Abstract
Dermatophytes cause the majority of superficial mycoses in humans and animals. However, little is known about the pathogenicity of this specialized group of filamentous fungi, for which molecular research has been limited thus far. During experimental infection of guinea pigs by the human pathogenic dermatophyte Arthroderma benhamiae, we recently detected the activation of the fungal gene encoding malate synthase AcuE, a key enzyme of the glyoxylate cycle. By the establishment of the first genetic system for A. benhamiae, specific ΔacuE mutants were constructed in a wild-type strain and, in addition, in a derivative in which we inactivated the nonhomologous end-joining pathway by deletion of the A. benhamiae KU70 gene. The absence of AbenKU70 resulted in an increased frequency of the targeted insertion of linear DNA by homologous recombination, without notably altering the monitored in vitro growth abilities of the fungus or its virulence in a guinea pig infection model. Phenotypic analyses of ΔacuE mutants and complemented strains depicted that malate synthase is required for the growth of A. benhamiae on lipids, major constituents of the skin. However, mutant analysis did not reveal a pathogenic role of the A. benhamiae enzyme in guinea pig dermatophytosis or during epidermal invasion of the fungus in an in vitro model of reconstituted human epidermis. The presented efficient system for targeted genetic manipulation in A. benhamiae, paired with the analyzed infection models, will advance the functional characterization of putative virulence determinants in medically important dermatophytes.
INTRODUCTION
Dermatophytes represent a group of specialized filamentous fungi which account for the majority of superficial fungal infections. Millions of so-called dermatophytoses, which in many cases are long lasting and difficult to eradicate, are recorded for humans and animals every year (29). As a peculiarity, dermatophytes specifically infect keratinized host structures, such as stratum corneum, hair, and nails. At the molecular level, however, little is known of the nature of the pathogenicity mechanisms in dermatophytes (17, 30). This drawback might be related to the fact that these fungi, which grow comparatively slowly under laboratory conditions, have so far not intensively been studied genetically, in contrast to other medically important fungi, such as Candida albicans, Aspergillus fumigatus, and Cryptococcus neoformans. Full genome sequence information for dermatophytes have been available since only very recently (http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative/MultiHome.html), and genetic tools have hardly been established. As a consequence, in dermatophyte species only a few genes have to date been analyzed by targeted inactivation, i.e., pacC and MDR2 in Trichophyton rubrum (8, 9), Ku80, areA, and Trim4 in Trichophyton mentagrophytes (Arthroderma vanbreuseghemii) (32), and areA in Microsporum canis (31). Specifically constructed dermatophyte mutants have, to our knowledge, not yet been tested in animal models.
In a recent study, we monitored a broad-scale gene expression profile in the human pathogenic dermatophyte Arthroderma benhamiae during keratin degradation and also during cutaneous infection of guinea pigs (25). By this approach, the in vivo activation of genes encoding key enzymes of the glyoxylate cycle, i.e., malate synthase and isocitrate lyase, was detected. The glyoxylate cycle, which is absent in mammals, is known to permit the synthesis of glucose from lipids and other alternative carbon sources in microorganisms. A role of this metabolic pathway for microbial pathogenicity has previously been unraveled for the bacterial pathogen Mycobacterium tuberculosis (16), the opportunistic yeast C. albicans (15), and the plant pathogenic fungus Leptosphaeria maculans (11). In contrast, a pathogenic function of the glyoxylate cycle could not be assigned for C. neoformans-induced cryptococcosis (21) or invasive aspergillosis by the mold A. fumigatus (23). Since dermatophytes infect specifically host epidermal structures rich in keratin and lipids, the functional characterization of putative glyoxylate cycle genes in these fungi as possible contributors to virulence is of interest.
