Abstract
Saccharomyces cerevisiae Skn7p is a stress response transcription factor that undergoes aspartyl phosphorylation by the Sln1p histidine kinase. Aspartyl phosphorylation of Skn7p is required for activation of genes required in response to wall stress, but Skn7p also activates oxidative stress response genes in an aspartyl phosphorylation-independent manner. The presence of binding sites for the Yap1p and Skn7p transcription factors in oxidative stress response promoters and the oxidative stress-sensitive phenotypes of SKN7 and YAP1 mutants suggest that these two factors work together. We present here evidence for a DNA-independent interaction between the Skn7 and Yap1 proteins that involves the receiver domain of Skn7p and the cysteine-rich domains of Yap1p. The interaction with Yap1p may help partition the Skn7 protein to oxidative stress response promoters when the Yap1 protein accumulates in the nucleus.
INTRODUCTION
Approximately half of the 71 proteins induced by oxidative stress depend on Yap1p, a well-characterized oxidative stress-responsive B-ZIP transcription factor whose subcellular localization is under redox control (9, 20), while the other half, including TRX2, TRR1, GPX2, and CCP1, for example, are regulated by both Yap1p and a second oxidative stress response transcription factor, Skn7p (24). The codependence of this large set of oxidative stress response (OXR) genes on the activity of both transcription factors has been studied in some detail (4, 14, 24, 30, 41). In contrast, only two OXR genes (DNM1 and OLA1) are reported to depend on Skn7p alone (24). This raises the question of whether the role of Skn7p in oxidative stress is distinct from that of Yap1p.
Two types of Skn7p response elements and two types of Yap1p response elements have been identified (14, 25, 26, 31). The preferred Yap1p response element (YRE) is T(T/G)ACTAA consisting of a pair of inverted TTA half-sites or a TGA/TTA pair (11, 21, 49), but we identified an alternative YRE [T(T/G)ACAAA] in YAP1-dependent genes such as TSA1 that lack the preferred sequence (14). The function of the alternate YRE was confirmed for the TSA1 gene and others using reporter assays and DNA-binding studies (14, 24). In the present study we consider whether Yap1p is also able to associate indirectly with OXR promoters via interactions with other DNA-binding proteins such as Skn7p.
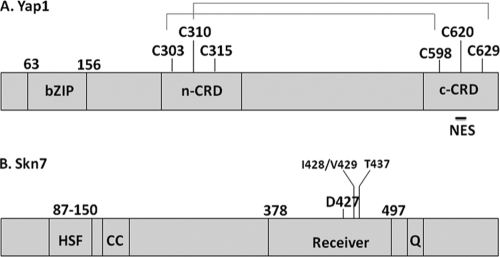
The Yap1 protein is regulated by subcellular localization. Under non-stress conditions the export receptor, Crm1p, binds to the Yap1p nuclear export signal and escorts Yap1p from the nucleus to the cytoplasm (20, 22, 50). Hydrogen peroxide stress leads to disulfide bond formation between cysteines within the C-terminal cysteine-rich domain (c-CRD; C598, C620, and C629) and the N-terminal cysteine-rich domain (n-CRD; C303, C310, and C315) (9, 50), causing conformational changes that prevent the interaction with Crm1p and result in nuclear accumulation of Yap1p (9, 10, 20, 35, 46). Both C303-C598 and C310-C629 disulfides have been documented in cells challenged with hydrogen peroxide (32, 46, 47) (Fig. 1A). More recently, proper folding of Yap1p in the presence of hydrogen peroxide has also been shown to be required for transcriptional activation via recruitment of the mediator protein, Rox3 at some promoters (13).
Fig. 1.
Domain organization of the Skn7p and Yap1p transcription factors. (A) Schematic diagram of major domains of the Yap1 protein, including the positions of the two cysteine-rich domains (n-CRD and c-CRD) and the nuclear export sequence (NES). Disulfide bonds formed in response to hydrogen peroxide stress are also shown. (B) Domain organization of the Skn7 transcription factor, showing the heat shock factor like DNA-binding domain (HSF), the coiled-coil domain (CC), the receiver domain (Receiver), and the glutamine-rich (Q) domains. Amino acid positions of domains and of critical residues mentioned in the text are indicated above both drawings.
The hydrogen peroxide signal is initially sensed by the Gpx3p peroxidase (10). Gpx3p transduces the signal to Yap1p via formation of a transient Yap1p-Gpx3p disulfide intermediate that involves Yap1p C598. The Yap1p-Gpx3p disulfide resolves into intramolecular Yap1p disulfides (10). In some strain backgrounds, the Yap1p binding protein Ybp1p is also required for hydrogen peroxide activation of Yap1p. The Gpx3p-Yap1p intermediate cannot be formed in the absence of Ybp1p (42).
In contrast to the redox-based regulation of Yap1p localization, current data indicate that Skn7p is constitutively nuclear (2, 28, 34) and able to associate with OXR promoters independent of oxidative stress. The Skn7 protein has two highly conserved domains, an N-terminal DNA-binding domain that is related to the DNA-binding domain of heat shock factor and a C-terminal receiver domain (Fig. 1B). The receiver domain contains the aspartic acid residue (D427) required for carrying out the aspartate-based phosphoacceptor function of Skn7p in SLN1-SKN7 His-Asp-based signal transduction (2, 3, 18, 25). Sln1p-dependent aspartyl phosphorylation of Skn7p is triggered by cell wall stress (36) and is distinct from the OXR function of Skn7p. The skn7 D427N mutation, which abolishes the aspartyl phosphoaccepting activity of the receiver domain, does not affect expression of OXR genes, nor does it cause the oxidative stress sensitivity seen in the skn7Δ mutant (15, 30).
Although the aspartate residue is dispensable, both the DNA-binding and the receiver domains are required for the OXR function of Skn7p. Certain receiver domain mutants exhibit sensitivity to oxidants (OxS), including hydrogen peroxide and tert-butyl hydroperoxide (t-BOOH) (15) and SKN7 constructs encoding fusions of the receiver domain to heterologous DNA-binding domains, are capable of mediating an oxidative stress response (4, 14, 19, 33).
We previously used electrophoretic mobility shift assays (EMSAs) to show Skn7p and Yap1p codependent complexes at various OXR gene promoters (14, 15). The Yap1p+Skn7p+DNA complex is disrupted by specific mutations in the receiver domain of SKN7 that do not affect the Skn7p+DNA complex but do cause an OxS phenotype (14, 15). These observations suggest that Skn7p+Yap1p complex formation requires more than both transcription factors binding to the promoter in close proximity and lead to the hypothesis that the two proteins interact directly.
We address here two important questions pertaining to the activation of OXR genes by the Skn7p and Yap1p transcription factors. First, does the joint activity of the Skn7p and Yap1p transcription factors depend on the presence of DNA containing both Skn7p and Yap1p response elements? Second, which domains of Skn7p and Yap1p are required for activating OXR genes that depend on both factors? In brief, we demonstrate that joint regulation of OXR genes by the two transcription factors requires only one binding site because the two factors are able to interact. We find that this interaction requires the receiver domain of Skn7p and the cysteine-rich domains of Yap1p.
MATERIALS AND METHODS
Media and growth conditions.
