Abstract
Errors during gene expression from DNA to proteins via transcription and translation may be deleterious for the functional maintenance of cells. In this paper, extensive genetic studies of the misreading of a GA repeat introduced into the lacZ gene of Escherichia coli indicate that in this bacteria, errors occur predominantly by a +2 translational frameshift, which is controlled by a tRNA modification involving the MnmE and GidA proteins. This ribosomal frameshift results from the coincidence of three events: (1) decreased codon–anticodon affinity at the P-site, which is caused by tRNA hypomodification in mnmE− and gidA− strains; (2) a repetitive mRNA sequence predisposing to slippage; and (3) increased translational pausing attributable to the presence of a rare codon at the A-site. Based on genetic analysis, we propose that GidA and MnmE act in the same pathway of tRNA modification, the absence of which is responsible for the +2 translational frameshift. The difference in the impact of the mutant gene on cell growth, however, indicates that GidA has at least one other function.
Keywords: Translational fidelity, 5-methylaminomethyl-2-thio-uridine, gidA, mnmE, thdF, argU
Processes ensuring the fidelity of DNA replication have been studied extensively, presumably because of the overwhelming importance of gene mutations in evolution, hereditary diseases and cancer (Friedberg et al. 1995). In contrast, less is known about the fidelity of transcription and translation, which must be crucial for the functional maintenance of cells and organisms. Functional degeneration of nondividing cells limits the lifespan of organs such as the heart or brain. Therefore, decreasing fidelity of gene expression may be an underlying cause of cellular aging, degenerative diseases, and death. Accurate gene expression is dependent on the fidelity of both transcription of DNA and translation of mRNA.
Fidelity of transcription depends on proofreading by a 3′ → 5′ exonuclease activity found in a number of eukaryotic and prokaryotic RNA polymerases (Thomas et al. 1998). The Escherichia coli RNA polymerase exhibits error rates ranging from 1 to 10 misincorporations per 105 synthesized nucleotides (for review, see Libby and Gallant 1991). Aberrant RNAs also include the presence of altered nucleotides in the mRNA caused either by direct alteration of the mRNA or by misincorporation of altered mutagenic nucleotides, such as 8-oxo-GTP, by the RNA polymerase (Taddei et al. 1997). The presence of 8-oxo-G in cellular RNA was shown to be restricted to vulnerable neurons in dementia such as Alzheimer's disease (AD) (Nunomura et al. 1999) or Parkinson's disease (Zhang et al. 1999) and to be correlated with the synthesis of altered proteins (Dukan et al. 2000). The rate of transcriptional frameshifting is unknown but it was proposed that it may lead to some neurodegenerative diseases (van Leeuwen et al. 1998b). In patients with AD, the accumulation of aberrant protein species shifted to the +1 frame with no related mutation in the DNA blueprint has been reported. The appearance of those proteins was shown to be caused by a molecular misreading of a coding GA repeat resulting in the deletion of a dinucleotide GA in the mRNA (Hol et al. 1998; van Leeuwen et al. 1998a,b).
Translational errors have been characterized in E. coli and measured in vivo and/or in vitro (for review, see Kurland 1992). These include missense errors resulting in the substitution of one amino acid for another, or incorporation of extra amino acids caused by read-through of termination codons. Overall, translational errors occur at a frequency of 10−3 to 10−4 (Loftfield and Vanderjagt 1972; Edelmann and Gallant 1977; Ellis and Gallant 1982; Parker 1989) and are limited by the ability of the ribosome to discriminate between correct and incorrect tRNAs entering the A-site (Thompson 1988; Nierhaus 1990; Czworkowski and Moore 1996). When induced by the addition of some antibiotics (e.g., streptomycin), translational misreading leads to increased concentrations of oxidized proteins (Dukan et al. 2000), which were found to accumulate in aging cells (Stadtman 1992). Other translational errors include premature termination (Menninger 1977; Kurland et al. 1996) and translational frameshifting. The latter generally gives rise to a shorter peptide because of the subsequent encounter of stop codons in the shifted frame. Spontaneous frameshifts resulting from translational errors occur at a frequency of <10−5 per codon (Kurland 1992), but were shown to increase in E. coli cells entering stationary phase (Barak et al. 1996).
Basic mechanisms involved in DNA replication, recombination, transcription, and translation are often conserved phylogenetically (Lewin 1994). Therefore, we set out to study in E. coli the source of errors brought about by the presence of a GA repeat within an engineered frameshifted allele of the lacZ gene (Table 1). The characterization of E. coli mutants expressing an intermediate level of β-galactosidase led to the identification of three genes that are involved in the faithful translation of the GA repeat. Two of these genes, mnmE and gidA, are involved in tRNA modification and the third, argU, encodes the tRNA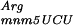 . The analysis of the mechanism of this unusual +2 translational frameshift at this sequence is consistent with the recently proposed +1/−1 translational frameshift dual error model (Qian et al. 1998; Björk et al. 1999; Farabaugh and Björk 1999), which can be extended to explain larger ribosomal frameshifting and short tRNA hopping. Measurements of the growth rate of strains carrying a mutation in the gidA or mnmE genes showed that the impact of tRNA hypomodification varies with nutritional conditions and is not equivalent for both strains (Table 2). These observations suggest that GidA may also be involved in other pathways.
. The analysis of the mechanism of this unusual +2 translational frameshift at this sequence is consistent with the recently proposed +1/−1 translational frameshift dual error model (Qian et al. 1998; Björk et al. 1999; Farabaugh and Björk 1999), which can be extended to explain larger ribosomal frameshifting and short tRNA hopping. Measurements of the growth rate of strains carrying a mutation in the gidA or mnmE genes showed that the impact of tRNA hypomodification varies with nutritional conditions and is not equivalent for both strains (Table 2). These observations suggest that GidA may also be involved in other pathways.
Table 1.
Sequences of alleles used and their corresponding β-galactosidase activities
| Allele
|
Sequencea
|
Stop codon positionb
|
β-Galactosidase activityc
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lacZ+ | A | A | T | G | A | A | T | C | A | G | G | C | 3073 | 12400 ± 10 | |||||||
| A | s | n | G | l | u | S | e | r | G | l | y | ||||||||||
| lacZ (+GA) | A | A | T | G | A | G | A | G | A | G | T | G | G | C | 1603 | 0.10 ± 0.01 | |||||
| A | s | n | G | l | u | A | r | g | V | a | l | ||||||||||
| lacZ+ revertants | A | A | T | G | A | G | A | G | T | G | G | C | 3073 | 12600 ± 240 | |||||||
| A | s | n | G | l | u | S | e | r | G | l | y | ||||||||||
| lacZ (+ Arg462) | A | A | T | G | A | G | A | G | A | A | G | T | G | G | C | 3076 | 0.01 ± 0.001 | ||||
| A | s | n | G | l | u | A | r | g | S | e | r | G | l | y | |||||||
All sequences are shown from codon 460 of the lacZ gene.
Nucleotide number of the first base of the following stop codon.
Miller units ± standard error.
Table 2.
Generation time of strains in various growth media at 40°C
| Relevant genotypes
|
Mediaa
|
|
|---|---|---|
| LB
|
Mm + 0.4% glucose
|
|
| Wild type (NECB1) | 32.9 ± 0.7 | 75.7 ± 3.7 |
| gidA∷Tn10 | 38.0 ± 1.2b | 91.1 ± 3.4e |
| mnmE∷Tn10 | 38.2 ± 0.4c | 77.7 ± 2.5 |
| gidA∷Tn10, mnmE∷kan | 45.2 ± 0.7d | 95.8 ± 2.0f |
Numbers represent the average generation time of 5 cultures given in minutes ± SEM.
b–fGeneration times that are significantly different from that of the wild type in the same medium. T-test comparison between WT and mutants grown in the same media have the following P-values: b, 0.0054; c, 0.0002; d, <0.0001; e, 0.0154; f, 0.0013.
Results
The reporter system
We studied the control of error rates of expression (transcription or translation) of a gene containing a coding GA repeat in E. coli. A reporter system for such errors was constructed using the lacZ gene encoding the β-galactosidase enzyme.