A. benhamiae serves as an appropriate model organism for the molecular analysis of the biology and pathogenicity of dermatophytes. The species is zoophilic, and inflammatory cutaneous infections are produced in both humans and guinea pigs, which allowed the recent establishment of an animal model (25). The comparatively fast in vitro growth of A. benhamiae, paired with the production of abundant microconidia and the ability to undergo sexual development, further facilitates the establishment of genetic methodologies. As a major additional advantage, the genome of our A. benhamiae strain, a clinical isolate from a patient suffering from highly inflammatory tinea faciei (10), has recently been sequenced (4) (http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative/MultiHome.html). In light of these benefits, it appeared timely to establish a genetic system for A. benhamiae, which allows isogenic strain construction and gene targeting, fundamental prerequisites for functional gene analysis. Basic improvements in dermatophyte genetics have recently been implemented by Yamada et al., who demonstrated the reproducible transformation of T. mentagrophytes and M. canis (32–34). Most filamentous fungi are known to only poorly support site-specific integration of linear DNA constructs in the genome via homologous recombination. The increased frequency of targeted insertion, however, has been observed with mutants of different filamentous fungi which lack enzymes of the Ku recombinase complex, e.g., in Neurospora crassa (18), Aspergillus spp. (6, 12), and recently also T. mentagrophytes (32). Consequently, such derivatives are commonly used for genetic manipulations in these microorganisms, thereby allowing major molecular insights into biological and/or virulence-associated traits. Nevertheless, by using parental strains lacking the nonhomologous end-joining (NHEJ) recombination pathway, an altered risk for secondary, unforeseen genetic variations and/or even an altered virulence in infection models cannot be excluded (14).
In the present study we set out to establish for the first time a genetic manipulation system for A. benhamiae. Efficient targeted gene deletion is achieved in both the wild type and the Δku70 mutant background, thereby demonstrating the applicability of two dominant selection markers in this species. A comparison of A. benhamiae wild-type and Δku70 mutant cells during infection revealed that deletion of AbenKU70 had no detectable effect on the virulence of the fungus, a benefit for future genetic studies. Addressing the pathogenicity of A. benhamiae, the gene encoding malate synthase AbenAcuE was deleted. Mutants of this enzyme showed a particular growth defect on lipids and other selected carbon sources in vitro, yet inflammatory infection of guinea pigs was not impaired by the lack of AbenACUE. To further study the process of epidermal invasion by dermatophytes, an in vitro model of reconstituted human epidermis (RHE) was applied for A. benhamiae.
MATERIALS AND METHODS
Strains and growth conditions.
A. benhamiae strains used in this study are listed in Table 1. A. benhamiae Lau2354-2 was used as a wild-type strain and is deposited in the Belgian Coordinated Collections of Microorganisms (BCCM/IHEM) under IHEM20163 (Scientific Institute of Public Health, Brussels, Belgium) and under CBS 112371 in the Netherlands culture collection Centraalbureau voor Schimmelcultures (CBS). The strain was isolated as the causative agent of highly inflammatory tinea faciei in a patient at the University hospital of Lausanne, Switzerland (10). Lau2354-2 and transformant derivatives were stored as frozen stocks with 15% glycerol at −80°C. For routine growth, the strains were cultivated on Sabouraud glucose agar (1% peptone, 2% glucose, 1.5% agar; here referred to as Sabouraud) or potato dextrose agar (PDA) (0.4% potato extract, 2% glucose, 1.5% agar [Carl Roth]) at 30°C. Liquid Sabouraud cultures were inoculated with a plug of fresh mycelium in 100 ml medium and incubated for 5 days at 30°C without shaking.
Table 1.