Rich medium (yeast extract-peptone-dextrose [YPD]) and synthetic complete medium lacking one or more amino acids (e.g., leucine) were prepared as previously described (37). t-BOOH (Acros Organics catalog no. 180342500) was a 70% (wt/vol) solution. All yeast strains were grown at 30°C.
Strains and plasmids.
Strains (Table 1) designated JF are isogenic derivatives of S288C from the Fassler laboratory collection. The YBP1 and GPX3 deletion strains were obtained from the BY4742 MATα deletion collection (Research Genetics) (16), and the SKN7-TAP-tagged strain was obtained from the C-terminally TAP-tagged collection in the BY4741 MATa background (Open Biosystems) (12). Bacterial strain BL21 Star (DE3) (Invitrogen) was used for the in vitro protein coprecipitation assay.
Table 1.
Strains used in this study
| Strain | Genotype | Notes | Source or reference |
|---|---|---|---|
| JF1565 | MATα canRcyhRhis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | 40 | |
| JF1904 | MATα skn7Δ::TRP1 canRcyhRhis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | skn7Δ::TRP1 one-step disruption in JF1565 | |
| JF2081 | MATayap1Δ canRcyhRhis3Δ200 leu2Δ1 ura3-52 trp1Δ63 | yap1Δ disruption in JF1562 | |
| JF2312 | MATα skn7Δ::TRP1 yap1Δ canRcyhRhis3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | skn7Δ::TRP1, yap1Δ derivative of JF2217 with URA3 removed with 5-FOA | |
| JF2413 | MATayap1Δ::KanMX trp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ lys2::GAL1-HIS3 met2::GAL7-lacZ ade2::GAL2-ADE2 | yap1Δ::KanMX one-step disruption in pJ69-4A | 17 |
| YHR206W (SKN7)-TAP | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YHR206W-TAP | TAP-tagged collection | Open Biosystems |
The plasmids used in the present study are listed in Table 2. pXH1853 contains SKN7 (−1293/+1866)–2×MYC–SKN7 3′ untranslated region (3′UTR; +1870/+2173) in the CEN plasmid, pRS316 (CEN, LEU2) (15, 38). This plasmid and its derivatives were constructed by using NotI and XhoI sites to clone SKN7 into the pRS316 backbone. The SKN7 sequence was amplified from JF1565 (Table 1). The myc epitope tag was cloned at the 3′ end of SKN7 by amplifying the myc sequence from pU6 H2myc (7). The SKN7 3′UTR sequence was amplified from the JF1565 genome. The sequences of the primers used to amplify these and other sequences are available upon request. pXH1854 (skn7 T437A), pXH1856 (skn7 T449A), pXH1858 (skn7 D427N), and pXH1928 (skn7 I428A/V429A) and pXH1939 (skn7 T449A in pRS425, 2μ, LEU2) have been described previously (15).
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pXH1853 | SKN7 (−1293/+1866)–MYC–3′UTR in pRS315 (CEN, LEU2) | 15 |
| pXH1854 | Skn7 T437A (−1293/+1866)–MYC–3′UTR in pRS315 (CEN, LEU2) | 15 |
| pXH1855 | Skn7 T455A (−1293/+1866)–MYC–3′UTR in pRS315 (CEN, LEU2) | 15 |
| pXH1928 | Skn7 I428A/V429A (−1293/+1866)–MYC–3′UTR in pRS315 (CEN, LEU2) | 15 |
| pXH1939 | Skn7 T449A (−1293/+1866)–MYC–3′UTR in pRS425 (2μ, LEU2) | 15 |
| pXH1941 | skn7 I428A/V429A (−1293/+1866)–MYC–3′UTR in pRS425 (2μ, LEU2) | 15 |
| pXH1957 | pGBD-C(2)-SKN7 +1051/+1572–MYC | Vector (17) |
| pSD2012 | 3×MYC in pET28(+) | 8 |
| pSD2013 | SKN7 (−1 to +582)–3×MYC His6 in pET28(+) | 8 |
| pSD2014 | SKN7 (−1 to +1045)–3×MYC His6 in pET28(+) | 8 |
| pSD2015 | SKN7 (−1 to +1870)–3×MYC His6 in pET28(+) | 8 |
| pSD2016 | skn7 D427N (−1 to 1870)–3×MYC His6 in pET28(+) | 8 |
| pSD2017 | skn7 D427E (−1 to 1870)–3×MYC His6 in pET28(+) | 8 |
| pSEY18-R2.5 | YAP1 in pRS426 (2μ, URA3) | W. S. Moye-Rowley (48) |
| pSM58wt | YAP1 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (44) |
| pJAW1 | yap1 Δ220-335 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (44) |
| pJAW76-2 | yap1 Δ322-469 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (43) |
| pJAW6 | yap1 Δ220-430 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (43) |
| p951 | yap1 Δ220-243 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (5) |
| p952 | yap1 Δ220-307 in pRS316 (CEN, URA3) | W. S. Moye-Rowley (5) |
| p976 | yap1 C303A in pRS316 (CEN, URA3) | W. S. Moye-Rowley (5) |
| pTYH6 | HA-YAP1 in YCp50 (CEN, URA3) | W. S. Moye-Rowley (39) |
| pSL26 | HA-YAP1 in YEp351 (2μ, LEU2) | W. S. Moye-Rowley |
| pSMS38 | HA-yap1 CSE598AAA in YEp351 (2μ, LEU2) | W. S. Moye-Rowley |
| pSL-C629A | HA-yap1 C629A in pRS315 (CEN, LEU2) | S. Lee |
The one-hybrid plasmid, pXH1957, was constructed by ligating PCR-amplified SKN7 (+1051/+1572)−2×MYC from the pXH1853 template into BamHI and HindIII sites downstream of GAL4-DBD in the plasmid pGBD-C2 (17).
The His6 plasmids pSD2013 to pSD2017 were constructed by introducing PCR-amplified SKN7 fragments (−1/582 [HSF domain only]; −1/1045 [HSF and CC domains]; and −1/1870 [full length]) using the JF1565 genomic template with added BamHI and SalI sites into BamHI/SalI-digested pSD2012, a pET-28a(+) vector with a 3×MYC tag inserted at unique HindIII and XhoI sites. The resulting plasmids express Skn7–3× myc–His6 fusion genes under the control of a T7 promoter.
Northern blot analysis.
Yeast strains JF1565, JF1904, JF2081, and JF2312 were grown in YPD at 30°C to 1 × 107 to 2 × 107 cells/ml. Log-phase cultures were treated with 0.5 mM t-BOOH for 10, 20, 30, and 40 min at 30°C. Cells were harvested by centrifugation at 5,000 rpm for 5 min, and the resulting pellets were frozen at −80°C. RNA was prepared using hot acidic phenol (1). A portion (10 μg) of each RNA sample was analyzed on a 1.2% agarose–1× morpholinepropanesulfonic acid (MOPS)–3.7% formaldehyde gel and transferred to a Nytran membrane in 10× saline sodium citrate (SSC). Hybridization was performed in Perfect Plus Pre-Hybridization solution (Sigma) at 68°C overnight in the presence of α-32P-labeled DNM1 (+929/+1348), OLA1 (+878/+1205), TRX2 (−160/+324), or PGK1 probes. The blot was then washed twice in 2× SSC–0.1% sodium dodecyl sulfate (SDS) and twice in 0.5× SSC–0.1% SDS before being exposed to a PhosphorImager screen. Values were normalized to PGK1, and induction was calculated relative to the value of the untreated culture.
