Abstract
The conserved multifunctional Paf1 complex is important for the proper transcription of numerous genes, and yet the exact mechanisms by which it controls gene expression remain unclear. While previous studies indicate that the Paf1 complex is a positive regulator of transcription, the repression of many genes also requires the Paf1 complex. In this study we used ARG1 as a model gene to study transcriptional repression by the Paf1 complex in Saccharomyces cerevisiae. We found that several members of the Paf1 complex contribute to ARG1 repression and that the complex localizes to the ARG1 promoter and coding region in repressing conditions, which is consistent with a direct repressive function. Furthermore, Paf1 complex-dependent histone modifications are enriched at the ARG1 locus in repressing conditions, and histone H3 lysine 4 methylation contributes to ARG1 repression. Consistent with previous reports, histone H2B monoubiquitylation, the mark upstream of histone H3 lysine 4 methylation, is also important for ARG1 repression. To begin to identify the mechanistic basis for Paf1 complex-mediated repression of ARG1, we focused on the Rtf1 subunit of the complex. Through an analysis of RTF1 mutations that abrogate known Rtf1 activities, we found that Rtf1 mediates ARG1 repression primarily by facilitating histone modifications. Other members of the Paf1 complex, such as Paf1, appear to repress ARG1 through additional mechanisms. Together, our results suggest that Rtf1-dependent histone H2B ubiquitylation and H3 K4 methylation repress ARG1 expression and that histone modifications normally associated with active transcription can occur at repressed loci and contribute to transcriptional repression.
INTRODUCTION
The organization of eukaryotic DNA into chromatin presents a significant obstacle to transcription by RNA polymerase II (Pol II). To allow proper gene expression, a multitude of accessory factors associate with RNA Pol II to facilitate the transcription of a chromatin template. A conserved, multifunctional protein complex that enables proper RNA Pol II transcription is the Paf1 complex. In Saccharomyces cerevisiae, the Paf1 complex consists of Paf1, Ctr9, Cdc73, Rtf1, and Leo1 (36, 45, 64, 66). Many physical and genetic interactions and phenotypes implicate the Paf1 complex in regulating the elongation stage of transcription. Specifically, strains lacking Paf1 complex members exhibit phenotypes associated with transcription elongation defects, such as sensitivity to 6-azauracil and mycophenolic acid (12, 66). During transcription elongation, the Paf1 complex associates with RNA Pol II on open reading frames (ORFs) (36, 54), where it orchestrates modifications to the chromatin template (11, 35, 49, 50, 78) and influences the phosphorylation state of the RNA Pol II carboxy-terminal domain (CTD) (46, 51). In addition, the Paf1 complex genetically and physically interacts with elongation factors such as the Spt4-Spt5 (yDSIF) and Spt16-Pob3 (yFACT) complexes, suggesting coordinated functions of these elongation factors during transcription (12, 36, 66).
Appropriate transcription by RNA Pol II depends on the dynamic regulation of chromatin structure, which is modulated by histone modifications. Members of the Paf1 complex are required for the establishment of several histone modifications that are associated with active genes. Specifically, Paf1 and Ctr9 are required for histone H3 lysine (K) 36 trimethylation by the histone methyltransferase Set2 (11), and Paf1 and Rtf1 are needed for methylation of histone H3 K4 and K79 by the histone methyltransferases Set1 and Dot1, respectively (35, 49, 50). Di- and trimethylation of histone H3 K4 and K79 is dependent on the monoubiquitylation of histone H2B K123 by the ubiquitin conjugating enzyme Rad6 and the ubiquitin ligase Bre1 (7, 68). Because Paf1 and Rtf1 are also required for histone H2B ubiquitylation, the Paf1 complex most likely regulates histone H3 K4 and K79 methylation indirectly through histone H2B ubiquitylation (49, 78). Both histone H3 K4 methylation and H2B ubiquitylation correlate with active transcription. These modifications are enriched on the coding regions of active genes (5, 60, 80), and the necessary histone-modifying enzymes are recruited to active genes in a Paf1 complex-dependent manner (35, 50, 80). Importantly, Rad6 and Bre1 are evolutionarily conserved, and the interconnections between the Paf1 complex, histone H2B ubiquitylation, and gene expression observed in yeast extend to other eukaryotes, including humans (87).
Due to its multiple roles during transcription elongation, it is not surprising that the Paf1 complex is required for the proper expression of many genes (53). However, it is unclear how the Paf1 complex regulates the expression of most genes or whether the complex uses similar mechanisms to effect gene activation and repression. To investigate the repressive function of the Paf1 complex, we focused on ARG1, a gene whose expression is negatively regulated by Paf1 (53) and whose cis- and trans-regulatory factors are well characterized. ARG1 transcription is repressed by the ArgR/Mcm1 complex in rich media (2, 4, 13, 14, 17, 19, 20, 55) and induced by Gcn4 in conditions of amino acid starvation (17, 24). Interestingly, although histone H2B ubiquitylation is generally associated with active transcription, this modification has been implicated in ARG1 repression. Deletion of RAD6, mutation of the Rad6 ubiquitin conjugation site, or mutation of histone H2B K123 results in derepression of an ARG1-lacZ reporter construct (73). Consistent with these observations, both gene-specific and genome-wide studies found increased ARG1 expression in htb1-K123R cells (38, 48, 84). Therefore, it is possible that the Paf1 complex may promote transcriptional activation and repression through the very same histone modifications. However, it is unknown whether the Paf1 complex or Paf1 complex-dependent modifications are enriched at repressed loci such as ARG1 or contribute to their repression. Therefore, we investigated the role of the Paf1 complex in transcriptional repression, with a particular focus on characterizing the contributions of Paf1 complex-dependent histone modifications to repression. Our results indicate that the Paf1 complex associates with and determines the histone modification state at ARG1 under repressing conditions, that Rtf1-dependent histone H2B ubiquitylation can both activate and repress transcription, and that Paf1 has roles in ARG1 repression beyond its known roles in facilitating histone modifications.
MATERIALS AND METHODS
Yeast strains and media.
Rich (yeast extract-peptone-dextrose [YPD]) and synthetic complete (SC) media were prepared as described previously (56). Yeast strains used in these studies are isogenic with FY2, a GAL2+ derivative of S288C, and listed in Table 1 (77). Because certain cellular auxotrophies influence the level of ARG1 repression (E. Crisucci and K. Arndt, unpublished observations), experiments were performed with prototrophic strains where possible. Mating types of prototrophic strains were assigned through visual examination of mating with MATa and MATα tester strains. Gene disruptions were created through PCR-mediated gene replacement via transformation and/or mating, sporulation, and tetrad dissection and confirmed by PCR or Southern analysis (3, 56). PCR fragments for gene replacement with KanMX were generated by amplification of the KanMX cassette on pRS400 (6). Strains containing an integrated copy of htb1-K123R as the only source of H2B were constructed and verified as described previously (72). Strains containing rtf1 internal deletion mutations were created through a two-step gene replacement method in which constructs encoding the N-terminally triple hemagglutinin (HA)-tagged Rtf1 derivatives were integrated to replace endogenous RTF1 (57). Comparisons between strains expressing HA-tagged and untagged Rtf1 derivatives revealed that the HA tag did not alter Rtf1 function or interfere with ARG1 repression. A yeast strain containing an integrated, tagged copy of RPB1, RPB1-13xMYC::KanMX, was constructed as previously described and generously provided by Joe Martens (22).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| KY1302 | MATα RPB1-13×MYC::KanMX |
| KY1698 | MATa |
| KY1699 | MATα |
| KY1700 | MATα paf1Δ::KanMX |
| KY1703 | MATartf1Δ::KanMX |
| KY1704 | MATα rtf1Δ::KanMX |
| KY1705 | MATactr9Δ::KanMX |
| KY1706 | MATα cdc73Δ::KanMX |
| KY1709 | MATα arg80Δ::KanMX |
| KY1711 | MATarad6Δ::KanMX |
| KY1713 | MATabre1Δ::KanMX |
| KY1714 | MATabre1Δ::KanMX ura3Δ0 |
| KY1715 | MATaset1Δ::KanMX |
| KY1716 | MATaset2Δ::KanMX |
| KY1717 | MATadot1Δ::KanMX |
| KY1721 | MATα 3×HA-PAF1 |
| KY1722 | MATa3×HA-rtf1Δ1 |
| KY1723 | MATa3×HA-rtf1Δ3 |
| KY1724 | MATa3×HA-rtf1Δ4 |
| KY1725 | MATα 3×HA-rtf1Δ7 |
| KY1726 | MATa3×HA-rtf1Δ12 |
| KY1727 | MATa3×HA-rtf1Δ13 |
| KY1731 | MATα HTA1-htb1-K123R (hta2-htb2)Δ::KanMX paf1Δ::KanMX |
| KY1732 | MATα HTA1-htb1-K123R (hta2-htb2)Δ::KanMX ura3Δ0 |
| KY1755 | MATα set1Δ::KanMX |
| KY1805 | MATα leo1Δ::KanMX |
| KY1821 | MATaset1Δ::KanMX set2Δ::KanMX |
| KY1826 | MATα set1Δ::KanMX dot1Δ::KanMX |
| KY1832 | MATα set2Δ::KanMX dot1Δ::KanMX |
| KY1847 | MATaset1Δ::KanMX set2Δ::KanMX dot1Δ::KanMX |
| KY1980 | MATa3×HA-rtf1Δ5 |
| KY1981 | MATa3×HA-rtf1-102-104A |
| KY1982 | MATα 3×HA-rtf1-E104K |
| KY1983 | MATa3×HA-rtf1-108-110A |
| KY1984 | MATa3×HA-rtf1-F80V,F123S |
| KY2074 | MATaHTA1-htb1-K123R (hta2-htb2)Δ::KanMX rtf1Δ::KanMX |
| KY2082 | MATα 3×HA-RTF1 leu2Δ1 trp1Δ63 lys2-128Δura3-52 |
Northern analysis.