A. benhamiae strains used in this study
| Strain(s) | Parent | Description or genotype | Reference or source |
|---|---|---|---|
| Lau2354-2 | Wild-type strain | 10 | |
| AbenKU70M1A and AbenKU70M1B | Lau2354-2 | Δku70::PACT1-neo | This study |
| AbenACUEM1A and AbenACUEM1B | Lau2354-2 | ΔacuE::Pgpd-hph-TtrpC | This study |
| AbenACUEM2A and AbenACUEM2B | AbenKU70M1A | Δku70::PACT1-neo ΔacuE::Pgpd-hph-TtrpC | This study |
| AbenACUEK1A | AbenACUEM1A | ΔacuE::ACUE-TACUE-PACT1-neo | This study |
| AbenACUEK1B | AbenACUEM1B | ΔacuE::ACUE-TACUE-PACT1-neo | This study |
For the analysis of hair perforation and growth on hair, blond human hair was autoclaved and inoculated with a plug of fresh mycelium from Sabouraud plate cultures. The cultures were incubated at 25°C for 50 days, before growth was evaluated macroscopically and hair perforation of single hair inspected by light microscopy. To test for the growth of A. benhamiae on various carbon sources, Aspergillus minimal medium (http://www.fgsc.net/methods/anidmed.html) was used and supplemented with 2% glucose, 1% peptone, 0.5% olive oil, 2% ethanol, 100 mM sodium acetate, 0.5% myristic acid, or 0.5% palmitic acid. For the production of A. benhamiae microconidia, the fungal cells were grown for 5 days on MAT agar (0.1% peptone, 0.2% glucose, 0.1% MgSO4, 0.1% KH2PO4) at 30°C (27).
Plasmid constructions.
A hygromycin resistance cassette containing the Escherichia coli hygromycin B phosphotransferase (hph) gene under the control of the Aspergillus nidulans glyceraldehyde-3-phosphate dehydrogenase (gpd) promoter (Pgpd), and followed by the A. nidulans trpC terminator sequences (TtrpC), was amplified by PCR with primers HYG-1 and HYG-2 (all primers are listed in Table 2). As a template, plasmid pSKHPH1T23 was used. Plasmid pSKHPH1T23 carries the NotI fragment of p123 (28) consisting of the hygromycin resistance cassette inserted into the NotI site of pBluescript SK+ (Stratagene). The PCR product was digested at the introduced BamHI and HindIII sites and cloned in the BamHI/HindIII-digested vector pBluescript II KS (Stratagene) to result in plasmid pHPH1. Sequence information for genes AbenACUE (locus ARB_07638) and AbenKU70 (locus ARB_06096) were obtained from the annotated A. benhamiae genome (http://www.broadinstitute.org/annotation/genome/dermatophyte_comparative/MultiHome.html). An ApaI-HindIII fragment containing AbenACUE upstream sequences from positions −490 to −3, with respect to the start codon, was obtained by PCR with the primers AbenACUE-1 and AbenACUE-2. A BamHI-XbaI fragment with AbenACUE downstream sequences from positions + 1966 to +2890 was amplified by PCR with the primers AbenACUE-3 and AbenACUE-4. Genomic DNA from the wild-type A. benhamiae strain Lau2354-2 was used as a template. The AbenACUE upstream and downstream sequences were successively cloned via the introduced restriction sites in plasmid pHPH1 to result in pAbenACUEM1 and pAbenACUEM2, respectively. In plasmid pAbenACUEM2, the hygromycin resistance marker is flanked by AbenACUE sequences for deletion of the entire coding region of the AbenACUE gene (Fig. 