Abstract
Eukaryotic heat shock transcription factors (HSF) regulate an evolutionarily conserved stress-response pathway essential for survival against a variety of environmental and developmental stresses. Although the highly similar HSF family members have distinct roles in responding to stress and activating target gene expression, the mechanisms that govern these roles are unknown. Here we identify a loop within the HSF1 DNA-binding domain that dictates HSF isoform specific DNA binding in vitro and preferential target gene activation by HSF family members in both a yeast transcription assay and in mammalian cells. These characteristics of the HSF1 loop region are transposable to HSF2 and sufficient to confer DNA-binding specificity, heat shock inducible HSP gene expression and protection from heat-induced apoptosis in vivo. In addition, the loop suppresses formation of the HSF1 trimer under basal conditions and is required for heat-inducible trimerization in a purified system in vitro, suggesting that this domain is a critical part of the HSF1 heat-stress-sensing mechanism. We propose that this domain defines a signature for HSF1 that constitutes an important determinant for how cells utilize a family of transcription factors to respond to distinct stresses.
Keywords: HSF, HSP70, stress response, apoptosis
A critical response to cellular stress is the rapid production of heat shock proteins (Hsps). The Hsps, or molecular chaperones, function to stabilize the folded state of proteins and prevent aggregation of damaged or unfolded proteins (for reviews, see Parsell and Lindquist 1993; Craig et al. 1994; Morimoto et al. 1994). In all eukaryotes an evolutionarily conserved protein, heat shock transcription factor (HSF), activates stress-inducible transcription of Hsp genes. HSF binds as a homotrimer to its cognate regulatory site, the heat shock element (HSE), composed of a minimum of two inverted repeats of the consensus 5-base pair sequence 5′-nGAAn-3′. The HSF protein is modular, composed of an amino-terminal DNA-binding domain (DBD), an adjacent coiled–coil trimerization domain, a central regulatory domain, a second coiled–coil domain, and a transcriptional activation domain at the carboxyl terminus. The structures of the DNA-binding and trimerization domains are highly conserved among all characterized HSFs (for reviews, see Lis and Wu 1993; Voellmy 1994; Wu 1995; Morimoto 1998; Morano and Thiele 1999).
In yeast and Drosophila, a single gene encodes HSF, but in mammals and higher plants, distinct genes encode multiple HSF isoforms (Sorger and Pelham 1988; Wiederrecht et al. 1988; Sarge et al. 1991; Nover et al. 1996; Jedlicka et al. 1997; Nakai et al. 1997). The existence of multiple HSF isoforms is consistent with observations that these proteins are differentially activated and may regulate distinct target genes and cellular responses. Mammalian HSF1, the archetype stress-inducible HSF, is activated in response to many types of physical, chemical, pharmacological, and physiological stresses. Indeed cells derived from hsf1 null mice lack activation of several Hsp genes in response to heat shock and are exquisitely sensitive to thermal stress-induced apoptosis (McMillan et al. 1998). In addition, hsf1−/− mice show developmental defects and female infertility, indicating a broader requirement for HSF1 in specific developmental pathways beyond its role in stress response (Xiao et al. 1999; Christians et al. 2000). While HSF1 is activated by a variety of physiological stresses, much less is known about the activators and targets for HSF2 and HSF4. It is now clear from studies of hsf1−/− mice that these two highly related transcription factors are not redundant with HSF1 and must be regulated differently or activate distinct target genes.
The activation of HSF1 is controlled at multiple levels. Under nonstress conditions, mammalian HSF1 and the orthologous Drosophila HSF (dHSF) appear as monomeric forms with low DNA-binding activity (Zimarino and Wu 1987; Sarge et al. 1991). Molecular and genetic studies of these HSFs by deletion and mutagenesis analysis have revealed a number of important intramolecular interactions between multiple domains, including the amino- and carboxy-terminal hydrophobic heptads and a linker domain, that restrain HSF in an inactive state (Rabindran et al. 1993; Zuo et al. 1994; Orosz et al. 1996; Farkas et al. 1998; Liu and Thiele 1999). Genetic and biochemical evidence also supports a role for HSP70 and HSP90 in modulating the activation and recovery of HSF1 in response to stress (Abravaya et al. 1992; Ali et al. 1998; Duina et al. 1998; Shi et al. 1998; Zuo et al. 1998; Bharadwaj et al. 1999). Activation of HSF1 leads to the formation of homotrimeric complexes in which monomers associate through the formation of a three-stranded coiled–coil by the trimerization domain (Sorger and Nelson 1989; Peteranderl and Nelson 1992). The effect of trimerization is to orient the individual DNA-binding domains such that they bind to inverted repeats of the HSE pentamer in a cooperative manner, thereby increasing both the affinity and specificity for target gene promoters. The underlying mechanisms by which HSF1 senses thermal stress to switch from the monomer to the homotrimeric form are poorly understood.
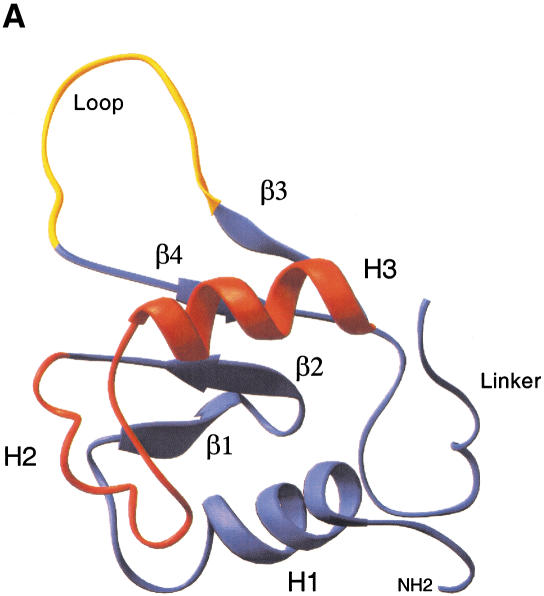
HSF belongs to the winged helix-turn-helix (wHTH) family of DNA-binding proteins that also includes the mammalian ETS and HNF-3/forkhead proteins. A defining feature of this family is the central helix-turn-helix fold, composed of α-helices 2 and 3, where α-helix3 is the DNA recognition motif that docks within the major groove of the DNA helix (Fig. 1A; Harrison et al. 1994; Vuister et al. 1994a,b; Damberger et al. 1995). The cocrystal structure of the Kluyveromyces lactis HSF DBD bound to an HSE revealed an important distinction between HSF and related wHTH proteins (Littlefield and Nelson 1999). The loop of the yeast HSF DBD does not contact DNA like the wHTH protein PU.1 does; rather, structural analysis indicates that the loop is juxtaposed against the DBD of an adjacent HSF molecule. Thus the loop may form a potential protein–protein interaction surface at the interface between adjacent subunits of the HSF homotrimer or between adjacent DNA-bound trimers and may facilitate high-affinity binding to HSEs through enhancing cooperativity (Littlefield and Nelson 1999).