The constructed lacZ(+GA) frameshifted allele contains a repetition of the dinucleotide GA starting from the first base of codon 461 to the first base of codon 463 (Table 1). These codons normally encode amino acids involved in the active site of β-galactosidase (Cupples and Miller 1988). This insertion results in a sequence shifted to the +2 reading frame when compared with the wild-type sequence and the emergence of a premature stop codon 210 nucleotides downstream, thereby generating a mRNA encoding a 534-amino-acid polypeptide. The β-galactosidase activity of NECB1 cells, which carry this new allele, is 125,000 times lower than the activity of the isogenic lacZ+ cells (Table 1). As a result, NECB1 cells are not able to grow on minimal media containing lactose as the sole source of carbon, and form white colonies on LB plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside).
Only deletion of a dinucleotide GA can restore an active β-galactosidase
To study errors on this GA repeat, we first wanted to establish the spectrum of mutants that could restore partially or totally the β-galactosidase activity. This question was addressed by selecting for spontaneous reversion mutants at the DNA level, which restore an active β-galactosidase.
NECB1 cells were plated on minimal medium containing lactose and Lac+ revertants were scored. The frequency of appearance of Lac+ colonies was about 10−9 per viable cell. The lacZ gene of 20 of those clones taken from three different cultures was sequenced and all of them had a deletion of a GA dinucleotide that restores the wild-type sequence of the protein (Table 1). The β-galactosidase activity of those revertants was measured and is comparable with the activity of the isogenic lacZ+ strain. With this screen we found no single nucleotide insertions that could potentially have restored the reading frame and generated a polypeptide with an extra amino acid. A mutant we constructed containing an extra A downstream to the GA repeat had a β-galactosidase activity about 106-fold lower than the lacZ+ strain (Table 1). The expected protein encoded by this lacZ (+Arg462) mutant contains an extra arginine in position 462 (Table 1). This indicates that only a mutation restoring the wild-type frame of the protein can result in an active β-galactosidase, consistent with the central position of these amino acids in the enzyme active site (Cupples and Miller 1988). As a consequence, mutation rate being very low, production of an active β-galactosidase should result mostly from a deletion of a dinucleotide GA at the transcription level or +2 frameshift at the translation level.
Isolation of mutants affected in the phenotypic expression of lacZ(+GA)
The low level of β-galactosidase activity of the lacZ(+GA) strain suggests that this type of misreading (−GA) at the level of transcription or translation, producing occasional active proteins, are very rare in wild-type E. coli. Therefore, a cell with a mutation that increases these error rates is expected to express only a low to intermediate level active β-galactosidase. Following mutagenic treatments, mutants were scored for their ability to form pale blue colonies on plates containing X-Gal.
The first mutagenesis of the NECB1 strain was a random mini-Tn10 insertion using the λNK1323 phage (Kleckner et al. 1991). Following mutagenesis, about 20,000 tetracycline-resistant colonies were screened and 10 of them presented a light blue coloration. Nine of the 10 were stable and eight of those conferred an intermediate level of β-galactosidase when the Tn10 was retransduced by P1 phage in NECB1 cells. For these clones, the transduction of the tetracycline resistance into Lac+ cells does not detectably affect the β-galactosidase-specific activity (data not shown), making it unlikely that the increased β-galactosidase level is attributable to a mutation affecting the level of transcription or translation of the lacZ gene.
As transposon mutagenesis will not reveal essential genes that may be involved in the control of gene expression, we performed a random point mutagenesis using 2-aminopurine (2-AP). This molecule is an adenine analog, which can mispair with cytosine and generates mostly transitions. Approximately 30,000 colonies from three different cultures of NECB1 cells grown with 2-AP were screened and 133 mutants harboring a stable pale blue coloration were isolated. These mutants are described below.
Characterization of mutants containing a Tn10 insertion
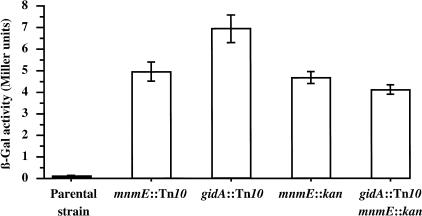
The eight mutants containing a Tn10 insertion that was associated with increased β-galactosidase activity were identified by inverse PCR (Triglia et al. 1988) and sequencing of the amplified fragments. Mutants 3, 4, 8, and 10 contained a Tn10 insertion in the gidA gene at position 707, 1025, 5, and 51, respectively. The function of this gene, the closest to the origin of replication oriC in E. coli, is as yet unidentified but has been associated with a glucose inhibited division phenotype (von Meyenburg and Hansen 1980). In all cases, β-galactosidase activity returned to the level of the parental strain when transformed with a plasmid containing the gidA gene (data not shown), suggesting that all insertions result in the lack of production of an active GidA protein. Mutant 3, hereafter designated as gidA::Tn10, having a β-galactosidase activity ∼70-fold higher than the parental strain (Fig. 1) was chosen for further analysis.
Figure 1.
Error rate measurement with β-galactosidase assays. β-galactosidase assays were performed on cells grown overnight in LB containing the proper antibiotic. Bars represent the average of β-galactosidase measurements ± standard errors. The activity of mnmE::kan is not statistically different from mnmE::Tn10 (t-test = −0.526, P = 0.604). Double mutant gidA::Tn10, mnmE::kan activity is not different from the activity of mnmE::kan (t-test = −1.556, P = 0.134) but is different from gidA::Tn10 (t-test = 4.451, P = 0.0001) suggesting that mnmE is epistatic on gidA.
Mutants 2, 6, and 7 had Tn10 insertions in the mnmE gene, at positions 798, 707, and 43, respectively. The mnmE gene was previously called trmE, because of its involvement in tRNA modification (Elseviers et al. 1984). For the three mutants, transformation with a plasmid containing the mnmE gene decreased the β-galactosidase activity to the level of the parental strain (data not shown). Mutant 2, hereafter designated as mnmE::Tn10, has a β-galactosidase activity ∼50-fold higher than the parental strain (Fig. 1). This mutant was used as a representative of mnmE::Tn10 mutants for further analysis.
Mutant 9 has a Tn10 insertion in the ebgR gene, known to be the repressor of the evolved β-galactosidase operon. When this operon is derepressed, the EbgA enzyme is produced, resulting in a weakly positive lactose utilization phenotype (Campbell et al. 1973; Arraj and Campbell 1975). This mutant was not analyzed further.
Characterization of mutants obtained after 2-AP mutagenesis
As the majority of the insertion mutants were in mnmE and gidA, we first identified which of the point mutants were complemented with plasmids expressing these genes. Transformed cells were scored for the disappearance of the pale blue coloration. Of the 133 mutants, 36 were complemented by mnmE and 56 were complemented with gidA. This accounted for ∼70% of all the clones, confirming the importance of these two genes in the fidelity of gene expression. The β-galactosidase activity of gidA mutants has an average of 5.7 ± 1.4 Miller units, whereas that for mnmE mutants is 3.7 ± 0.3, both of which are comparable with the activities in the strains with Tn10 disruptions of those genes.
To determine if the 41 mutants not complemented by mnmE or gidA contained mutations altering phenotypic expression, we eliminated mutations in cis in the lacZ gene and those causing derepression of genes coding for β-galactosidase-like activities. These two types of mutants were identified by transduction using P1 stocks made on lacI42::Tn10 lacZ(+GA) and lacI42::Tn10 lacZΔM15 strains, respectively. When the lacZ(+GA) allele was introduced into its genome, one mutant lost the blue coloration showing that the intermediate β-galactosidase activity was attributable to a cis mutation of the lacZ(+GA) allele (not sequenced). Six mutants remained constantly blue when their lacZ gene was replaced by a deletion of lacZ, which is the hallmark of a strain expressing β-galactosidase-like activity.