One-hybrid assays.
Yeast strain JF2413 containing a UASG-lacZ reporter gene was transformed with pXH1957 (Skn7-RD+1051/+1572) and individual YAP1 plasmids. Transformants were grown at 30°C in synthetic complete medium lacking the appropriate amino acids to 1 × 107 to 2 × 107cells/ml. Log-phase cultures were treated with 0.5 mM t-BOOH at 30°C for 30 min. Treated and untreated cells were harvested by centrifugation at 5,000 rpm for 5 to 10 min, and the resulting pellets were frozen at −80°C. The beta-galactosidase activity was measured in modified Miller units (29) and normalized to the protein concentration. The data represent average values ± the standard deviations for triplicate assays. Oxidative stress induction was determined by comparing the average value for the treated samples to the average value for the untreated samples of the same strain.
Oxidative stress spot assays.
Yeast strain JF1904 was transformed with pXH1853 (SKN7+), pXH1854 (skn7 T437A), or pXH1928 (skn7 I428A/V429A) and various YAP1 plasmids. Cells were grown at 30°C in synthetic complete medium lacking the appropriate amino acids to 107cells/ml. Serial dilutions of each culture were then spotted onto plates with YPD plus 0, 0.375, 0.5, 0.75, or 1 mM t-BOOH, starting with 106 cells/ml. All plates were incubated at 30°C for 3 days with growth scanned after 2 and 3 days. The best images are shown.
Coimmunoprecipitation.
Yeast strain JF1904 transformed with pXH1853 (SKN7+), pXH1854 (skn7 T437A), pXH1941 (skn7 I428A/V429A), or pXH1939 (skn7 T449A) and pSEY18-R2.5 (YAP1, 2μ), and yeast strain JF2117 (yap1Δ) transformed with pXH1853 were grown at 30°C in synthetic complete medium lacking the appropriate amino acids to 1 × 107 to 2 × 107 cells/ml. Protein extracts were prepared as described previously (27) and precleared by incubation with 50% Sepharose G-beads (4 Fast Flow; GE Healthcare) at 4°C for 20 min prior to incubation with α-Myc antibody (UI Hybridoma; 1:100) overnight at 4°C. After the addition of a 50% Sepharose G-bead slurry, the beads were collected by centrifugation, washed four times in HNTG (20 mM HEPES, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol), incubated at 95°C for 5 min in 2× Laemmli buffer (0.125 M Tris [pH 6.8], 3% SDS, 20% sucrose, 0.02% bromophenol blue, 20% bovine serum albumin), and loaded on SDS–10% PAGE gels for Western blot analysis with α-Myc (1:1,000 to 1:2,500 in 5% milk) and α-Yap1p (a gift from S. Moye-Rowley; 1:2,000 in 5% milk). The secondary antibodies were horseradish peroxidase (HRP)-conjugated goat anti-mouse (Jackson Immunoresearch; 1:3,000 in 5% milk) and goat anti-rabbit (Sigma catalog no. A4914; 1:3,000 in 5% milk). The signal was detected by chemiluminescence using a SuperSignal West Pico kit (Pierce).
For immunoprecipitation experiments with TAP-tagged Skn7, the SKN7-TAP strain (from the TAP strain collection) and the skn7Δ yap1Δ strain JF2312 were transformed with the HA-YAP1 (CEN) plasmid, and cultures were treated with 2 mM t-BOOH for 20 min. Cells were collected by centrifugation, and pellets were stored at −80°C prior to being resuspended in 30 mM Tris-HCl (pH 8.0), 5 mM EDTA, 3 mM dithiothreitol, and 5% glycerol supplemented with phosphatase and protease inhibitors (sodium fluoride, phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail (PIC), and sodium orthovanadate) for extract preparation by glass bead lysis. Precleared extracts were mixed with α-TAP antibody (0.5 μg/μl; Open Biosystems) and incubated at 4°C overnight prior to addition of a 50% Sepharose bead-protein G slurry and 1 h of further incubation. Beads were collected by centrifugation, washed once in HNTG plus 1 M NaCl and three times in HNTG buffer, resuspended in 2× Laemmli buffer (23) with 20% BSA, and heated to 95°C prior to being loaded on long-format (to resolve hemagglutinin [HA]-Yap1p from a nonspecifically associated protein) 10% SDS-polyacrylamide gels. After transfer, the blots were blocked in 1× Tris-buffered saline–Tween with 5% milk and incubated overnight with α-TAP (Open Biosystems, 1:2,500 in 5% milk) or α-HA antibody (12CA5; 1:1,500 in 0.5% milk) and for 2 h with goat anti-rabbit (Sigma catalog no. A4914; 1:3,000 in 5% milk) (TAP) or goat α-mouse secondary antibodies (Jackson Immunoresearch; 1:3,000 in 5% milk) (HA).
Induction and purification of Skn7-His6 protein from Escherichia coli.
Recombinant Skn7-His6 protein was expressed from plasmid pSD2015 in BL21 Star (DE3) cells after treatment with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 16°C for 20 h. Cultures were harvested by centrifugation and stored at −20°C. The pellets were resuspended in 1× bind buffer (Novagen His-Bind kit, catalog no. 70239-3) supplemented with 1 mM PMSF, PIC, and RNase. Lysozyme was added to 100 μg/ml, and the cells were incubated for 15 min at 30°C, followed by 10 freeze-thaw cycles in a dry ice-ethanol bath. Lysates were cleared by centrifugation prior to incubation with the His-Bind resin for 30 min at room temperature. Washes were performed by using the batch method described in the manufacturer's instructions. The extent and quality of the recovered protein was evaluated by SDS–10% PAGE, followed by Coomassie blue staining (1).
Skn7p-His6 pull-down experiments.
Portions (0.5 mg) of cleared yeast protein extract in coprecipitation buffer (50 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 140 mM NaCl, 0.1% Triton X-100, 5 mM EDTA, 1 mM dithiothreitol, and 1 mM PMSF plus PIC [Roche]) were diluted with cold coprecipitation buffer plus 1% bovine serum albumin to a volume of 500 μl and then mixed with 15 μl of Skn7p-His6-bound His-Bind resin. Mixtures were incubated with rocking in the cold for 12 to 24 h and then washed twice in coprecipitation buffer, twice in phosphate-buffered saline (PBS) plus 0.1% Tween 20, and twice in PBS plus 0.2% Tween 20 with 5 min on ice between washes. The resin was then mixed with 5× LSB (15% SDS, 0.575 M sucrose, 0.325 M Tris-HCl [pH 6.8], 5% β-mercaptoethanol, 0.002% bromophenol blue) loading buffer, and the proteins were separated by SDS–10% PAGE. Western blot analysis was conducted using α-Yap1 primary antibody (a gift from S. Moye-Rowley; 1:2,000 in 5% milk) and HRP-conjugated goat anti-rabbit secondary antibody (Sigma catalog no. A4914; 1:3,000 in 5% milk).