Unless stated otherwise, 10 μg of total RNA, isolated from cells grown in YPD at 30°C to a density of 1 × 107 to 2 × 107 cells/ml, were subjected to Northern analysis with random-prime-labeled, PCR-amplified DNA probes for ARG1 (+34 to +1201), SNZ1 (+79 to +890), GAP1 (+133 to +1213), and SCR1 (−242 to +283) as described previously (70). Signals were quantified by using phosphorimager and ImageQuant software. ARG1 signals were normalized to the loading control SCR1. To facilitate comparisons between samples and avoid introducing errors from the very low ARG1 transcript levels in wild-type strains, normalized ARG1 transcript levels in experimental samples are presented relative to normalized ARG1 transcript levels in an arg80Δ control strain, which was processed in parallel. The normalized ARG1 transcript levels in arg80Δ samples (not shown) were set equal to 1. Relative signals for at least three independent samples were averaged and plotted with the standard deviation.
Western analysis.
Whole-cell extracts were prepared by a rapid boiling method as described previously (67). Briefly, cells were grown in YPD to a density of approximately 4 × 107 cells/ml. A 1.5-ml portion of culture was harvested by centrifugation and resuspended in 20-μl sample buffer (80 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 10% glycerol, 2 mM phenylmethylsulfonyl fluoride, and 0.1% bromophenol blue) and immediately boiled for 2 min at 100°C. After glass bead lysis, an additional 80-μl sample buffer was added, and 20 μl of this lysate was separated on an SDS–10% polyacrylamide gel. Membranes were probed with a 1:2,500 dilution of anti-HA antibody (Roche catalog no. 11666606001), followed by a 1:5,000 dilution of sheep anti-mouse horseradish peroxidase-coupled secondary antibody (GE Healthcare). As a loading control, membranes were probed with a 1:100,000 dilution of anti-glucose-6-phosphate dehydrogenase antibody (G6PDH; Sigma catalog no. A9521), followed by a 1:5,000 dilution of donkey anti-rabbit horseradish peroxidase-coupled secondary antibody (GE Healthcare).
Chromatin immunoprecipitation (ChIP) assays.
Cells were grown in YPD to a density of ∼107 cell/ml and harvested or washed and resuspended in minimal media and then incubated for an additional 30 min. Chromatin was prepared as described previously (65). Sonicated chromatin was incubated with antibodies at 4°C overnight. Agarose-conjugated anti-HA (Santa Cruz Biotechnology sc-7392 AC) or anti-MYC (Santa Cruz Biotechnology sc-40 AC) was used to precipitate HA-Paf1 or Rpb1-Myc, respectively. Polyclonal anti-Rtf1 antibody (66), anti-H3 trimethyl K36 (Abcam catalog no. ab9050), anti-H3 trimethyl K4 (Active Motif catalog no. 39159), anti-H3 dimethyl K4 (Millipore catalog no. 07-030), or anti-H3 (Abcam catalog no. ab1791), followed by incubation with protein A-coupled Sepharose beads (GE Healthcare catalog no. 17-5280-01), were used to precipitate Rtf1 or the appropriate histone protein. Precipitated DNA was purified by using PCR purification columns (Qiagen). For HA-Paf1 and Rtf1 ChIP assays, two dilutions of input and immunoprecipitated (IP) DNA from three independent chromatin preparations were amplified by PCR in the presence of [α-32P]dATP. PCR products were separated on 6% native polyacrylamide gels, and signals were quantified by using a phosphorimager and ImageQuant software. After signals were multiplied by their dilution factor, the average input was divided by the average IP signal. IP/input signals for ARG1 were normalized to a subtelomeric control region on chromosome VI (75). For ChIP assays examining histone modification levels, immunoprecipitated DNA from three independent chromatin preparations was used in quantitative real-time PCR with SYBR green detection (Fermentas). IP/input values for the histone modifications were normalized to those for total histone H3. Error bars represent the standard error of the mean.
RESULTS
Members of the Paf1 complex repress ARG1 transcription.
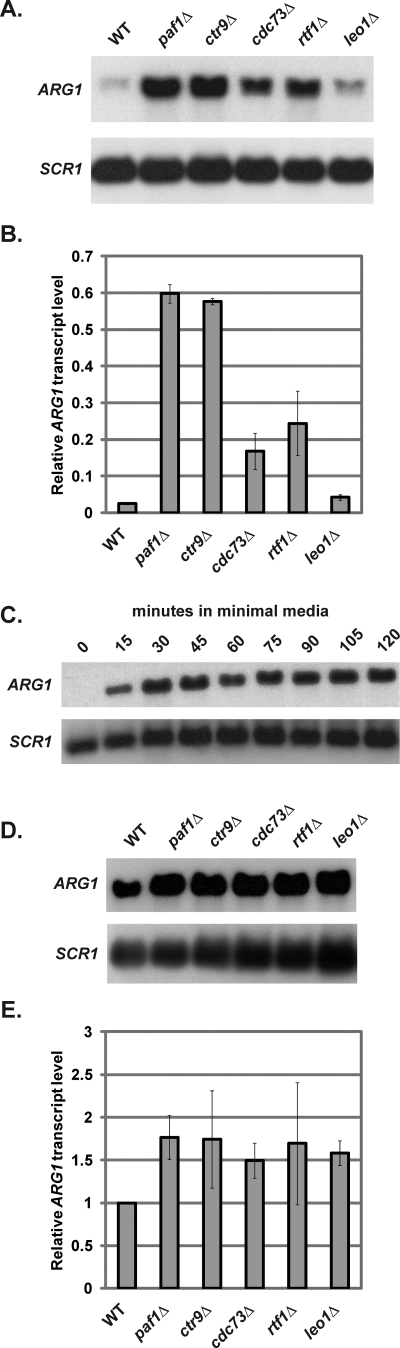
The Paf1 complex was first implicated in ARG1 repression by microarray analyses investigating changes in gene expression in paf1Δ cells (53). To determine whether other members of the Paf1 complex are required for ARG1 repression in rich media (YPD), ARG1 transcript levels were examined by Northern analysis in wild-type strains and strains lacking individual members of the Paf1 complex (Fig. 1A and B). Early in our analysis, we discovered that certain cellular auxotrophies influence the degree of ARG1 repression in otherwise wild-type cells, even when grown in rich media (data not shown). Therefore, to eliminate any effects of auxotrophies on our measurements, we analyzed ARG1 transcript levels in prototrophic strains wherever possible. Under these conditions, wild-type cells had very low ARG1 transcript levels (Fig. 1A and B). In contrast, strains lacking Paf1, Ctr9, Cdc73, or Rtf1 exhibited high levels of ARG1 expression relative to the isogenic wild-type control strain. Of these strains, paf1Δ and ctr9Δ mutants were most defective in ARG1 repression. These results indicate that although Leo1 does not appear to have a very significant role, Paf1, Ctr9, and to a lesser extent Cdc73 and Rtf1 each contribute to ARG1 repression in nutrient-rich conditions.
