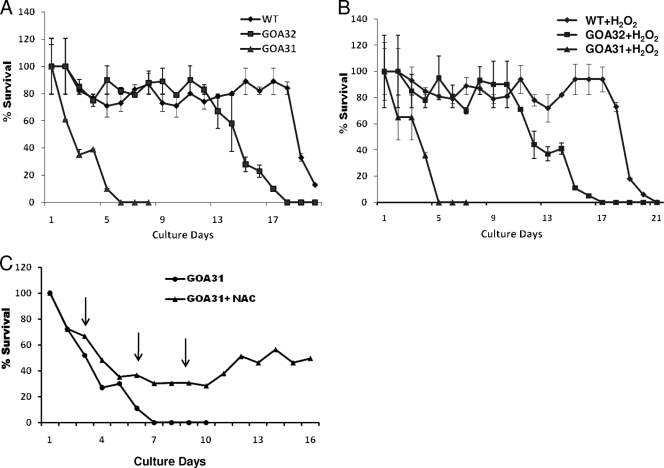
Chronological life span is decreased in GOA31.
Increased amounts of cell ROS as a result of defective CI induces apoptosis in mammalian and fungal cells (2, 11, 15, 25, 36, 37, 54, 56). To explore this possibility in C. albicans, a suspension of 106 cells/ml in SC broth was prepared and incubated with or without 5 mM H2O2. In treated cultures, we maintained high levels of H2O2 content by daily replenishing the medium with this oxidant. At daily intervals for 21 days, aliquots of cells were counted and plated on YPD medium to determine the number of CFU of each strain. In untreated cells, within 2 to 3 days, the viability of GOA31 was reduced by ∼50%, while that of GOA32 and WT cells remained high, at ∼80 to 90% (Fig. 6 A). At 6 to 7 days of incubation, we could not detect any viable cells of GOA31, in contrast to control strains, which retained a viability of ∼80 to 90%. Viability of GOA32 began to decrease significantly at about 11 to 12 days while wild-type cells remained about 80% viable until 18 days, and then this proportion steadily decreased. These data indicate that loss of chronological life span (decreased viability) of GOA31 is associated with an increased level of cell ROS (Fig. 5). Interestingly, loss of viability in all strains treated with H2O2 was equal to that in untreated cells (Fig. 6B). Our interpretation of these data is that cell levels of ROS were already high in GOA31 compared to levels in WT and GOA32 cells so that the addition of H2O2 did not cause a change in chronological life span profiles. Further, WT and GOA32 cells adapted to added peroxide. To directly demonstrate that the association of ROS increases with a decrease in chronological life span, we treated cells of GOA31 with the antioxidant N-acetyl cysteine ([NAC] 30 mM), which was added several times during the experiment. We found that compared to survival of untreated cells, survival of GOA31 persisted at about 30% through ∼10 days and then began increasing thereafter (Fig. 6C).
Fig. 6.
Chronological life span of GOA31 (goa1Δ/goa1Δ) cells is significantly less than that of WT or GOA32 (GOA1/goa1Δ) cells. All strains were grown in YPD medium overnight, washed, and then incubated in fresh SC medium in the absence (A) or presence (B) of 5 mM H2O2. Samples were removed daily for 3 weeks, and viability was determined by plating on YPD medium again. After 48 h of incubation, the number of CFU was determined. (C) GOA31 was untreated or treated with the antioxidant N-acetyl cysteine (NAC; 30 mM). NAC was added at the time points indicated (arrows). The presence of NAC increased survival of GOA31.
Apoptosis is increased in GOA31.
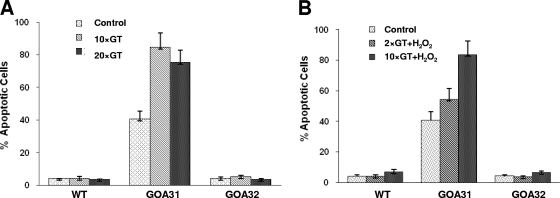
To determine if the decrease in viability in GOA31 is related to programmed cell death, we measured caspase activity (Fig. 7). For untreated cells, caspase activity was determined in cell populations at 0 (control), 10, or 20 generation times (GT) (8). Our data indicate that the levels of caspase were increased in GOA31 at all time points compared to levels in the WT and GOA32 strains, which produced negligible caspase activity (Fig. 7A). The difference in caspase activity of GOA31 at 10 and 20 GT was not statistically significant. The addition of H2O2 to cultures did not influence the level of caspase at 2 and 10 GT (Fig. 7B). It also appears that significant apoptosis as determined by caspase activity had already occurred in overnight washed cultures of GOA31 (zero time; control cultures).