RESULTS
Expression of candidate SKN7-dependent, YAP1-independent OXR genes.
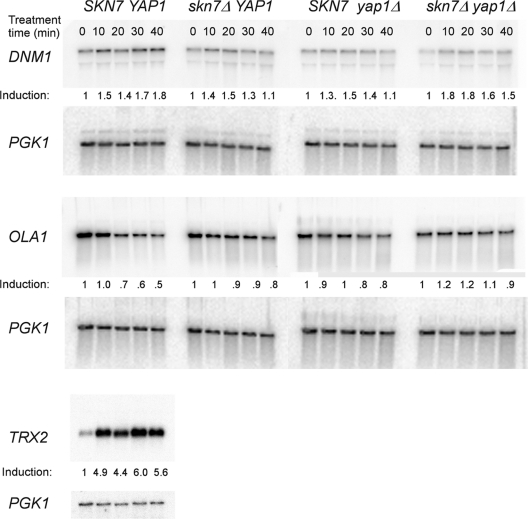
Oxidative stress induction of many OXR genes is nearly equally sensitive to the loss of SKN7 or YAP1 and is virtually the same in a skn7Δ yap1Δ double mutant as in either single mutant (14, 15, 19, 30, 41). Hence, the prevailing idea has been that the two transcription factors work together. To specifically investigate whether any OXR genes are regulated by SKN7 but not by YAP1, we examined two genes (DNM1 and OLA1) previously identified in proteomic scale two-dimensional polyacrylamide gel experiments, whose oxidative stress response appeared to depend on SKN7 but not YAP1 (24). A time course analysis of DNM1 and OLA1 expression in wild-type strains showed modest (1.5-fold for DNM1) or no (0.5-fold OLA1) induction after treatment with 0.5 mM t-BOOH. As a control, we found that the same cultures exhibited normal 5-fold induction of the SKN7- and YAP1-dependent TRX2 gene (Fig. 2) (30). The 1.5-fold induction of DNM1 expression seen at all treatment times was not affected by deletion of skn7 or yap1 or by simultaneous deletion of both genes. The expression of OLA1 was slightly increased by the mutation of both skn7 and yap1. We conclude that even if DNM1 and OLA1 were OXR genes with subtle induction properties, neither qualifies as a gene whose expression is dependent on SKN7 alone. Thus far, no known OXR genes exhibit dependence on SKN7 but not on YAP1.
Fig. 2.
Expression of the DNM1 and OLA1genes is largely unaffected by oxidative stress and by SKN7. RNA isolated from yeast strains JF1565 (SKN7 YAP1), JF1904 (skn7Δ), JF2081 (yap1Δ), and JF2312 (skn7Δ yap1Δ) was analyzed by Northern blot analysis for DNM1, OLA1, TRX2, and PGK1 gene expression. All strains were treated with 0.5 mM t-BOOH for 0, 10, 20, 30, or 40 min. The probes were as follows: DNM1 (+929/+1348), OLA1 (+878/+1205), and TRX2 (−160/+324). Values were normalized to PGK1 expression. Induction, shown below each panel, was calculated using the normalized value for the untreated (0 min) lane in the denominator.
SKN7 does not regulate expression of Yap1p activators.
SKN7 activation of Yap1p or genes encoding Yap1p activators is one possible explanation for the requirement for Skn7p in Yap1p-mediated activation of genes in response to oxidative stress. No differences in Yap1 protein levels have been observed in SKN7 versus skn7Δ extracts. Thus, regulation of Yap1p expression by SKN7 is unlikely (J. S. Fassler and K. E. Mulford, unpublished results). However, regulation of the Yap1p regulators, GPX3 and YBP1 has not been tested. To examine the potential for indirect regulation of Yap1p activity by SKN7, the expression of the Yap1p regulators, GPX3 and YBP1, which are required for the Yap1p response to hydrogen peroxide (10, 13, 42), was examined. Wild-type, skn7, yap1, and skn7 yap1 double-mutant strains were treated with t-BOOH, and the levels of GPX3 and YBP1 RNA were analyzed in Northern hybridization experiments. No significant effect of SKN7 on GPX3 or YBP1 expression was observed in t-BOOH-treated or untreated cultures. Likewise, no effect was seen on their expression by YAP1. Finally, there was no evidence that the expression of either GPX3 or YBP1 was inducible by oxidative stress (data not shown).
The Skn7 and Yap1 proteins interact.
The formation of complexes on the promoters of OXR genes that are codependent on Skn7p and Yap1p was previously established by using EMSAs (14, 15, 24). Such complexes are sensitive to mutations in the receiver domain of Skn7p (e.g., skn7 T437A and skn7 I428A/V429A) that confer an OxS phenotype but that do not affect DNA binding or the nuclear localization of the Skn7 protein (15).
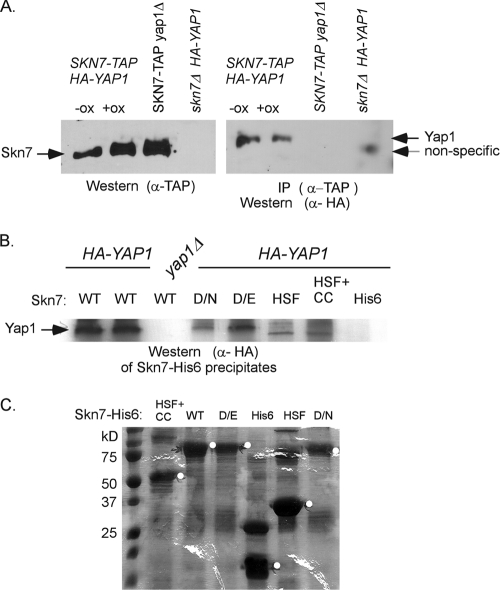
To examine the Skn7p-Yap1p interaction in cells in which neither protein is highly overexpressed, a strain carrying a genomic SKN7-TAP allele driven by the native SKN7 promoter was used in conjunction with a low-copy-number plasmid expressing YAP1-HA under the control of the YAP1 promoter. α-TAP antibody was used to precipitate Skn7p from whole-cell extracts. Immunoprecipitates were then analyzed by using both α-TAP antibody to detect Skn7p and α-HA antibody to detect Yap1p (Fig. 3A). Yap1p was detected in α-TAP precipitates of Skn7p-containing extracts whether the extracts were prepared from oxidant treated or untreated cultures and was not detected in extracts prepared from skn7Δ strains or from strains lacking the YAP1-HA plasmid. This suggests that Yap1p is specifically associated with Skn7p even when Skn7p is present at physiological levels.
Fig. 3.