To characterize mutations in genes not identified previously, the seven mutants out of the 34 remaining that showed the highest β-galactosidase level (140-fold higher than the parental strain on average) were transformed with a genomic DNA library made from the parental lacZ(+GA) strain. Plasmids that suppressed the blue color all contained argU, which encodes tRNAArg. To confirm whether this gene was deficient in these seven mutants, a plasmid containing the argU gene was constructed and used to transform cells of those seven clones. The β-galactosidase activity of all clones transformed with this plasmid was reduced to a level comparable with that of the NECB1 parental strain. The presence of mutations in the argU structural gene or in its promoter was confirmed by sequencing.
The 27 mutants harboring the lowest β-galactosidase activity (sevenfold higher than the parental strain on average) have not been characterized further.
GidA and MnmE may be in the same pathway
The mnmE gene product is known to be involved in the biosynthesis of the hypermodified nucleoside 5-methylaminomethyl-2-thio-uridine (mnm5s2U), which is found in the wobble position of bacterial tRNAs specific for glutamate, lysine, glutamine, arginine, and leucine (Björk 1996). It is unclear how many enzymatic reactions are involved in the first step of this process (Hagervall et al. 1987). There is no clear evidence for the function of GidA, but thermosensitive mutants of that gene were found in a tRNA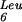 (supP) mutant background (Nakayashiki and Inokuchi 1998), which we interpret as a role for this protein in tRNA biogenesis.
(supP) mutant background (Nakayashiki and Inokuchi 1998), which we interpret as a role for this protein in tRNA biogenesis.
To determine whether GidA is involved in the same pathway as MnmE, we transduced a mutant allele of mnmE interrupted with a kanamycin cassette (o454::kan1, called mnmE::kan hereafter; Cabedo et al. 1999) into lacZ(+GA) parental and derivative gidA::Tn10 strains. The β-galactosidase activity of mnmE::kan was not different from that of mnmE::Tn10, and the activity of the double mutant (gidA::Tn10, mnmE::kan) is not different from that of the single mnmE::kan mutant (Fig. 1) but is significantly lower than the activity of the single gidA::Tn10 mutant. This implies that the gene products from gidA and mnmE are likely to be in the same biosynthetic pathway, and furthermore, that the MnmE protein activity precedes the activity of the GidA protein.
+2 Ribosomal frameshifting caused by a less efficient codon–anticodon recognition
The +2 translational frameshifting, observed in mnmE::Tn10 or gidA::Tn10, could be explained by an extension of the recently described +1/−1 translational frameshift dual error model (Qian et al. 1998), which postulates such a slipping of the ribosome when a hypomodified tRNA present in the P-site results in a weaker codon–anticodon interaction. The tRNA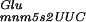 , which decodes the first codon of the GA repeat, is one of the tRNAs that carry an MnmE-dependent mnm5s2U34 modification (Björk 1996), which allows it to decode either GAG or GAA codons with similar efficiencies (Kruger et al. 1998). In the mnmE::Tn10 strain, and likely in the gidA::Tn10 strain, the U34 is hypomodified resulting in tRNA with a less efficient base-pairing in the wobble position (Kruger et al. 1998). The observed frameshifting could result from the presence of this hypomodified tRNA
, which decodes the first codon of the GA repeat, is one of the tRNAs that carry an MnmE-dependent mnm5s2U34 modification (Björk 1996), which allows it to decode either GAG or GAA codons with similar efficiencies (Kruger et al. 1998). In the mnmE::Tn10 strain, and likely in the gidA::Tn10 strain, the U34 is hypomodified resulting in tRNA with a less efficient base-pairing in the wobble position (Kruger et al. 1998). The observed frameshifting could result from the presence of this hypomodified tRNA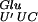 (U′ is a hypomodified form of the uridine) in the P-site of the ribosome.
(U′ is a hypomodified form of the uridine) in the P-site of the ribosome.
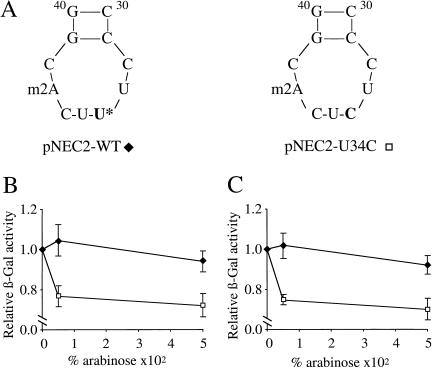
To test the effect of the pairing stability of this tRNA on frameshift errors, we engineered gltT to have a CUC anticodon. This mutant and the gltT+ gene were cloned into the pBAD33 vector under the control of the arabinose inducible promoter (Guzman et al. 1995), generating the pNEC2 plasmid series (Fig. 2; Table 3). When coexpressed with the four wild-type copies on the chromosome, the tRNA harboring the CUC anticodon reduced the translational frameshifting by 30% in mnmE::Tn10 and gidA::Tn10 strains (Fig. 2B,C) as well as in the wild-type strain (t-test induced vs. non-induced = −2.8, P = 0.02; data not shown). This mutated tRNA therefore appears to be able to increase the codon–anticodon interaction, thereby reducing the frameshifting of the ribosome. The selective competition for translational machinery (e.g., aminoacylation) between the tRNA produced by the plasmid and the wild-type tRNA produced from the four genomic genes could explain why the error rate was not reduced to the level of the parental lacZ(+GA) strain.
Figure 2.
Expression of a tRNA with an exact anticodon reduces the errors. Mutant gltT tRNAGlu was created to determine the role of the anticodon loop sequence in translational errors. (A) Sequence from nucleotides 30–40 (anticodon loop) of the tRNA encoded by gltT cloned in pNEC2-WT. The pNEC2–U34C plasmid carries the mutation U34 to C (bold letters, U* designates the modified uridine), thereby generating a CUC anticodon, which is an exact match to the GAG codon. (B,C) Transformed NECB10 (mnmE::Tn10 Δara leu::Tn5) and NECB11 (gidA::Tn10 Δara leu::Tn5) cells were grown overnight in LB containing 0%, 0.005%, or 0.05% arabinose to induce expression of the gltT gene that is under the control of the arabinose promoter in pBAD33 (see Table 3). For each strain and plasmid pair, the β-galactosidase values were normalized to the value at 0% arabinose induction. The decreased β-galactosidase activity observed with the expression of the gltT allele carried by pNEC2–U34C (□) is significant in mnmE::Tn10 (t-test induced vs. noninduced = −4.2, P = 0.002) (B) and in gidA::Tn10 (t-test induced vs. noninduced = −4.8, P = 0.001) (C). In both strains the expression of the gltT allele carried by the pNEC2-WT (♦) has no effect on ribosomal frameshifting. These results suggest that translational errors in mnmE::Tn10 and gidA::Tn10 are attributable to a less efficient hypomodified tRNA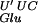 (U′ represents the hypomodified form of the uridine). Each point represents the average of normalized β-galactosidase activity ± standard error.
(U′ represents the hypomodified form of the uridine). Each point represents the average of normalized β-galactosidase activity ± standard error.
Table 3.