1A). A DNA cassette which contains the neomycin resistance gene (neo) under the control of the A. benhamiae actin promoter (PACT1) was constructed as follows. An A. benhamiae PACT1 fragment was amplified by PCR with the primers AbenACT-1 and AbenACT-2, using genomic DNA from strain Lau2354-2 as a template. The PCR product was cloned via the introduced BamHI and NcoI restriction sites in pEGFP (Clontech). The resulting pactinEGFP1 was digested at NcoI/NotI to replace the GFP gene by a BspHI-NotI fragment of pEBFP (Clontech) carrying the neo gene, generating pacG418RY58. The PACT1-neo fragment was amplified by PCR with the primers NEO-1 and NEO-2, using plasmid pacG418RY58 as a template. The PCR product was digested at the introduced BamHI and HindIII sites and cloned in the BamHI/HindIII-digested vector pBluescript II KS to result in plasmid pNEO1. For reinsertion of the ACUE gene at its original locus in the two ΔacuE mutants, AbenACUEM1A and AbenACUEM1B, the AbenACUE gene, including its upstream and downstream sequences (positions −493 to +2890) was obtained by PCR using primers AbenACUE-9 and AbenACUE-4, and genomic DNA of strain Lau2354-2 as a template. The resulting PCR product carries a ClaI site at position +2840. The SalI-ClaI fragment was cloned in the SalI-ClaI-digested plasmid pNEO1 to result in pAbenACUEK1. The BamHI-XbaI downstream ACUE fragment from pAbenACUEM2 was cloned in pAbenACUEK1, resulting in plasmid pAbenACUEK2 (Fig. 1B). An ApaI-HindIII fragment with AbenKU70 upstream sequences from positions −1454 to +2 was amplified by PCR with the primers AbenKU70-1 and AbenKU70-2. A BamHI-SacII fragment containing AbenKU70 downstream sequences from positions +2123 to +3646 was obtained by PCR with the primers AbenKU70-3 and AbenKU70-4. Genomic DNA from A. benhamiae strain Lau2354-2 was used as a template. The AbenKU70 upstream and downstream sequences were successively cloned via the introduced restriction sites in plasmid pNEO1 to result in pAbenKU70M1 and pAbenKU70M2, respectively. In plasmid pAbenKU70M2, the neomycin resistance cassette is flanked by AbenKU70 sequences for deletion of the entire coding region of the AbenKU70 gene (Fig. 2).
Table 2.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| HYG-1 | ATATAGGATCCTTGGTTGAATTTAGAACGTGGCACTATTGATC |
| HYG-2 | ATATAAAGCTTTGAAGGCGTTACTAGGTTGCAGTCAATGC |
| AbenACT-1 | CGggATccCATCAGCGGCCGGAGTG |
| AbenACT-2 | CCTCCATGGTGGCTGATAGTAC |
| AbenACUE-1 | CGGTGGCTTCGATGGgcccGATGGAATGTGTAC |
| AbenACUE-2 | CTTCCACCGAGGTaagCTTGCGTTGGTGCTAG |
| AbenACUE-3 | CCATAATGTAAATggatCCTAACATTTTGAACGCGC |
| AbenACUE-4 | CGTGGCTCTAGATAAGCTGAC |
| AbenACUE-9 | CGGTGGCTTCGATGGTCgAcATGGAATGTGTAC |
| NEO-1 | CTCTAGAGGATCCCATCAGC |
| NEO-2 | TATAAAGCTTTTTCAGAAGAACTCGTCAAGAAGGCG |
| AbenKU70-1 | CCGTATCACTGGTCTGGGCCCCGAGCCAGGC |
| AbenKU70-2 | CGCGCACCCAGTTAGAAGCttTGACAGAAGC |
| AbenKU70-3 | CGAATTATCAAGCCTCTAACTCGGatccTAATATAGCTC |
| AbenKU70-4 | GGACAACGCGGGCcGCGGCGGGCAGCGTTACCC |
The lowercase letters represent nucleotide exchanges introduced to create the underlined restriction sites.
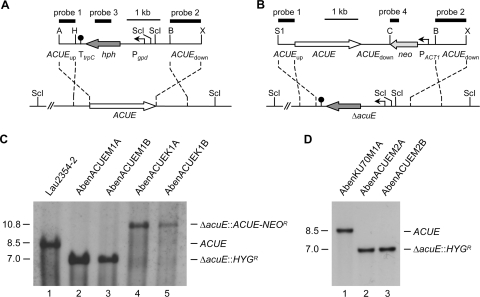
Fig. 1.