Figure 1.
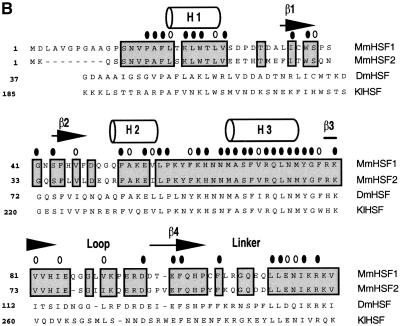
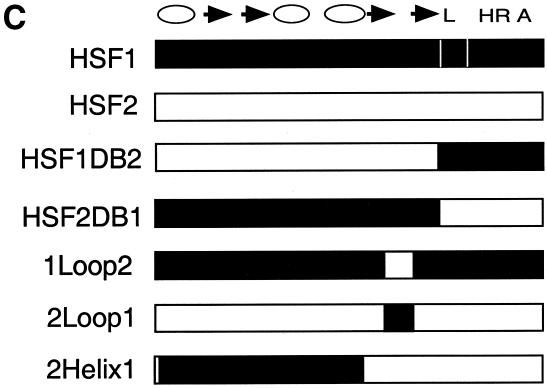
Structure of the heat shock transcription factor (HSF) DNA-binding domain. (A) The central helix-turn-helix motif (red) is composed of α-helix2 and α-helix3, where the latter is the DNA recognition helix. The structure was modeled using the Ribbons program from the coordinates of NMR structure of the Drosophila DBD (Vuister et al. 1994a). The loop (yellow) was determined to be solvent exposed. (B) Alignment of the amino acid sequences for the mouse HSF1, HSF2, Drosophila HSF1, and Kluyveromyces lactis HSF DBDs. Predicted secondary structure for the mouse HSFs is based on homology to the solved K. lactis and Drosophila HSF DBD structures (Harrison et al. 1994; Vuister et al. 1994a), where cylinders represent α-helicies and arrows represent β-sheets. Residues conserved between HSF1 and HSF2 are boxed; residues identical among all four sequences are indicated by a solid oval; those positions that have conserved substitutions are indicated by an open oval. (C) Diagram of specific chimeras used in this study. HSF1 sequences (white); HSF2 sequences (black). (L) Linker; (HR A) trimerization domain.
Although distinct patterns of mammalian target gene activation by different HSF isoforms have not yet been shown, studies of DNA binding by mouse HSF proteins to the hsp70 promoter have revealed intriguing differences between HSF1 and HSF2. Footprinting studies have shown that HSF1 protected all five hsp70 HSE repeats, but HSF2 failed to protect site 1 (Kroeger et al. 1993). These differences in the HSF1 and HSF2 DNA-binding patterns were attributed to the greater ability of HSF1 to bind DNA cooperatively. Furthermore, DNA binding to randomized oligonucleotides using a SELEX procedure showed that sequences composed of long arrays of HSE units were preferentially selected by HSF1 and shorter HSE arrays were selected by HSF2 (Kroeger and Morimoto 1994). Consistent with this idea, human HSF1 more potently activated transcription of an SSA3 HSE–lacZ reporter gene in yeast, containing five HSE repeats than did human HSF2 (Liu et al. 1997). The region that determines differential activation of SSA3–lacZ did not require either the oligomerization or linker domains (Liu and Thiele 1999), suggesting that the DNA-binding domain itself dictates the selectivity of binding. Collectively, these data suggest that HSF1 and HSF2 might activate distinct subsets of target genes depending upon the architecture of the HSEs and the differential interaction between HSF isoforms and HSEs.
In this study, we have investigated the mechanistic basis for differences in DNA binding between HSF1 and HSF2 to the hsp70 HSEs and its effect on transcription. We map the determinant for DNA-binding specificity for HSF1 and HSF2 to a loop region in the DNA-binding domain (DBD) and show that this region is sufficient to confer isoform-specific patterns of DNA binding and target gene transcription. We further show that the HSF1 loop plays an important role in thermal sensing by purified HSF proteins in vitro, in heat shock induced gene transcription in vivo and in protecting mouse cells from heat shock induced apoptosis. These results reveal a signature domain within the HSF1 DBD that is critical for stress-responsive gene activation.
Results
DNA-binding specificity of HSF1 maps to the loop
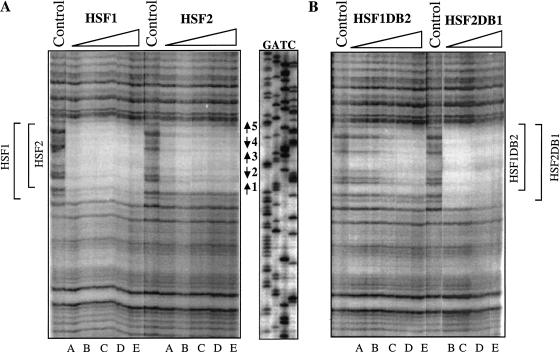
To delineate what determines the DNA-binding specificity between HSF1 and HSF2, Dnase I footprinting was used to analyze interactions between mouse HSF derivatives and the hsp70 promoter. Sequences encoding HSF1 and HSF2 and several chimeras between the two isoforms (Fig. 1B,C) were cloned into a T7 expression system with a carboxy-terminal His-tag as described in Materials and Methods. As shown previously (Kroeger et al. 1993), HSF1 protected the entire series of HSEs in the hsp70 promoter, from nucleotides −19 to −86, even at the lowest protein concentration. In contrast the region of protection by HSF2 was smaller, extending from nucleotides −112 to −86 (Fig. 2A). Protection by HSF1 was ∼70% throughout the HSE at the lowest concentration of the protein. The protection by HSF2 was nearly the same except in the region of site 1, where the level of protection was ∼25% and did not increase significantly with additional protein.
Figure 2.
Differential binding to the hsp70 heat shock element (HSE) maps to the heat shock transcription factor (HSF) DNA-binding domain. (A) DNase I footprinting by mHSF1 and mHSF2 to the hsp70 HSE. The hsp70 HSE probe was labeled at the 5′ end of the coding strand, and the concentration of the probe in all reaction mixtures was 0.1 nM. The amounts of bacterial lysates in lanes A–E were 0.5, 1, 2, 4, and 8 μL, respectively. Control reaction shows DNase I footprinting reaction in the absence of protein. The extent of HSF1 and HSF2 protection are indicated at the left with brackets. The positions of HSEs 1 to 5 are marked by arrows. (B) Specificity in DNA-binding maps to the DNA-binding domain. Chimeras in which the DBD was interchanged were expressed and purified as in Materials and Methods and used for DNase I footprinting. Lane assignments are as described for panel A. The extent of protein binding is delimited by the brackets.
