Abstract
Conidiation is an asexual sporulation pathway that is a response to adverse conditions and is the main mode of dispersal utilized by filamentous fungal pathogens for reestablishment in a more favorable environment. Heterotrimeric G proteins (consisting of α, β, and γ subunits) have been shown to regulate conidiation in diverse fungi. Previous work has demonstrated that all three of the Gα subunits in the filamentous fungus Neurospora crassa affect the accumulation of mass on poor carbon sources and that loss of gna-3 leads to the most dramatic effects on conidiation. In this study, we used 1H nuclear magnetic resonance (NMR) to profile the metabolome of N. crassa in extracts isolated from vegetative hyphae and conidia from cultures grown under conditions of high or low sucrose. We compared wild-type and Δgna-3 strains to determine whether lack of gna-3 causes a significant difference in the global metabolite profile. The results demonstrate that the global metabolome of wild-type hyphae is influenced by carbon availability. The metabolome of the Δgna-3 strain cultured on both high and low sucrose is similar to that of the wild type grown on high sucrose, suggesting an overall defect in nutrient sensing in the mutant. However, analysis of individual metabolites revealed differences in wild-type and Δgna-3 strains cultured under conditions of low and high sucrose.
INTRODUCTION
The filamentous fungus Neurospora crassa is a model organism that has been extensively studied for over a hundred years. N. crassa possesses a predominantly haploid life cycle, with 28 different cell types and three sporulation pathways (3). The genome has been sequenced (3), and knockout mutants are available for most of its ∼10,000 genes (10). Since N. crassa has simple nutritional requirements, being able to synthesize all cellular constituents (except for the vitamin biotin) from a medium containing simple salts, trace elements, and a carbon and nitrogen source, it is an ideal system for analysis of biochemical pathways and metabolic flux (17).
N. crassa produces a type of asexual spore, the macroconidium (referred to as a conidium) for dissemination in the environment (66). Conidiation initiates with the production of aerial hyphae that rise perpendicular to the substratum (65). The tips of the aerial hyphae form constrictions that eventually develop into complete crosswalls (septa) and finally sever, leading to release of the mature macroconidia. Since conidiation is a major mode of dispersal utilized by fungal pathogens (7), it also plays an important role in pathogenesis in these organisms.
Several factors have been shown to cause conidiation in N. crassa, including carbon or nitrogen deprivation, desiccation, heat shock, and blue-light exposure. Lowering the sucrose concentration from 1.5% to 0.15% (wt/vol) or imposing nitrogen limitation will cause the wild type to form conidiophores in shaking submerged cultures (55, 68). Desiccation-induced conidiation is observed when N. crassa is cultured on solid medium or in standing liquid cultures (65). Heat shocking N. crassa submerged liquid cultures at 46°C followed by incubation at 25°C causes inappropriate conidiogenesis (67). Finally, exposing N. crassa to blue light induces conidiation (44). From these findings, it is evident that conidiation is a common biological response to adverse conditions and a means by which the fungus can reestablish itself in a more favorable environment.
A major signaling pathway that detects and responds to external signals in fungi and other eukaryotes is mediated by heterotrimeric GTP-binding proteins (consisting of α, β, and γ subunits) (38, 42). Briefly, the binding of a ligand to a G protein-coupled receptor (GPCR) leads to a conformational change that causes the associated Gα subunit to exchange GDP for GTP. Consequently, the βγ-heterodimer dissociates from the Gα protein, allowing both to interact with downstream effectors that influence cell growth and development.
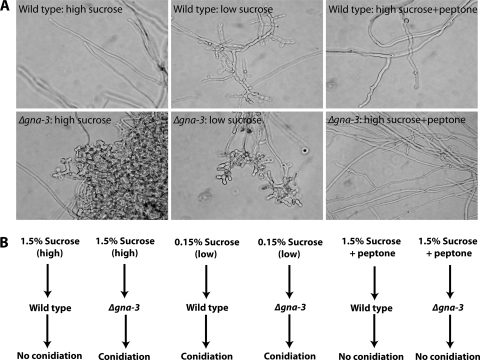
Three Gα (GNA-1, GNA-2, and GNA-3), one Gβ (GNB-1), and one Gγ (GNG-1) subunit have been identified in N. crassa (36, 42). Loss of gna-3 has dramatic effects on conidiation, leading to the production of short aerial hyphae and premature conidiation in plate cultures and inappropriate conidiation in submerged cultures (Fig. 1) (37). The phenotypes of Δgna-3 mutants are consistent with GNA-3 as a negative regulator of conidiation in N. crassa. Δgna-3 mutants have low levels of adenylyl cyclase protein (CR-1) and cyclic AMP (cAMP), and Δcr-1 mutants produce conidiophores in submerged cultures, revealing a link between low cAMP levels and conidiation. Interestingly, supplementation of submerged cultures of Δgna-3 and Δcr-1 mutants with 2% peptone reverses the submerged conidiation phenotype (37), hinting at a nutrient-sensing function for GNA-3 and CR-1. These observations are also supported by work showing that loss of gna-3 or cr-1 leads to reduced mass accumulation on poor carbon sources (41).
Fig. 1.
Submerged-culture phenotypes. (A) Morphology of submerged cultures. Cultures were grown in high or low sucrose with or without peptone as indicated in the figure. Photographs were taken after 16 h of growth. (B) Summary of treatments used for submerged cultures. The strains and media used to grow cultures, as well as the final conidiation phenotype, are indicated.
Previous work has identified metabolites present in N. crassa tissues under various conditions. Most of the amino acids have been detected in cell extracts from conidia and hyphal cultures using high-pressure liquid chromatography (HPLC) followed by ninhydrin staining (61, 62). Alanine, glutamine, and glutamate have also been quantified in intact hyphal cells using 15N nuclear magnetic resonance (NMR) (34). Several sugars/carbon sources have been detected in hyphal cultures, including trehalose (enzymatic detection) (20), glucose (enzymatic detection) (11), glycerol (enzymatic detection) (54), and tricarboxylic acid intermediates and organic acids (HPLC) (52).
The goals of this research were 2-fold. First, we sought to determine the metabolite profile of wild-type N. crassa hyphae grown under high- and low-carbon (sucrose) conditions and of conidia cultured in high sucrose. Second, we wanted to explore whether lack of a single gene (gna-3) causes a significant metabolite shift in conidia or hyphae under the same conditions used for the wild type. The latter objective addresses whether the Δgna-3 mutant conidiates in submerged conditions due to interruption of a signal transduction relay from outside stimuli or because of a metabolic block that leads to nutrient deprivation. A metabonomics approach using 1H NMR (33, 45, 46, 59, 71, 73) was used to create a metabolite fingerprint of tissue extracts to compare levels of amino acids and other metabolites present in the wild type and the Δgna-3 mutant. 1H NMR was chosen as the method for recording metabolite profiles because it is rapid and direct and the quantitative information from multiple metabolites in a single experiment has been shown to correlate well with the data from quantitative enzymatic assays in other systems (5, 46).
MATERIALS AND METHODS
Cell growth and extraction of metabolites.
The N. crassa strains used in this study were wild-type ORS-SL6a (74a, mat a, Fungal Genetics Stock Center no. 4200; Fungal Genetics Stock Center, Kansas City, MO) and Δgna-3 strain 43c2 (37). Tissue was collected from a total of six biological replicates per strain for each treatment. Each strain was grown in three different types of medium: Vogel's minimal medium (VM) with 1.5% sucrose (wt/vol), VM with 0.15% sucrose (wt/vol), and VM with 1.5% sucrose (wt/vol) plus 2% peptone (wt/vol). Solid medium contained 1% agar.
Conidia were propagated as follows. A small amount of conidia from a slant culture was used to inoculate 50 ml of VM agar medium in a 250-ml foam-stoppered flask. Flasks were then incubated at 30°C in the dark for 2 days before being moved to 25°C in constant light for an additional 5 days. Conidia that were used to inoculate liquid cultures were collected by filtration using water (17) and used immediately, while those to be used directly for metabonomics studies were collected using Soltrol (see below).
Glass Erlenmeyer flasks (25-ml total volume) to be used for liquid cultures were treated with dichlorodimethylsilane (5% [vol/vol] in chloroform; TCI America) in order to prevent hyphae from adhering to the inside walls of the flask, possibly leading to desiccation-induced conidiation. Cultures containing 6 ml of liquid VM were inoculated with water-harvested conidia to a final concentration of 1 × 106 conidia/ml and then incubated at 30°C with shaking at 200 RPM in the dark for 16 h. Cultures were collected by vacuum filtration using filter paper (Whatman #3), transferred to 2-ml screw-cap tubes, and stored at −80°C until extraction.