Construction of A. benhamiae ΔacuE mutants and complemented strains. (A) Structure of the AbenACUE deletion cassette from plasmid pAbenACUEM2 (top), containing the hygromycin resistance marker (HYGR), and genomic structure of the AbenACUE locus in the wild-type strain A. benhamiae Lau2354-2 (bottom). The ACUE coding region is represented by the white arrow, and the upstream and downstream regions are represented by the solid lines. The hygromycin resistance gene (hph) is shown as a dark gray arrow, the gpd promoter (Pgpd) as a bent arrow, and the transcription termination sequence TtrpC as a filled circle. (B) Structure of the DNA fragment from plasmid pAbenACUEK2 (top), containing the neomycin resistance marker (NEOR), which was used for reinsertion of the ACUE gene into its original locus in the ΔacuE mutants (bottom). The neomycin resistance gene (neo) is shown as a light gray arrow, and the A. benhamiae ACT1 promoter (PACT1) as a bent arrow. The probes which were used for Southern analysis of the transformants are indicated by the black bars. Only the following relevant restriction sites are given in panels A and B: A, ApaI; B, BamHI; C, ClaI; H, HindIII; ScI, SacI; S1, SalI; X, XbaI. (C) Southern hybridization of SacI-digested genomic DNA of the wild-type strain A. benhamiae Lau2354-2 (lane 1), ΔacuE mutants in wild-type AbenACUEM1A (lane 2) and AbenACUEM1B (lane 3), and complemented strains AbenACUEK1A (lane 4) and AbenACUEK1B (lane 5) with ACUE-specific probe 1. (D) Southern hybridization of SacI-digested genomic DNA of the Δku70 mutant AbenKU70M1A (lane 1) and ΔacuE mutants generated in AbenKU70M1A, i.e., AbenACUEM2A (lane 2) and AbenACUEM2B (lane 3), with ACUE-specific probe 1. The sizes of the hybridizing fragments (in kilobases) are given on the left, and their identities on the right.
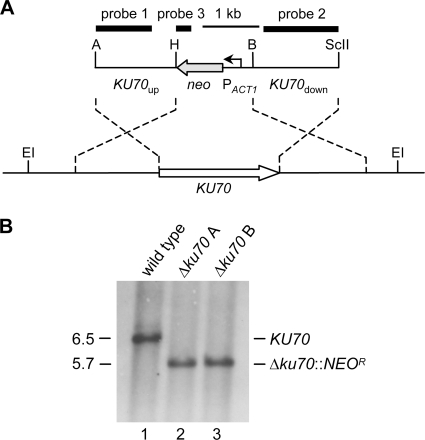
Fig. 2.
Construction of A. benhamiae Δku70 mutants. (A) Structure of the deletion cassette from plasmid pAbenKU70M2 (top), containing the neomycin resistance marker (NEOR), and genomic structure of the KU70 locus in wild-type A. benhamiae Lau2354-2 (bottom). The KU70 coding region is represented by the white arrow, and the upstream and downstream regions are represented by the solid lines. The neomycin resistance gene (neo) is shown as a gray arrow, and the A. benhamiae ACT1 promoter (PACT1) as a bent arrow. The probes which were used for Southern analysis of the transformants are indicated by the black bars. Only the following relevant restriction sites are given: A, ApaI; B, BamHI; EI, EcoRI; H, HindIII; ScII, SacII. (B) Southern hybridization of EcoRI-digested genomic DNA of the wild-type strain A. benhamiae Lau2354-2 (lane 1) and Δku70 mutants AbenKU70M1A (lane 2) and AbenKU70M1B (lane 3) with KU70-specific probe 1. The sizes of the hybridizing fragments (in kilobases) are given on the left, and their identities on the right.