To test whether the difference in occupancy of the hsp70 promoter was due solely to the DBD, we generated HSF hybrid molecules in which the DBD was interchanged. Substituting the HSF2 DBD into HSF1 (HSF1DB2) resulted in a decrease in the size of the protected area: the HSF1DB2 chimera showed no protection of site 1, even with titration of increasing protein amounts (Fig. 2B). In contrast, the DNA-binding pattern of HSF2 harboring the DBD of HSF1 (HSF2DB1) was qualitatively similar to HSF1. As the concentration of the chimeric protein increased, all five HSE repeats were protected. These results indicate that differences in the DNA-binding properties of mouse HSF1 and HSF2 are due to the differences in their DBDs.
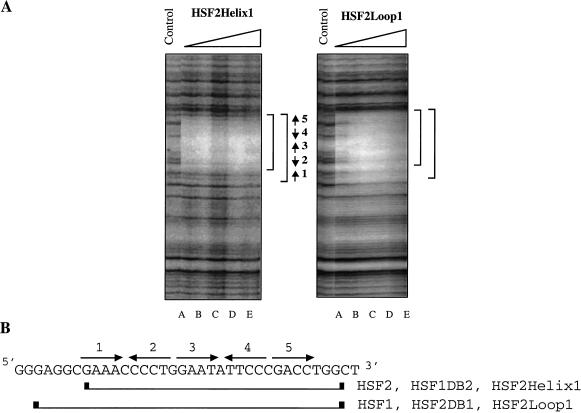
Although the DBD structure has not been solved for mammalian HSFs, sequence comparison among HSF proteins for which structures have been solved allow us to predict the position of structural elements such as α-helixes, β-strands, and an extended loop (Fig. 1A; Harrison et al. 1994; Vuister et al. 1994a). An alignment between HSF1 and HSF2 reveals only minor differences (70% identity, 80% similarity) in amino acid sequence over the entire DBD (Fig. 1B). Because residues within the recognition helix (α3) are identical between HSF1 and HSF2, we focused on divergent regions flanking α-helix 3. We expressed and purified a number of hybrid proteins chimeric within the HSF1 and HSF2 DBD. Substitution of HSF1 sequences encompassing β-strands 1 and 2 and α-helicies 1 and 2 into corresponding sequences of HSF2 did not change the DNA-binding pattern of HSF2 (Fig. 3A, HSF2Helix1). Strikingly, a chimera composed of HSF2 with the HSF1 loop bound to the hsp70 HSE with HSF1-like specificity (Fig. 3A, HSF2Loop1). The HSF2Loop1 footprint exhibited increased protection of repeat 1 at the 5′ end of the HSE, like that observed for wild-type HSF1. These results suggest the HSF1 loop, encompassing amino acids 83 to 98 but not regions amino-terminal of α-helix 3, is a critical determinant for the specific pattern of binding to the extended arrays of HSE that are present in the hsp70 promoter. Attempts to introduce specific point mutations or small clusters of mutations in the HSF2 loop did not alter DNA-binding specificity (data not shown). Thus it is likely that sequences comprising the entire HSF1 loop are necessary for extended occupancy of the hsp70 HSE. The footprint results summarized in Figure 3B show that the loop chimera (2Loop1) is the minimal domain that interconverts the pattern of DNA binding between HSF1 and HSF2.
Figure 3.
Extended occupancy of the hsp70 heat shock element (HSE) requires the HSF1 loop. (A) DNase I footprinting by chimeric heat shock transcription factor (HSF) proteins to the hsp70 HSE. HSF2Helix1 (mHSF2 in which aa 18–43 were substituted with aa 26– 51 from mHSF1) and HSF2Loop1 (mHSF2 aa 73–91 were substituted with aa 81–99 from mHSF1) were purified and used for DNase I footprint experiments, as described in Figure 2. The positions of the HSEs are shown by arrows. (B) Sequence of the hsp70 HSE with the five inverted HSE repeats are indicated with arrows. The boundaries of protection by HSF1 and HSF2 and the chimeras are indicated by the brackets.
The HSF1 loop imparts preferential activation of yeast HSP70
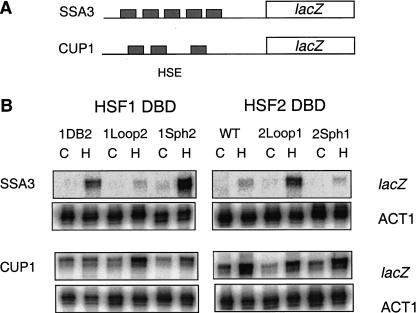
To ascertain whether the loop could alter selective target gene activation by HSF in vivo, we expressed mouse HSF1, HSF2, and specific chimeras in yeast lacking the endogenous yeast HSF gene and assayed for inducible activation of two HSE-lacZ reporter genes. As shown previously, two yeast HSF target genes, CUP1 encoding copper metallothionein and SSA3 encoding a member of the cytosolic Hsp70 chaperone family, can be activated in yeast by mammalian HSFs in response to heat shock (Liu et al. 1997; Liu and Thiele 1999). The CUP1 promoter contains a short, gapped HSE (Fig. 4A); whereas the SSA3 HSE is composed of five contiguous, inverted repeats of the pentameric recognition sequence similar to the mammalian hsp70 HSE arrangement. Consistent with previous results with human HSF1 and HSF2, mouse HSF1 activated the SSA3–lacZ reporter gene more strongly than the CUP1–lacZ fusion gene, whereas HSF2 failed to significantly activate the SSA–lacZ reporter (Fig. 4B). Chimeras in which HSF1–HSF2 fusions were made at a conserved SphI site within the trimerization domain followed the activation pattern according to the DBD, in agreement with DNA-binding specificity in vitro for these chimeras (Kroeger and Morimoto 1994). When sequences encoding the HSF2 loop were replaced by the corresponding sequence for the HSF1 loop, this HSF2Loop1 fusion protein activated the SSA3–lacZ reporter, as well as proteins containing the complete HSF1 DBD (HSF2DB1, HSF1Sph2). Thus the HSF1 loop is sufficient, when placed in the context of the full-length HSF2, for dictating the selective activation from the SSA3 promoter. In contrast, the HSF1Loop2 chimera, like HSF2, activated the CUP1 reporter in response to stress but did not significantly activate the SSA3 promoter. These data show that the loop of HSF1 is a critical determinant for selective activation from extended HSEs and is sufficient for interchanging the target gene specificity between mammalian HSF1 and HSF2 in this yeast transcription assay.