Coimmunoprecipitation of Yap1p depends on the Skn7p receiver domain. (A) Coimmunoprecipitation of HA-Yap1p in extracts expressing genomic SKN7-TAP. The presence of the TAP-tagged Skn7p in oxidant-treated and untreated extracts is shown in the α-TAP Western blot on the left. HA-Yap1p that immunoprecipitated with Skn7p-TAP is shown in the α-HA Western blot on the right. A nonspecific band is sometimes seen in extracts prepared from skn7Δ extracts. However, prolonged electrophoresis (shown) reveals that it migrates faster than HA-Yap1p. (B) Coprecipitation of HA-Yap1p with recombinant full-length Skn7p-His6 (WT), full-length Skn7p-D427N-His6 (D/N), full-length Skn7p-D427E-His6 (D/E), and truncated Skn7p-His6 constructs lacking the receiver domain but including the HSF DNA binding and the coiled-coil domains (HSF+CC) or including the HSF DNA-binding domain only (HSF). (C) SDS–10% PAGE and Coomassie blue staining of equivalent bead volumes (equal to 1/5 of the total) associated with each Skn7p-His6 protein. The remaining (4/5 total volume) beads were mixed with 0.5 mg of each yeast extract for the pull-down experiment shown in panel B except for wild-type beads, which were divided, with 2/5 being added to YAP1+ extract and 2/5 added to yap1Δ extract (first and third lanes in panel B). The white circles on the gel are to the right of bands migrating at the expected molecular mass of each Skn7p-His6 species.
To investigate the Skn7p determinants required for the interaction, His6-tagged Skn7 proteins were expressed and purified from bacteria and then added to yeast whole-cell extracts. Full-length His6-tagged Skn7p precipitated Yap1p as expected (Fig. 3B). Yap1p was also found in association with recombinant Skn7D427N-His6 and Skn7D427E-His6 proteins (Fig. 3B). The skn7 D427N and skn7 D427E mutations are known to affect the SLN1-SKN7 pathway but not the oxidative stress pathway (15, 19, 26, 30). The association of Yap1p with recombinant Skn7 proteins was dependent on the presence of Skn7p since Yap1p was not present in His6 vector control reactions (Fig. 3B, “His6”). To further examine the role of the receiver domain in the interaction with Yap1p, two truncated proteins were used in the same type of experiment. One lacked the receiver domain (HSF and CC), and the other truncation protein lacked both the receiver domain and the coiled-coil domain (HSF). Neither truncated protein precipitated detectable levels of Yap1p (Fig. 3B). These results are consistent with the need for the Skn7p receiver domain in an interaction with Yap1p.
Finally, α-Myc immunoprecipitation was used to investigate the effect of three OxS skn7 receiver domain mutations on the Yap1p-Skn7p association. Protein extracts were prepared from strains expressing untagged YAP1 from a high-copy-number plasmid and various skn7-MYC alleles from a CEN plasmid. Immunoprecipitation of Yap1p was significantly less efficient in extracts prepared from skn7 T437A, skn7 T449A, and skn7 I428A/V429A mutants than in the wild-type SKN7 strain. The average recovery of Yap1p relative to that in the SKN7 strain was 23% in the skn7 T437A mutant, 11% in the skn7 T449A mutant, and 17% in the skn7 I428A/V429A mutant (Table 3). Since the OxS phenotype of skn7 receiver domain mutations correlates with the absence of a Skn7p-Yap1p complex in EMSA experiments and is partially suppressed by high-copy YAP1 in some cases (15), these numbers may be underestimates of the effects of the skn7 mutations on the interaction with the Yap1 protein.
Table 3.
Coimmunoprecipitation of Yap1p with Skn7p
| Parameter | SKN7+a | skn7Δ | T455A | T437A | T449A | I428A/V429A | SKN7+yap1Δ |
|---|---|---|---|---|---|---|---|
| Avg Yap1/Skn7b | 3.08 | NDd | 1.39 | 0.59* | 0.58* | 0.50* | ND |
| SD | 1.34 | – | 0.18 | 0.2 | 0.06 | 0.04 | – |
| % Recoveryc | 100 | – | 45.1 | 19.2 | 18.8 | 16 | – |
Protein extracts were prepared from the skn7Δ strain, JF1904, carrying the SKN7-MYC plasmids pXH1853 (SKN7+), pXH1855 (skn7 T455A), pXH1854 (skn7 T437A), pXH1939 (skn7 T449A), or pXH1941 (skn7 I428A/V429A), as well as the YAP1 expression plasmid pSEY18-R2.5.
Average Yap1/Skn7 values represent the average intensity of the Yap1p band from the immunoprecipitate relative to the intensity of the Skn7p band from the beads in each reaction mixture in three experiments. Asterisks indicate that the ratio for the mutant is significantly different (P<0.05) from the wild-type ratio. The skn7 T455A mutant, which is not oxidative stress sensitive, is included as a control.
The percent Yap1p recovered is the relative amount of Yap1 protein in the immunoprecipitate of skn7 mutant extracts compared to that in SKN7 wild-type extracts.
ND, not detectable.
Identifying Yap1p determinants needed for interaction with Skn7p: one hybrid analysis.
Previously, we found that oxidative stress activation of a UASG-lacZ reporter with a GAL4DBD-SKN7 (bp +1051/+1866) fusion protein depended on the presence of Yap1p, as well as certain amino acid residues in the receiver domain of Skn7p (15). Since the lacZ reporter gene does not contain Yap1p binding sites and a GAL4DBD construct lacking Skn7 sequences exhibited no activation that could be attributed to fortuitous binding of Yap1p to the reporter (15), these results suggest that oxidative stress activation of the reporter requires an interaction between the receiver domain of Skn7p and Yap1p.
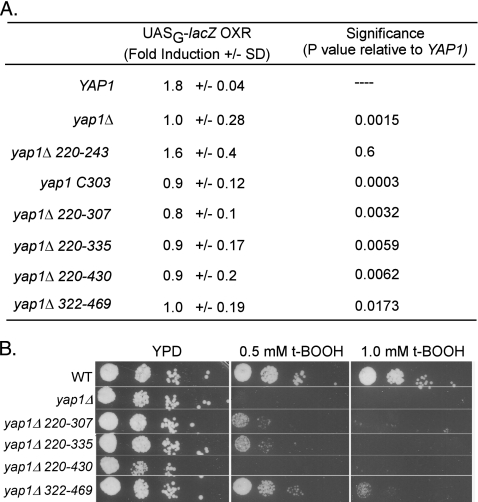
In the present study, the features of Yap1 protein required for oxidative stress activation were investigated by conducting the assay in a yap1Δ strain into which plasmids bearing different yap1 alleles were introduced. In the presence of a plasmid carrying a wild-type YAP1 allele, expression of the lacZ reporter gene was induced 1.8-fold by oxidant treatment, while strains lacking a YAP1 plasmid showed no induction (Fig. 4A). A collection of YAP1 expression plasmids having small deletions or point mutations (Δ220-243, Δ220-307, Δ220-335, Δ220-430, Δ322-469, and C303A) were also assayed. These mutant Yap1 proteins are expressed at normal levels and exhibit robust activation in response to diamide (5, 43, 44). All but the Δ220-243 and Δ322-469 alleles were previously found to confer sensitivity to hydrogen peroxide (5, 43). The t-BOOH phenotypes (Fig. 4B) for these mutants were consistent with previous reports of H2O2 sensitivity with the exception of the Δ322-469 mutant, which exhibits modest sensitivity to t-BOOH but not to H2O2 (43). In the one-hybrid test, the OxR yap1 Δ220-243 mutant was induced normally in oxidant-treated cultures, indicating that this region is not essential for the Skn7p-Yap1p interaction (Fig. 4A). In contrast, the yap1 C303A, Δ220-307, Δ220-335, and Δ220-430 mutants were not induced upon oxidative stress (Fig. 4A). Thus, amino acids 243 to 307 and, in particular, residue C303, are required for the Skn7p-Yap1p interaction.