Fig. 7.
Caspase activity in strains of C. albicans. Cells of each strain were grown overnight, standardized, and inoculated in YPD medium in the absence (A) or presence (B) of 10 mM H2O2. Caspase activity was measured using a poly-caspase (FAM-VAD-FMK) apoptosis detection kit (Immunochemistry Technologies, LLC). At designated times, FAM-VAD-FMK was added to cell suspensions. After unbound FAM-VAD-FMK was washed away, anti-FAM-VAD-FMK antibody was added. The number of cells with green fluorescence is a direct measurement of caspase-positive cells.
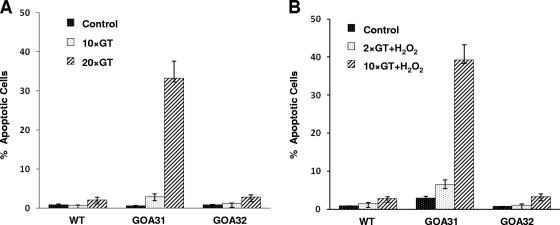
TUNEL assays were also established to verify that apoptosis had increased in GOA31 (Fig. 8). In unstressed cells (0, 10, and 20 GT), in WT or GOA32, TUNEL assays verified a low level of apoptosis in untreated or treated cultures, while that of GOA31 exhibited high levels by 10 and 20 cell generations (Fig. 8A). In cells treated with H2O2, again only strain GOA31 showed a marked increase in TUNEL activity at 10 generations (Fig. 8B). Importantly, propidium iodide staining, which measures necrotic cells, was quite low, in the range of 1 to 2%.
Fig. 8.
TUNEL assays in strains of C. albicans. In this assay, a terminal deoxynucleotidyltransferase (TdT) adds Br-dUTP (bromolated dUTP nucleotides) to DNA breaks. The incorporation of Br-dUTP results in a green-fluorescent (FITC) signal once anti-BrdU antibody is added. In situ DNA fragmentation was assayed in all strains. The cells were either left untreated or were treated with 10 mM H2O2 and collected at the time points indicated on the graph. The DNA labeling solution was added for 1 h, and fluorescence was measured by flow cytometry.
DISCUSSION
Mitochondrial dysfunctions: general considerations.
In WT cells of C. albicans, C. parapsilosis, and other fungi, the primary respiratory pathway in regard to energy formation and O2 consumption appears to be the CRC (28, 29, 46). The AOX and PAR pathways consume less oxygen and generate less ATP. Although the AOX pathway has been suggested as a compensatory mechanism in case of CRC failure, other data indicate that both the CRC and at least AOX1 are constitutively expressed in fungi (28, 29, 31, 46). S. cerevisiae has three alternative NADH dehydrogenases (NdeI, Nde2, and Ndi1) that oxidize cytosolic NADH and participate in NADH-dependent mitochondrial respiration but do not translocate protons and are only partially sensitive to rotenone (42). These proteins are also described in Yarrowia lipolytica (27). Therefore, our thought was that the orthologs in C. albicans (NDE1 and YMX6) might account for the compensatory increase in respiration in SHAM-treated GOA31 cells. We now show that transcription levels of both NDE1 and YMX6 are lower in GOA31 cells than in control strains when cells are treated with SHAM but that flavone, an inhibitor of these dehydrogenases, did reduce the compensatory respiration in GOA31. The characterization of growth mutants in different organisms has demonstrated that mitochondria defective in the CRC pathway are able to compensate for these impairments by a process termed retrograde regulation, which includes the induction of certain genes that under normal conditions are not expressed (48). We are left with the observation that the compensatory increase in GOA31 respiration in SHAM-treated cells is due to the PAR pathway and external NADH dehydrogenases.
Our data support the concept that a major defect has occurred in the respiration of GOA31 (loss of oxygen consumption, membrane potential, and ATP formation) that is associated with a dysfunctional CRC (8; also this study). We now demonstrate that CI is dysfunctional in the GOA31 strain. Two lines of proof were used to validate this conclusion: in vitro activity and in situ gel analysis as well as drop plate and respiration assays with rotenone. Further, transcription of AOX genes (AOX1 in untreated cells and AOX1 and AOX2 in peroxide-treated cells) is downregulated in GOA31, such that only the PAR mitochondrial pathway apparently remains intact, similar to what has been shown in C. parapsilosis (46). A reduction in CI activity is especially detrimental to cells since it provides ∼50% of the energy generated by oxidative phosphorylation. The reduction in CI activity results in a deficiency in membrane potential (ΔψM) in mitochondria since CI is the major donor to the proton gradient.