Strains, plasmids, and primers
|
|
Description
|
Source
|
|---|---|---|
| Strains | ||
| FR576 | NR3835 F−: Δ(proAB-lac) ara−thi−trpE997 | Taddei et al. 1997 |
| FR575 | NR3835 F′ [proAB-lacZ+] | Schaaper and Dunn 1987 |
| NECB1 | FR576 F′[proAB-lacZ(+GA)] | This study |
| NECB2 | NECB1 mnmE∷Tn10 | This study |
| NECB3 | NECB2 gidA∷Tn10 | This study |
| IC3652 | recB21 recC22 sbcB15 sbcC201 Sal−o454-1∷kan | Cabedo et al. 1999 |
| NECB4 | NECB1 o454-1∷kan (or mnmE∷kan) | This study |
| NECB5 | NECB3 o454-1∷kan (or mnmE∷kan) | This study |
| SS14 | trp31 his1 argG6 rpsl 104 tonA2 Δ(lacZ) supE44 | Vulic et al. 1999 |
| xyl7 mtl2 metB1 serB28 specR F42[finP301 lacI42∷Tn10 lacZ+] | ||
| NECB6 | NECB1 lacI42∷Tn10 | This study |
| GC3560 | thr1 leu6 proA2 his4 thi1 argE3 galK2 ara14 xyl5 | R. D’Ari's collection |
| mtl1 tsx33 str31 supE44 lacIq lacZΔM15 λJFL100 lacY+ | ||
| NECB7 | rph1 lacI42∷Tn10 lacZΔM15 | This study |
| NECB8 | rph1 Δara leu∷Tn5 lacZ101 | Lab collection |
| NECB9 | NECB1 Δara leu∷Tn5 | This study |
| NECB10 | NECB2 Δara leu∷Tn5 | This study |
| NECB11 | NECB3 Δara leu∷Tn5 | This study |
| NECB13 | FR576 F′[proAB-lacZ(+Arg462)] | This study |
| Plasmids | ||
| pNEC1 | pACYC184 containing argU at BamHI site | This study |
| pNEC2-WT | pBAD33 containing gltT between KpnI and PstI | Guzman et al. 1995 |
| sites under the control of the arabinose promoter | and this study | |
| pNEC2-U34C | pNEC2-WT with U34 to C34 mutation in gltT | This study |
| pNEC3 | pUC18 containing gidA between EcoRI and SphI sites | This study |
| pNEC4 | pUC18 containing mnmE between EcoRI and PstI sites | This study |
| Primers (5′ → 3′) | ||
| LacUp | CCCGAAACTGTGGAGCGCCGAA | |
| LacDwn | CCGACCACGGGTTGCCGTTTTC | |
| LacZGAGA | AGCGCCGTGGCCACTCTCTCATTCCCCAG | |
| ArgUB1Ua | CGGGATCCCGCAGGACGGGTTCGGAGATAb | |
| ArgUB1La | CGGGATCCCGACATACCAAGGCGGCTAAGb | |
| GltTK1Ua | GGGGTACCCCGAAGCGTACTTTGTAGTGCb | |
| GltTP1La | AACTGCAGAACCAATGCATTGGCCTCACAACCCGAAGATGTb | |
| GltTDir | GCCCTCTCACGGCGGTAACAGGGGTTCG | |
| GltTRev | GCCGTGAGAGGGCGGTGTCCTGGGCCTC | |
| MnmEE1Ua | CGGAATTCAAATCCTGATTCGGTGAGTb | |
| MnmEP1Da | TTCTGCAGAGTCAGAATGCGGCTTCGTb | |
| GidAE1Ua | TGGTCGTGAATTCTTTTAATACCCAGGATCCCAb | |
| GidAS1Da | TGGTCGTGCATGCTTGCCTGGTAAGCGGGTGCTb | |
| Tn10U | ACGATGCCCATTTTGTTGA | |
| Tn10L | ACGGCGTTCACTCCAATCT |
Primers used for cloning. These primers contain restriction site B1, BamHI; K1, Kpn1; P1, Pst1; E1, EcoR1; and S1, Sph1.
The restriction sites for cloning of the PCR product are underlined.
+2 Translational frameshift caused by the presence of a rare codon in the A-site
The +1/−1 translational frameshift dual error model also proposes that frameshifting could occur at a pausing site during translation, which could be caused by the presence of a slowly translated codon in the A-site. The extent of vacancy of the A-site has been shown to correlate to a +1 translational frameshift (Curran and Yarus 1989; Sipley and Goldman 1993). In the case of lacZ(+GA), the AGA codon (codon 462; Table 1) is one of the rarest codons in E. coli (Hénaut and Danchin 1996; Nakamura et al. 2000) and is decoded by a minor tRNA encoded by argU (Ikemura 1981; Dong et al. 1996). The AGA codon may therefore constitute a pausing site and lead to frameshifting.
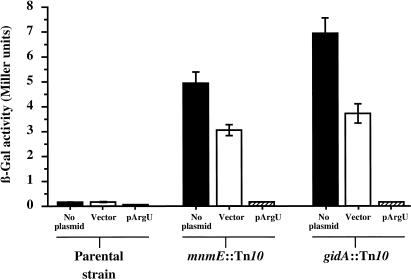
To test this hypothesis, the lacZ(+GA) and the derivatives gidA::Tn10 and mnmE::Tn10 were transformed with a plasmid containing the argU gene. The overexpression of the tRNA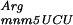 encoded by argU in gidA::Tn10 or mnmE::Tn10 strains abolishes the error phenotype observed in those strains (Fig. 3), as the β-galactosidase activities for transformed gidA::Tn10 and mnmE::Tn10 strains are comparable with the parental strain. It should be noted that the presence of the vector alone slightly reduced ribosomal frameshifting. This decrease may be attributable to the chloramphenicol, which was added to the growth media to select for the plasmid and is known to be a translation inhibitor (Bergmann and Sicher 1952). The error rate is significantly reduced by the overproduction of argU, even in the transformed parental strain (Fig. 3), showing that this same +2 frameshift occurs in wild-type cells (NECB1) by pausing of the ribosome caused by the scarcity of tRNA
encoded by argU in gidA::Tn10 or mnmE::Tn10 strains abolishes the error phenotype observed in those strains (Fig. 3), as the β-galactosidase activities for transformed gidA::Tn10 and mnmE::Tn10 strains are comparable with the parental strain. It should be noted that the presence of the vector alone slightly reduced ribosomal frameshifting. This decrease may be attributable to the chloramphenicol, which was added to the growth media to select for the plasmid and is known to be a translation inhibitor (Bergmann and Sicher 1952). The error rate is significantly reduced by the overproduction of argU, even in the transformed parental strain (Fig. 3), showing that this same +2 frameshift occurs in wild-type cells (NECB1) by pausing of the ribosome caused by the scarcity of tRNA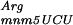 . These results show that the abolition of the ribosome frameshifting in parental, mnmE::Tn10, and gidA::Tn10 strains is caused by the filling of the A-site with the overproduced tRNA
. These results show that the abolition of the ribosome frameshifting in parental, mnmE::Tn10, and gidA::Tn10 strains is caused by the filling of the A-site with the overproduced tRNA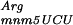 .
.
Figure 3.
Overexpression of argU abolishes errors. To know whether the presence of a rare AGA codon could be involved in these translational errors, parental mnmE::Tn10 and gidA::Tn10 strains were transformed with pNEC1, a pACYC184 derivative that carries the argU gene. The overexpression of the tRNAArg (paired hatched bars) decoding the AGA codon in mnmE::Tn10 and in gidA::Tn10 reduces translational errors to the level of the parental strain. It should be noted that even in the parental strain, the overexpression of argU significantly decreases the β-galactosidase activity (t-test = 5.62, P = 0.002 when compared with activity from the parental strain carrying pNEC1 or the control plasmid). For comparison, β-galactosidase activities of plasmid-less strains are presented by solid bars. As a control, strains were also transformed with pACYC184 (open bars), which by itself reduces translational frameshifting (P = 0.04 in mnmE::Tn10; P = 0.01 in gidA::Tn10).
The absence of the mnm5s2U modification and the cell growth rate
The gidA gene was first isolated in association with a glucose-inhibited division phenotype (von Meyenburg and Hansen 1980) and is located next to the origin of replication (oriC). It was therefore proposed to have a role in the control of cell division. As it is likely to belong to the same pathway as the mnmE gene in the control of the +2GA frameshift, we have tested the effect of mutant alleles of these genes on growth rate.
To estimate the growth rate of these strains defective for the mnm5s2U modification, we measured the generation time of the parental strain, gidA::Tn10, mnmE::Tn10, and the double mutant (gidA::Tn10 mnmE::kan) in LB and in minimal medium at 40°C. Comparison of generation times showed that gidA::Tn10 mnmE::Tn10 and (gidA::Tn10 mnmE::kan) strains had a growth rate lower than that of the parental strain during growth in LB (Table 2) or in LB containing glucose (data not shown). Under conditions of slower growth (minimal medium), we failed to detect a significant difference between the parental and the mnmE::Tn10 strains, but the gidA::Tn10 and (gidA::Tn10 mnmE::kan) strains had a generation time 16% and 20% longer, respectively, than that of the parental or mnmE::Tn10 strains. Unlike in the β-galactosidase assays showing the translational error rate of the +2GA frameshift, the double mutant did not have the same phenotype as the mnmE::Tn10 strain. These results indicate that mutations in gidA or mnmE have an independent impact on growth rate.