To determine the metabolite profile of pure conidia, conidia were collected from the agar flasks described above using Soltrol 170 isoparaffin, a nonaqueous solvent that has previously been shown to prevent germination of conidia (4). Approximately 50 ml of Soltrol was poured into the flask, and the contents were vortexed and then filtered, using Handi-wipe towels, into a sterile flask. The conidial suspension was then collected on a Whatman #3 filter paper using vacuum filtration. The conidia were scraped off the filter paper using a spatula and transferred to a 2-ml screw-cap tube before being frozen at −80°C.
Similar to recent work in the filamentous fungus Fusarium graminearum (46), the metabolite extraction protocol used in this study was developed using Arabidopsis (32). To extract the metabolites, the frozen tissues in 2-ml tubes were pulverized, using a glass rod, after the addition of liquid nitrogen. After the tissue reached the consistency of a fine powder, 750 μl of extraction buffer was added to each tube. The extraction buffer consisted of 1:1 (vol/vol) acetonitrile-d3 (CD3CN):deuterium oxide (D2O) containing 50 mM sodium acetate-d3 (CD3COOD) and 100 μM 3-trimethylsilylpropionic acid-d4 sodium salt (TMSP) as a chemical shift reference (0.000 ppm). Acetonitrile (50%) has been shown to precipitate proteins without small-molecule loss (2). Tissue homogenization in the presence of the extraction buffer was carried out at room temperature for 4 min.
The extracts were clarified by centrifugation at 2,300 × g for 5 min and the supernatants transferred to a new tube, while cellular debris was discarded. We found that a simple evaporation and reconstitution step provided selectivity for hydrophilic metabolites, removing broad resonances contributed by long hydrocarbon chains (which obscure the region around 0.9 ppm), alleviating spectral crowding, and allowing accurate integration of resonances from aliphatic amino acids without appreciable loss of amino acids, sugars, and nitrogen-containing metabolic intermediates (72). Extracts were evaporated using a speed vacuum overnight (16 h) at room temperature and reconstituted in 700 μl of D2O containing 100 mM CD3COOD, 100 μM TMSP, and 500 μM NaN3 as a biocide.
Hydrophobic interferents were removed by the addition and removal of 100 μl of deuterated chloroform (CDCl3) at room temperature. The D2O/CDCl3 phases were vortexed for 60 s, and the emulsion was broken by centrifugation at 2,300 × g for 5 min. This step further improved our ability to quantify organic acids (occupying the region around 1.3 ppm) by removing endogenous polar lipids (43). This protocol is summarized in Kaiser et al. (32). The aqueous portion, in the amount of 600 μl, was transferred to a microcentrifuge tube wherein the pD of each extract was adjusted to 7.40 ± 0.08 (mean ± standard deviation) using deuterium chloride and sodium deuteroxide, where pD = pH meter reading +0.4 to correct for isotope effects in glass pH electrodes calibrated with aqueous buffers. All deuterated reagents were from Cambridge Isotope Laboratories (Andover, MA). The solution was stored at −80°C until measurement of metabolites.
1H NMR.
Each sample was thawed and then transferred to a 5-mm NMR tube (Wilmad, Buena, NJ) for analysis. Spectra were acquired for bioreplicate samples with selective saturation of the solvent resonance using a Bruker Avance NMR spectrometer operating at 600.06 MHz. Free induction decays (FIDs) were acquired into 25,860 time points and zero filled to 131,072 points. A spectral width of 7,716 Hz was excited using a 90° pulse. A relaxation delay of 1.5 s was used, and 640 scans were coadded following 16 dummy scans, for a total experiment time of 34 min. The temperature of the sample was maintained at 298 K. FIDs were apodized by multiplication by an exponential function equivalent to 1.0-Hz line broadening prior to Fourier transformation. Manual shimming was performed for each sample, and the TMSP line width at half-height after the application of 1.0-Hz exponential line broadening was 2.43 ± 0.16 Hz.
Metabonomics and metabolite profiling.
Spectra were processed using Topspin 2.0 (Bruker, Billerica, MA); phasing was applied automatically, while baseline correction was applied by manual fitting with a sine function. The conidia spectra were baseline corrected using a cubic spline function with approximately 100 user-defined baseline points utilized in the operation. This was necessary due to residual contributions from Soltrol, which could not be completely removed by chloroform treatment, in the aliphatic region of the spectra. Prior to integration, each spectrum was aligned such that the chemical shift reference (TMSP) was at 0.00 ppm. Spectra were integrated for the purpose of metabonomics using equidistant integral regions of 0.02 ppm width over the range of 0.50 to 9.00 ppm, excluding the regions containing the resonances of the solvent constituents acetate and acetonitrile (1.96 to 2.04 ppm) and residual water (4.28 to 4.72 ppm). For metabolic profiling, a second set of integration regions were manually defined for the quantitation of well-resolved peaks. The relative amounts of metabolites present were determined by normalizing each integral to the constant sum of the NMR spectrum (47, 74). We found the method of normalization to a constant sum to be superior to mass normalization (data not shown), even though approximately equal tissue weights were used in these experiments. In our hands, normalization to a constant sum better compensates for variability in the recovery of metabolites by extraction.
Confirmation of analyte resonance assignments was made by comparison to standard solutions. The NMR spectra of standards were measured using the same NMR parameters as for the extracts, except that fewer scans were required.
Statistical analysis.
Our goal in this work was to record metabolic fingerprints and compare treatments for their effect on the global metabolome; therefore, principal components analysis (PCA) (58) was selected as the appropriate technique for dimensionality reduction and data visualization. PCA, an unsupervised pattern recognition method, was performed with the statistical analysis program Minitab, version 15 (Minitab, Inc., State College, PA), using the equidistant integrals obtained from the NMR spectra. These calculations were performed in accordance with the recommendations of Broadhurst and Kell (6) to avoid false discoveries through aggressive cross-validation. Mean centering was applied in this study (2, 70). No scaling was applied prior to PCA.
One of our objectives was to determine whether loss of GNA-3 and/or sucrose limitation directly influences metabolite profiles. Some of our growth conditions were conidiogenic, while others supported a predominantly hyphal mycelium (Fig. 1). In order to study metabolites specifically accumulated in conidia, Soltrol 170 isoparaffin was added to a set of conidial suspensions to prevent germination. However, residual Soltrol remaining after our extraction procedure interfered with routine baseline correction. Because of the resonances due to residual Soltrol and the different modes of baseline correction, the conidia spectra are not directly compared with the submerged-culture profiles represented in Fig. 3A and B. Importantly, the presence of Soltrol did not affect the integration and univariate analysis of many conidial metabolites in regions of the spectrum unaffected by the Soltrol resonances.
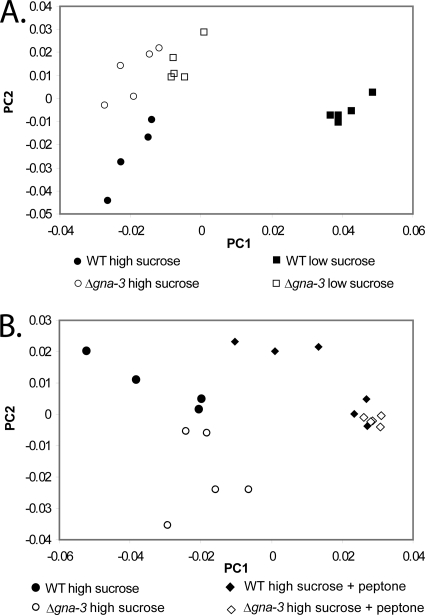
Fig. 3.
Principal component analysis (PCA) score plot. (A) Analysis of results for low- and high sucrose-cultures. PCA analysis of relative integrals from 1H NMR spectra measured for replicate samples of both strains grown under high-sucrose and low-sucrose conditions. In this plot of PC1 versus PC2, the x axis is the value of the first principal component, explaining 55.3% of the variance, while the y axis indicates the second principal component, explaining 24.9% of the variance. (B) Analysis of results for high-sucrose cultures with or without peptone. PCA analysis of 1H NMR data was performed for the strains described for panel A except that high-sucrose cultures with or without peptone were compared. x and y axes are as described for panel A; the first principal component explains 55.4% of the variance, while the second principal component accounts for 18.3% of the variance.
Univariate statistical analyses for metabolite profiling were carried out in Excel 2008 (Microsoft, Redmond, WA). Each integral region was treated as an independent variable with six replicates per treatment per strain. The Q-test was conducted to exclude up to one replicate per treatment per strain that was considered an outlier.
Identification of metabolites.