Fig. 1.
Members of the Paf1 complex are required for repression of ARG1 in both repressing and inducing conditions. Representative Northern analysis (A) and quantitation (B) of ARG1 transcript levels in wild-type (KY1699), paf1Δ (KY1700), ctr9Δ (KY1705), cdc73Δ (KY1706), rtf1Δ (KY1704), and leo1Δ (KY1805) strains were performed. SCR1 served as a loading control. Transcript levels were quantified and normalized to the levels detected in an arg80Δ (KY1709) control strain (not shown) as described in Materials and Methods. The values shown are the means of three independent experiments. Error bars represent one standard deviation of the mean. (C) Northern analysis of ARG1 transcript levels in cells shifted from YPD to minimal media for various times. Wild-type (KY1699) cells were grown to log phase in YPD, washed in water, resuspended in minimal media, and harvested at various time points. Representative Northern analysis (D) and quantitation (E) of ARG1 transcript levels in wild-type (KY1699), paf1Δ (KY1700), ctr9Δ (KY1705), cdc73Δ (KY1706), rtf1Δ (KY1704), and leo1Δ (KY1805) strains that were grown to log phase in YPD then shifted to minimal medium for 30 min prior to harvesting for RNA was performed. Relative signal in wild-type cells was set equal to one. The means of three independent experiments are shown. Error bars represent one standard deviation of the mean.
To determine whether the repressive functions of the Paf1 complex are specific to nutrient-rich conditions or whether the Paf1 complex also negatively regulates ARG1 expression in nutrient-limiting conditions, when ARG1 is activated, we examined the effects of deleting Paf1 complex members on ARG1 transcript levels in minimal media. First, we examined the timing of ARG1 induction in wild-type cells and found that ARG1 expression was fully induced 30 min after the cells were transferred to minimal media from rich media (Fig. 1C). Consequently, we measured ARG1 expression in Paf1 complex mutant strains that were grown in YPD and shifted to minimal media for 30 min. We found that deletion of genes encoding Paf1 complex members resulted in slightly higher ARG1 expression in inducing conditions (Fig. 1D and E). Consistent with our results, it was shown previously that paf1Δ strains exhibit more ARG1 expression than wild-type cells when ARG1 transcription is induced with sulfometuron methyl, which increases cellular levels of the ARG1 activator Gcn4 (69). These results suggest that the Paf1 complex acts as a transcriptional repressor of ARG1 in both repressing and inducing conditions.
Paf1 and Rtf1 are present at ARG1 in repressing conditions.
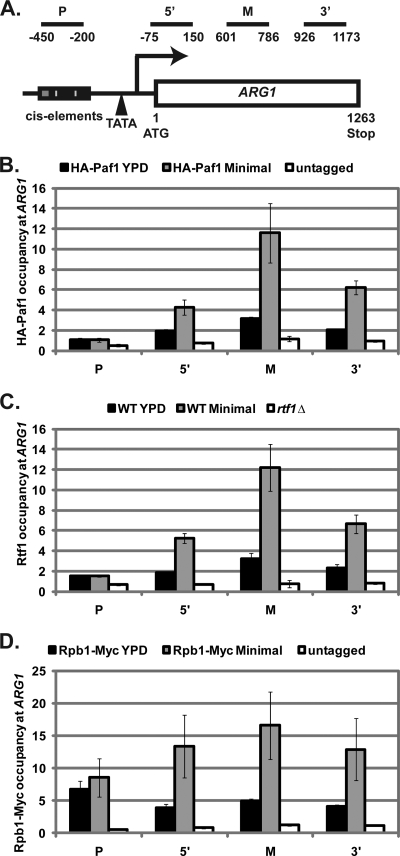
While it is known that the Paf1 complex associates with RNA Pol II during transcription elongation (36, 54), it is unclear whether the Paf1 complex localizes to repressed genes. Therefore, we examined whether the Paf1 complex localizes to ARG1 in repressing conditions by performing ChIP analysis using PCR primers that amplify the promoter, 5′, middle, and 3′ coding regions of ARG1 (Fig. 2A). Relative to the untagged control strain, we reproducibly detected a low level of HA-Paf1 occupancy at the ARG1 coding region in repressing conditions (Fig. 2B). Rtf1 occupancy, detected with polyclonal antisera against Rtf1, mirrored that of HA-Paf1 and was enriched over an rtf1Δ control strain (Fig. 2C). These results indicate that members of the Paf1 complex localize to the ARG1 coding region in repressing conditions. When cells were shifted to minimal media, HA-Paf1 and Rtf1 occupancy increased across the ARG1 coding region indicating that, similar to other active genes, Paf1 complex occupancy correlates with gene expression levels at ARG1 (Fig. 2B and C) (43).
Fig. 2.
The Paf complex localizes to the ARG1 promoter and coding region in repressing conditions. (A) Locations of PCR products used for ChIP analysis of the ARG1 locus. (B) ChIP analysis of HA-Paf1 occupancy at promoter (P), 5′, middle (M), and 3′ regions of ARG1 in cells expressing HA-tagged Paf1 (KY1721) or untagged Paf1 (KY1699) grown in rich medium (YPD) or shifted to minimal medium for 30 min (Minimal). (C) ChIP analysis of Rtf1 association with ARG1 in wild-type (KY1699) and rtf1Δ (KY1704) strains grown in rich medium (YPD) or shifted to minimal medium for 30 min (Minimal). (D) ChIP analysis of Rpb1-Myc occupancy at the ARG1 locus in an untagged control strain (KY1699) and cells expressing Myc-tagged Rpb1 (KY1302) grown in YPD or shifted to minimal medium for 30 min. ChIP data were quantified and normalized as described in Materials and Methods. Shown are the means of three independent experiments. Error bars represent the standard error of the mean.
Since the Paf1 complex associates with RNA Pol II during transcription elongation (36, 54), Paf1 complex occupancy correlates with RNA Pol II levels on active genes (43). To determine whether Paf1 complex occupancy also correlates with RNA Pol II occupancy on a Paf1 complex-repressed gene, ChIP analysis was performed to examine Rpb1-Myc levels at ARG1 in strains grown in repressing or inducing conditions (Fig. 2D). In repressing conditions, Rpb1-Myc was enriched at the ARG1 promoter and coding region compared to the untagged control strain, indicating that low levels of RNA Pol II are present at ARG1 in repressing conditions (Fig. 2D). This is consistent with our finding that long exposures of Northern blots revealed low levels of ARG1 transcription in wild-type cells grown in rich media (data not shown). As expected, Rpb1-Myc occupancy increased across the ARG1 coding region when cells were shifted to minimal media (Fig. 2D). Therefore, similar to its association with activated genes, the Paf1 complex likely associates with ARG1 through its interaction with RNA Pol II even under repressing conditions.
Histone H3 methylation contributes to Paf1 complex-mediated ARG1 repression.