These results suggest that GidA may be acting in another pathway, such as a different tRNA modification path, or in DNA replication as suggested previously (Ogawa and Okazaki 1994).
Discussion
We studied the potential causes of errors during expression of a GA repeat sequence inserted out of frame in the lacZ gene of E. coli. Mutants having an increased error rate of gene expression were isolated based on the production of a moderate amount of active β-galactosidase. Approximately 83% of all these mutants (generated by both insertional and point mutagenesis) were alleles of mnmE and gidA, both apparently involved in the same tRNA modification pathway. The preponderance of these two genes suggests that they are the predominant genetic targets for the increased frameshift (−GA) misreading in gene expression of E. coli. The impact of the observed ribosomal errors on cell growth rate is largely dependent on nutritional conditions and is different in both strains.
Misreading of a GA repeat in E. coli mostly occurs by translational frameshifting
The elucidation of the observed (−GA) misreading suggests that the strong increase in +2GA translational frameshifting results from the coincidence of three events (see Fig. 4). The first repeat-containing codon to be decoded by the ribosome is a GAG glutamate codon normally read by tRNA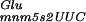 , which in mnmE and gidA mutants is probably present only in the tRNA
, which in mnmE and gidA mutants is probably present only in the tRNA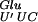 hypomodified form. This hypomodified form of the tRNA is less efficient for the decoding of the GAG codon. In addition, the GAG codon cannot be decoded by a near-cognate tRNA because only one species of tRNAGlu exists in E. coli (Komine et al. 1990). Therefore, when the tRNA
hypomodified form. This hypomodified form of the tRNA is less efficient for the decoding of the GAG codon. In addition, the GAG codon cannot be decoded by a near-cognate tRNA because only one species of tRNAGlu exists in E. coli (Komine et al. 1990). Therefore, when the tRNA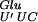 is in the P-site of the ribosome, there is a weaker base pairing between the tRNA and the mRNA. The second element affecting this +2 frameshift is the repetitive GAG-AGA mRNA sequence, which seems to predispose the translational complex to slippage. In addition, the next codon to be read at the frameshift site is an AGA arginine codon. This codon was shown previously to be decoded by a minor tRNA (Ikemura 1981; Dong et al. 1996) and to be one of the rarest codons in the E. coli genome (Hénaut and Danchin 1996; Nakamura et al. 2000). The presence of this codon in the A-site probably results in a pause during translation. Therefore, the conjunction of these three events — the presence of the hypomodified tRNA
is in the P-site of the ribosome, there is a weaker base pairing between the tRNA and the mRNA. The second element affecting this +2 frameshift is the repetitive GAG-AGA mRNA sequence, which seems to predispose the translational complex to slippage. In addition, the next codon to be read at the frameshift site is an AGA arginine codon. This codon was shown previously to be decoded by a minor tRNA (Ikemura 1981; Dong et al. 1996) and to be one of the rarest codons in the E. coli genome (Hénaut and Danchin 1996; Nakamura et al. 2000). The presence of this codon in the A-site probably results in a pause during translation. Therefore, the conjunction of these three events — the presence of the hypomodified tRNA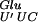 in the P-site, the repetitive sequence of the mRNA and the rare AGA codon in the A-site allows the ribosome to shift to another frame, as described in Figure 4. Therefore on the lacZ(+GA) mRNA, the ribosome slips over the 2 bases (GA) toward the 3′ end on the mRNA, producing a +2 frameshift that restores the normal reading frame for an active β-galactosidase.
in the P-site, the repetitive sequence of the mRNA and the rare AGA codon in the A-site allows the ribosome to shift to another frame, as described in Figure 4. Therefore on the lacZ(+GA) mRNA, the ribosome slips over the 2 bases (GA) toward the 3′ end on the mRNA, producing a +2 frameshift that restores the normal reading frame for an active β-galactosidase.
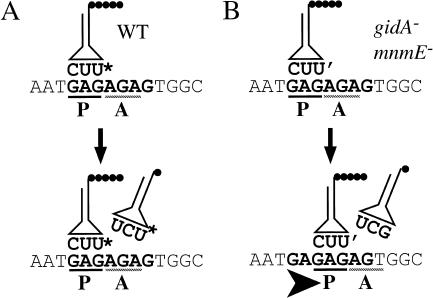
Figure 4.
Model for the observed +2 frameshift. (A) The normal process of translation in NECB1 lacZ(+GA) begins with the peptidyl-tRNA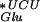 (U* is the modified uridine at the wobble position, which could be either mnm5-s2-U or mnm5-U) bound to the mRNA in the P-site of the ribosome. The empty A-site is normally filled with a tRNA
(U* is the modified uridine at the wobble position, which could be either mnm5-s2-U or mnm5-U) bound to the mRNA in the P-site of the ribosome. The empty A-site is normally filled with a tRNA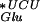 and the translational complex can proceed to the next step of elongation by transferring the peptide and translocating the accepted tRNA in the P-site. (B) In mnmE mutants, and probably in gidA mutants, the uridine at the wobble position of the tRNAGlu is hypomodified (U′: s2-U or some other hypomodified form of the mnm5-s2-U). This hypomodified nucleotide is less efficient at pairing with a guanine in the wobble position. The presence of a rare AGA codon at the A-site results in a pause during the translation process. Therefore, the conjunction of a less efficient pairing between the tRNA and the mRNA at the P-site and the presence of a rare codon at the A-site renders the ribosome prone to shifting. This translation complex moves to (or possibly scans for) the next GAG cognate codon of the tRNA in the P-site, which is easy on this repetitive sequence and then resumes translation, but in a different frame.
and the translational complex can proceed to the next step of elongation by transferring the peptide and translocating the accepted tRNA in the P-site. (B) In mnmE mutants, and probably in gidA mutants, the uridine at the wobble position of the tRNAGlu is hypomodified (U′: s2-U or some other hypomodified form of the mnm5-s2-U). This hypomodified nucleotide is less efficient at pairing with a guanine in the wobble position. The presence of a rare AGA codon at the A-site results in a pause during the translation process. Therefore, the conjunction of a less efficient pairing between the tRNA and the mRNA at the P-site and the presence of a rare codon at the A-site renders the ribosome prone to shifting. This translation complex moves to (or possibly scans for) the next GAG cognate codon of the tRNA in the P-site, which is easy on this repetitive sequence and then resumes translation, but in a different frame.
The +2(GA) translational frameshifting is counteracted by tRNA modification
The product of the mnmE gene encodes an evolutionarily conserved GTPase (Cabedo et al. 1999). It is involved in the modification of the uridine residue in the wobble position (nucleotide 34) of tRNA that reads split codon boxes (a codon box in which the first two nucleotides are the same, while the third nucleotide determines which of the two alternative amino acids are encoded). In those tRNAs, the modification is of the type xm5U (or a thiolated derivative xm5s2U) where x can be an amino (m), methylamino (mn), or carboxymethylamino (cmn) group. The first reaction in this process, catalyzed by MnmE, is the addition of the cmnm group in position 5 of the uridine. For the tRNAGlu of E. coli, which decodes GAA and GAG codons, it was shown that 5-methylaminomethyl group on the uracil facilitates its base-pairing with G while decreasing the base-pairing with A. This results in rates of translation of GAG and GAA that approach the average speed of the ribosome (Kruger et al. 1998). Therefore, in the case of lacZ(+GA) in mnmE::Tn10 strain, this tRNAGlu is less efficiently paired with the first codon of the GA repeat, which is likely to facilitate the observed ribosomal frameshifting (see above; Fig. 4).