In human cells, CI is the major ROS-generating site in mitochondria, whereas CII, CIII, and CIV could be inhibited by about 70% without an increase in ROS production (11, 40). During optimum respiration, referred to as state 3 respiration, when oxygen consumption is coupled to ATP production, ROS levels are low. However, in state 4 respiration, when the CRC slows down or is dysfunctional at CI, the production and release of ROS increase (11). An imbalance in ROS is thought to initiate programmed cell death (PCD) as in several types of neurodegenerative diseases. In structurally and functionally intact mitochondria, a large antioxidant defense capacity negates ROS that is formed; therefore, there is little net cellular ROS in the matrix of active mitochondria. However, once this balance is disturbed, the accumulated ROS will further damage mitochondria, causing more free-radical molecules to form. The ROS free radicals are also able to damage biomolecules such as nucleic acids, lipids, and proteins. Usually, young cells have very small amounts of net ROS, but as mitochondria age and become more dysfunctional, larger amounts of ROS are generated, and cell viability decreases. Loss of cell viability is also accompanied by the ROS-induced inactivation of complex protein Fe-S clusters, which is followed by a release of iron that induces the formation of hydroxyl radicals (39). Therapies to counter neurodegenerative diseases and even cancer are currently being sought that will alter carbohydrate metabolism to either attenuate or heighten mitochondrial activity in a disease-specific manner (11, 19).
Our hypothesis is that Goa1p maintains CI activity and that in its absence, cells have less membrane potential, with an increase in ROS accumulation and cell death by apoptosis. This is also reminiscent of data in Neurospora Crassa, where treatment with phytosphingosine targets complex I (17). Subsequently, cell ROS increases, followed by cell death, which is very similar to what has been described in Podospora anserina and S. cerevisiae (48).
Complex I.
The C. albicans CI is believed to consist of 39 proteins, 2 of which are unique to Candida species (22). Comparative genomics indicates that CI shares structural similarity with the complex I of other eukaryotes (22). Eukaryotic CI consists of a conserved core of 14 subunits, present in bacterial NADH:ubiquinone oxidoreductases, plus a variable number of supernumerary subunits, e.g., 31 in mammals (24, 36) and 26 in the yeast Y. lipolytica (1, 22, 47). This species is more closely related to C. albicans, and we have found that all of the Y. lipolytica subunits except one are conserved in the C. albicans genome (unpublished results). The core subunits include seven peripheral proteins that serve as binding sites for NADH, flavin mononucleotide (FMN), and iron-sulfur clusters and seven membrane-spanning subunit proteins that collectively form an L-shaped structure, with the peripheral arm located perpendicular to the mitochondrial inner membrane (22). Many of the supernumerary subunits are conserved across all species. Fungal CI has also been investigated in N. crassa (6, 44). In that fungus, the lack of a single subunit protein in knockout mutants caused a complete block in the assembly pathway of CI (44). It is important to recognize that following the evolutionary recruitment of the core proteobacterial and lower eukaryotic proteins to CI, subsequently, the complex diversified along the different eukaryotic lineages (mammals, fungi, insects, and nematodes) with new and continuous recruitment of subunits (22). Thus, significant differences exist among these lineages to warrant study of the C. albicans CI. Presently, we are pursuing the identification of complex I proteins in GOA31 and control strains. Since clearly CI activity is decreased and there is less CI, we hypothesize that Goa1p regulates complex activity and assembly or is required for its activity as a part of CI. These defects have been observed in human CI-specific disorders (36, 49).
AOX.
All indications are that Goa1p is required for transcription of the AOX pathway genes in untreated (AOX1) and peroxide-treated cells (AOX1 and AOX2). However, we have not determined whether the relationship of the AOX pathway and AOX1 and AOX2 transcription with Goa1p is direct or indirect. Our hypothesis is that Goa1p and Aox1p/Aox2p are each critical to peroxide adaptation. Additional experiments are planned to examine the response of both proteins to other forms of cell stress.
Apoptosis.