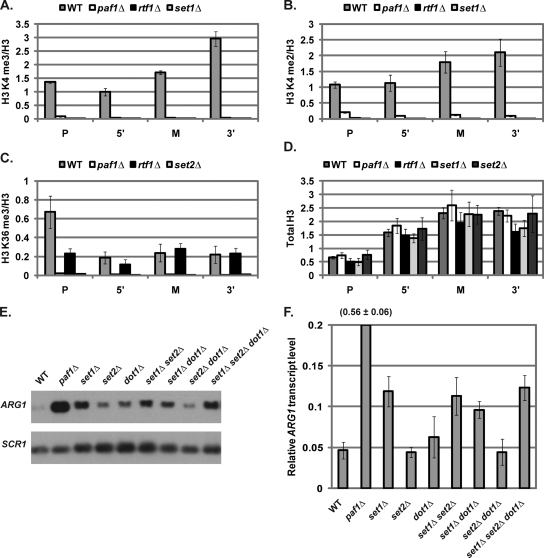
The presence of the Paf1 complex at the ARG1 locus in repressing conditions suggests that the Paf1 complex may regulate the histone modification state at ARG1 under these conditions. To determine whether histone H3 methylation is present at the promoter, 5′, middle, or 3′ coding region of ARG1 in repressing conditions, we performed ChIP assays using antibodies that detect histone H3 K4 trimethylation, H3 K4 dimethylation, and H3 K36 trimethylation. While total histone H3 levels were similar in all strains examined (Fig. 3D), changes in the histone modification pattern at the ARG1 locus were detected in the absence of Paf1 complex members. Specifically, both histone H3 K4 di- and trimethylation marks were detected in wild-type strains at all four regions examined but were lost in strains deleted for Paf1, Rtf1, or Set1 (Fig. 3A and B). Similarly, whereas histone H3 K36 trimethylation was detected at all four locations in the wild-type strain, H3 K36 trimethylation was undetectable in the paf1Δ and set2Δ strains (Fig. 3C). Furthermore, histone H3 K36 trimethylation was specifically reduced at the promoter in the absence of Rtf1 (Fig. 3C). These results indicate that the histone H3 K4 and K36 methylation marks are present at the ARG1 locus in repressing conditions in a Paf1 complex-dependent manner.
Fig. 3.
Paf1 complex-dependent histone modifications are present at ARG1 in repressing conditions and contribute to transcriptional repression. ChIP analysis of relative H3 K4 trimethylation (me3) (A), H3 K4 dimethylation (me2) (B), H3 K36 me3 (C), and total H3 (D) levels in wild-type (KY1699), paf1Δ (KY1700), rtf1Δ (KY1704), set1Δ (KY1755), and set2Δ (KY1716) strains. ChIP data were quantified and normalized as described in Materials and Methods. Histone H3 K4 me3, K4 me2, and K36 me3 levels are presented relative to total H3 levels. The means of three independent experiments are shown. Error bars represent the standard error of the mean. Representative Northern analysis (E) and quantitation (F) of ARG1 mRNA levels in wild-type (KY1699), set1Δ (KY1715), set2Δ (KY1716), dot1Δ (KY1717), set1Δ set2Δ (KY1821), set1Δ dot1Δ (KY1826), set2Δ dot1Δ (KY1832), and set1Δ set2Δ dot1Δ (KY1847) strains were performed. Values shown are the means of three independent experiments, quantified and normalized to the levels detected in an arg80Δ (KY1709) control strain (not shown) as described in Materials and Methods. Error bars represent one standard deviation of the mean. The y axis was cropped to allow for comparisons between lower values. The value for paf1Δ is indicated above the appropriate bar.
To determine to what extent Paf1 complex-dependent histone H3 methylation is required for ARG1 repression, we performed Northern analysis of ARG1 transcript levels in strains lacking Set1, Set2, or Dot1. While previous work showed that deletion of SET2 or DOT1 caused increased expression of an ARG1-lacZ reporter construct (48), these mutations did not lead to a change in repression of the native ARG1 gene that was statistically different from wild-type (Fig. 3E and F). The differing results may be due to increased sensitivity of the ARG1 expression reporter or the presence of auxotrophies in the previously analyzed strains. In contrast, set1Δ strains exhibited an increase in ARG1 expression (Fig. 3E and F). This result is consistent with the finding that loss of Bre2 or Swd3, components of the Set1-containing COMPASS complex, results in increased expression of an ARG1-lacZ reporter construct (48). However, the increase in endogenous ARG1 transcript levels in set1Δ cells was not as high as in paf1Δ cells (Fig. 3E and F). Together, these results suggest that none of the methyltransferases examined individually are as important for ARG1 repression as Paf1. Since the Paf1 complex is important for multiple methylation marks, we tested whether the combined loss of multiple methyltransferases might derepress ARG1 to the same degree as deleting PAF1. Surprisingly, no combination of double or triple mutations caused any more than an ∼2.6-fold increase in ARG1 transcript levels, whereas deletion of PAF1 resulted in an ∼12-fold increase in ARG1 transcript levels (Fig. 3E and F). Together, these results demonstrate that Paf1-dependent histone H3 K4 and K36 methylation are present at ARG1 in repressing conditions and histone H3 K4 methylation contributes to ARG1 repression; however, the Paf1 subunit has repressive functions in addition to facilitating histone H3 methylation.
Rtf1 represses ARG1 by promoting histone modifications.
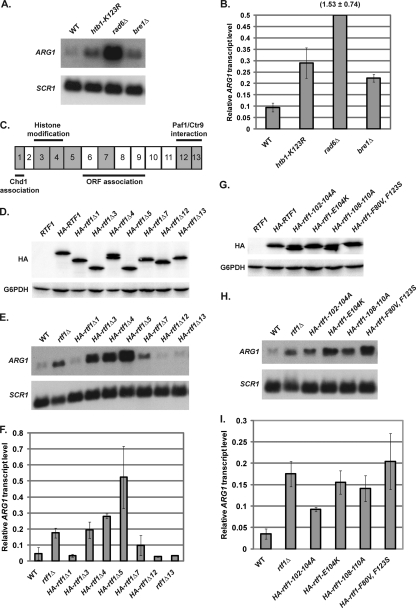
Our results demonstrate that histone H3 K4 methylation is present at ARG1 in cells grown in rich media and can contribute to ARG1 repression. Because histone H3 K4 methylation is dependent on histone H2B ubiquitylation (68, 78), we tested whether the Paf1 complex mediates ARG1 repression by promoting histone H2B K123 ubiquitylation. Indeed, as previously reported, we found that rad6Δ and bre1Δ strains and strains in which the histone H2B ubiquitylation site is mutated (htb1-K123R) exhibited ARG1 derepression (Fig. 4A and B) (26, 38, 48, 73, 84). Note that rad6Δ cells exhibited higher levels of ARG1 derepression than either bre1Δ or htb1-K123R strains. These results are consistent with reports that Rad6 may function with another ubiquitin ligase that is required for ARG1 repression and further suggest that Rad6 has additional targets important for ARG1 repression (73). Together, these results suggest that histone H2B ubiquitylation and downstream histone H3 K4 methylation are important for ARG1 repression. We previously identified a region within Rtf1 that is essential for these histone modifications (76); therefore, we decided to further examine the role of Rtf1 in ARG1 repression.
Fig. 4.