GidA protein may be involved in the mnm5s2U34 modification pathway
There is no clearly identified function for the gidA gene, which has been described to have the phenotype of glucose inhibited division (von Meyenburg and Hansen 1980) and is located next to the chromosomal origin of replication, oriC, of E. coli. Results obtained in this study with the mnmE::kan gidA::Tn10 double mutant show that the products of both genes are likely to be involved in the same tRNA modification pathway and, furthermore, that the MnmE activity precedes that of the GidA. The synthesis of mnm5s2U could occur in the following steps:
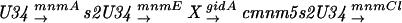 |
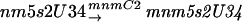 |
It was recently suggested that the genetic symbols for genes involved in this pathway should be revised and that trmU (or asuE) be replaced by mnmA, trmE (or thdF) by mnmE, and trmC by mnmC (Leung et al. 1998). The mnmA gene product is responsible for the thiolation of position 2 of the uridine (Sullivan et al. 1985). No mutant of this gene has been isolated with our screen, probably because the 2-thio group added by this enzyme is important for aminoacylation and does not have a significant role for the decoding of the GAG codon (Kruger et al. 1998). MnmC is composed of two activities (Hagervall et al. 1987), which transform the cmnm intermediate into the final mnm modification. The fact that we did not isolate mutants of this gene and the fact that a cmnm intermediate is used as the final form of one tRNALeu, suggest that this step may not change the decoding properties of the modified tRNA. The mnmE gene product was shown to be responsible for the first modification of the 5-position of the uracil, but it is unclear how many steps precede the formation of cmnm5 (Elseviers et al. 1984; Hagervall et al. 1987).
The role we propose for GidA in this modification pathway could explain the recent results of Nakayashiki and others. Thermosensitive lethal mutants of this gene have been isolated in a strain harboring the amber suppressor mutant gene tRNA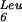 (supP) (Nakayashiki and Inokuchi 1998), which changes the anticodon of this dispensable tRNA from CAA to CUA. Normally, tRNA
(supP) (Nakayashiki and Inokuchi 1998), which changes the anticodon of this dispensable tRNA from CAA to CUA. Normally, tRNA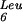 recognizes the UUG codon, whereas the mutated supP tRNA recognizes the UAG amber codon. The UUG codon is also decoded by tRNA
recognizes the UUG codon, whereas the mutated supP tRNA recognizes the UAG amber codon. The UUG codon is also decoded by tRNA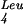 , which has a cmnm5m2UAA (m2: methyl group in position 2) in its anticodon such that it can recognize both UUG and UUA (Horie et al. 1999). By analogy with tRNAGlu, one could speculate that the absence of cmnm5 at nonpermissive temperature may decrease the base pairing of the tRNA
, which has a cmnm5m2UAA (m2: methyl group in position 2) in its anticodon such that it can recognize both UUG and UUA (Horie et al. 1999). By analogy with tRNAGlu, one could speculate that the absence of cmnm5 at nonpermissive temperature may decrease the base pairing of the tRNA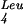 with the UUG codon, thereby requiring the tRNA
with the UUG codon, thereby requiring the tRNA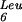 under these conditions for normal translation. Furthermore, the first attempt at isolating mutants affected in the biosynthesis of mnm5s2U allowed the characterization of two loci, named trmE and trmF. The phenotype of these two mutants was not clearly different but they were mapped to two different locations on the E. coli chromosome. The trmE gene was shown to be the same gene as mnmE (Cabedo et al. 1999) and trmF was mapped near 83 min on the E. coli map between uncA and bglR in the region of oriC (Elseviers et al. 1984). The map location of gidA is 84.5 min, close to oriC. We therefore propose that trmF is an allele of gidA and that this locus may be renamed mnmG as suggested previously (Leung et al. 1998).
under these conditions for normal translation. Furthermore, the first attempt at isolating mutants affected in the biosynthesis of mnm5s2U allowed the characterization of two loci, named trmE and trmF. The phenotype of these two mutants was not clearly different but they were mapped to two different locations on the E. coli chromosome. The trmE gene was shown to be the same gene as mnmE (Cabedo et al. 1999) and trmF was mapped near 83 min on the E. coli map between uncA and bglR in the region of oriC (Elseviers et al. 1984). The map location of gidA is 84.5 min, close to oriC. We therefore propose that trmF is an allele of gidA and that this locus may be renamed mnmG as suggested previously (Leung et al. 1998).
The growth rate of both gidA::Tn10 and mnmE::Tn10 strains is reduced in LB, but, in minimal medium, this decrease was only observed for the gidA::Tn10 strain. This difference of phenotype may suggest that the unknown product of MnmE (indicated by an X in the modification pathway presented above), which is likely to be accumulated in a gidA− strain, could be toxic for cells under this slow growth condition. At this stage, however, we can not exclude that GidA may be involved in other tRNA modification pathways or in DNA replication as suggested previously (Ogawa and Okazaki 1994).
Extension of the dual error model may explain tRNA hopping
Most of the described nonprogrammed ribosomal frameshifts are thought to slip in either +1 or −1 frame (Farabaugh and Björk 1999, and references therein). The +1/−1 dual error model proposed previously (Qian et al. 1998) suggests several explanations for nonprogrammed +1 and −1 frameshifts that could occur, especially at CCCN and GGGN sites (N, any nucleotide). This model assumes that slipping of the ribosome would happen at a precise stage after the three nucleotide translocation. At this step, the ribosome is prone to slip on the mRNA, in particular when a near-cognate tRNA or a hypomodified tRNA is present in the P-site. In this case, the shift is attributable to a weaker codon–anticodon pairing at the wobble position. Even if a normal cognate tRNA is in the P-site, the ribosome could slip when a translational pause is induced because of the presence of a rare codon in the A-site.
An extended version of the +1/−1 dual error model could also explain the tRNA hopping described by Weiss et al. (1987). The mutation they studied is a leaky −1 frameshift allele trpE91 in Salmonella (Riyasaty and Atkins 1968). The low-level translational frameshifting that occurs at this site was studied by transferring the region surrounding the mutation into a lacZ expression vector (Weiss et al. 1987). The sequence of the expressed peptide revealed that the frameshift occurred by a translational hopping, creating a +2 rather than a −1 frameshift, via a tRNAVal hopping by +2. Two other sequences were identified on which the ribosome is prone to slip (or hop) over 5 or 6 nucleotides. Protein sequencing confirmed that tRNAAsn hops 5 nucleotides between cognate codons and that tRNALeu hops over an in-frame stop codon between near-cognate codons. The presence of an in-frame stop codon for both +2 and +6 hopping apparently creates a pausing site during translation, a favorable condition for the frameshifting (Farabaugh 1996). The +5 event, however, occurred at an in-frame UCA codon that is not predicted to cause a translational pause. The strain used by Weiss and colleagues was CSH26 with a deletion of the pro–lac region, where the strC and trmB genes have been mapped, respectively, by P1 co-transduction (Roberts and Reeve 1970) and conjugation (Marinus et al. 1975), which may have an influence on this +5 frameshift. The strC gene is known to be involved in streptomycin resistance and to modify the ribosome structure (Roberts and Reeve 1970). The TrmB protein is a tRNA methyltransferase, which catalyzes the formation of methyl-7-guanosine (m7G) in some tRNAs, including the tRNAAsn hopping over 5 nucleotides. Whether this modification is important to stabilize the translation complex remains to be tested. All observations of ribosome frameshifting with hypomodified tRNA or in presence of a pausing site during translation are therefore in favor of the extended version of the dual error model described above.
An evolutionarily conserved modification pathway
The mnmE and gidA gene products display significant similarities with proteins of Saccharomyces cerevisiae and Caenorhabditis elegans as well as with several expressed sequence tags from Arabidopsis thaliana, mouse and human. In S. cerevisiae, homologs of MnmE and GidA are respectively Mss1p and Mto1p and have been shown to be nuclear-encoded mitochondrial genes (Decoster et al. 1993; Colby et al. 1998). Therefore, MSS1 and MTO1 genes are likely to have evolved from mitochondrial genes translocated into the nucleus.