Resonances were assigned to individual metabolites by comparison with spectra recorded for approximately 100 authentic metabolite standards (Sigma-Aldrich, St. Louis, MO) and with chemical shifts and multiplicities reported in the published literature and also in the Madison Metabolomics Consortium Database (http://mmcd.nmrfam.wisc.edu/) (14). In cases of ambiguity, a pure standard was spiked into the fungal extract and the resulting NMR spectra were recorded (see Table S1 in the supplemental material). For the metabolites listed in Table S1, good agreement was obtained for all the resolved resonances of a given metabolite.
RESULTS
The global metabolome of the Δgna-3 mutant is similar to that of the wild-type strain.
In this study, we analyzed the endometabolome, or intracellular metabolites (31), of N. crassa. The levels of extractable, water-soluble metabolites (lipids were removed) were measured under several conditions using 1H NMR. We assessed liquid cultures from the wild-type and Δgna-3 strains in the presence of high (1.5%) sucrose, low (0.15%) sucrose, and 1.5% sucrose plus 2% peptone (Fig. 1A and B). As previously reported, wild-type strains produce only hyphae in high-sucrose submerged cultures but form hyphae with associated conidia in low-sucrose submerged cultures (Fig. 1A and B) (37). In contrast, the Δgna-3 mutant forms hyphae and conidia under both high- and low-sucrose conditions in submerged cultures (Fig. 1A and B) and peptone reverses the submerged conidiation phenotype of the Δgna-3 mutant in high sucrose (Fig. 1A and B) (37). Thus, analysis of submerged cultures under these varied conditions could reveal a possible contribution of altered metabolite levels in inducing conidiation, as well as a role for the G protein GNA-3 in regulating metabolite levels in N. crassa. We also recorded metabolite profiles of purified conidia harvested from wild-type and Δgna-3 strain cultures grown in solid medium containing 1.5% sucrose. Together with the results for high-sucrose submerged cultures, these studies produce a baseline metabolome for purified conidia and vegetative hyphae from submerged cultures under high-carbon growth conditions.
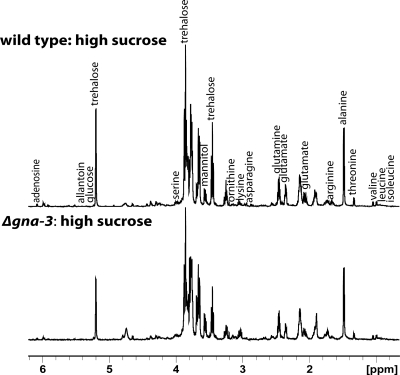
Six biological replicates were used to generate the initial data, with up to one outlier discarded per treatment (see Materials and Methods). Representative spectra of extracts obtained from submerged cultures of wild-type and Δgna-3 strains grown in high sucrose (Fig. 2) reveal both similarities and differences between the two strains; this pattern was repeated with the other treatments (see below). We examined the spectra for the presence of peaks corresponding to 102 metabolites, with a total of 21 being unambiguously identified (see Table S1 in the supplemental material).
Fig. 2.
Representative 1H NMR spectra for extracts of wild-type and Δgna-3 strain submerged cultures. Strains were cultured in high-sucrose medium for 16 h, and metabolites extracted as described in Materials and Methods. The major peaks identified are labeled.
The data were subjected to PCA, an unsupervised statistical method that can be used to compare large data sets, reducing multidimensional data to two principal component axes which represent the differences between samples in a two-dimensional graphical format (58). We initially conducted PCA (Fig. 3A) with the peptone medium data omitted, since peptone is a rich source of metabolites that could influence the observed clustering. The analysis revealed that principal component 1 (PC1) is responsible for 55.3% of the variance, while PC2 accounts for 24.9%. The PCA score plot (Fig. 3A) revealed two major groupings, with separation of all samples. The grouping on the left side of the plot consisted of submerged cultures from the wild type cultured on high sucrose and the Δgna-3 mutant grown on high and low sucrose. The second grouping on the right side of the plot was the wild type cultured on low sucrose. These results suggest several conclusions. First, there is a large shift in the metabolite profile produced in wild-type N. crassa when cultured under high- versus low-sucrose conditions. Second, the observation that the Δgna-3 strain cultured in high and low sucrose is more similar to high-sucrose than low-sucrose wild-type cultures suggests that loss of gna-3 does not have a global effect on the metabolome of N. crassa in adequate carbon and that the Δgna-3 mutant is relatively “blind” to reduced carbon levels in submerged cultures. Third, the closer grouping between the conidiating Δgna-3 strain and nonconidiating (high-sucrose) rather than conidiating (low-sucrose) wild-type cultures suggests that any effect of the Δgna-3 mutation on conidiation does not involve large changes in the metabolome.
The results from the PCA score plot that included peptone-supplemented cultures (Fig. 3B) reinforced the conclusions of the initial analysis. This plot revealed two major groupings; the first, on the left side of the plot, contained separable clusters for wild-type and Δgna-3 strain high-sucrose submerged cultures. The second grouping, on the right side of the plot, consisted of both strains cultured in high sucrose plus peptone. The results from peptone-supplemented cultures suggest that, analogous to the observations in high-sucrose medium, the Δgna-3 strain has a metabolome similar to that of the wild type and/or that small molecules in the nutrient-rich peptone have a dominant effect on the metabolic profile that obscures any effect due to the Δgna-3 mutation.
The Soltrol solvent used to isolate conidia was present as an interference in the aliphatic region (0.8 ∼ 1.1 ppm) of the NMR spectra of conidia extracts. Therefore, the conidial samples were not directly compared with other treatments by multivariate analysis (PCA), and only univariate statistical analyses of metabolites from conidia were performed.
It has been postulated that “metabolic network robustness” in a gene knockout results from gene redundancy and alternate metabolic pathways (39). The PCA score plots (Fig. 3) are consistent with this premise, as the overall global metabolome does not change significantly with loss of gna-3. This could be due to the presence of the other two Gα subunits (GNA-1 and GNA-2) in the N. crassa genome. Loss of gna-1 also leads to inappropriate conidiation, although a higher cell density is required (3 × 106 conidia/ml). Furthermore, mutation of gna-2 in the Δgna-3 background leads to more severe effects on submerged conidiation and overall growth (36). Thus, the observed overlapping roles of Gα subunits may explain the overall similarity in the metabolomes of Δgna-3 and wild-type strains.
Levels of individual metabolites differ in the Δgna-3 strain and the wild type.
Although PCA analysis showed that the metabolomes were comparable in the wild-type and Δgna-3 strains, visual inspection of the NMR spectra revealed significant changes in several metabolites between treatments and also between strains (Fig. 2; also see Fig. S1 in the supplemental material). Stacking of the spectra was used to identify changes in metabolite levels and to assess the reproducibility of replicates (Fig. 2; also see Fig. S1). Taking into account all sample preparation steps, relative standard deviations of ∼10% were observed between bioreplicates, which is comparable to the most widely used gas chromatography-mass spectrometry (GC-MS) metabolomics method of Fiehn et al. (23), who reported a relative standard deviation of 8%. The work of Dieterle et al. (21) demonstrates that with a sample size of 4,000-plus bioreplicates, relative standard deviations on the order of 7% could be achieved for creatinine, a proposed “housekeeping” metabolite, using NMR-based metabolomics in small mammals. A majority of the amino acids and a number of sugars were cataloged, as well as other metabolites (see below). In many cases, supplementation of cultures with peptone led to elevation of amino acid levels, presumably because peptone is a rich source of amino acids (Fig. 4).
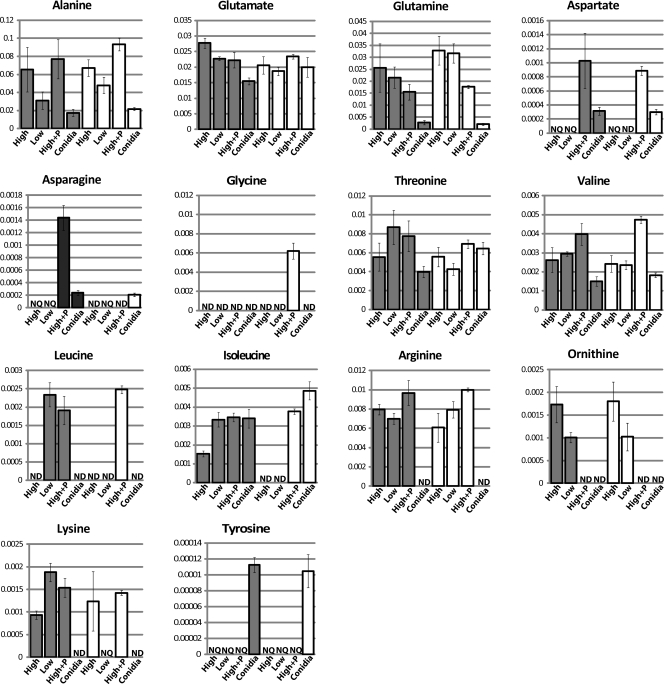
Fig. 4.