Rtf1 mediates ARG1 repression primarily through histone H2B ubiquitylation. Representative Northern analysis (A) and quantitation (B) measuring ARG1 transcript levels in wild-type (KY1698), htb-K123R (KY1732), rad6Δ (KY1711), and bre1Δ (KY1713) strains were performed. The y axis was shortened to facilitate comparison between lower values. The value for rad6Δ is indicated above the bar. (C) Schematic of the 13 regions of Rtf1 defined by internal deletion mutations and their associated function (62). Regions that were examined for effects on ARG1 repression are indicated in gray. (D) Western analysis of wild-type and mutant Rtf1 protein levels using an anti-HA antibody in strains expressing untagged Rtf1 (KY1698), HA-Rtf1 (KY2082), HA-rtf1Δ1 (KY1722), HA-rtf1Δ3 (KY1723), HA-rtf1Δ4 (KY1724), HA-rtf1Δ5 (KY1980), HA-rtf1Δ7 (KY1725), HA-rtf1Δ12 (KY1726), or HA-rtf1Δ13 (KY1727). G6PDH serves as a loading control. Note that a faster-migrating band observed for HA-rtf1Δ4 is likely a degradation product, which has been reproducibly observed with several forms of Rtf1 (72, 76). Representative Northern analysis (E) and quantitation (F) of relative ARG1 transcript levels in wild-type (KY1698), rtf1Δ (KY1703), HA-rtf1Δ1 (KY1722), HA-rtf1Δ3 (KY1723), HA-rtf1Δ4 (KY1724), HA-rtf1Δ5 (KY1980), HA-rtf1Δ7 (KY1725), HA-rtf1Δ12 (KY1726), and HA-rtf1Δ13 (KY1727) strains were performed. The means of three independent experiments are shown, quantified and normalized to the levels detected in an arg80Δ (KY1709) control strain (not shown) as described in Materials and Methods. Error bars represent one standard deviation of the mean. (G) Western analysis of wild-type and mutant Rtf1 protein levels using an anti-HA antibody in strains expressing untagged Rtf1 (KY1698), HA-Rtf1 (KY2082), HA-rtf1-102-104A (KY1981), HA-rtf1-E104K (KY1982), HA-rtf1-108-110A (KY1983), and HA-rtf1-F80V,F123S (KY1984). G6PDH serves as a loading control. The faster-migrating band for HA-rtf1-102-104A and HA-rtf1-108-110A has been previously observed and is likely a product of proteolysis (72, 76). Representative Northern analysis (H) and quantitation (I) of relative ARG1 transcript levels in wild-type (KY1698), rtf1Δ (KY1703), HA-rtf1-102-104A (KY1981), HA-rtf1-E104K (KY1982), HA-rtf1-108-110A (KY1983), and HA-rtf1-F80V,F123S (KY1984) strains were performed. Graphs depict the means of three independent experiments, quantified and normalized to the levels detected in an arg80Δ (KY1709) control strain (not shown) as described in Materials and Methods. Error bars represent one standard deviation of the mean.
In addition to defining the region of Rtf1 required for histone modifications, we assigned other Rtf1 functions, including ORF association, Paf1 complex assembly, and interaction with the chromatin remodeling factor Chd1, to specific regions of the Rtf1 protein using deletion analysis (76). To determine which region and thus which function of Rtf1 is important for ARG1 repression, ARG1 transcript levels were examined in rtf1 deletion strains that define different functional classes (Fig. 4C). Mutations were chosen because they delete a region of Rtf1 with a known function (rtf1Δ1, rtf1Δ3, rtf1Δ4, rtf1Δ7, rtf1Δ12, and rtf1Δ13) or because they cause a phenotype that indicates a defect in transcription (rtf1Δ5). While Western analysis confirmed that the internal Rtf1 deletion mutant proteins were expressed to similar levels as full-length Rtf1 (Fig. 4D), Northern analysis indicated that the internal rtf1 deletions had differential effects on ARG1 transcript levels. Deletion of Rtf1 region 1 (amino acids 3 to 30), which is required for an interaction between Rtf1 and Chd1 (76), did not cause ARG1 derepression, suggesting that Rtf1-dependent recruitment of Chd1 is not required for ARG1 repression (Fig. 4E and F). Consistent with this result, a chd1Δ mutation did not alter ARG1 repression (data not shown). Similarly, cells lacking Rtf1 region 7 (amino acids 251 to 300), which is required for the association of Rtf1 with ORFs (76), showed only a slight increase in ARG1 transcription under repressing conditions (Fig. 4E and F). This result suggests that stable association with the ARG1 coding region may not be required for full repression of ARG1 by Rtf1. Furthermore, deletion of Rtf1 regions 12 (amino acids 491 to 535) or 13 (amino acids 536 to 558), which are required for the interaction between Rtf1 and other Paf1 complex members, Paf1 and Ctr9 (76), did not result in ARG1 derepression, suggesting that a stable interaction between Rtf1 and other Paf1 complex members is not required for ARG1 repression (Fig. 4E and F).
Rtf1 regions 3 (amino acids 62 to 109) and 4 (amino acids 112 to 152) are required for Rtf1-dependent histone modifications, leading us to define these regions collectively as the Rtf1 histone modification domain (HMD) (72, 76). Interestingly, deletion of Rtf1 region 3 or 4 resulted in significant ARG1 derepression (Fig. 4E and F). Furthermore, disruption of the Rtf1 HMD derepressed ARG1 to the same degree as completely deleting the RTF1 gene, suggesting that Rtf1 mediates ARG1 repression primarily through promoting histone modifications (Fig. 4E and F). We recently identified a set of specific amino acid substitutions within the Rtf1 HMD that impair its histone modification functions (72). Therefore, we examined whether these substitutions, which greatly diminish histone H2B K123 ubiquitylation, also result in ARG1 derepression. While Western analysis demonstrated that wild-type Rtf1 and Rtf1 point mutant proteins were expressed to similar levels (Fig. 4G), cells expressing the Rtf1 point mutants, Rtf1-102-104A, Rtf1-E104K, Rtf1-108-110A, and Rtf1-F80V,F123S, exhibited ARG1 derepression similar to strains lacking Rtf1 entirely (Fig. 4H and I). Our results strongly suggest that Rtf1 mediates ARG1 repression by promoting histone H2B ubiquitylation and subsequent H3 K4 methylation.
Complete deletion of RTF1 causes a suppressor-of-Ty (Spt−) phenotype, indicating that deletion of RTF1 suppresses defects in transcription caused by the insertion of Ty transposons or their long terminal repeats within the promoters or 5′ ends of genes (67). Strains lacking any of Rtf1 regions 3 to 9 (spanning amino acids 62 to 395) individually have an Spt− phenotype, suggesting that regions 3 to 9 are each important for transcriptional regulation (76). Of these regions, only region 5 has yet to be assigned a specific function. We found that rtf1Δ5 strains exhibited levels of ARG1 derepression that were higher than an rtf1Δ strain (Fig. 4E and F), indicating that region 5 may have a negative effect on the function of the rest of the protein. Because our current data have not defined an obvious role for region 5 in regulating histone modifications or other known activities of Rtf1, future analysis of this region may reveal new insights on the regulation or functions of the Paf1 complex.
Histone H2B K123 is required for full derepression in paf1Δ cells.
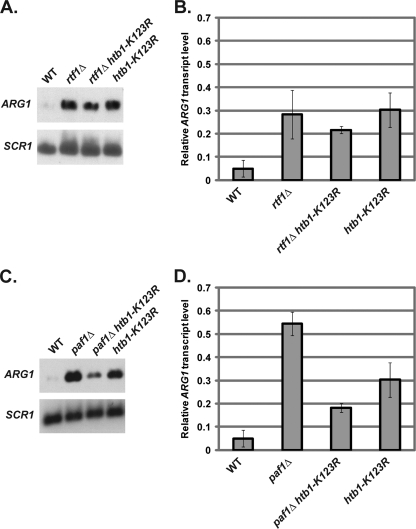
Our results suggest that Rtf1 mediates ARG1 repression primarily through histone H2B ubiquitylation and H3 K4 methylation. To test this hypothesis, we examined the effect of mutating the histone H2B ubiquitylation site, alone or in combination with deletion of RTF1. We found that rtf1Δ and rtf1Δ htb1-K123R cells had similar levels of ARG1 derepression (Fig. 5A and B), consistent with Rtf1 and histone H2B ubiquitylation functioning in the same pathway for ARG1 repression. However, rtf1Δ cells reproducibly showed significantly lower levels of ARG1 derepression than paf1Δ cells, suggesting that Paf1 has repressive functions aside from its role in promoting histone H2B ubiquitylation (Fig. 1A and B). To test whether Paf1 and H2B ubiquitylation have independent roles in ARG1 repression, we performed Northern analysis on paf1Δ htb1-K123R double-mutant cells. If Paf1 and H2B ubiquitylation have completely independent effects on ARG1 repression, paf1Δ htb1-K123R double-mutant strains should exhibit an elevated level of ARG1 derepression compared to paf1Δ and htb1-K123R single-mutant strains. In contrast to this prediction, htb1-K123R significantly reduced the level of ARG1 derepression in paf1Δ cells (Fig. 5C and D). This result suggests that histone H2B ubiquitylation is required for full ARG1 derepression in paf1Δ cells and argues that this modification can have both positive and negative effects on the same gene.