Mutants of Mto1p and Mss1p were reported to be implicated in a respiratory deficiency phenotype only when the 15S rRNA gene carries a mutation that confers resistance to paromomycin (Decoster et al. 1993; Colby et al. 1998). This mutation is localized to helix 47, which is part of the A-site of the ribosome (Li et al. 1982; De Stasio and Dahlberg 1990). Those mutant ribosomes were also shown to have a decreased level of natural frameshifting, suggestive of a more stringent proofreading (Weiss-Brummer and Huttenhofer 1989). It is significant that mitochondrial tRNALeu and tRNATrp contain a modified U at the wobble position (Björk 1998). This modification is of the type cmnm5Um and allows these tRNA to translate codons terminating in either A or G. We propose that Mss1p and Mto1p are responsible for this modification and that hypomodified tRNAs are less efficient for the decoding of the codon ending in G. This deficiency could be stronger with the more stringent paromomycin-resistant ribosome, thereby causing the respiratory defect observed in mss1 and mto1 mutant cells. Furthermore, the products of these two genes in S. cerevisiae were shown to be in the same complex, and are therefore likely to act in the same pathway (Colby et al. 1998). Whether GidA and MnmE are also in the same protein complex, which may counteract the previously proposed toxicity of the unknown MnmE product (X), remains to be determined in E. coli.
The alteration of growth rate in both gidA::Tn10 and mnmE::Tn10 strains and the respiratory deficient phenotype of yeast mutants could be linked to errors made during translation, which are likely to be processivity errors. As this pathway seems to be conserved in many organisms, mutation of these genes may be involved in genetic disorders such as mitochondrial diseases.
Materials and methods
Growth conditions
The cell growth media are liquid LB supplemented with the following antibiotics, when appropriate, at the following concentrations: tetracycline 12.5 μg/mL, chloramphenicol 30 μg/mL, rifampicin 100 μg/mL, kanamycin 100 μg/mL, and ampicillin 100 μg/mL. For strain constructions, clones were selected on LB plates supplemented with the appropriate antibiotic and/or X-Gal (200 μg/mL) and 20 mM sodium citrate. Selections for auxotrophy, or carbon source, were made on minimal M9 plates (1.5% agar) supplemented with 0.4% of the relevant carbon source and 0.1% of the suitable amino acids. Growth rates were measured by optical density at 530 nm in 96-well plates.
Strain constructions
The genotypes and sources of the strains of E. coli used in this study are listed in Table 3. The NECB1 was constructed as described in Cupples and Miller (1988) using LacZGAGA as mutagenic primer. The introduced mutation was verified by sequencing.
Derivative strains were constructed by P1-mediated transduction as described by Miller (1972). Conjugations were made by mixing 500 μL of overnight cultures of donor and recipient cells in 5 mL of LB and placed at 37°C without shaking for 2 h. Transconjugants were then selected by plating on the appropriate media.
Mutagenic treatments and identification of transposon insertion site
The random insertion mutants were created by transposition of mini-Tn10 from λNK1323 (Kleckner et al. 1991). Phage stocks and infections were made as described in (Miller 1992). Transfected cells were plated on LB plates containing tetracycline and X-Gal to screen for light blue colonies. The Tn10 insertion sites were identified by inverse PCR in bacterial clones showing intermediate levels of β-galactosidase activity. Genomic DNA (500 ng) from such clones was digested with PstI and purified with a QIAquick PCR purification kit (QIAGEN). Fragments were then incubated overnight at 12°C with 40,000 units of T4 DNA ligase in a final volume of 1 mL. DNA was precipitated with ethanol and resuspended in 20 μL of water. The DNA fragments were then amplified by PCR with VentR(exo−) DNA polymerase (New England Biolabs) using Tn10U and Tn10L primers (Genset). Comparison of the sequence of the PCR product with the sequence of the E. coli genome allowed us to determine the insertion site for each clone.
The point mutations were made by growing NECB1 cells overnight in LB containing 700 μg/mL of 2-AP as described by Miller (1992). Mutagenesis was monitored by determining the frequency of rifampicin resistant mutants as compared with nontreated culture. After the treatment, cells were plated on LB plates containing X-Gal, and mutants having a pale blue coloration were re-streaked on the same type of medium.
Plasmid construction
All plasmids and their constructs were transformed into the appropriate E. coli cells by electrotransformation. Ligations were carried out by T4 DNA ligase (New England Biolabs) according to the manufacturer’s instructions and the Electromax DH10B cells (Life Technologies) were used for primary transformations. For gene amplifications, a single colony of NECB1 was resuspended in 50 μL of water, and 1 μL of the suspension was added to the PCR mix. Cells were lysed during the first cycle of PCR by 5-min incubation at 95°C.
To construct the pNEC1 plasmid, the region of argU was amplified under standard conditions with the Pwo polymerase, using ArgUB1U and ArgUB1L as primers. The amplified fragment was digested with BamHI and inserted into pACYC184 at the BamHI site. Positive clones were checked to be chloramphenicol-resistant and tetracycline-sensitive and the presence of the insert was monitored by digestion with BamHI.
The gltT gene was amplified in the same manner as argU by using GltTK1U and GltTP1L as primers for the construction pNEC2-WT. The linear fragment was digested with KpnI and PstI and inserted into pBAD33 (Guzman et al. 1995) between KpnI and PstI sites. Plasmids from transformed clones were extracted using a QIAquick Miniprep kit (QIAGEN) and digested with KpnI and PstI to monitor the presence of the inserted fragment. Linear fragments for the construction of pNEC2-U34C were generated by sequential PCR steps using GltTK1U, GltTDir, GltTP1L, and GltTRev (Table 3).
Linear fragments for the cloning of gidA (pNEC3) or mnmE (pNEC4) were obtained by amplification with Pwo DNA polymerase (Roche) using GidAE1U and GidAS1D, respectively, or MnmEE1U and MnmEP1D as primers. Fragments were then digested with the appropriate enzyme (Table 3) and inserted into pUC18. Positive clones were checked for the lack of α-complementation (Yanisch-Perron et al. 1985) and were verified by restriction.
For the genomic library, 2 μg of genomic DNA from NECB1 prepared with a Wizard Genomic DNA Preparation kit (Promega) was partially digested with Sau3AI. Fragments ranging from 3–4 kb were eluted from agarose with a QIAquick gel extraction kit (QIAGEN). Fragments were then cloned into the BamHI site of pACYC184 using a fragment:plasmid ratio of 10:1 for the ligation and the transformed cells were selected on LB plates containing chloramphenicol. Plasmid DNA of 30 single colonies was isolated and digested with EcoRV and SalI to estimate the efficiency of ligation, which was ∼90%. Plasmids were then extracted from 20,000 colonies resuspended in MgSO4 10 mM with a QIAfilter Maxiprep kit (QIAGEN).
β-Galactosidase activity
For all β-galactosidase assays, 200 μL of an overnight culture was used. Activities were measured and were expressed as described by Miller (1972). The OD530 was not measured as all samples were centrifuged before the reading of the OD420. To measure low levels of β-galactosidase activities, longer incubation times were necessary (up to 1 d, always controlling for spontaneous hydrolysis of O-nitrophenyl-β-D-galactoside). All β-galactosidase activities are given in Miller units ± standard error.
Acknowledgments
We thank P. Leroy, M.F. Bredeche, and M. Kobayashi for technical assistance and R. D'Ari and M.E. Armengod for providing strains. We thank I. Matic and O. Tenaillon for constant encouragement. We thank E. Stewart, F.W. Stahl, M.S. Fox, C.G. Kurland, G.R. Björk, T.G. Hagervall, and J.A. Gallant for critical reading of the manuscript and F. VanLeeuwen, H. Grosjean, and P. Farabaugh for their interest in this work. This work was supported by grants from ACI blanche, MENRT, INSERM and La Ligue contre le cancer.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bregeon@necker.fr; FAX 33-1-40-61-53-22.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.207701.