The consequence of heightened ROS levels associated with CI dysfunction is apparently due to programmed cell death in GOA31. That dysfunctional CI is a source of heightened ROS and PCD has been demonstrated in N. crassa (17). Phytosphingosine-treated conidia caused increased ROS, evidence of PCD, and reduced viability similar to our current observations of GOA31. However, a CI mutant of N. crassa was more resistant to phytosphingosine than wild-type cells, indicating that CI is a target for this compound (17). An important point of this study is, therefore, that CI defects in a fungus are associated with ROS and PCD. In GOA31, an increase in caspase and TUNEL activity accompanies the loss of viability. Our hypothesis that GOA31 cells undergo apoptosis is based upon the combined application of several assays, as suggested by others (16). In S. cerevisiae, PCD and apoptosis are now thought to have several subroutines, including necrosis, which phenotypically includes a ruptured plasma membrane (15). PCD generally occurs through two types of mechanisms, extrinsic, as described above, with inducers forced upon cells, and intrinsic, associated with mitochondrial dysfunction and ROS production. Both pathways overlap to a great degree, making analysis of defects somewhat complex. Carmona-Gutierrez et al. (15, 16) differentiate between replicative aging of cells, defined as the number of divisions an individual mother cell has before death occurs, and chronological aging, which is distinguished by viability over time of cells in stationary phase. Based upon our life span experiments (Fig. 6), we believe that GOA31 cells have reduced chronological aging, which means that Goa1p is essential for cell survival.
In summary, our data demonstrate that Goa1p is required for mitochondrial functions, especially CI activity, and, as a consequence, is critically important in maintaining nontoxic levels of ROS. As a result, chronological aging is extended by weeks in comparison to cells that lack GOA1. Although there are certainly other additional factors that could contribute to the observed growth defects in the goa1 mutant, it is clear that CI dysfunction results in the overproduction of ROS that impacts survival of C. albicans.
ACKNOWLEDGMENTS
We thank the flow cytometry facility at the Georgetown University Center Lombardi Cancer Center for helping with the fluorescence-activated cell sorter analysis. We also thank Robert Balaban, Stephanie French, Susham S. Ingavale, Quentin Li, John Bennett, and Kyung J. Kwon-Chung at NHLBI and NIAID, NIH, for their advice with the oxygen consumption assays.
Footnotes
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Abdrakhmanova A., et al. 2004. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1658:148–156 [DOI] [PubMed] [Google Scholar]
- 2. Aerts A., et al. 2009. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 583:113–117 [DOI] [PubMed] [Google Scholar]
- 3. Alby K., Schaefer D., Sherwood R., Jones S., Bennett R. 2010. Identification of a cell death pathway in Candida albicans during the response to pheromone. Eukaryot. Cell 9:1690–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Dhaheri R. S., Douglas L. J. 2010. Apoptosis in Candida biofilms exposed to amphotericin B. J. Med. Microbiol. 59:149–157 [DOI] [PubMed] [Google Scholar]
- 5. Alonso-Monge R., Cavaheiro S., Nombela C., Rial E., Pla J. 2009. The Hog1 MAPK controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology 155:413–423 [DOI] [PubMed] [Google Scholar]
- 6. Alves P., Videra A. 1998. The membrane domain complex I is not assembled in the stopper mutant E35 of Neurospora. Biochem. Cell Biol. 76:139–143 [PubMed] [Google Scholar]
- 7. Askew C., et al. 2009. Transcriptional regulation of carbohydrate metabolism in Candida albicans. PLoS Pathog. 5:e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bambach A., et al. 2009. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot. Cell 8:1706–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barrelle C., et al. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 6:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrientos A. 2002. In vivo and in organello assessment of OXPHOS activities. Methods 26:307–316 [DOI] [PubMed] [Google Scholar]