Relative amino acid levels. Metabolites extracted from both the wild-type (dark bars) and the Δgna-3 (white bars) strain grown under various conditions were subjected to 1H NMR analysis, and the relative resonance intensities were determined. On the x axis, “High” indicates metabolites extracted from tissue grown in high sucrose medium, “Low” refers to low-sucrose medium, “High+P” denotes high-sucrose with the addition of 2% peptone, and “Conidia” represents conidia collected from the strain grown on high-sucrose solid medium. “ND” indicates that the metabolite was not detected in that treatment. “NQ” indicates that the metabolite was detected but was not quantifiable due to low concentration. The y axis represents the quantity of a metabolite that has been normalized to the total pool of metabolites present in that particular sample. Error bars show standard errors.
Amino acids and related metabolites. (i) Alanine, glutamate, and glutamine.
Alanine serves as a convenient reservoir of amino groups and pyruvate during nitrogen and carbon sufficiency. Alanine is made from glutamate by reverse transamination (34) and can be converted back to glutamate by alanine transaminase. Glutamine is synthesized from glutamate by glutamine synthetase (16).
The results from our analysis validate earlier reports that alanine is the most abundant amino acid in N. crassa hyphal cultures, followed by glutamate and glutamine (34, 62). In the wild type, the NMR spectra show that alanine and glutamate levels are elevated in high versus low sucrose (Fig. 4). In the Δgna-3 strain, the same trend is observed for alanine, while glutamate levels are relatively similar under high- and low-carbon conditions. For glutamine, the levels in both strains are not appreciably influenced by carbon availability. However, the relative glutamine amount is higher in the Δgna-3 mutant than in the wild type cultured on low-sucrose medium. This effect cannot be explained by the greater conidiation of the Δgna-3 strain, as conidia from the Δgna-3 mutant have the lowest relative levels of glutamine detected in the six biological treatments.
On a relative basis, conidia have much lower levels of alanine and glutamine than hyphae. The glutamate amounts in Δgna-3 conidia are similar to those in hyphal cultures, whereas the levels in wild-type conidia are significantly less. The relative levels of these three amino acids roughly correlate with those measured in conidia in a previous study (61). Furthermore, our results demonstrating that alanine, glutamate, and glutamine are some of the most abundant amino acids in conidia support the results of earlier work showing that the levels of these three compounds constituted nearly 70% of free amino acids in conidia (Fig. 4) (61).
(ii) Aspartate and asparagine.
Aspartate can be synthesized from the citric acid cycle intermediate oxaloacetate using aspartate aminotransferase (49). Evidence for synthesis of aspartate in a pathway involving catabolism of glutamate using glutamic acid decarboxylase during conidial germination has been obtained (9). Asparagine is produced by transamination of aspartate (49). The levels of aspartate and asparagine were very low or could not be detected in the high- and low sucrose-submerged cultures of both strains (Fig. 4). With the exception of high-sucrose-plus-peptone cultures of the Δgna-3 mutant, where asparagine could not be detected, the relative levels of aspartate and asparagine are similar in wild-type and Δgna-3 strains and are much greater in peptone-supplemented cultures than in conidia (Fig. 4).
(iii) Serine and glycine.
Serine is formed from 3-phospho-hydroxypyruvate, which in turn is derived from glycerate-3-phosphate, a metabolic intermediate of glycolysis (1). Glycine is then produced from serine by serine hydroxymethyltransferase (13). We detected serine in the wild-type strain grown in high sucrose, low sucrose, or the presence of peptone and in the Δgna-3 mutant cultured in high and low sucrose (data not shown). However, it was not possible to reliably quantify serine levels because overlap of the serine resonance with the shoulder of the much more intense trehalose peak at 3.88 ppm made integration unreliable. Serine could not be detected in conidia isolated from wild-type and Δgna-3 strains or in peptone-treated cultures of the Δgna-3 strain (data not shown).
Glycine could only be detected and quantified in the Δgna-3 strain grown in peptone-supplemented medium (Fig. 4).
(iv) Threonine, valine, leucine, and isoleucine.
Threonine is derived from glycine and serine and, along with pyruvate, is a direct precursor to isoleucine (16, 57). Pyruvate is a precursor of valine, and leucine is formed from the last intermediate in the valine biosynthetic pathway (16, 57). Although they are formed from different initial substrates, the pathways for synthesis of isoleucine and valine share four enzymes (16, 57).
Under high-sucrose conditions, the relative levels of threonine are comparable in wild-type and Δgna-3 strain submerged cultures (Fig. 4). However, the relative threonine amount is greater in the wild-type but not in the Δgna-3 strain under low-sucrose conditions. This suggests that gna-3 is required for increased threonine in response to poor carbon availability. Peptone supplementation did not significantly affect the relative threonine levels. Finally, the threonine levels are much greater in conidia from the Δgna-3 mutant than in those from the wild type (Fig. 4).
The relative valine levels are not affected by sucrose in submerged cultures from the two strains (Fig. 4). Similar to the results for threonine, the valine amounts are slightly higher in conidia from the Δgna-3 mutant than in those from the wild type. The relative valine levels are highest in peptone-supplemented cultures (Fig. 4).
Leucine was only detected in low-sucrose wild-type cultures and peptone-supplemented cultures from both strains (Fig. 4). Interestingly, although isoleucine could not be detected in high- or low-sucrose submerged cultures from the Δgna-3 strain, Δgna-3 conidia possessed the highest relative levels of isoleucine detected in our study (Fig. 4). In the wild type, the relative isoleucine amount was higher in low-sucrose submerged cultures and conidia than in high-sucrose submerged cultures (Fig. 4). Finally, peptone-supplemented cultures from Δgna-3 and wild-type strains had comparable amounts of isoleucine. The results for the Δgna-3 strain suggest that GNA-3 is required for isoleucine production in submerged cultures but not in conidia.
(v) Arginine and ornithine.
Ornithine is produced in the mitochondrion and then transported into the cytoplasm to serve as a precursor for arginine and polyamine synthesis (16). In N. crassa cultured on minimal medium, >98% of cellular arginine and ornithine are stored in the vacuole. Arginine or nitrogen starvation results in the release of stored arginine and ornithine into the cytoplasm, where these compounds can be used for protein synthesis and/or as nitrogen sources (69).
Comparison of high- versus low-sucrose cultures demonstrates that the arginine level decreases slightly in the wild type, while the Δgna-3 mutant shows the opposite trend (Fig. 4). The levels of arginine are highest in peptone-supplemented cultures. The relative levels of ornithine are similar in wild-type and Δgna-3 strain submerged cultures (Fig. 4), with lesser amounts in low versus high sucrose. Ornithine could not be detected in peptone-supplemented cultures (Fig. 4). The results for ornithine are consistent with positive regulation of this amino acid by the sucrose levels in the growth medium in both strain backgrounds.
Arginine and ornithine could not be detected in conidia from either strain. The observation of lower arginine levels in conidia versus the levels in hyphae is consistent with results from previous work (61) and also supported by the hypothesis (69) that arginine is metabolized to glutamate during conidiation and then eventually converted back into arginine after conidial germination.
(vi) Lysine.
Lysine is synthesized using the α-aminoadipate pathway in N. crassa (53), with the majority of lysine being utilized for protein synthesis (30). Our results demonstrate that sucrose deprivation leads to an almost 2-fold increase in lysine levels in wild-type submerged cultures (Fig. 4). The lysine amounts are similar in peptone-supplemented cultures of both strains. Lysine could not be detected in low-sucrose Δgna-3 strain cultures or in conidia from either the wild-type or the Δgna-3 strain (Fig. 4).
(vii) Tyrosine.
In N. crassa, tyrosine, phenylalanine, and tryptophan are produced from the shikimate pathway (15). Tyrosine is synthesized starting with the conversion of chorismate to prephenate by chorismate mutase, and then prephenate is converted to tyrosine by prephenate dehydrogenase (15, 16). Tyrosine is a precursor to the secondary metabolite 3,4-dihydroxyphenylalanine (DOPA) melanin, through a pathway involving the enzyme tyrosinase (40). In our study, tyrosine levels were very low in submerged cultures and this amino acid could only be quantified in conidia of both wild-type and Δgna-3 strains (Fig. 4). The relatively high levels of tyrosine in mature conidia suggest a function for this amino acid during conidial biogenesis and/or germination.