Fig. 5.
ARG1 derepression in paf1Δ cells partially requires histone H2B ubiquitylation. Representative Northern analysis (A) and quantitation (B) examining ARG1 transcript levels in wild-type (KY1699), rtf1Δ (KY1703), rtf1Δ htb1-K123R (KY2074), and htb1-K123R (KY1732) strains were performed. Representative Northern analysis (C) and quantitation (D) examining ARG1 transcript levels in wild-type (KY1699), paf1Δ (KY1700), paf1Δ htb1-K123R (KY1731), and htb1-K123R (KY1732) strains were also performed. The means of three independent experiments are shown, quantified and normalized to the levels detected in an arg80Δ (KY1709) control strain (not shown) as described in Materials and Methods. Error bars represent one standard deviation of the mean.
The Paf1 complex uses similar mechanisms to repress other genes.
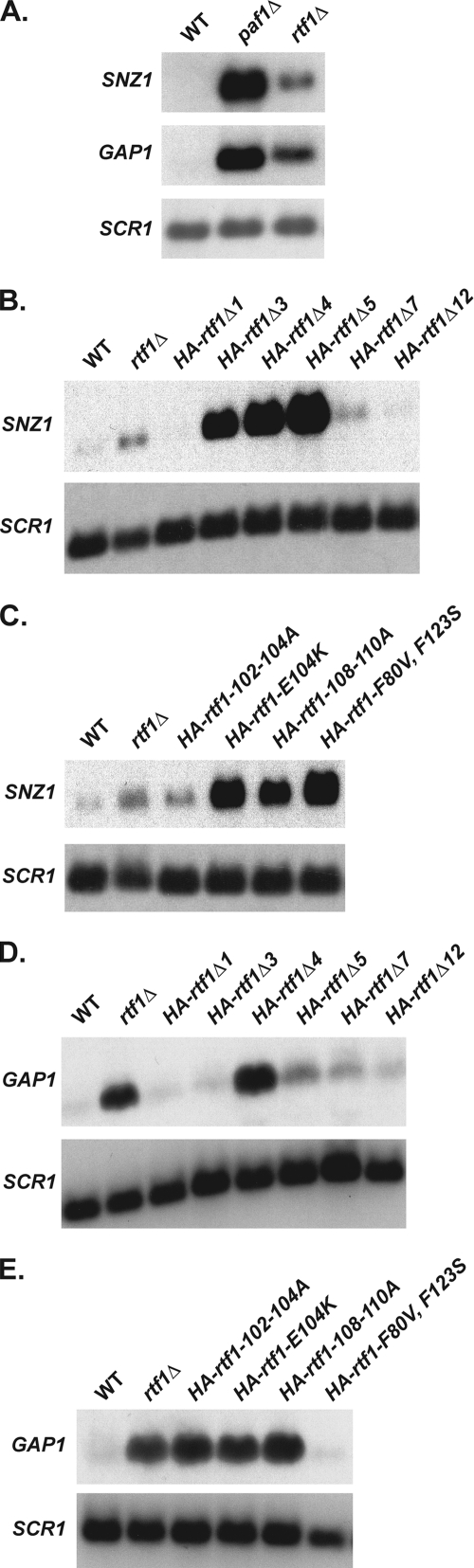
To determine whether the manner in which the Paf1 complex mediates repression of ARG1 extends to other genes, we examined the effects of deleting PAF1 and RTF1 on the expression of SNZ1 and GAP1, which encode a protein involved in vitamin B biosynthesis and a general amino acid permease, respectively. We chose to examine these genes because, like ARG1, GAP1 and SNZ1 have been shown by genome-wide expression studies to be derepressed in paf1Δ and htb1-K123R strains (48, 53, 84). Using Northern analysis, we found that SNZ1 and GAP1 were repressed in wild-type cells and derepressed in the absence of Paf1 or Rtf1 (Fig. 6A). Similar to ARG1, paf1Δ cells exhibited higher derepression of these genes than rtf1Δ cells, suggesting that Paf1 and Rtf1 may function in a similar manner at all three genes.
Fig. 6.
The Paf1 complex has similar repression mechanisms at other genes. Northern analysis of SNZ1 and GAP1 transcript levels in wild-type (KY1699), paf1Δ (KY1700), and rtf1Δ (KY1704) strains. Northern analysis of SNZ1 (B and C) or GAP1 (D and E) in wild-type (KY1698), rtf1Δ (KY1703), HA-rtf1Δ1 (KY1722), HA-rtf1Δ3 (KY1723), HA-rtf1Δ4 (KY1724), HA-rtf1Δ5 (KY1980), HA-rtf1Δ7 (KY1725), HA-rtf1Δ12 (KY1726), HA-rtf1-102-104A (KY1981), HA-rtf1-E104K (KY1982), HA-rtf1-108-110A (KY1983), and HA-rtf1-F80V,F123S (KY1984) strains was performed. The Northern blots are representative of at least two independent experiments.
To further test the requirements for SNZ1 and GAP1 repression, we performed Northern analyses of these genes in strains expressing the rtf1 internal deletion mutations. We found that the expression profile of SNZ1 mirrored that of ARG1 with rtf1Δ3, rtf1Δ4, and rtf1Δ5 cells exhibiting high levels of SNZ1 derepression (Fig. 6B). Consistent with a requirement for the Rtf1 HMD in repressing SNZ1 transcription, rtf1 point mutations within the HMD-coding region also caused SNZ1 derepression (Fig. 6C). Furthermore, the levels of SNZ1 derepression that occurred in these mutants closely mimicked the effects we observed at ARG1 (Fig. 4H and I), with rtf1-102-104A cells exhibiting the least dramatic derepression and rtf1-F80V,F123S cells exhibiting the most dramatic derepression (Fig. 6C).
Similar to both ARG1 and SNZ1, repression of GAP1 requires a functional Rtf1 HMD, since rtf1Δ4 cells or strains expressing the rtf1 HMD point mutations showed significant derepression of GAP1 (Fig. 6D and E). However, unlike ARG1 and SNZ1, high levels of GAP1 derepression did not occur in rtf1Δ3 or rtf1Δ5, suggesting that Rtf1 regions 3 and 4 are not equivalent in all cases (Fig. 6D and E). Furthermore, while amino acid substitutions within the HMD resulted in GAP1 derepression, the relative levels of derepression caused by these substitutions differed from those observed at ARG1 and SNZ1. Specifically, rtf1-102-104A cells exhibited a high level of GAP1 derepression and rtf1-F80V,F123S cells exhibited a low level of GAP1 derepression (Fig. 6E). While the differences between GAP1 and the other genes examined will likely enrich further studies of the functions of the Paf1 complex, the overall similarities point toward a common mechanism of gene repression by the Paf1 complex in which Rtf1-dependent histone modifications play a prominent role.
DISCUSSION
In this study, we investigated the mechanisms by which the yeast Paf1 complex negatively regulates transcription, using the well-characterized ARG1 gene as a framework for our studies. Although genome-wide expression patterns indicate that the repressive effects of the Paf1 complex are widespread (53), an analysis of how the Paf1 complex mediates gene repression has not been previously described. Here, we report that the Paf1, Rtf1, Ctr9, and Cdc73 subunits of the Paf1 complex contribute to ARG1 repression. Consistent with a direct repressive role, the Paf1 complex is present at the ARG1 coding region when cells are grown in conditions that strongly repress ARG1 transcription. Under these conditions, histone modifications primarily controlled by Rtf1 are present at ARG1 and contribute to repression. Interestingly, Paf1 appears to have repressive functions beyond its role in mediating known Paf1 complex-dependent histone modifications. Finally, an analysis of two additional genes, SNZ1 and GAP1, indicates that the characteristics of Paf1 complex-mediated transcriptional repression observed at ARG1 extend to other genes.