- 11. Bayir H., Kagan V. 2008. Bench-to-bedside review: mitochondrial injury, oxidative stress and apoptosis—there is nothing more practical than a good theory. Crit. Care 12:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Camougrand N., Zniber S., Guerin M. 1991. The antimycin-A insensitive respiratory pathway of Candida parapsilosis: evidence for a second quinone involved specificity in its functioning. Biochim. Biophys. Acta 1057:124–130 [DOI] [PubMed] [Google Scholar]
- 13. Cao Y., et al. 2009. Candida albicans cells lacking CaMCA1-encoded metacaspase show resistance to oxidative stress-induced death and change in energy metabolism. Fungal Genet. Biol. 46:183–189 [DOI] [PubMed] [Google Scholar]
- 14. Carman A., Vylkova S., Lorenz M. 2008. Role of acetyl coenzyme A synthesis and breakdown in alternative carbon source utilization in Candida albicans. Eukaryot. Cell 7:1733–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carmona-Gutierrez D., et al. 2010. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 17:763–773 [DOI] [PubMed] [Google Scholar]
- 16. Carmona-Gutierrez D., Jungwirth H., Eisenberg T., Madeo F. 2010. Cell cycle control of death in yeast. Cell Cycle 9:4046. [DOI] [PubMed] [Google Scholar]
- 17. Castro A., Lemos C., Falco A., Glass N., Videira A. 2008. Increased resistance of complex I mutants to phytosphingosine-induced programmed cell death. J. Biol. Chem. 283:19314–19321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cavalheiro R., et al. 2004. Respiration, oxidative phosphorylation and uncoupling protein in Candida albicans. Braz. J. Med. Biol. Res. 37:1455–1461 [DOI] [PubMed] [Google Scholar]
- 19. Derdak Z., et al. 2008. The mitochondrial uncoupling protein-2 promotes chemoresistance in cancer cells. Cancer Res. 68:2813–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forsmark-Andree P., Lee C.-P., Dallner G., Ernster L. 1997. Lipid peroxidation and changes in the ubiquinone content and the respiratory chain enzymes of submitochondrial particles. Free Rad. Biol. Med. 22:391–400 [DOI] [PubMed] [Google Scholar]
- 21. Fradin C., et al. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56:397–415 [DOI] [PubMed] [Google Scholar]
- 22. Gabaldon T., Rainey D., Huynen M. 2005. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 348:857–870 [DOI] [PubMed] [Google Scholar]
- 23. Gallagher S. 2010. Digital image processing and analysis with Image J. Curr. Protoc. Essent. Lab. Tech. 3:A.3C.1–A.3C.24 doi: 10.1002/9780470089941.eta03cs03 [Google Scholar]
- 24. Garbian Y., Ovadia O., Dadon S., Mishmar D. 2010. Gene expression patterns of oxidative phosphorylation complex I subunits are organized as clusters. PLoS One 5:e9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gogvadze V., Orrenius S. 2006. Mitochondrial regulation of apoptotic cell death. Chem. Biol. Interact. 163:4–14 [DOI] [PubMed] [Google Scholar]
- 26. Guerin M., Camougrand N. 1994. Partitioning of electron flux between the respiratory chains of the yeast Candida parapsilosis: parallel working of two chains. Biochim. Biophys. Acta 1184:111–117 [DOI] [PubMed] [Google Scholar]
- 27. Guerro-Castillo S., Vazquez-Acevedo M., Gonzalez-Halphen D., Uribe-Carajal S. 2009. In Yarrow lipolytica mitochondria, the alternative NADH dehydrogenase interacts specifically with the cytochrome complexes of the classic respiratory pathway. Biochim. Biophys. Acta 1787:75–85 [DOI] [PubMed] [Google Scholar]
- 28. Helmerhorst E., Stan M., Murphy M., Sherman F., Oppenheim F. 2005. The concomitant expression and availability of conventional and alternative, cyanide-insensitive, respiratory pathways in Candida albicans. Mitochondrion 5:200–211 [DOI] [PubMed] [Google Scholar]
- 29. Helmerhorst E., Murphy M., Troxler R., Oppenheim F. 2002. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim. Biophys. Acta 1556:73–80 [DOI] [PubMed] [Google Scholar]
- 30. Huh W.-K., Kang S. 2001. Characterization of a gene family encoding alternative oxidases from Candida albicans. Biochem. J. 356:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huh W.-K., Kang S. 1999. Molecular cloning and functional expression of an alternate oxidase from Candida albicans. J. Bacteriol. 181:4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ingavale S., et al. 2008. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 4:e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joseph-Horne T., Hollomon D., Wood P. M. 2001. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504:179–195 [DOI] [PubMed] [Google Scholar]
- 34. Juarez O., Guerra G., Martinez F., Pardo J. 2004. The mitochondrial respiratory chain of Ustilago maydis. Biochim. Biophys. Acta 1658:244–251 [DOI] [PubMed] [Google Scholar]