(viii) Amino acids that could not be quantitated and/or detected.
The amino acids that could not be detected and/or quantitated in our studies were serine (as mentioned above), cysteine, histidine, methionine, phenylalanine, proline, and tryptophan.
Sugars and related metabolites. (i) Trehalose and glucose.
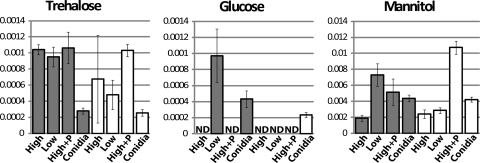
Previously published studies have shown that metabolites that can be readily used as carbon sources, such as glutamate and trehalose, are found in dormant spores of N. crassa (61). A role for trehalose as a thermal protectant has been established for N. crassa (56). This previous work also demonstrated that although trehalose is a large component of the metabolite pool in conidia, the relative levels of trehalose are similar in conidia and hyphae of wild-type strains.
In our study, we observed that the relative levels of trehalose are greater in high-sucrose submerged cultures than in all other submerged cultures of the wild type (Fig. 5). In contrast, the relative trehalose amounts are similar in high- and low-sucrose submerged cultures of the Δgna-3 strain, with lower levels under conditions of peptone supplementation (Fig. 5). The results for high- and low-sucrose submerged cultures are consistent with a defect in nutrient sensing for the Δgna-3 strain.
Fig. 5.
Relative levels of sugar metabolites. Metabolites were extracted and analyzed and x and y axes labeled as indicated in the legend to Fig. 4.
The relative glucose levels are higher in low-sucrose than in high-sucrose submerged cultures for both the wild-type and the Δgna-3 strain (Fig. 5). Still-lower relative amounts are found in peptone-supplemented cultures (Fig. 5).
Our attempts to measure relative amounts of trehalose and glucose in conidial extracts were unsuccessful due to residual enzymatic activity of trehalase, which we did not observe in hyphal extracts (data not shown). Trehalase is known to be extremely stable in solution, at a range of temperatures and within a wide pH range (22). The levels of trehalose are high in fungal conidia, and activation of trehalase has been shown to accompany conidial germination in numerous fungi (18, 19, 29). Previous studies have demonstrated that ungerminated conidia contain the highest levels of trehalase detected during N. crassa development (27), effectively priming conidia for germination and colonization.
(ii) Fumarate.
Fumarate is an intermediate of the citric acid cycle. In our study, it could not be detected in high- or low-sucrose submerged cultures from either strain or in peptone-supplemented cultures from the Δgna-3 mutant. There was wide variation in the amounts of fumarate in the biological replicates for peptone-supplemented cultures of the wild type. The fumarate levels were similar in conidia from both the Δgna-3 and wild-type strain (data not shown).
(iii) Mannitol.
Mannitol is an abundant sugar alcohol in fungal tissue that is derived from fructose (64). In the wild type, the relative mannitol levels are greater in low-sucrose than in high-sucrose submerged cultures (Fig. 5). In contrast, the relative mannitol amounts in the Δgna-3 mutant are similar in high- and low-sucrose cultures and greater than the levels detected in wild-type submerged cultures (Fig. 5). Conidia of both the wild-type and Δgna-3 strain contain the highest relative levels of mannitol detected in our studies (Fig. 5). Taken together, these results reveal a positive correlation between the proportion of conidia in a culture and the relative level of mannitol.
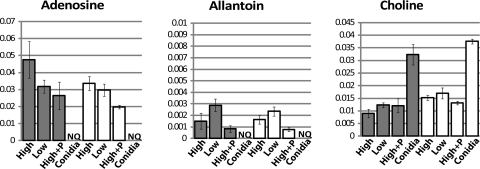
Miscellaneous metabolites. (i) Adenosine.
Adenosine is a precursor of AMP, ADP, and ATP and the second messenger cAMP (51). We observed that the adenosine levels were similar in all three submerged cultures of the wild type (Fig. 6). The relative adenosine levels exhibited high variability in high-sucrose cultures of the Δgna-3 mutant. The levels in low-sucrose cultures of the Δgna-3 mutant were lower than those observed in the wild type, while the amount of adenosine in peptone-supplemented cultures was similar to the amount in the wild type (Fig. 6). The relative adenosine levels were similar in conidia from the wild-type and Δgna-3 strain and were greatly reduced relative to the levels in submerged cultures.
Fig. 6.
Relative levels of adenosine, allantoin, and choline. Metabolites were extracted and analyzed and x and y axes labeled as indicated in the legend to Fig. 4.
(ii) Allantoin.
As a small molecule (158.12 molecular weight) with four nitrogen atoms, allantoin is an ideal nitrogen storage compound (12). Allantoin is an intermediate in purine catabolism and can be utilized as a nitrogen source after sequential degradation to urea and then ammonium (48). We could not detect allantoin in high-sucrose cultures of either strain with or without peptone or in low-sucrose Δgna-3 cultures (Fig. 6). The relative allantoin amounts were the highest in low-sucrose submerged cultures and conidia from the wild-type strain, with lower levels observed in Δgna-3 conidia (Fig. 6).
In Saccharomyces cerevisiae, allantoin is stored as a nitrogen reserve during starvation (48). Our results are also consistent with a storage function in N. crassa conidia, with allantoin being sequestered in conidia and possibly serving as a nitrogen reserve for germination. The presence of lower relative allantoin levels in Δgna-3 samples than in the wild type suggests that the loss of GNA-3 disrupts the normal storage mechanism, particularly in submerged cultures that produce conidia.
(iii) Choline.
Choline is an important component of cellular membranes (28). We noted that choline was present at similar levels in wild-type and Δgna-3 strains cultured on high sucrose (Fig. 6). However, under low-sucrose conditions, the relative levels of choline are much higher in the wild-type strain. The relative choline levels in conidia from both strains are similar and are higher than those in high-sucrose submerged cultures. Peptone supplementation results in marked elevation of choline levels in Δgna-3 cultures (Fig. 6).
Data from previous mRNA profiling experiments support the metabolite profiling results for several amino acids.
An earlier study by Kasuga et al. (35) used long-oligomer microarray profiling to detect transcript levels for 3,366 predicted genes in wild-type N. crassa during a time course of conidial germination. Among the genes analyzed are numerous loci encoding amino acid biosynthetic and degradative enzymes. We took advantage of this data set by comparing the levels of amino acids that could be detected and quantified in wild-type conidia and high-sucrose submerged cultures to the available corresponding mRNA levels for amino acid metabolic genes in conidia and at various times during germination. Within these constraints, we were able to analyze pathways for 10 amino acids: glutamate, glutamine, asparagine, threonine, valine, isoleucine, ornithine, arginine, lysine, and tyrosine (Table 1). In general, the mRNA and metabolic profiling data correlate. In most instances, the transcript levels for amino acid biosynthetic genes increase during conidial germination (usually within 0.5 to 1 h), and this is reflected in higher levels of the relevant amino acid in hyphae relative to the levels in ungerminated conidia (Table 1). The exceptions are asparagine, isoleucine, and tyrosine, whose levels are lower in hyphae than in conidia. The metabolism of isoleucine is complicated by its sharing of four enzymes with valine biosynthesis, as well as inhibition of the first common enzyme (acetolactate synthase) by valine but not isoleucine (16). In the case of tyrosine, this amino acid shares several intermediates with phenylalanine, tryptophan, and the vitamin p-aminobenzoic acid (24–26) and is a precursor for DOPA melanin (40). Asparagine is interconverted with aspartate (57). Finally, transcripts for genes that metabolize glutamate, glutamine, and arginine to other compounds are also upregulated during conidial germination (Table 1).
Table 1.