The correlation between Paf1 complex occupancy and gene activity (43) raises the question of how the Paf1 complex is recruited to a gene in repressing conditions. Our data indicate a modest but significant occupancy of both the Paf1 complex and RNA Pol II at the ARG1 coding region in nutrient-rich media. In these conditions, a very low level of transcriptional activity can be detected by our Northern blot assays. Therefore, consistent with its known association with RNA Pol II during transcription elongation (36, 54), we hypothesize that the low levels of transcription occurring in repressing conditions are sufficient to result in enrichment of the Paf1 complex across the ARG1 locus. Interestingly, an antisense transcript traversing the ARG1 coding region was detected by Steinmetz and coworkers (16, 82), raising the possibility that antisense transcription could contribute to RNA Pol II occupancy at ARG1. Consistent with transcriptional activity in the antisense direction, histone H3 K4 methylation and K36 methylation at ARG1 were highest at 3′ and 5′ locations, respectively, a histone methylation pattern that is opposite of the typical distribution (34, 35, 37, 40, 50, 61, 79). A reversed histone modification pattern has been observed at GAL10 (27), one of several genes recently shown to be regulated by antisense transcription (25, 27, 74, 81). Whether the Paf1 complex and its associated histone modifications repress ARG1 expression by impacting antisense transcription at the ARG1 locus remains to be determined.
In accordance with the localization of the Paf1 complex to ARG1 in repressing conditions, ChIP analysis demonstrated that histone H3 K4 and K36 methylation are significantly enriched at ARG1 in a Paf1 complex-dependent manner. Both histone H3 K4 and K36 methylation have been shown to impact the levels of histone acetylation on genes through several established pathways of histone cross talk. In one well-studied pathway, histone H3 K36 dimethylation is required for the activity of the Rpd3S histone deacetylase complex (HDAC), which reduces histone acetylation on transcribed genes and inhibits transcription from cryptic promoters within coding regions (8, 31, 39, 41). We found that eliminating histone H3 K36 methylation by deleting SET2 had little impact on ARG1 expression in repressing conditions, suggesting that Set2-dependent histone deacetylation is unlikely to be involved in maintaining ARG1 repression. We also found no indications that the histone H3 K79 methyltransferase, Dot1, plays an important role in ARG1 repression.
In contrast to the effects of deleting SET2 and DOT1, deletion of SET1, the gene encoding the histone H3 K4 methyltransferase, caused a significant reduction in ARG1 repression. Interestingly, histone H3 K4 methylation has been implicated in pathways that direct either the acetylation or deacetylation of histones. By recruiting the NuA3 histone acetyltransferase (HAT) complex, histone H3 K4 methylation increases histone H3 K14 acetylation levels and gene activation (42, 71). By activating the Set3 HDAC, histone H3 K4 dimethylation lowers histone acetylation levels at the 5′ ends of genes (21, 33). Because histone deacetylation has well-established links to gene repression, including the silencing of genes near telomeres (reviewed in reference 62), it is possible that histone deacetylation driven by histone H3 K4 methylation and the Set3 HDAC could be involved in repressing ARG1 and other loci. However, we did not observe a loss of ARG1 repression in set3Δ cells (data not shown). Therefore, although we cannot rule out the possibility that histone H3 K4 methylation leads to the recruitment of other HDACs, we currently have no experimental support for a model in which this modification represses ARG1 through activation of the Set3 complex.
Because Rtf1 is essential for histone H3 K4 di- and trimethylation, we chose to investigate further the role of this Paf1 complex subunit in gene repression. We previously showed that disruption of the Rtf1 HMD, either through deletion or substitution of conserved residues, dramatically reduces global levels of histone H2B K123 ubiquitylation and histone H3 K4 tri- and dimethylation (72, 76). Because these same rtf1 mutations alleviate ARG1 repression to approximately the same degree as an rtf1-null allele, we conclude that Rtf1 mediates ARG1 repression primarily through its histone modification functions. In support of this idea, a comparison of rtf1Δ cells and rtf1Δ htb1-K123R cells revealed approximately the same levels of ARG1 depression, strongly suggesting that Rtf1 and histone H2B ubiquitylation function in the same pathway for ARG1 repression. Therefore, we conclude that Rtf1 mediates repression by promoting histone H2B ubiquitylation and downstream H3 K4 methylation. Similar effects of the rtf1 mutations were obtained for two other genes, SNZ1 and GAP1, suggesting that Rtf1 can repress a subset of genes through similar mechanisms.
Microarray analysis of transcript levels in htb1-K123R cells revealed that the majority of affected genes exhibited increased expression, indicating that the repressive functions of histone H2B ubiquitylation are required at many genes (48). Providing a possible mechanism for gene repression by H2B K123 ubiquitylation, a recent study revealed that this modification enhances nucleosome stability at the promoters of repressed genes (9). Although we did not detect a reduction in histone H3 occupancy at the ARG1 promoter or coding region in rtf1Δ cells, it remains possible that our ChIP assays lacked the sensitivity to detect subtle changes in nucleosome stability. In addition to its role in nucleosome stability, histone H2B ubiquitylation is required for proper telomeric silencing (28, 68). Consequently, complete deletion of RTF1 (35, 49) or disruption of the Rtf1 HMD results in telomeric silencing defects (72, 76). The genome-wide loss of histone H3 K4 and K79 methylation in these cells has been proposed to cause a redistribution of telomeric silencing factors from their normal sites of action (reviewed in reference 59). Whether similar mechanisms can influence the occupancy of regulatory factors at genes such as ARG1 remains to be determined.
In addition to its repressive role, histone H2B K123 also positively regulates ARG1 expression under certain circumstances. For example, derepression of ARG1 in a paf1Δ strain is partially suppressed by the htb1-K123R substitution (Fig. 5). The histone H2B K123 residue itself may be important for full ARG1 derepression in paf1Δ cells through effects on nucleosome structure. Alternatively, the finding of an effect of htb1-K123R in a paf1Δ cells suggests that in paf1Δ cells, a low level of histone H2B ubiquitylation occurs that is required for full levels of ARG1 derepression. In support of this idea, a bre1Δ mutation also partially suppresses ARG1 transcription in paf1Δ strains (data not shown). Another possibility is that histone H2B ubiquitylation and subsequent deubiquitylation, which is important for full expression of inducible genes, such as GAL1 and SUC2 (15, 23, 30), may be required for full ARG1 expression in the absence of Paf1. Consistent with this possibility, the loss of Ubp8, which deubiquitylates histone H2B, somewhat reduces ARG1 expression in inducing conditions (38). Histone H2B ubiquitylation in humans has also been shown to have both positive and negative influences on transcription. For example, histone H2B ubiquitylation facilitates transcription elongation in vitro (52) and preferentially associates with sites of active transcription in vivo (44). However, removal of histone H2B ubiquitylation by the deubiquitylating enzyme Usp22 inhibits heterochromatic silencing and facilitates gene activation (85, 86). Importantly, histone H2B ubiquitylation in human cells promotes transcription of tumor suppressor genes and represses several proto-oncogenes, indicating that both the negative and the positive transcriptional effects of histone H2B ubiquitylation are critical for cancer prevention (63).
In contrast to our observations on histone modifications, our data do not indicate strong repressive roles for other Rtf1 functions, including Chd1 interaction, ORF association, and Paf1 complex association. In agreement with previous studies (46, 76), these observations suggest that members of the Paf1 complex retain some functionality when their stable interactions with each other or elongating RNA Pol II are disrupted. Differing reports on whether human Rtf1 is absent from (58, 83, 87) or present in (32) the human Paf1 complex has led to the conclusion that, like Drosophila Rtf1 (1), human Rtf1 is a less stably associated member of the complex. However, despite its less stable association with the Paf1 complex, human Rtf1 retains its effects on gene expression (18, 47). Therefore, it may not be surprising that, in yeast, repression of a subset of genes by Rtf1 does not require stable association with other Paf1 complex members.
Although Rtf1 mediates repression primarily through histone H2B ubiquitylation and its downstream modifications, our results suggest that Paf1 has repressive functions aside from histone H2B ubiquitylation and other known Paf1-dependent histone modifications. Since most known roles for the Paf1 complex are intimately connected to histone modifications (reviewed in reference 29), it will be important to explore histone modification-independent functions of the complex. Interestingly, in a recent study, the human Paf1 complex was shown to stimulate in vitro transcription of a chromatin template independently of histone modifications (32). The extensive functional conservation between the yeast and human Paf1 complexes (reviewed in reference 29) strongly suggests that mechanistic studies of Paf1 complex-mediated gene repression in yeast will yield insights on the human complex, defects in which are associated with cancers (reviewed in reference 10) and the loss of stem cell identity (18).