- 35. Kim S. Y., Kim J. 2010. Roles of dihydrolipoamide dehydrogenase Lpd1 in Candida albicans filamentation. Fungal Genet. Biol. 47:782–788 [DOI] [PubMed] [Google Scholar]
- 36. Lazarou M., Thorburn D. R., Ryan M. T., McKenzie M. 2009. Assembly of mitochondria complex I and defects in disease. Biochim. Biophys. Acta 1793:78–88 [DOI] [PubMed] [Google Scholar]
- 37. Lee Y.-I., et al. 2004. Human hepatitis B virus-X protein alters mitochondria function and physiology in human liver cells. J. Biol. Chem. 279:15460–15471 [DOI] [PubMed] [Google Scholar]
- 38. Lenaz G., et al. 1995. Under evaluation of complex I activity by the direct assay of NADH-coenzyme Q. reductase in rat liver mitochondria. FEBS Lett. 366:119–121 [DOI] [PubMed] [Google Scholar]
- 39. Lill R. 2009. Function and biogenesis of iron-sulphur clusters. Nature 460:831–838 [DOI] [PubMed] [Google Scholar]
- 40. Lin M., Beal M. 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–798 [DOI] [PubMed] [Google Scholar]
- 41. Lorenz M., Bender J., Fink G. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luttik M., et al. 1998. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 273:24524–24534 [DOI] [PubMed] [Google Scholar]
- 43. Magnani T., et al. 2007. Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stress. FEMS Microbiol. Lett. 271:230–238 [DOI] [PubMed] [Google Scholar]
- 44. Marques I., Duarte M., Assunção J., Ushakova A., Videira A. 2005. Composition of complex I from Neurospora crassa and disruption of two accessory subunits. Biochim. Biophys. Acta 1707:211–220 [DOI] [PubMed] [Google Scholar]
- 45. McDonough J., Bhattacherjee V., Sadlon T., Hostetter M. 2002. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Genet. Biol. 36:117–127 [DOI] [PubMed] [Google Scholar]
- 46. Milani G., et al. 2001. Respiratory chain network in mitochondria of Candida parapsilosis: ADP/O appraisal of the electron pathways. FEBS Lett. 508:231–235 [DOI] [PubMed] [Google Scholar]
- 47. Morgner N., et al. 2008. Subunit mass fingerprinting of mitochondria complex I. Biochim. Biophys. Acta 1777:1384–1391 [DOI] [PubMed] [Google Scholar]
- 48. Osiewacz H., Scheckhuber C. 2006. Impact of ROS on ageing of two model systems: Saccharomyces cerevisiae and Podospora anserina. Free Radic. Res. 40:1350–1358 [DOI] [PubMed] [Google Scholar]
- 49. Papa S., Sardanelli A., Capitanio N., Piccoli C. 2009. Mitochondrial respiratory dysfunction and mutations in mitochondrial DNA in PINK1 familial parkinsonism. J. Bioenerg. Biomembr. 41:509–516 [DOI] [PubMed] [Google Scholar]
- 50. Ramierz M., Lorenz M. 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 6:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rodaki A., et al. 2009. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol. Biol. Cell 20:4845–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruy F., Vercesi A., Kowaltowski A. 2006. Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. J. Bioenerg. Biomembr. 38:129–135 [DOI] [PubMed] [Google Scholar]
- 53. Sabar M., Balk J., Leaver C. 2005. Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J. 44:893–901 [DOI] [PubMed] [Google Scholar]
- 54. Semighini C., Hornby J., Dumitru P., Nickerson K., Harris D. 2006. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 59:753–764 [DOI] [PubMed] [Google Scholar]
- 55. Seo B., et al. 1998. Molecular remedy of complex I defects: rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 95:9167–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shirtliff M., et al. 2009. Farnesol-induced apoptosis in Candida albicans. Antimicrob. Agents Chemother. 53:2392–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Velazquez I., Pardo J. 2001. Kinetic characterization of the rotenone-insensitive internal NADH:ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 389:7–14 [DOI] [PubMed] [Google Scholar]
- 58. Vellucci V., Gygax S., Hostetter M. 2007. Involvement of Candida albicans pyruvate dehydrogenase complex protein X (Pdx1) in filamentation. Fungal Genet. Biol. 44:979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vogel R., Smeitink J., Nijtmans L. 2007. Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim. Biophys. Acta 1776:1215–1227 [DOI] [PubMed] [Google Scholar]
- 60. Wu X. Z., Chang W. Q., Cheng A. X., Sun L. M., Lou H. X. 2010. Plagiochin E, an antifungal active macrocyclic bis(bibenzyl), induced apoptosis in Candida albicans through a metacaspase-dependent apoptotic pathway. Biochim. Biophys. Acta 1800:439–447 [DOI] [PubMed] [Google Scholar]