Comparison of relative metabolite and transcript levels for amino acid metabolic pathways
| Amino acid pathway | Broad gene no.a | EC no. | Enzyme(s) | Gene nameb | Relative mRNA expression inc: |
Relevant metabolite(s)e | Relative metabolite level [(mean ± SD) × 103] inf: |
||
|---|---|---|---|---|---|---|---|---|---|
| Conidia | Hyphae (hd) | Conidia | Hyphae | ||||||
| Threonine synthesis | NCU04118 | 2.7.2.4 | Aspartate kinase | 0.68–1.66 | (0.5) 2.32–5.87 | Threonine | 4.0 ± 0.6 | 5.6 ± 1.5 | |
| NCU03935 | 1.1.1.3 | Homoserine dehydrogenase | 0.92–1.23 | (0.5) 6.79–8.88 | |||||
| NCU04277 | 2.7.1.39 | Homoserine kinase | thr-4 | 0.44–0.79 | (0.5) 3.44–6.18 | ||||
| NCU03425 | 4.2.3.1 | Threonine synthase | thr-2 | 0.64–1.34 | (0.5) 1.53–3.12 | ||||
| Glutamate synthesis | NCU01195 | 1.4.1.4 | Glutamate dehydrogenase (NADP+) | am | 0.51–1.08 | (1) 3.15–4.30 | Glutamate | 16.0 ± 1.1 | 27.0 ± 1.6 |
| NCU01744 | 1.4.1.14, 1.4.1.13 | Glutamate synthase (NADPH) GOGAT | en (am)-2 | 0.99–3.44 | (0.5) 10.40–22.38 | ||||
| Glutamate degradation | NCU00461 | 1.4.1.2 | Glutamate dehydrogenase (NAD+) | gdh | 0.61–1.46 | (0.5) 2.26–2.16 | |||
| Glutamine synthesis | NCU06724 | 6.3.1.2 | Glutamine synthetase, β subunit | gln-1 | 0.34–0.52 | (1) 1.32–1.52 | Glutamine | 2.7 ± 0.9 | 25.6 ± 10.0 |
| NCU04856 | 6.3.1.2 | Glutamine synthetase, α subunit | gln-2 | 0.95–2.58 | (0.5) 1.51–2.60 | ||||
| Glutamine degradation | NCU04216 | 2.4.2.14 | Amidophosphoribosyl transferase | ad-7 | 1.04–2.48 | (0.5) 3.89–7.42 | |||
| Asparagine synthesis | NCU04303 | 6.3.5.4 | Asparagine synthase | asn-1 | 0.49–1.10 | (0.5) 1.43–2.71 | Asparagine | 0.24 ± 0.04 | NQg |
| Threonine synthesis | NCU04118 | 2.7.2.4 | Aspartate kinase | 0.68–1.66 | (0.5) 2.32–5.87 | Threonine | 4.0 ± 0.6 | 5.6 ± 1.5 | |
| NCU03935 | 1.1.1.3 | Homoserine dehydrogenase | 0.92–1.23 | (0.5) 6.79–8.88 | |||||
| NCU04277 | 2.7.1.39 | Homoserine kinase | thr-4 | 0.44–0.79 | (0.5) 3.44–6.18 | ||||
| NCU03425 | 4.2.3.1 | Threonine synthase | thr-2 | 0.64–1.34 | (0.5) 1.53–3.12 | ||||
| Valine and isoleucine | NCU07982 | 2.2.1.6 | Acetolactate synthase, large subunit | ilv-3g | 1.62–2.95 | (1) 2.73–3.74 | Valine/isoleucine | 1.5 ± 0.3/3.4 ± 0.5 | 2.6 ± 0.7/1.5 ± 0.2 |
| synthesis | NCU01666 | 2.2.1.6 | Acetolactate synthase, small subunit | ilv-4 | 0.36–0.70 | (1) 6.44–6.85 | |||
| NCU03608 | 1.1.1.86 | Ketol acid reductoisomerase | ilv-2 | 0.47–1.19 | (1) 23.37–27.84 | ||||
| NCU04579 | 4.2.1.9 | Dihydroxy acid dehydratase | ilv-1 | 1.22–2.62 | (0.5) 6.02–12.57 | ||||
| NCU04754 | 2.6.1.42 | Branched-chain amino acid aminotransferase | 0.85–1.86 | (0.5) 3.67–7.13 | |||||
| Ornithine synthesis | NCU01295/NCU10468 | 2.3.1.1 | Glutamate N-acetyltransferase | arg-14 | 0.49–0.99 | (0.5) 3.23–6.56 | Ornithine | NQ | 1.7 ± 0.4 |
| NCU00567 | 2.7.2.8, 1.2.1.38 | Acetylglutamate kinase and N-acetylglutamyl phosphate reductase | arg-6 | 0.41–0.80 | (0.5) 1.22–2.57 | ||||
| NCU05410 | 2.6.1.11 | Acetylornithine aminotransferase | arg-5 | 0.63–1.27 | (0.5) 1.68–2.62 | ||||
| Arginine synthesis | NCU07732 | 6.3.5.5 | Carbamoyl phosphate synthase, small chain, arginine specific | arg-2 | 0.69–1.46 | (1) 8.18–8.71 | Arginine | NQ | 8.0 ± 0.6 |
| NCU01667 | 2.1.3.3 | Ornithine carbamoyl transferase | arg-12 | 0.71–1.83 | (0.5) 3.66–7.59 | ||||
| Arginine degradation | NCU02333 | 3.5.3.1 | Arginase | aga | 1.31–2.09 | (0.5) 5.11–3.82 | |||
| Lysine synthesis | NCU05526 | 2.3.3.14 | Homocitrate synthase (mitochondrial precursor) | lys-5 | 0.50–1.33 | (0.5) 2.28–4.81 | Lysine | NQ | 0.9 ± 0.1 |
| NCU08898 | 4.2.1.36 | Homoaconitase | lys-6 | 0.57–1.61 | (1) 3.33–3.44 | ||||
| NCU02954 | 1.1.1.87 | Homoisocitrate dehydrogenase | lys-7 | 0.40–0.70 | (0.5) 4.01–7.43 | ||||
| NCU03010 | 1.2.1.31 | l-Aminoadipate-semialdehyde dehydrogenase, large subunit | lys-3 | 0.55–1.04 | (0.5) 2.27–3.63 | ||||
| NCU03748 | 1.5.1.10 | Saccharopine dehydrogenase | lys-2 | 1.23–1.85 | (0.5) 7.26–8.22 | ||||
| Tyrosine synthesis | NCU01632 | Pentafunctional arom polypeptide | aro-1 | 0.63–1.62 | (0.5) 2.78–5.44 | Tyrosine | 0.11 ± 0.01 | NQ | |
| NCU05420 | Chorismate synthase/flavin reductase (NADPH) | aro-3 | 0.49–1.39 | (0.5) 2.07–5.20 | |||||
All Broad gene numbers are from annotation version 1.
mRNA data are from Kasuga et al. (35). Ranges of relative values are shown; complete data are available in supplementary table gki953_S2_Expression_Data of Kasuga et al. (35).
First time point with increased value after onset of germination.
Amino acid produced or consumed by indicated enzyme(s).
Data obtained from experiments whose results are shown in Fig. 4 for wild-type conidia and high-sucrose submerged hyphal cultures. NQ, not quantifiable; amino acid was detected, but levels were too low to quantitate.
Reannotated as ilv-3 based on linkage to met-2 in genetic and physical maps.
The results from a study that used Northern analysis to measure mRNA expression further validate our metabolite profiling results for arginine and ornithine levels in wild-type conidia and hyphae (60). This work showed that mRNA levels for arg-2 (encoding the small subunit of arginine-specific carbamoyl phosphate synthetase, required for arginine and ornithine biosynthesis) (16) are low in conidia and then become elevated during germination (50).
DISCUSSION
To our knowledge, this is the first report profiling N. crassa metabolites using 1H NMR. The advantage of 1H NMR is that an entire cellular “snapshot” of metabolites can be produced from a single experiment, thus allowing comparative global analysis of multiple strains and growth conditions. We first determined whether there were large differences in the metabolomes in a nontargeted approach using PCA analysis and then turned to the identification of specific metabolites using relative integrals of resolved resonances. To our knowledge, we are the first to report relative quantitation of several metabolites, including allantoin and mannitol, in N. crassa.
Schmit and Brody previously conducted a metabolic study with N. crassa in which they measured the levels of amino acids and other compounds in conidia and hyphae of the wild type using column chromatography and detection with ninhydrin (61). Overall, our results are consistent with this earlier work, validating the accuracy of the two methods for metabolite profiling of N. crassa. However, there were some differences in the results obtained using the two methodologies. Proline, methionine, and cysteine could not be detected in conidia and their levels were very low in hyphae in the Schmit and Brody study. We could not detect these three amino acids in either conidia or hyphae. Schmit and Brody reported that the predominant amino acids in conidia were glutamate, alanine, and glutamine, with glutamate in the greatest abundance. Similar results were obtained in our experiments, except that glutamate and alanine were present at similar levels in conidia. Schmit and Brody also found that the amounts of ornithine and arginine were lower in conidia than in hyphal cultures; this trend was also noted in our study, except that ornithine and arginine could not be detected in conidia.