Fig. 4.
t-BOOH sensitivity of YAP1 alleles and their effect on Gal4DBD-Skn7RD hybrid protein activity. (A) t-BOOH-mediated induction of the Gal4DBD-Skn7RD hybrid protein in the presence of various YAP1 alleles. YAP1 alleles were transformed into skn7Δ yap1Δ strain JF2413 carrying the Gal4-Skn7p fusion on pXH1957. The YAP1 plasmids were as follows: YAP1+, pSM58; yap1Δ (vector); yap1 Δ220-243, p951; yap1 Δ220-307, p952; yap1 Δ220-335, pJAW1; yap1 Δ220-430, pJAW6; yap1 Δ322-469, pJAW76-2; and yap1 C303, p976. Fold induction values are the averages ± the standard deviations of three to six measurements made using different transformants. Significance was determined using an unpaired Student t test and reflects the probability that the average induction value for the mutant yap1 allele differs from the average induction value for wild-type YAP1. (B) A plate assay showing t-BOOH sensitivity of yap1 alleles relative to YAP1+ (WT) and null (yap1Δ) strains.
The yap1 Δ322-469 deletion also abolished induction (Fig. 4A), which is consistent with its t-BOOH-sensitive phenotype (Fig. 4B). This deletion should not affect cysteine bond formation since neither cysteine-rich domain is eliminated. Taken together, the data suggest that the n-CRD and amino acids 322 to 469 located between the two CRDs are important for full Skn7p-Yap1p-mediated oxidative stress induction.
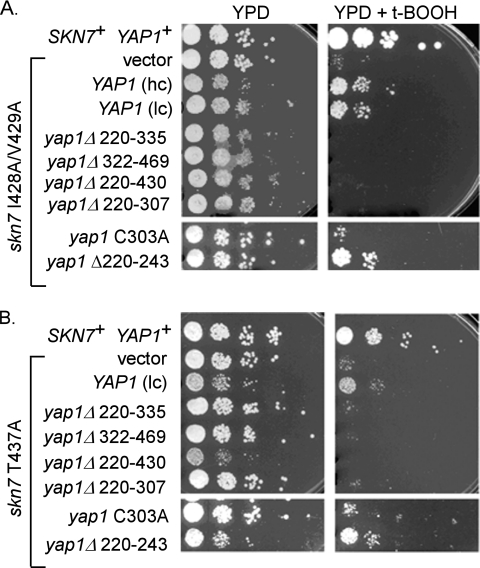
Overexpression of YAP1 suppresses the oxidative stress sensitivity of skn7 I428A/V429A and skn7 T437A mutants.
The oxidative stress-sensitive phenotype of the skn7 I428A/V429A and skn7 T437A receiver domain mutations, visible as reduced viability on oxidant plates, is suppressed by modest overexpression of YAP1 (15). We therefore used this assay to examine the requirement for the C303 residue and the 322-469 region of Yap1p (Fig. 5). YAP1 overexpression plasmids (both low copy and high copy [lc and hc in Fig. 5]) containing wild-type and mutant forms of YAP1 were introduced into YAP1+ skn7 T437A and skn7 I428A/V429A mutant strains and the oxidative stress phenotype of the transformants was evaluated. The oxidative stress-sensitive phenotype of the skn7 I428A/V429A and skn7 T437A mutants was suppressed by overexpression of the wild-type YAP1 and the yap1 Δ220-243 alleles (Fig. 5, right panel). Overexpression of other YAP1 deletion alleles or the yap1 C303A allele did not appreciably suppress the oxidative stress phenotype of the skn7 mutants. These results support the conclusion from the one-hybrid assays that C303 and the 322-469 region of Yap1p each play a role in the interaction of Yap1p with Skn7p.
Fig. 5.
Overexpression of YAP1 but not yap1 C303 suppresses oxidative stress sensitivity of skn7 I429A/V429A (A) and skn7 T437A (B) mutants. Oxidative stress assays of the yeast strain JF1904 containing plasmids carrying SKN7 (pXH1853), skn7 I428A/V429A (pXH1928), or skn7 T437A (pXH1854) with or without YAP1+ plasmids (pSEY18-R25, 2μ [high copy, hc], or pSM58, CEN [low copy, lc]) or yap1 mutant plasmids (pJAW1, pJAW76-2, pJAW6, p952, p976, or p951). Tenfold serial dilutions were spotted on YPD (untreated) or YPD plus 0.75 or 1 mM t-BOOH (treated) plates starting at 106cells/ml. The plates were incubated at 30°C for 2 to 3 days.
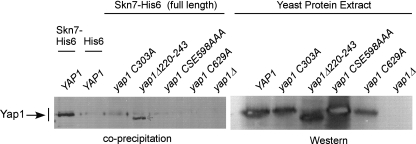
Yap1p association with Skn7p depends on both cysteine rich domains.
The one-hybrid analysis described above implicated the n-CRD of Yap1p and residues located between the CRDs in the Skn7p-Yap1p interaction. To further investigate the involvement of CRDs, the in vitro association of CRD mutant Yap1 proteins with full-length recombinant Skn7p-His6 was examined. As a control, the Yap1Δ220-243 mutant protein was efficiently precipitated in this experiment (Fig. 6), which was expected from the one-hybrid (Fig. 4) and high-copy (Fig. 5) suppression assays. In contrast, the Yap1 C303A mutant protein exhibited substantially reduced coprecipitation with Skn7p-His6, supporting the conclusion that the n-CRD is important for the Skn7p interaction, perhaps through disulfide bond formation. Furthermore, neither the Yap1CSE598AAA nor the Yap1C629A c-CRD mutant proteins were efficiently precipitated by Skn7p-His6. Taken together, these results indicate that both the c-CRD and the n-CRD are important for a normal interaction between Yap1p and Skn7p.
Fig. 6.
Yap1p coprecipitation with Skn7p depends on residues within the n- and c-CRDs of Yap1p. Shown are the results of an in vitro coprecipitation assay in which bacterially expressed Skn7p-His6 (pSD2015) or His6 alone (pSD2012) was mixed with yeast protein extracts prepared from a yap1Δ skn7Δ strain (JF2312) carrying plasmids expressing various YAP1 alleles, including YAP1 (pSM58), yap1 C303A (p976), yap1 Δ220-243 (p951), yap1 CSE598A (pSMS38), and yap1 C629A (pSL-C629A). α-Yap1 antibody (a gift from S. Moye-Rowley) was used to detect Yap1p in the precipitate (left) and to examine Yap1 protein levels in the extracts (right).