Figure 4.
The HSF1 loop dictates preferential activation from an extended (heat shock element) HSE in yeast. (A) Cartoon illustration of the arrangement of HSE repeats in the SSA3 and CUP1 gene promoters. (B) RNA levels for lacZ or ACT1 were measured by RNase protection assays using RNA isolated from control (C) or heat-shocked (HS; 40°C for 15 min) yeast expressing the indicated mouse heat shock transcription factor (HSF). The abbreviations for the HSF chimeras are identical to those described in the Figure 1 legend.
The HSF1 loop restrains activity of the inactive monomer
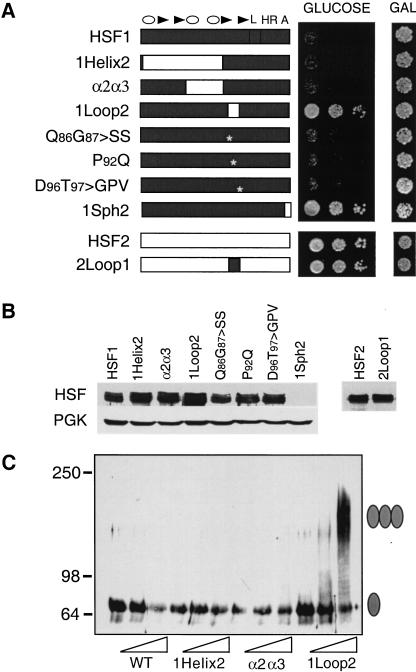
An interesting observation arose from the studies on target gene activation by mouse HSFs in yeast that suggested a second function for the loop. As we have shown previously, HSF1 fails to support growth of hsf1Δ yeast (Liu et al. 1997; Liu and Thiele 1999); however, a chimera in which the HSF1 loop was replaced by sequences from the HSF2 loop (HSF1Loop2) complemented the viability defect of hsfΔ yeast (Fig. 5A). This effect was specific because several other DBD chimeras between HSF1 and HSF2 all failed to functionally replace the endogenous yeast HSF protein. Expression of HSF1 proteins harboring small clustered point mutations within the loop also failed to support yeast growth. The inability of these other chimeras to complement was not due to a lack of protein expression as immunoblot analysis revealed approximately equivalent levels of fusion proteins (Fig. 5B). Human HSF1 fails to support growth of yeast cells because it is unable to convert from the inactive monomer to an active trimeric form;whereas HSF1 derivatives that could complement the viability defect all are capable of trimerization (Liu et al. 1997; Liu and Thiele 1999). We reasoned that the HSF1Loop2 might also trimerize constitutively in yeast and performed EGS cross-linking with extracts prepared from yeast expressing wild-type HSF1, HSF1Helix2, and HSF1Loop2 to test this hypothesis. As shown in Figure 5C, only extracts from yeast expressing HSF1Loop2 showed high molecular weight bands corresponding to the trimeric forms of HSF. Neither HSF1 nor the HSF1Helix2 chimera showed significant oligomerization in the extracts, consistent with their inability to function in yeast cells. These data suggest that the HSF1 loop may modulate activation of the HSF1 monomer under nonstress conditions by suppressing trimer formation.
Figure 5.
The HSF1 loop contributes to suppression of HSF1 activity in yeast. (A) Complementation assay for the ability of mouse HSF1, HSF2, and chimeras to substitute for yeast (heat-shock transcription factor) HSF. Cells harboring a chromosomal disruption of yeast hsf1 and bearing a plasmid with (heat stress transcription factor) HSF under the control of the GAL1 promoter were transformed with another plasmid expressing the indicated mouse HSF from a constitutive promoter. Transformants were diluted serially (from left to right, OD650, 0.1, 0.01, 0.001) and spotted onto glucose- or galactose-containing medium, incubated for 3 d at 30°C and photographed. (B) Western analysis for expression of mouse HSF isoforms in yeast using antibodies specific to HSF1 or HSF2. Levels of PGK are shown for normalization of loading. The 1Sph2 isoform is not detected with the HSF1 antibody because the primary epitopes recognized by this antibody map to the carboxy-terminal regions of the protein, which have been replaced by HSF2 in the chimera. (C) Crosslinking of HSF reveals that the HSF1 loop suppresses trimerization. Whole-cell extracts were prepared from yeast cells expressing the indicated forms of mouse HSF and subjected to in vitro cross-linking with 0, 0.5, or 2 mM EGS (indicated by wedge). HSF proteins were detected by immunoblotting with HSF1-specific antibody. The monomeric and trimeric HSF species are depicted with a single or triple oval, respectively.
Temperature-dependent trimerization of HSF in vitro depends on the HSF1 loop
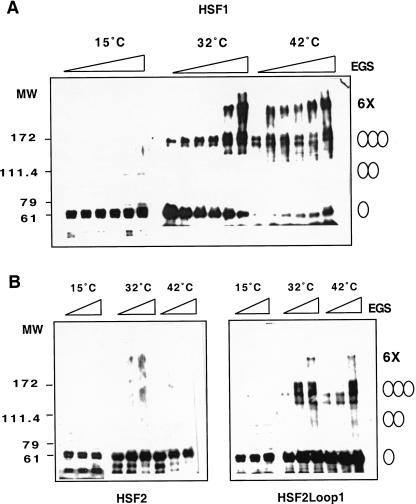
Because the HSF1 loop contributes to the monomer to trimer transition in yeast, we asked whether the loop might also be directly involved in the heat-inducible monomer-to-trimer conversion. We purified HSF1, HSF2 and HSF2loop1 proteins from Escherichia coli to >95% purity as ascertained by coomassie blue staining of SDS-PAGE (data not shown) and determined the oligomerization states of each isoform in vitro at 15°C, 32°C, and 42°C by EGS cross-linking, SDS-PAGE, and immunoblotting. As shown in Figure 6A, purified HSF1 was mostly monomeric at low temperature (15°C); however, incubation at 32°C or 42°C resulted in the formation of higher molecular weight species that correlated with the predicted size of the dimer and trimer upon titration with EGS. An additional band of lower mobility, estimated to be comprised of six HSF1 monomers, was detected at high concentrations of EGS at 32°C and at virtually all EGS doses at 42°C. In contrast, HSF2 was detected as a monomer at all temperatures and EGS concentrations (Fig. 6B). The HSF2loop1 chimera, like HSF1, was monomeric at 15°C but could be inducibly trimerized upon shift to 32°C and 42°C (Fig. 6B). These experiments were performed using purified HSF proteins, indicating that HSF1 is capable of sensing heat stress directly, consistent with previous studies (Goodson and Sarge 1995; Farkas et al. 1998; Zhong et al. 1998); significantly, the HSF1 loop is sufficient to transpose this thermal-sensing ability to HSF2.