ACKNOWLEDGMENTS
We thank Joe Martens for yeast strains used in this study. We are grateful to Joe Martens, Margaret Shirra, Brett Tomson, and Kristin Klucevsek for the critical reading of the manuscript and Alan Hinnebusch for helpful discussions.
This study was supported by NIH grant GM52593 to K.M.A.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Adelman K., et al. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 26:250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amar N., Messenguy F., El Bakkoury M., Dubois E. 2000. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 20:2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ausubel F. M., et al. (ed.). 1988. Current protocols in molecular biology, vol. 1 John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 4. Bechet J., Greenson M., Wiame J. M. 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 12:31–39 [DOI] [PubMed] [Google Scholar]
- 5. Bernstein B. E., et al. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U. S. A. 99:8695–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brachmann C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132 [DOI] [PubMed] [Google Scholar]
- 7. Briggs S. D., et al. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 8. Carrozza M. J., et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592 [DOI] [PubMed] [Google Scholar]
- 9. Chandrasekharan M. B., Huang F., Sun Z. W. 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc. Natl. Acad. Sci. U. S. A. 106:16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaudhary K., Deb S., Moniaux N., Ponnusamy M. P., Batra S. K. 2007. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene 26:7499–7507 [DOI] [PubMed] [Google Scholar]
- 11. Chu Y., Simic R., Warner M. H., Arndt K. M., Prelich G. 2007. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26:4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costa P. J., Arndt K. M. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crabeel M., et al. 1995. Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast 11:1367–1380 [DOI] [PubMed] [Google Scholar]
- 14. Crabeel M., Lavalle R., Glansdorff N. 1990. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol. Cell. Biol. 10:1226–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniel J. A., et al. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867–1871 [DOI] [PubMed] [Google Scholar]
- 16. David L., et al. 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U. S. A. 103:5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delforge J., Messenguy F., Wiame J. M. 1975. The regulation of arginine biosynthesis in Saccharomyces cerevisiae: the specificity of argR− mutations and the general control of amino-acid biosynthesis. Eur. J. Biochem. 57:231–239 [DOI] [PubMed] [Google Scholar]
- 18. Ding L., et al. 2009. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4:403–415 [DOI] [PubMed] [Google Scholar]
- 19. Dubois E., Bercy J., Messenguy F. 1987. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol. Gen. Genet. 207:142–148 [DOI] [PubMed] [Google Scholar]
- 20. El Bakkoury M., Dubois E., Messenguy F. 2000. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Mol. Microbiol. 35:15–31 [DOI] [PubMed] [Google Scholar]
- 21. Govind C. K., et al. 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39:234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A. 2010. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Henry K. W., et al. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hinnebusch A. G. 1986. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. CRC Crit. Rev. Biochem. 21:277–317 [DOI] [PubMed] [Google Scholar]
- 25. Hongay C. F., Grisafi P. L., Galitski T., Fink G. R. 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735–745 [DOI] [PubMed] [Google Scholar]
- 26. Hossain M. A., Claggett J. M., Nguyen T., Johnson T. L. 2009. The cap binding complex influences H2B ubiquitination by facilitating splicing of the SUS1 pre-mRNA. RNA 15:1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32:685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang H., Kahana A., Gottschling D. E., Prakash L., Liebman S. W. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaehning J. A. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799:379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kao C. F., et al. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keogh M. C., et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605 [DOI] [PubMed] [Google Scholar]
- 32. Kim J., Guermah M., Roeder R. G. 2010. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140:491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim T., Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137:259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kizer K. O., et al. 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25:3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krogan N. J., et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721–729 [DOI] [PubMed] [Google Scholar]
- 36. Krogan N. J., et al. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krogan N. J., et al. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee K. K., Florens L., Swanson S. K., Washburn M. P., Workman J. L. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 25:1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li B., et al. 2007. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316:1050–1054 [DOI] [PubMed] [Google Scholar]
- 40. Li B., Howe L., Anderson S., Yates III J. R., Workman J. L. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897–8903 [DOI] [PubMed] [Google Scholar]
- 41. Li B., et al. 2009. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 284:7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin D. G., et al. 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26:7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mayer A., et al. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17:1272–1278 [DOI] [PubMed] [Google Scholar]
- 44. Minsky N., et al. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 10:483–488 [DOI] [PubMed] [Google Scholar]
- 45. Mueller C. L., Jaehning J. A. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447–456 [DOI] [PubMed] [Google Scholar]
- 47. Muntean A. G., et al. 2010. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell 17:609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mutiu A. I., Hoke S. M., Genereaux J., Liang G., Brandl C. J. 2007. The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics 277:491–506 [DOI] [PubMed] [Google Scholar]
- 49. Ng H. H., Dole S., Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625–33628 [DOI] [PubMed] [Google Scholar]
- 50. Ng H. H., Robert F., Young R. A., Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709–719 [DOI] [PubMed] [Google Scholar]
- 51. Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. 2008. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7:1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pavri R., et al. 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125:703–717 [DOI] [PubMed] [Google Scholar]
- 53. Penheiter K. L., Washburn T. M., Porter S. E., Hoffman M. G., Jaehning J. A. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20:213–223 [DOI] [PubMed] [Google Scholar]
- 54. Pokholok D. K., Hannett N. M., Young R. A. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- 55. Qiu H. F., Dubois E., Broen P., Messenguy F. 1990. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Gen. Genet. 222:192–200 [DOI] [PubMed] [Google Scholar]
- 56. Rose M. D., Winston F., Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 57. Rothstein R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281–301 [DOI] [PubMed] [Google Scholar]
- 58. Rozenblatt-Rosen O., et al. 2005. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rusche L. N., Kirchmaier A. L., Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481–516 [DOI] [PubMed] [Google Scholar]
- 60. Santos-Rosa H., et al. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411 [DOI] [PubMed] [Google Scholar]
- 61. Schaft D., et al. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31:2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shahbazian M. D., Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100 [DOI] [PubMed] [Google Scholar]
- 63. Shema E., et al. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22:2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shi X., et al. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shirra M. K., Rogers S. E., Alexander D. E., Arndt K. M. 2005. The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169:1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Squazzo S. L., et al. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stolinski L. A., Eisenmann D. M., Arndt K. M. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun Z. W., Allis C. D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108 [DOI] [PubMed] [Google Scholar]
- 69. Swanson M. J., et al. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Swanson M. S., Malone E. A., Winston F. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Taverna S. D., et al. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24:785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tomson B. N., Davis C. P., Warner M. H., Arndt K. M. Identification of a role for histone H2B ubiquitylation in non-coding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 73. Turner S. D., et al. 2002. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22:4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Uhler J. P., Hertel C., Svejstrup J. Q. 2007. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U. S. A. 104:8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vogelauer M., Wu J., Suka N., Grunstein M. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495–498 [DOI] [PubMed] [Google Scholar]
- 76. Warner M. H., Roinick K. L., Arndt K. M. 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27:6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Winston F., Dollard C., Ricupero-Hovasse S. L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55 [DOI] [PubMed] [Google Scholar]
- 78. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739–34742 [DOI] [PubMed] [Google Scholar]
- 79. Xiao T., et al. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xiao T., et al. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu Z., et al. 2011. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 7:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu Z., et al. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yart A., et al. 2005. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 25:5052–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang X., et al. 2005. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J. Biol. Chem. 280:2759–2770 [DOI] [PubMed] [Google Scholar]
- 85. Zhang X. Y., Pfeiffer H. K., Thorne A. W., McMahon S. B. 2008. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 7:1522–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao Y., et al. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29:92–101 [DOI] [PubMed] [Google Scholar]
- 87. Zhu B., et al. 2005. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell 20:601–611 [DOI] [PubMed] [Google Scholar]