Our extraction method produced hyphal extracts that were stable for more than 105 min of incubation at room temperature prior to acquiring spectra. The same was true for conidial extracts, with the exception of trehalose, glucose, and citrate (data not shown). The citrate levels were too low to quantitate in any sample in our analysis. We were unable to quantitate the other two sugars in conidia, due to apparent metabolism of trehalose to glucose, which resulted in large errors for these two compounds in our univariate analysis (data not shown). Future experiments to measure trehalose and glucose levels in conidia will require an alternative extraction method, possibly that recently reported by Lowe et al. (46).
PCA analysis revealed somewhat surprising results in that, although the Δgna-3 mutant produced conidia under all growth conditions (with the exception of peptone), they did not group with low-sucrose, conidiating wild-type cultures on the score plot but instead clustered near wild-type high-sucrose cultures. This suggests that the Δgna-3 mutation does not have a global effect on the metabolome in high-sucrose conditions. However, major differences were observed in the PCA analysis for the wild type and the Δgna-3 mutant cultured in limiting sucrose. Taken together, these data and those from previous work in our group (41) suggest that GNA-3 regulates carbon sensing and conidiation using different pathways. One interpretation is that without GNA-3, N. crassa will initiate conidiation but at the same time will not be able to sense available carbon and so will metabolically resemble a strain cultured in abundant carbon.
As mentioned above, conidiation and conidial germination in N. crassa are influenced by nutrient availability (62), and changes in the levels of several intracellular metabolites have been reported during these processes. We inspected our data for metabolite(s) that correlated with (i) the sucrose level in the growth medium, (ii) the Δgna-3 mutation, or (iii) the presence of conidia. There were three metabolites that were associated with low or high sucrose in submerged cultures of both the wild-type and the Δgna-3 strain: alanine, ornithine, and glucose. Alanine and ornithine are positively correlated, while the amount of glucose varies inversely with sucrose levels in the growth medium.
There were several metabolites that could be associated with the Δgna-3 mutation, but not under all growth conditions. The relative threonine levels are higher in the wild-type than in the Δgna-3 strain under low-sucrose conditions. Isoleucine cannot be detected in high- or low-sucrose submerged cultures from the Δgna-3 mutant. Leucine cannot be detected and the levels of adenosine and choline are low in low-sucrose Δgna-3 strain submerged cultures. The observation of lower metabolite levels in the Δgna-3 mutant cultured on limiting carbon hints at a requirement for gna-3 in the response to poor carbon availability.
Regarding metabolites that were associated with conidiation, we noted that the sugar alcohol mannitol was elevated in tissues that contain conidia, with the highest levels in ungerminated conidia (Fig. 5). Mannitol has been proposed to play roles as a carbohydrate reserve in environmental stress tolerance and during conidiation in the wheat pathogen fungus Stagonospora nodorum (63, 64). In the insect pathogenic fungus Metarhizium anisopliae, mannitol is the most abundant metabolite in conidial extracts (8).
A reduced carbon level signals to the fungus that the immediate environment is no longer suitable for sustained vegetative hyphal growth and that a new location must be sought to ensure survival in more favorable surroundings. In the absence of a mating partner, this is accomplished in many species of filamentous fungi by conidiation and the subsequent spread of mature spores by animals or wind currents. The Δgna-3 mutant appears to possess two defects. First, the Δgna-3 strain is deficient in a negative-control mechanism for conidiation that is independent of major changes in metabolite levels. Second, the Δgna-3 mutant is lacking in the ability to detect abundant sucrose in growth media. These malfunctions suggest that GNA-3 plays separate roles in the signal transduction cascades responsible for conidiation and nutrient sensing in N. crassa.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Borkovich and Larive laboratories for comments on the manuscript and many helpful discussions.
We acknowledge support for this work by National Science Foundation Integrative Graduate Education Research and Training Program fellowships DGE-0504249 to K.K. and J.D.K. and Public Health Service grants GM048626 and GM086565 from the National Institute of General Medical Sciences to K.A.B. C.K.L. gratefully acknowledges support from National Science Foundation grant CHE 0848976.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 1 April 2011.
REFERENCES
- 1. Abelson P. H., Vogel H. J. 1954. Amino acid biosynthesis in Torulopsis utilis and Neurospora crassa. J. Biol. Chem. 213:355–364 [PubMed] [Google Scholar]
- 2. Boernsen K. O., Gatzek S., Imbert G. 2005. Controlled protein precipitation in combination with chip-based nanospray infusion mass spectrometry. An approach for metabolomics profiling of plasma. Anal. Chem. 77:7255–7264 [DOI] [PubMed] [Google Scholar]
- 3. Borkovich K. A., et al. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brambl R. 1975. Presence of polyribosomes in conidiospores of Botryodiplodia theobromae harvested with nonaqueous solvents. J. Bacteriol. 122:1394–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Branco-Price C., Kaiser K. A., Jang C. J. H., Larive C. K., Bailey-Serres J. 2008. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56:743–755 [DOI] [PubMed] [Google Scholar]
- 6. Broadhurst D. I., Kell D. B. 2006. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2:171–196 [Google Scholar]
- 7. Brown J. K. M., Hovmoller M. S. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537–541 [DOI] [PubMed] [Google Scholar]
- 8. Carollo C. A., et al. 2010. Fungal tyrosine betaine, a novel secondary metabolite from conidia of entomopathogenic Metarhizium spp. fungi. Fungal Biol. 114:473–480 [DOI] [PubMed] [Google Scholar]
- 9. Christensen R. L., Schmit J. C. 1980. Regulation and glutamic acid decarboxylase during Neurospora crassa conidial germination. J. Bacteriol. 144:983–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colot H. V., et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colvin H. J., Sauer B. L., Munkres K. D. 1973. Glucose utilization and ethanolic fermentation by wild type and extrachromosomal mutants of Neurospora crassa. J. Bacteriol. 116:1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cooper T. G., Chisholm V. T., Cho H. J., Yoo H. S. 1987. Allantoin transport in Saccharomyces cerevisiae is regulated by two induction systems. J. Bacteriol. 169:4660–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cossins E. A., Pang S. H. Y., Chan P. Y. 1980. Glycine synthesis in a Neurospora mutant deficient in serine hydroxymethyltransferase. Plant Cell Physiol. 21:719–729 [Google Scholar]
- 14. Cui Q., et al. 2008. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotech. 26:162–164 [DOI] [PubMed] [Google Scholar]
- 15. Davis R. H. 2000. Neurospora: contributions of a model organism. Oxford University Press, New York, NY [Google Scholar]
- 16. Davis R. H. 2010. Amino acids and polyamines: polyfunctional proteins, metabolic cycles, and compartmentation, p. 339–358 In Borkovich K. A., Ebbole D. J. (ed.), Cellular and molecular biology of filamentous fungi. ASM press, Washington, DC [Google Scholar]
- 17. Davis R. H., de Serres F. J., Tabor H., Tabor C. W. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17:79–143 [Google Scholar]
- 18. d'Enfert C., Bonini B. M., Zapella P. D. A., Fontaine T., da Silva A. M., Terenzi H. F. 1999. Neutral trehalases catalyse intracellular trehalose breakdown in the filamentous fungi Aspergillus nidulans and Neurospora crassa. Mol. Microbiol. 32:471–483 [DOI] [PubMed] [Google Scholar]
- 19. d'Enfert C., Fontaine T. 1997. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol. Microbiol. 24:203–216 [DOI] [PubMed] [Google Scholar]
- 20. de Pinho C. A., de Lourdes M., Polizeli T. M., Jorge J. A., Terenzi H. F. 2001. Mobilisation of trehalose in mutants of the cyclic AMP signalling pathway, cr-1 (CRISP-1) and mcb (microcycle conidiation), of Neurospora crassa. FEMS Microbiol. Lett. 199:85–89 [DOI] [PubMed] [Google Scholar]
- 21. Dieterle F., Ross A., Schlotterbeck G., Senn H. 2006. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78:4281–4290 [DOI] [PubMed] [Google Scholar]
- 22. Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology 13:17R–27R [DOI] [PubMed] [Google Scholar]