Figure 6.
The HSF1 loop confers heat shock inducible multimerization in vitro. (A) Temperature-dependent trimerization of HSF1. Purified HSF1 (4 μM) was incubated at 15°C, 32°C, or 42°C for 30 min and then subjected to EGS cross-linking (EGS concentration indicated by the wedge 0, 0.1, 0.2, 0.5, 1.0, 2.5 mM from left to right) and detection by Western blotting using a HSF1-specific antibody. The predicted monomer, dimer, trimer, and putative hexamers are indicated by the ovals. (B) The HSF1 loop confers heat inducible trimerization to HSF2. Purified HSF2 and HSF2loop1 proteins, incubated at 15°C, 32°C, or 42°C, were subjected to EGS cross-linking (0, 0.5, 2.5 mM left to right) and analyzed as in A.
The HSF1 loop is required for heat inducible HSP70 expression and protection from heat-induced apoptosis in mouse cells
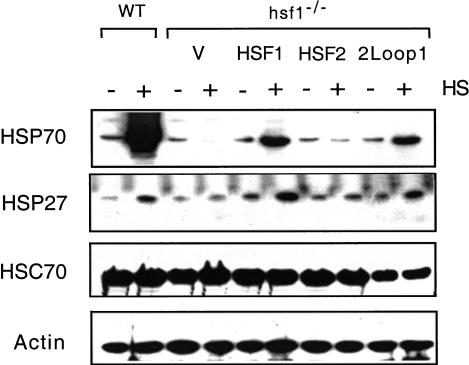
To understand the potential role of the HSF1 loop in stress-inducible activation and target gene selectivity in mouse cells, we transfected hsf1−/− mouse embryonic fibroblasts (MEFs) with plasmids expressing HSF1, HSF2, or the loop chimeras and assayed for steady-state protein levels of several HSF target genes. In agreement with previous results, hsf1−/− MEFs fail to accumulate HSP70 protein following heat shock (McMillan et al. 1998); whereas MEFs from wild-type mice give robust induction of HSP70, indicating an absolute dependence upon HSF1 for heat-inducible transcription of the hsp70 gene (Fig. 7). Accordingly, reintroduction of HSF1, but not HSF2 or vector alone, into the knockout cells restored heat-inducible expression of HSP70. Transfection with the chimera of HSF2 harboring the loop from HSF1 (HSF2Loop1) also led to heat-inducible accumulation of HSP70. The level of HSP70 induction by HSF2Loop1 was reproducibly lower compared to wild-type HSF1 and may reflect either a weaker activation domain (data not shown) or less responsiveness by this chimera to heat shock. Thus consistent with the in vitro trimerization data, the HSF1 loop when transposed into HSF2 can confer heat-induced target gene activation in cells. A nearly identical pattern of expression was observed for HSP27, a stress-inducible member of the small HSP family, suggesting that the loop is important for expression of at least a small group of HSPs. Immunoblot analysis using antibodies specific to HSF1 or HSF2 revealed that the chimeric proteins were expressed at comparable levels to ectopically expressed wild-type proteins (data not shown). These data show that the HSF1 loop is required for robust activation of hsp70 and hsp27 in vivo and is sufficient, when transplanted into HSF2, to allow heat-inducible activation from a promoter harboring an extended HSE arrangment.
Figure 7.
The HSF1 loop confers heat-inducible activation of Hsp gene expression in hsf1−/− mouse embryonic fibroblast (MEF) cells. Immunoblot analyses for levels of HSP70, HSP27, HSC70, and actin were performed on extracts prepared from control (C) or heat-shocked (HS; 42°C for 1 h, then at 37°C for 3 h ) hsf1−/− MEF cells transfected with pcDNA3.1 (V) or vector containing the indicated mouse HSF isoform. Extracts from isogenic wild-type (WT) MEFs are shown for comparison.
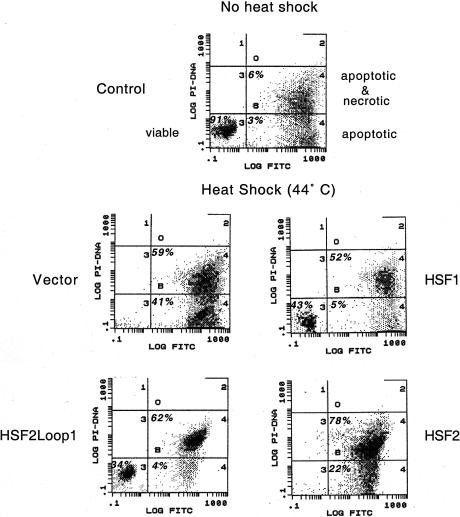
The heat shock response and HSPs protect cells from injury and death during cellular stress. One consequence of disrupting the mouse hsf1 gene is an increase in heat-induced apoptosis because these cells fail to accumulate anti-apoptotic Hsps, such as Hsp70 and Hsp27 (McMillan et al. 1998). As the HSF2Loop1 chimera has the DNA-binding and target activation properties of HSF1, we tested whether HSF2Loop1 could physiologically compensate for loss of HSF1 in the protection of hsf1−/− fibroblasts from heat-induced apoptosis. hsf1−/− MEFs transfected with vector alone or containing HSF1, HSF2, or the HSF2Loop1 chimera were exposed to a 44°C heat shock and assessed for viability by annexin V–FITC binding. To quantify this protection from heat stress, we analyzed cells by flow cytometry and scored them for annexinV and propidium iodide staining. As shown in Figure 8, hsf1−/− MEFs transfected with HSF1 and HSF2Loop1 had 43% and 34% viable cells, respectively; whereas vector and HSF2-transfected cell populations underwent nearly quantitative cell death in response to heat stress. Likewise replacing the HSF1 loop with the loop from HSF2 (HSF1loop2) also failed to protect transfected hsf1 knockout cells from heat-induced apoptosis (data not shown). Thus the HSF1 loop is not only a signature for HSF1 in recognizing preferred HSEs in target genes like hsp70, but plays a physiologically relevant role in the response of the molecule to stress and protection of cells from heat-induced death.
Figure 8.
Expression of HSF2Loop1 in hsf1−/− cells prevents heat-induced apoptosis. hsf1−/− mouse embryonic fibroblasts (MEFs) were transfected with pcDNA3 alone (Vector) or harboring the indicated HSF sequence. Cells were exposed for 1 h to 44°C or kept at 37°C (control) and then cultured for another 24 h at 37°C prior to detection of apoptosis by flow cytometry of annexin V-FITC and propidium iodide (PI)-stained cells. The percentage of cells for each population of viable, apoptotic or apoptotic and necrotic, falling within each quadrant is indicated. Viable cells sort to the lower left quadrant.