DISCUSSION
A role for a Skn7p-Yap1p protein-protein interaction at oxidative stress responsive promoters was previously hypothesized based on EMSA data in which Skn7p and Yap1p codependent complexes found at OXR promoters were disrupted by OxS mutations in the Skn7 receiver domain (15). That the two transcription factors can function collaboratively is clear from the similar OxS phenotypes conferred by skn7Δ and yap1Δ mutations in plate assays, the effects of single deletion mutations on OXR gene expression in Northern analysis, and the lack of additivity in the response of the double mutant (4, 14, 24, 30, 41). The presence of individual binding sites for each protein in many OXR gene promoters suggests that the two proteins may sit shoulder to shoulder on the DNA. The question we addressed here was whether an interaction between the two proteins contributes to the OXR.
Two candidate SKN7-dependent YAP1-independent genes (DNM1 and OLA1) were identified in two-dimensional gel analysis of the oxidative stress proteome (24). However, our analysis revealed no oxidant or SKN7-dependent regulation of DNM1 or OLA1 expression. One possible explanation for the discrepancy between our results and the proteome study is that the Dnm1 and Ola1 proteins are stabilized by Skn7p under oxidative conditions. Stabilization of the Skn7p-associated Crz1 protein has been reported (45). A more comprehensive transcriptome analysis may reveal true Skn7-dependent Yap1-independent OXR candidate genes, but thus far, none have come to light.
Oxidative stress-mediated activation of a UASG-driven reporter gene by the Gal4-Skn7RD fusion protein required YAP1 (Fig. 4). Fortuitous binding of Yap1p to UASG is unlikely because there was no activation of this reporter by the Gal4-DBD fusion protein (no Skn7p) when measured in a skn7Δ YAP1+ strain (15). YAP1 independence was reported in similar experiments performed in another laboratory (4); however, the same authors reported that a 16-fold induction of a lacZ reporter gene driven by a synthetic YAP1 dependent promoter was eliminated in a skn7Δ strain (4).
The requirement for Yap1p in this assay could reflect recruitment of Yap1p by Skn7p to the promoter. It could also indicate a conformational change in Skn7p caused by a transient interaction with Yap1p. Alternatively, it could reflect posttranslational modification of Skn7p requiring Yap1p. We previously found that Yap1p does play a role in serine/threonine phosphorylation of Skn7p (15). However, we also found that the need for Skn7p phosphorylation in the oxidative stress response can be partially bypassed by overexpressing YAP1 (15). The simplest interpretation is that it is not phosphorylation per se but rather the recruitment of Yap1p to the promoter that is the key to activation of OXR genes. This view is supported by the observation that the Skn7 T437A receiver domain mutant protein is successfully phosphorylated but is nonetheless unable to form a Skn7p-Yap1p-codependent complex in EMSAs (15). In addition, the OxS phenotype of the skn7 T437A mutant is partially suppressed by overexpression of the YAP1 gene. A reasonable interpretation is that a weak Skn7p-Yap1p interaction can be partially overcome by increasing the concentration of Yap1p. In sum, the evidence is consistent with oxidative stress activation of the UASG-lacZ reporter due to the recruitment of Yap1p to the promoter by the Gal4-Skn7p protein.
Several types of coprecipitation assays provide additional support for an interaction between Yap1p and Skn7p. First, the Yap1-HA fusion protein was coimmunoprecipitated by the Skn7-TAP protein (Fig. 3). In this experiment artifactual interactions were minimized through the use of genomic SKN7 and low-copy-number plasmid based YAP1. Second, coimmunoprecipitation of Yap1p with the Skn7p-MYC protein from yeast extracts was shown to be compromised by OxS receiver domain mutations (Table 3). In this experiment SKN7-MYC and various individual alleles of YAP1 were expressed from pairs (SKN7, YAP1) of centromere-based low-copy vectors. Third, the association of Yap1p (from yeast extracts) with purified recombinant His6-tagged Skn7p was shown to be dependent on the receiver domain of Skn7p (Fig. 3B) and the cysteine-rich domains of Yap1p (Fig. 6).
Although these experiments do not explicitly address the role of DNA in the interaction, we deem the involvement of DNA to be unlikely based on the observation that receiver domain mutations that do not affect Skn7p binding to DNA (15) nonetheless render the cell sensitive to oxidant and simultaneously diminish Yap1p complex formation. In addition, our results showing that chimeric Gal4 DBD-Skn7 receiver domain proteins that bind to a UASG element are nonetheless sensitive to the presence of the Yap1 protein and to Skn7p receiver domain residues indicate that the Skn7p-Yap1p interaction does not require specific DNA sequences.
Our initial examination of yap1 mutants leads to the conclusion that the cysteine-rich domains and disulfide bond formation in Yap1p play a role in the Skn7p interaction. Deletions that eliminate the C303 residue needed for disulfide bond formation with C598 in response to hydrogen peroxide (9) failed to induce expression of the lacZ reporter gene upon oxidative stress (Fig. 4). Likewise, overexpression of these yap1 alleles failed to suppress the OxS phenotype of the skn7 T437A and skn7 I428A/V429A mutants (Fig. 5). Finally, the Yap1 C303A, CSE598AAA, and C629A proteins precipitated poorly with recombinant Skn7p-His6, confirming that these three cysteine residues are required for a normal interaction between Yap1p and Skn7p (Fig. 6). The failure of Yap1 proteins lacking C303, C598, or C629 to interact with Skn7p could be due to the direct involvement of these residues in the Skn7p-Yap1p interaction or to their effects on the oxidized conformation of Yap1. Like the cysteine mutants, the Yap1 Δ322-469 protein was also defective in the one-hybrid (Fig. 4) and high-copy suppression (Fig. 5) assays, indicating that the spacing between the two CRDs might be important. Alternatively, the region between the CRDs may be part of the protein interface needed for a robust interaction with Skn7p.
In vivo, the Skn7p-Yap1p complex forms only when conditions permit oxidized Yap1p to accumulate in the nucleus. The yap1 mutations used in the present study affect Yap1p localization in various ways. For example, the Yap1 Δ220-243 protein is constitutively nuclear (6). This protein exhibits normal resistance to hydrogen peroxide (6), interacts normally with Skn7p-His6 (Fig. 6), responds to oxidative stress in the one-hybrid assay (Fig. 4), and suppresses the oxidative stress sensitivity of skn7 T437A and skn7 I428A/V429A mutants when overexpressed (Fig. 5). The Yap1 Δ220-307 and the Yap1 C303A proteins are also constitutively nuclear (6). These mutants are sensitive to hydrogen peroxide and defective in their interaction with Skn7p. Finally, the Yap1 Δ220-335 protein translocates normally to the nucleus upon oxidative stress (5) but is nonetheless sensitive to hydrogen peroxide and defective in interacting with Skn7p. Hence, it is clear that the Skn7p interaction defects exhibited by various Yap1p cysteine mutants is not due to aberrant Yap1p localization.
The involvement of the CRDs in the Yap1p interaction with Skn7p, as well as for Yap1p recruitment of the Rox3p mediator protein in transcriptional activation of TRX2 (13), suggests a possible relationship between these two events. For example, the cysteine-rich domains and proper folding of Yap1p may be required for formation or stabilization of a Skn7p-Yap1p complex which may be, in turn, a prerequisite for Rox3p recruitment. The details of such a model require further testing.