- 23. Fiehn O., et al. 2000. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18:1157–1161 [DOI] [PubMed] [Google Scholar]
- 24. Halsall D. M., Catcheside D. E. 1971. Structural genes for DAHP synthase isoenzymes in Neurospora crassa. Genetics 67:183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Halsall D. M., Catcheside D. E., Doy C. H. 1971. Some properties of the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase isoenzymes from mutant strains of Neurospora crassa. Biochim. Biophys. Acta 227:464–472 [DOI] [PubMed] [Google Scholar]
- 26. Halsall D. M., Doy C. H. 1969. Studies concerning the biochemical genetics and physiology of activity and allosteric inhibition mutants of Neurospora crassa 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Biochim. Biophys. Acta 185:432–446 [DOI] [PubMed] [Google Scholar]
- 27. Hill E. P., Sussman A. S. 1964. Development of trehalase and invertase activity in Neurospora. J. Bacteriol. 88:1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hogg J. A., Richardson M. 1968. Biosynthesis of betaine in Neurospora crassa. Arch. Mikrobiol. 62:153–156 [DOI] [PubMed] [Google Scholar]
- 29. Horikoshi K., Ikeda Y. 1966. Trehalase in conidia of Aspergillus oryzae. J. Bacteriol. 91:1883–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horne D. W., Broquist H. P. 1973. Role of lysine and N-trimethyllysine in carnitine biosynthesis. I. Studies in Neurospora crassa. J. Biol. Chem. 248:2170–2175 [PubMed] [Google Scholar]
- 31. Jewett M. C., Hofmann G., Nielsen J. 2006. Fungal metabolite analysis in genomics and phenomics. Curr. Opin. Biotechnol. 17:191–197 [DOI] [PubMed] [Google Scholar]
- 32. Kaiser K. A., Barding G. A., Larive C. K. 2009. Metabolic profiling of plants by 1H-NMR: a comparison of metabolite extraction strategies using rosette leaves of the model plant Arabidopsis thaliana. Magn. Reson. Chem. 47:S147–S156 [DOI] [PubMed] [Google Scholar]
- 33. Kaiser K. A., Merrywell C. E., Fang F., Larive C. K. 2008. Metabolic profiling, p. 233–267 In Wawer I., Holzgrabe U., Diehl B. (ed.), NMR spectroscopy in pharmaceutical analysis. Elsevier, Oxford, United Kingdom [Google Scholar]
- 34. Kanamori K., Legerton T. L., Weiss R. L., Roberts J. D. 1982. Effect of the nitrogen source on glutamine and alanine biosynthesis in Neurospora crassa. An in vivo 15N nuclear magnetic resonance study. J. Biol. Chem. 257:14168–14172 [PubMed] [Google Scholar]
- 35. Kasuga T., et al. 2005. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 33:6469–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kays A. M., Borkovich K. A. 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G alpha proteins. Genetics 166:1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kays A. M., Rowley P. S., Baasiri R. A., Borkovich K. A. 2000. Regulation of conidiation and adenylyl cyclase levels by the G-alpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693–7705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koelle M. R. 2006. Heterotrimeric G protein signaling: getting inside the cell. Cell 126:25–27 [DOI] [PubMed] [Google Scholar]
- 39. Kuepfer L., Sauer U., Blank L. M. 2005. Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res. 15:1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lerch K. 1981. Metal ions in biological systems, vol. 13 Marcel Dekker, Inc., New York, NY [Google Scholar]
- 41. Li L., Borkovich K. A. 2006. GPR-4 is a Predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot. Cell 5:1287–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L., Wright S. J., Krystofova S., Park G., Borkovich K. A. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61:423–452 [DOI] [PubMed] [Google Scholar]
- 43. Lin C. Y., Wu H. F., Tjeerdema R. S., Viant M. R. 2007. Evaluation of metabolite extraction strategies from tissue samples using NMR metabolomics. Metabolomics 3:55–67 [Google Scholar]
- 44. Linden H., Ballario P., Macino G. 1997. Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 22:141–150 [DOI] [PubMed] [Google Scholar]
- 45. Lindon J. C., Holmes E., Nicholson J. K. 2003. So what's the deal with metabonomics? Anal. Chem. 75:384A–391A [DOI] [PubMed] [Google Scholar]
- 46. Lowe R. G., et al. 2010. A combined (1)H nuclear magnetic resonance and electrospray ionization-mass spectrometry analysis to understand the basal metabolism of plant-pathogenic Fusarium spp. Mol. Plant Microbe Interact. 23:1605–1618 [DOI] [PubMed] [Google Scholar]
- 47. Martin F. P., et al. 2007. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol. Syst. Biol. 3:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marzluf G. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Munkres K. D. 1968. Genetic and epigenetic forms of malate dehydrogenase in Neurospora. Ann. N. Y. Acad. Sci. 151:294–306 [DOI] [PubMed] [Google Scholar]
- 50. Orbach M. J., Sachs M. S., Yanofsky C. 1990. The Neurospora crassa arg-2 locus. Structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J. Biol. Chem. 265:10981–10987 [PubMed] [Google Scholar]
- 51. Pall M. L. 1981. Adenosine 3′,5′-phosphate in fungi. Microbiol. Rev. 45:462–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pall M. L., Robertson C. K. 1988. Regulation of lactate/pyruvate ratios by cyclic AMP in Neurospora crassa. Biochem. Biophys. Res. Commun. 150:365–370 [DOI] [PubMed] [Google Scholar]
- 53. Perkins D. D., Radford A., Sachs M. S. 2001. The Neurospora compendium: chromosomal loci. Academic Press, San Diego, CA [Google Scholar]
- 54. Pillonel C., Meyer T. 1997. Effect of phenylpyrroles on glycerol accumulation and protein kinase activity of Neurospora crassa. Pestic. Sci. 49:229–236 [Google Scholar]
- 55. Plesofsky-vig N., Light D., Brambl R. 1983. Paedogenetic conidiation in Neurospora crassa. Exp. Mycol. 7:283–286 [Google Scholar]
- 56. Plesofsky N., Brambl R. 1999. Glucose metabolism in Neurospora is altered by heat shock and by disruption of HSP30. Biochim. Biophys. Acta 1449:73–82 [DOI] [PubMed] [Google Scholar]
- 57. Radford A. 2004. Metabolic highways of Neurospora crassa revisited. Adv. Genet. 52:165–207 [DOI] [PubMed] [Google Scholar]
- 58. Ringner M. 2008. What is principal component analysis? Nat. Biotechnol. 26:303–304 [DOI] [PubMed] [Google Scholar]
- 59. Robertson D. G. 2005. Metabonomics in toxicology: a review. Toxicol. Sci. 85:809–822 [DOI] [PubMed] [Google Scholar]
- 60. Sachs M. S., Yanofsky C. 1991. Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev. Biol. 148:117–128 [DOI] [PubMed] [Google Scholar]
- 61. Schmit J. C., Brody S. 1975. Neurospora crassa conidial germination: role of endogenous amino acid pools. J. Bacteriol. 124:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmit J. C., Brody S. 1976. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol. Rev. 40:1–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Solomon P. S., Waters O. D. C., J.örgens C. I., Lowe R. G. T., Rechberger J., Trengove R. D., Oliver R. P. 2006. Mannitol is required for asexual sporulation in the wheat pathogen Stagonospora nodorum (glume blotch). Biochem. J. 399:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Solomon P. S., Waters O. D. C., Oliver R. P. 2007. Decoding the mannitol enigma in filamentous fungi. Trends Microbiol. 15:257–262 [DOI] [PubMed] [Google Scholar]
- 65. Springer M. L. 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays 15:365–374 [DOI] [PubMed] [Google Scholar]
- 66. Springer M. L., Yanofsky C. 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3:559–571 [DOI] [PubMed] [Google Scholar]
- 67. That T. C., Turian G. 1978. Ultrastructural study of microcyclic macroconidiation in Neurospora crassa. Arch. Microbiol. 116:279–288 [DOI] [PubMed] [Google Scholar]
- 68. Turian G., Bianchi D. E. 1972. Conidiation in Neurospora. Bot. Rev. 38:119–154 [Google Scholar]
- 69. Turner G. E., Weiss R. L. 2006. Developmental expression of two forms of arginase in Neurospora crassa. Biochim. Biophys. Acta 1760:848–857 [DOI] [PubMed] [Google Scholar]
- 70. van den Berg R. A., Hoefsloot H. C., Westerhuis J. A., Smilde A. K., van der Werf M. J. 2006. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Viant M. R., Rosenblum E. S., Tieerdema R. S. 2003. NMR-based metabolomics: a powerful approach for characterizing the effects of environmental stressors on organism health. Environ. Sci. Technol. 37:4982–4989 [DOI] [PubMed] [Google Scholar]
- 72. Villas-Boas S. G., Hojer-Pedersen J., Akesson M., Smedsgaard J., Nielsen J. 2005. Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast 22:1155–1169 [DOI] [PubMed] [Google Scholar]
- 73. Ward J. L., Baker J. M., Beale M. H. 2007. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 274:1126–1131 [DOI] [PubMed] [Google Scholar]
- 74. Zhang S., et al. 2009. Interdependence of signal processing and analysis of urine 1H NMR spectra for metabolic profiling. Anal. Chem. 81:6080–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.