Discussion
In mammals, three nonredundant HSF family members have been identified. The physiological and environmental signals that activate HSF2 and HSF4, as well as the respective roles of all three HSFs to developmental pathways, remain to be elucidated; however, detailed biochemical analyses of these proteins have revealed interesting and significant differences with respect to their activation and DNA binding characteristics. Previous genetic analyses of hsf1−/− cells established that HSF1 is the only heat-inducible HSF in mice, and in vitro biochemical studies have shown differences between HSF1 and HSF2 in the degree of cooperative DNA binding on the hsp70 promoter (Kroeger et al. 1993; Kroeger and Morimoto 1994). In this study, we have mapped a multifunctional region of the HSF1 DNA-binding domain, the loop, that allows discrimination between different HSE–HSF interactions and acts as a part of the sensing and regulatory mechanism for responding to thermal stress. The HSF1-specific DNA-binding property can be transposed to HSF2 by creating a hybrid of HSF2 with the loop of HSF1, and significantly, the HSF2Loop1 chimera is capable of activating transcription of an HSP70–lacZ reporter in yeast and the endogenous inducible hsp70 gene in mouse cells. Based on previous data that HSF1 binds to HSEs with greater cooperativity than HSF2 does (Kroeger et al. 1993; Kroeger and Morimoto 1994), we propose that the HSF1 loop facilitates interactions between HSF1 trimers, thereby promoting its binding to extended HSEs. Such cooperative DNA binding has also been established for lower eukaryotic HSFs from yeast and fly (Xiao et al. 1991). The HSF2 loop seems much less capable of promoting such cooperativity between trimers, as evidenced by its smaller footprint over the hsp70 HSE and weaker activation of the yeast HSP70–lacZ reporter. Therefore the loop domain may be a critical determinant in allowing HSF1 and HSF2 to activate target genes selectively based on the arrangement of the HSE.
The idea that different arrangements of response elements for a transciption factor may influence the pattern of gene regulation was proposed to explain how the glucocorticoid receptor could function as both a transcriptional activator and repressor depending upon the promoter context (Lefstin and Yamamoto 1998). The allosteric control hypothesis posited that distinct DNA recognition sequences influence the activity of a transcriptional activator by dictating the conformation of the DNA-binding protein bound to the site, which in turn specifies the protein surfaces available for interactions with other transcription factors. Similarly differences in the orientation of the HSE have been reported to promote distinct binding by yeast HSF. An arrangement of two repeats of the tail–tail sequence (nTTCnnGAAn) was found to bind two HSF trimers much more efficiently than two repeats of the head–head orientation (nGAAnnTTCn; Bonner et al. 1994). Furthermore, a comparison of yeast HSF binding to the gapped CUP1 promoter and a continuous HSE by proteolytic cleavage assays revealed differences in the sensitivity of HSF to the protease (Santoro et al. 1998). This difference in conformation was suggested to be one factor in determining the temperature threshold of HSF target gene activation. Although these studies show that yeast HSF is capable of adapting to different HSEs, our analysis of HSF1 and HSF2 suggest that the mammalian proteins exhibit more defined preferences for DNA binding and target gene activation. Although both proteins bind to the same core HSE sequences, differential binding and transactivation from shorter versus longer arrays of the pentameric HSE repeats by HSF2 and HSF1, respectively, would provide a basis for differential target gene activation in mammals. Interactions with other cis- and trans-acting factors may influence the protein–DNA interactions at some promoters. Although these data have addressed the molecular basis for discrimination for HSEs by HSF1 and HSF2, there may be overlapping targets for HSF1 and HSF2. For example, hsp70 can be activated in an HSF2-dependent manner during hemin-induced differentiation of K562 cells (Sistonen et al. 1992). However, it was observed that HSF1 and HSF2 differentially occupied the hsp70 HSE and activated the gene to significantly different levels in response to the heat shock and hemin, respectively. Thus, we propose that distinctions in the arrangement of the HSEs will provide an additional layer of selectivity among HSF-dependent target gene expression, especially under limiting levels of HSF1 and HSF2 protein concentrations, and may also modulate transcriptional potency. A full characterization of HSF2-preferred HSEs in vivo must await the identification of bona fide HSF2 target genes.
A question that is raised is how might the loop facilitate cooperative interactions. A potential answer may be found from the cocrystal structure of two K. lactis HSF DBDs bound to two inverted HSE repeats (Littlefield and Nelson 1999). Although disordered in that structural model, the loop was predicted to lie in the interface between the two DNA-bound HSF DBDs. The loop of one DBD was thought to be juxatoposed against the turn of the helix-turn-helix motif of an adjacent DBD. Although not encompassed by the structure, the loop-and-turn interface was also predicted to bridge interactions between adjacent trimers bound to an extended HSE. Thus the loop may comprise part of a critical protein–protein interaction surface that is responsible for stabilizing contacts between individual DBDs within a DNA-bound trimer and between adjacent trimers.
Even more striking, we find that the HSF1 loop is sufficient for conferring heat-inducible trimerization to purified HSF2 in vitro when transposed into this isoform, suggesting that the loop is part of the critical heat-sensing mechanism that distinguishes HSF1. We also show a physiological significance for the HSF1 loop in cells because the HSF2Loop1 protein, like HSF1 but not HSF2, is capable of rescuing transfected hsf1−/− MEFs from heat-induced apoptosis. Therefore, we propose that the HSF1 loop modulates the monomer-to-trimer transition in response to heat stress in vitro and in cells in addition to facilitating cooperative binding on extended HSEs. A potential model is that the loop undergoes a conformational change upon heat stress, which relieves intramolecular repression of the monomer and exposes the trimerization domain. Direct thermal sensing by HSF1 is reminiscent of the temperature-sensitive monomer to coiled–coil dimerization of the Salmonella DNA-binding protein TlpA (Hurme et al. 1997). A notable difference, however, is that HSF is multimerized and activated upon heat shock; whereas TlpA becomes unfolded and inactivated upon shift to elevated temperatures. Intriguingly the yeast HSF DBD was recently shown to affect transcriptional activation in vivo through a proposed mechanism involving changes in the structure of the DBD upon heat stress (Hardy et al. 2000). Yeast strains expressing HSF mutants that removed a conserved helical bulge in α-helix 2 of HSF exhibited improved survival at nonpermissive temperatures and increased thermotolerance against lethal heat shock, which correlated with higher basal levels of transcriptional activity, compared with cells expressing wild-type HSF. Using a similar yeast assay, we found that substitution of the HSF2 loop sequence into HSF1 resulted in constitutive trimerization and the ability to complement the viability defect of yeast lacking yeast HSF in contrast to HSF1. Together these data further support the idea that small structural changes within the DBD, either through mutagenesis or thermally induced conformational changes, can effect transitions from the inactive to the active states. These data are consistent with the idea that activation of HSF1 by heat proceeds through distinct steps involving conformational changes that unleash the fully activated molecule. We envision that in the HSF1 monomer the loop interacts with another region of HSF1 to repress the inactive state. Upon thermal stress, the loop or its interacting domain may undergo local conformational changes that allow oligomerization of HSF1 monomers. In the activated trimer state, the HSF1 loop promotes DNA-binding specificity by facilitating interactions between other HSF1 molecules to allow greater binding to and preferential gene expression from extended HSEs. In conclusion, a small flexible loop domain within the HSF1 DBD plays multiple roles in the regulation of HSF1 activation in response to heat stress and in the promotion of cooperative DNA binding, two mechanisms that may allow for functional specialization between similar transcription factors.