The oxidative stress-sensitive phenotypes of SKN7 and YAP1 mutants and the presence of binding sites for both the Yap1p and Skn7p transcription factors in many oxidative stress response promoters suggests that the factors work together in the oxidative stress response. We present evidence here for a protein-protein interaction between Skn7p and Yap1p that contributes to an efficient oxidative stress response. Since aspartyl phosphorylation of the Skn7p transcription factor in response to cell wall-activated SLN1-SKN7 signaling may cause redistribution of Skn7p to wall response genes, the interaction with Yap1p may guarantee the availability of the Skn7 protein when the redox-sensitive Yap1 protein accumulates in the nucleus.
ACKNOWLEDGMENTS
We acknowledge the technical support of G. Gingerich and the generosity of W. S. Moye-Rowley and members of his laboratory for plasmids, strains, α-Yap1 antibody, and helpful discussions. We also thank D. Weeks, R. Malone, and G. Gussin for critical review of the manuscript.
This study was supported by funding from the National Institutes of Health (GM056719 and GM068746) and by the Center for Biocatalysis and Bioprocessing at the University of Iowa to K.E.M.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Ausubel F. M., et al. 1989. Current protocols in molecular biology, vol. 1 John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 2. Brown J. L., Bussey H., Stewart R. C. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown J. L., North S., Bussey H. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charizanis C., Juhnke H., Krems B., Entian K. D. 1999. The oxidative stress response mediated via Pos9/Skn7 is negatively regulated by the Ras/PKA pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:740–752 [DOI] [PubMed] [Google Scholar]
- 5. Coleman S. T., Epping E. A., Steggerda S. M., Moye-Rowley W. S. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coleman S. T., Tseng E., Moye-Rowley W. S. 1997. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 272:23224–23230 [DOI] [PubMed] [Google Scholar]
- 7. De Antoni A., Gallwitz D. 2000. A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246:179–185 [DOI] [PubMed] [Google Scholar]
- 8. Dean S. 2004. Achieving specificity in yeast stress responses. Ph.D. thesis. University of Iowa, Iowa City, IA [Google Scholar]
- 9. Delaunay A., Isnard A. D., Toledano M. B. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471–481 [DOI] [PubMed] [Google Scholar]
- 11. Fernandes L., Rodrigues-Pousada C., Struhl K. 1997. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17:6982–6993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghaemmaghami S., et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 13. Gulshan K., Rovinsky S. A., Coleman S. T., Moye-Rowley W. S. 2005. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J. Biol. Chem. 280:40524–40533 [DOI] [PubMed] [Google Scholar]
- 14. He X. J., Fassler J. S. 2005. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 58:1454–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He X. J., Mulford K. E., Fassler J. S. 2009. Oxidative stress function of the Saccharomyces cerevisiae Skn7 receiver domain. Eukaryot. Cell 8:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes T. R., et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109–126 [DOI] [PubMed] [Google Scholar]
- 17. James P., Halladay J., Craig E. A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ketela T., Brown J. L., Stewart R. C., Bussey H. 1998. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259:372–378 [DOI] [PubMed] [Google Scholar]
- 19. Krems B., Charizanis C., Entian K. D. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327–334 [DOI] [PubMed] [Google Scholar]
- 20. Kuge S., et al. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuge S., Jones N. 1994. YAP1-dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuge S., Jones N., Nomoto A. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 24. Lee J., et al. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040–16046 [DOI] [PubMed] [Google Scholar]
- 25. Li S., et al. 1998. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 17:6952–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S., et al. 2002. The eukaryotic two-component histidine kinase Sln1p regulates OCH1 via the transcription factor, Skn7p. Mol. Biol. Cell 13:412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu J. M., Deschenes R. J., Fassler J. S. 2004. Role for the Ran binding protein, Mog1p, in Saccharomyces cerevisiae SLN1-SKN7 signal transduction. Eukaryot. Cell 3:1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J. M., Deschenes R. J., Fassler J. S. 2003. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30. Morgan B. A., et al. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moye-Rowley W. S., Harshman K. D., Parker C. S. 1989. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3:283–292 [DOI] [PubMed] [Google Scholar]
- 32. Okazaki S., Tachibana T., Naganuma A., Mano N., Kuge S. 2007. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol. Cell 27:675–688 [DOI] [PubMed] [Google Scholar]
- 33. Page N., Sheraton J., Brown J. L., Stewart R. C., Bussey H. 1996. Identification of ASK10 as a multicopy activator of Skn7p-dependent transcription of a HIS3 reporter gene. Yeast 12:267–272 [DOI] [PubMed] [Google Scholar]
- 34. Raitt D. C., et al. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ross S. J., Findlay V. J., Malakasi P., Morgan B. A. 2000. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol. Biol. Cell 11:2631–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shankarnarayan S., Malone C. L., Deschenes R. J., Fassler J. S. 2008. Modulation of yeast Sln1 kinase activity by the CCW12 cell wall protein. J. Biol. Chem. 283:1962–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sherman F., Fink G. R., Hicks J. B. 1986. Methods in yeast genetics, p. 163–167 Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Sikorski R. S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takeuchi T., Miyahara K., Hirata D., Miyakawa T. 1997. Mutational analysis of Yap1 protein, an AP-1-like transcriptional activator of Saccharomyces cerevisiae. FEBS Lett. 416:339–343 [DOI] [PubMed] [Google Scholar]
- 40. Tao W., Deschenes R. J., Fassler J. S. 1999. Intracellular glycerol levels modulate the activity of Sln1p, a Saccharomyces cerevisiae two-component regulator. J. Biol. Chem. 274:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsuzi D., Maeta K., Takatsume Y., Izawa S., Inoue Y. 2004. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 565:148–154 [DOI] [PubMed] [Google Scholar]
- 42. Veal E. A., Ross S. J., Malakasi P., Peacock E., Morgan B. A. 2003. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 278:30896–30904 [DOI] [PubMed] [Google Scholar]
- 43. Wemmie J. A., Steggerda S. M., Moye-Rowley W. S. 1997. The Saccharomyces cerevisiae AP-1 protein discriminates between oxidative stress elicited by the oxidants H2O2 and diamide. J. Biol. Chem. 272:7908–7914 [DOI] [PubMed] [Google Scholar]
- 44. Wemmie J. A., Wu A. L., Harshman K. D., Parker C. S., Moye-Rowley W. S. 1994. Transcriptional activation mediated by the yeast AP-1 protein is required for normal cadmium tolerance. J. Biol. Chem. 269:14690–14697 [PubMed] [Google Scholar]
- 45. Williams K. E., Cyert M. S. 2001. The eukaryotic response regulator Skn7p regulates calcineurin signaling through stabilization of Crz1p. EMBO J. 20:3473–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wood M. J., Andrade E. C., Storz G. 2003. The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry 42:11982–11991 [DOI] [PubMed] [Google Scholar]
- 47. Wood M. J., Storz G., Tjandra N. 2004. Structural basis for redox regulation of Yap1 transcription factor localization. Nature 430:917–921 [DOI] [PubMed] [Google Scholar]
- 48. Wu A., et al. 1993. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268:18850–18858 [PubMed] [Google Scholar]
- 49. Wu A. L., Moye-Rowley W. S. 1994. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14:5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan C., Lee L. H., Davis L. I. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]