References
- Arraj JA, Campbell JH. Isolation and characterization of the newly evolved ebg β- galactosidase of Escherichia coli K-12. J Bacteriol. 1975;124:849–856. doi: 10.1128/jb.124.2.849-856.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Z, Gallant J, Lindsley D, Kwieciszewki B, Heidel D. Enhanced ribosome frameshifting in stationary phase cells. J Mol Biol. 1996;263:140–148. doi: 10.1006/jmbi.1996.0565. [DOI] [PubMed] [Google Scholar]
- Bergmann ED, Sicher S. Mode of action of chloramphenicol. Nature. 1952;170:931–932. doi: 10.1038/170931a0. [DOI] [PubMed] [Google Scholar]
- Björk GR. Stable RNA modification. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low BK, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, DC.: American Society for Microbiology; 1996. pp. 861–886. [Google Scholar]
- ————— . Modified nucleosides at position 34 and 37 of tRNAs and their predicted coding capacities. In: Grosjean H, Benne R, editors. Modifiation and editing of RNA. Washington, DC.: American Society for Microbiology; 1998. pp. 577–581. [Google Scholar]
- Björk GR, Durand JM, Hagervall TG, Leipuviene R, Lundgren HK, Nilsson K, Chen P, Qian Q, Urbonavicius J. Transfer RNA modification: Influence on translational frameshifting and metabolism. FEBS Lett. 1999;452:47–51. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- Cabedo H, Macian F, Villarroya M, Escudero JC, Martinez-Vicente M, Knecht E, Armengod ME. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Lengyel JA, Langridge J. Evolution of a second gene for β-galactosidase in Escherichia coli. Proc Natl Acad Sci. 1973;70:1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby G, Wu M, Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- Cupples CG, Miller JH. Effects of amino acid substitutions at the active site in Escherichia coli β-galactosidase. Genetics. 1988;120:637–644. doi: 10.1093/genetics/120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J Mol Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Moore PB. The elongation phase of protein synthesis. Prog Nucleic Acid Res Mol Biol. 1996;54:293–332. doi: 10.1016/s0079-6603(08)60366-9. [DOI] [PubMed] [Google Scholar]
- De Stasio EA, Dahlberg AE. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J Mol Biol. 1990;212:127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- Decoster E, Vassal A, Faye G. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J Mol Biol. 1993;232:79–88. doi: 10.1006/jmbi.1993.1371. [DOI] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nystrom T. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci. 2000;97:5746–5749. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann P, Gallant J. Mistranslation in E. coli. Cell. 1977;10:131–137. doi: 10.1016/0092-8674(77)90147-7. [DOI] [PubMed] [Google Scholar]
- Ellis N, Gallant J. An estimate of the global error frequency in translation. Mol Gen Genet. 1982;188:169–172. doi: 10.1007/BF00332670. [DOI] [PubMed] [Google Scholar]
- Elseviers D, Petrullo LA, Gallagher PJ. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl- 2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ, Björk GR. How translational accuracy influences reading frame maintenance. EMBO J. 1999;18:1427–1434. doi: 10.1093/emboj/18.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: American Society for Microbiology; 1995. [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagervall TG, Edmonds CG, McCloskey JA, Björk GR. Transfer RNA(5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J Biol Chem. 1987;262:8488–8495. [PubMed] [Google Scholar]
- Hénaut A, Danchin A. Analysis and prediction from Escherichia coli sequences, or E. coli. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low BK, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Silico. In Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC.: American Society for Microbiology; 1996. pp. 2047–2066. [Google Scholar]
- Hol EM, Neubauer A, de Kleijn DP, Sluijs JA, Ramdjielal RD, Sonnemans MA, van Leeuwen FW. Dinucleotide deletions in neuronal transcripts: A novel type of mutation in non-familial Alzheimer's disease and Down syndrome patients. Prog Brain Res. 1998;117:379–395. doi: 10.1016/s0079-6123(08)64028-5. [DOI] [PubMed] [Google Scholar]
- Horie N, Yamaizumi Z, Kuchino Y, Takai K, Goldman E, Miyazawa T, Nishimura S, Yokoyama S. Modified nucleosides in the first positions of the anticodons of tRNA(Leu)4 and tRNA(Leu)5 from Escherichia coli. Biochemistry. 1999;38:207–217. doi: 10.1021/bi981865g. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Komine Y, Adachi T, Inokuchi H, Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990;212:579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kruger MK, Pedersen S, Hagervall TG, Sorensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J Mol Biol. 1998;284:621–631. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Kurland C, Hughes D, Ehrenberg M. Limitations of translational accuracy. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low BK, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, DC.: American Society for Microbiology; 1996. pp. 979–1004. [Google Scholar]
- Leung HCE, Hagervall TG, Björk GR, Winkler ME. Genetic locations and database accession numbers of RNA-modifying and -editing enzymes. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, DC.: American Society for Microbiology; 1998. pp. 561–568. [Google Scholar]
- Lewin B. Genes V. New York, NY: Oxford University Press; 1994. [Google Scholar]
- Li M, Tzagoloff A, Underbrink-Lyon K, Martin NC. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982;257:5921–5928. [PubMed] [Google Scholar]
- Libby RT, Gallant JA. The role of RNA polymerase in transcriptional fidelity. Mol Microbiol. 1991;5:999–1004. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Morris NR, Soll D, Kwong TC. Isolation and partial characterization of three Escherichia coli mutants with altered transfer ribonucleic acid methylases. J Bacteriol. 1975;122:257–265. doi: 10.1128/jb.122.1.257-265.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menninger JR. Ribosome editing and the error catastrophe hypothesis of cellular aging. Mech Ageing Dev. 1977;6:131–142. doi: 10.1016/0047-6374(77)90014-8. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- ————— . A short course in bacterial genetics. A laboratory manual for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki T, Inokuchi H. Novel temperature-sensitive mutants of Escherichia coli that are unable to grow in the absence of wild-type tRNA6Leu. J Bacteriol. 1998;180:2931–2935. doi: 10.1128/jb.180.11.2931-2935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH. The allosteric three-site model for the ribosomal elongation cycle: Features and future. Biochemistry. 1990;29:4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Li JN, Zhao H, Hagervall TG, Farabaugh PJ, Björk GR. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell. 1998;1:471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- Riyasaty S, Atkins JF. External suppression of a frameshift mutant in Salmonella. J Mol Biol. 1968;34:541–557. doi: 10.1016/0022-2836(68)90179-4. [DOI] [PubMed] [Google Scholar]
- Roberts LM, Reeve EC. Two mutations giving low-level streptomycin resistance in Escherichia coli K 12. Genet Res. 1970;16:359–365. doi: 10.1017/s0016672300002640. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Dunn RL. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: The nature of in vivo DNA replication errors. Proc Natl Acad Sci. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipley J, Goldman E. Increased ribosomal accuracy increases a programmed translational frameshift in Escherichia coli. Proc Natl Acad Sci. 1993;90:2315–2319. doi: 10.1073/pnas.90.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Sullivan MA, Cannon JF, Webb FH, Bock RM. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J Bacteriol. 1985;161:368–376. doi: 10.1128/jb.161.1.368-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Hayakawa H, Bouton MF, Cirinesi AM, Matic I, Sekiguchi M, Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- Thompson RC. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci. 1988;13:91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16:81–86. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen FW, Burbach JP, Hol EM. Mutations in RNA: A first example of molecular misreading in Alzheimer's disease. Trends Neurosci. 1998a;21:331–335. doi: 10.1016/s0166-2236(98)01280-6. [DOI] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RDJ, Salehi A, Martens GJM, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998b;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- von Meyenburg K, Hansen FG. Mechanistic studies of DNA replication and genetic recombination. New York, NY: Academic Press; 1980. The origin of replication, oriC, of the Escherichia coli chromosome: Genes near to oriC and construction of oriC deletion mutations; pp. 137–159. [Google Scholar]
- Vulic M, Lenski RE, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc Natl Acad Sci. 1999;96:7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss-Brummer B, Huttenhofer A. The paromomycin resistance mutation (parR-454) in the 15 S rRNA gene of the yeast Saccharomyces cerevisiae is involved in ribosomal frameshifting. Mol Gen Genet. 1989;217:362–369. doi: 10.1007/BF02464905. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson's disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–1429. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]