Materials and methods
Construction of chimeric HSFs
Mouse HSF1 and HSF2 were subcloned into plasmid pET21b (Novagen) to generate pHSF1-His and pHSF2-His, which have a 6-His epitope tag immediately preceding the stop codon, and DNA-binding domain chimeras were created by the overlapping extension recombinant PCR method (McClintock and Lerner 1997). The presence of desired substitutions and the absence of PCR-generated random mutations were confirmed by sequencing the entire DBD of the proteins. All HSF proteins were expressed in BL21-DE3 bacteria with isopropyl-β-D-thiogalactopyranoside induction. For DNA-binding studies, HSFs were purified from cells grown at 37°C, in which all HSFs are constitutively trimerized. For in vitro trimerization studies, HSFs were purified from cells grown at 20°C, conditions under which both HSF1 and HSF2Loop1 are present as monomers. Whole-cell extracts of induced bacteria were prepared as described previously (Studier et al. 1990). The amount of expressed HSF proteins affinity purified from E. coli extracts was estimated by Western blot analysis with anti-His6 specific antibodies (Novagen), and the volume of each extract was then adjusted with lysis buffer so that all extracts would contain the same relative amount of HSFs.
Analysis of DNA–protein interactions
A native gel shift assay was performed as described previously (Mosser et al. 1990) with a self-complimentary oligonucleotide harboring four consensus repeats of the HSE (5′-CTAGAA GCTTCTAGAAGCTTCTAG-3′). Equal amounts of HSF proteins were used for DNA-binding assays.
DNase I footprinting was done as described previously (Kroeger and Morimoto 1994), except that DNase I digestion was stopped by adding 55 μL of saturated NH4Ac. The HSP70 HSE oligonucleotide probe for footprinting studies was prepared as described, except that the single-end-labeled PCR product was gel purified. Typically, 1 μL of DNA (10,000–20,000 cpm) was used for each DNA-binding reaction. To confirm that the His-tag on the carboxyl terminus did not affect the DNA-binding properties of the proteins, HSF1, HSF2, HSF1DB2, and HSF2DB1 were subcloned into the pET3 plasmid (Novagen) to yield constructs that did not carry His-tag. DNA-binding analysis of these proteins resulted in footprints identical to those of His-tagged proteins (data not shown).
In vitro EGS cross-linking
His-tagged HSF proteins were affinity purified from E. coli extracts using Ni-agarose chromatography and the purity of the proteins determined by Coomassie blue staining of SDS-polyacrylamide gels. For cross-linking experiments, purified HSF proteins were diluted into phosphate-buffered saline to a final concentration of 4 μM and incubated for 30 min in 15°C, 32°C, or 42°C water baths. EGS (ethylene glycol bis[succinimidyl succinate]) was added to the desired final concentration from a 25 mM stock in DMSO and incubated for 30 min at 22°C and quenched with excess glycine. The samples were resolved by electrophoresis through a 6% SDS-polyacrylamide gel and analyzed by immunoblotting with polyclonal antibodies specific to HSF1 or HSF2 (Sarge et al.1991).
Yeast strains, growth conditions, and biochemical techniques
Yeast expression plasmids p424GPDHSF1 and p424GPDHSF2 harboring the complete mHSF1 and mHSF2 ORFs, respectively, under the control of constitutive yeast promoters were described previously (Liu et al. 1997). HSF chimeras were directly subcloned into these vectors. The recipient strain for all experiments was the W303 derivative PS145 (MATaade2-1 trp1 can1-100 leu2,3–112 his3-11,15 ura3 hsf::LEU2 YCpGAL1-yHSF) (Sorger and Pelham 1988). Plasmids harboring mHSFs were transformed according to the PEG-lithium acetate procedure and tested for the ability to complement hsfΔ cells as described (Liu et al. 1997). Cells were routinely grown at 30°C in selective synthetic complete medium and heat shocked for 15 min by submersing tubes into a shaking water bath equilibrated to 40°C.
Yeast cell extracts were prepared by alkaline lysis of cells in the presence of protease inhibitors, followed by TCA precipitation (Ooi et al. 1996). Immunoblotting was performed as described using antibodies specific to HSF1 (monoclonal antibody 10H8; Cotto et al. 1997) or HSF2. Procedures for EGS cross-linking, immunoblotting, and RNase protection assays have been described previously (Liu et al. 1997).
Mammalian tissue culture, transfection, and analysis
Mouse embryonic fibroblasts (MEFs) derived from hsf1−/− mice and congenic wild-type animals were a gift from Ivor Benjamin (University of Texas Southwestern Medical Center, Dallas) and cultured according to standard procedures (McMillan et al. 1998). MEF cells were transfected by electroportation using conditions described previously (Pirkkala et al. 2000) with only minor modifications: Specifically, 2 × 105 fibroblasts were transfected with 20 μg of plasmid DNA and used after 24-h recovery in complete medium. Heat shock treatments were performed by submerging sealed culture dishes into a water bath equilibrated to 42°C. Whole-cell lysates were prepared as described and resolved by denaturing SDS-PAGE, followed by Western blotting. Specific proteins were detected using the following antibodies: inducible HSP70 with anti-HSP70 (sc-24, Santa Cruz Biotechnology), HSP27 (sc-1049), HSC70 with rat anti-HSC70 antibody (StressGen SPA-815), and actin with anti-actin peptide antibody (C-11, Santa Cruz Biotechnology).
Apoptosis detection
Cell viability and apoptosis was assayed using annexin V-fluorescein and propidium iodide (Annexin-V-FLUOS kit, Roche) and visualized using a Nikon Eclipse E800 automated fluorescent microscope equipped with a digital camera or analyzed by flow cytometry. MEF cells were transfected by electroporation as described above and allowed to recover for 24 h. The plates were immersed in for 1 h in a 44°C water bath and then returned to 37°C 24 h prior to determination of cell death.
Acknowledgments
We thank Ivor Benjamin for providing the hsf1−/− MEF cells and Hillary Nelson for yeast strain PS145. We thank Lea Sistonen for advice with electroporation of MEFs, Jeanne Stuckey for assistance with the Ribbons software program for drawing the 3-D HSF DBD structure, and members of the Thiele and Morimoto laboratories, especially Sue Fox and Anu Mathew for help and useful discussions and Chen Kuang for technical support. P.C.C.L. and K.K. were supported by NRSA postdoctoral fellowships GM18858 and ES07284, respectively, from the National Institutes of Health. S.-G.A. was supported by Postdoctoral Fellowship 0120446Z from the American Heart Association. This work was supported by grants GM59911 to D.J.T. and GM38109 to R.M from the NIH and the Daniel F. and Ada L. Rice Institute for Biomedical Research and the Carol and Martin Gollob Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dthiele@umich.edu; FAX (734) 763-7799.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.894801.
References
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes & Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol Cell Biol. 1998;18:4949–4960. doi: 10.1128/mcb.18.9.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj S, Ali A, Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol Cell Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Ballou C, Fackenthal DL. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–508. doi: 10.1128/mcb.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. Maternal effect of Hsf1 on reproductive success. Nature. 2000;407:693–694. doi: 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- Cotto J, Fox S, Morimoto R. HSF1 granules: A novel stress-induced nuclear compartment of human cells. J Cell Sci. 1997;110:2925–2934. doi: 10.1242/jcs.110.23.2925. [DOI] [PubMed] [Google Scholar]
- Craig EA, Baxter BK, Becker J, Halladay J, Ziegelhoffer T. Cytosolic hsp70s of Saccharomyces cerevisiae: Roles in protein synthesis, protein translocation, proteolysis, and regulation. In: Morimoto RI, et al., editors. The biology of heat shock proteins and molecular chaperones. Vol. 26. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 31–52. [Google Scholar]
- Damberger FF, Pelton JG, Liu C, Cho H, Harrison CJ, Nelson HC, Wemmer DE. Refined solution structure and dynamics of the DNA-binding domain of the heat shock factor from Kluyveromyces lactis. J Mol Biol. 1995;254:704–719. doi: 10.1006/jmbi.1995.0649. [DOI] [PubMed] [Google Scholar]
- Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974. [DOI] [PubMed] [Google Scholar]
- Farkas T, Kutskov YA, Zimarino V. Intramolecular repression of mouse heat shock factor 1. Mol Cell Biol. 1998;18:906–918. doi: 10.1128/mcb.18.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Sarge KD. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Walsh ST, Nelson HC. Role of an alpha-helical bulge in the yeast heat shock transcription factor. J Mol Biol. 2000;295:393–409. doi: 10.1006/jmbi.1999.3357. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- Hurme R, Berndt KD, Normark SJ, Rhen M. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90:55–64. doi: 10.1016/s0092-8674(00)80313-x. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Morimoto RI. Selection of new HSF1 and HSF2 DNA-binding sites reveals differences in trimer cooperativity. Mol Cell Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger PE, Sarge KD, Morimoto RI. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Lis J, Wu C. Protein traffic on the heat shock promoter: Parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- Littlefield O, Nelson HC. A new use for the 'wing' of the 'winged' helix-turn-helix motif in the HSF–DNA cocrystal. Nat Struct Biol. 1999;6:464–470. doi: 10.1038/8269. [DOI] [PubMed] [Google Scholar]
- Liu PCC, Thiele DJ. Modulation of human heat shock factor trimerization by the linker domain. J Biol Chem. 1999;274:17219–17225. doi: 10.1074/jbc.274.24.17219. [DOI] [PubMed] [Google Scholar]
- Liu XD, Liu PCC, Santoro N, Thiele DJ. Conservation of a stress response: Human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 1997;16:6466–6477. doi: 10.1093/emboj/16.21.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Lerner MR. Functional analysis by imaging of melanophore pigment dispersion of chimeric receptors constructed by recombinant polymerase chain reaction. Brain Res Protoc. 1997;2:59–68. doi: 10.1016/s1385-299x(97)00030-5. [DOI] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expr. 1999;7:271–282. [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes & Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Tissieres A, Georgopoulos C. Cold Spring Harbor Monograph Series. Vol. 26. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. The biology of the heat shock proteins and molecular chaperones; p. 610. [Google Scholar]
- Mosser DD, Kotzbauer PT, Sarge KD, Morimoto RI. In vitro activation of heat shock transcription factor DNA-binding by calcium and biochemical conditions that affect protein conformation. Proc Natl Acad Sci. 1990;87:3748–3752. doi: 10.1073/pnas.87.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB. The Hsf world: Classification and properties of plant heat stress transcription factors. Cell Stress Chaperones. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CE, Rabinovich E, Dancis A, Bonifacino JS, Klausner RD. Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 1996;15:3515–3523. [PMC free article] [PubMed] [Google Scholar]
- Orosz A, Wisniewski J, Wu C. Regulation of Drosophila heat shock factor trimerization: Global sequence requirements and independence of nuclear localization. Mol Cell Biol. 1996;16:7018–7030. doi: 10.1128/mcb.16.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Peteranderl R, Nelson HC. Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled–coil. Biochemistry. 1992;31:12272–12276. doi: 10.1021/bi00163a042. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20:2670–2675. doi: 10.1128/mcb.20.8.2670-2675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol. 1998;18:6340–6352. doi: 10.1128/mcb.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Zimarino V, Holm K, Wu C, Morimoto RI. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes & Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes & Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Nelson HC. Trimerization of a yeast transcriptional activator via a coiled–coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Voellmy R. Transduction of the stress signal and mechanisms of transcriptional regulation of heat shock/stress protein gene expression in higher eukaryotes. Crit Rev Eukaryot Gene Expr. 1994;4:357–401. [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994a;1:605–614. [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Wu C, Bax A. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix-turn-helix and HNF-3/forkhead families of transcription factors. Biochemistry. 1994b;33:10–16. doi: 10.1021/bi00167a002. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G, Seto D, Parker CS. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell. 1988;54:841–853. doi: 10.1016/s0092-8674(88)91197-x. [DOI] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: Structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xiao H, Perisic O, Lis JT. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- Zimarino V, Wu C. Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. Nature. 1987;327:727–730. doi: 10.1038/327727a0. [DOI] [PubMed] [Google Scholar]
- Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled–coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]