A. benhamiae transformation.
A. benhamiae was transformed following a modified polyethylene glycol-mediated protoplast transformation protocol (34). In brief, microconidia were washed from MAT agar plates and incubated for germination at a concentration of 1.5 × 108 conidia in liquid Sabouraud medium overnight at 30°C with shaking (200 rpm). After filtering the cell suspension by the use of two layers of Miracloth (Calbiochem), the germinating cells were suspended in protoplast buffer (0.7 M KCl, 50 mM potassium phosphate buffer, pH 5.8) with 3.75 mg/ml lysing enzymes from Trichoderma harzianum (Sigma-Aldrich), kitalase (Wako Chemicals), and yatalase (Takara Bio Europe) and incubated at 30°C for 30 min with gentle shaking to digest the cell wall. Protoplast formation was monitored by microscopy. The protoplasts were collected by centrifugation at 1,200 × g for 5 min and resuspended in STC1 buffer (1.2 M sorbitol, 10 mM Tris-Cl [pH 7.5], 10 mM CaCl2). After a second centrifugation step at 1,200 × g for 5 min, the protoplast pellet was resuspended in STC2 buffer (1.2 M sorbitol, 10 mM Tris-Cl [pH 7.5], 50 mM CaCl2) at a final concentration of approximately 2 × 108 cells/ml. Two hundred microliters of the protoplast suspension was incubated on ice for 10 min with approximately 1 μg of a gel-purified, linear DNA fragment from plasmid pAbenACUEM2, pAbenACUEK2, or pAbenKU70M2. Three hundred microliters of 60% PEG 4000-TC (10 mM Tris-Cl [pH 7.5], 10 mM CaCl2) were gently mixed with the protoplasts and allowed to stand on ice for 10 min before the procedure was repeated with 600 μl PEG 4000-TC. The protoplasts were then centrifuged at 1,500 × g for 5 min, and the pellets were resuspended in STC1 buffer. The protoplast suspension was mixed with liquid Sabouraud-sorbitol-agar (Sabouraud with 1 M sorbitol and 0.7% agar) and poured on Sabouraud-sorbitol-agar (1.5% agar). These plates were incubated at 30°C and overlaid after 24 h with Sabouraud-sorbitol-agar containing the appropriate antibiotic. Depending on the used selection marker, hygromycin- or neomycin-resistant transformants were selected with 300 μg/ml hygromycin (ForMedium) or 200 μg/ml G418 (Carl Roth). For a comparison of site-specific integration frequencies in the A. benhamiae wild type and Δku70 mutant, three independent transformation experiments were carried out for each strain using the linear deletion cassette from plasmid pAbenACUEM2. The results were analyzed by the two-tailed unpaired Student's t test, and a P value of <0.05 was considered statistically significant.
Isolation of genomic DNA and Southern analysis.
Genomic DNA from A. benhamiae strains was isolated as follows. Mycelia from liquid Sabouraud cultures (containing 100 μg/ml hygromycin or G418 in the case of resistant transformants) were frozen in liquid nitrogen and ground into a powder with a mortar and pestle. The powder was resuspended in 800 μl proteinase K buffer (10 mM Tris-Cl [pH 7.5], 50 mM EDTA [pH 8.0], 0.5% SDS, 1 mg/ml proteinase K) and incubated at 60°C for 2 h with shaking at 1,400 rpm. After extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), the DNA was precipitated from the aqueous phase with 100% isopropanol. The DNA pellet was washed once with 70% ethanol, air dried, and dissolved in 70 μl H2O containing 1.4 μl RNase A (10-mg/ml stock).
Fifteen micrograms of DNA was digested with appropriate restriction enzymes and separated in 1% (wt/vol) agarose gels. After ethidium bromide staining, the gels were transferred by vacuum blotting onto nylon membranes and fixed by UV cross-linking. Southern hybridization using enhanced chemiluminescence-labeled probes was performed with the Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare) as recommended by the manufacturer.
In vivo experiments. (i) Infection of guinea pigs.
Specific-pathogen-free guinea pigs of the Hartley strain (Charles River Laboratories International, Wilmington, MA) were used to evaluate the virulence of the A. benhamiae strains AbenKU70M1A (12-month-old females, n = 3), AbenACUEM1A (2-month-old males, n = 3), and AbenACUEM1B (2-month-old males, n = 3). During the entire study, animals were housed separately. Infection was performed essentially as shown previously (1, 25). In brief, animals were anesthetized by intraperitoneal injection of medetomidine and ketamine. Both flanks of the animals (10-cm2 areas) were shaved with electric clippers and then dry shaved using a sterile scalpel blade to slightly break down the skin integrity. For each animal, the skin from one flank was infected with a specific mutant strain of A. benhamiae, while the skin from the opposite flank was infected with the control parental strain (A. benhamiae Lau2354-2). Inoculum consisted of mycelium and spores scraped from freshly grown 18-day-old Sabouraud plates suspended in 5% (wt/wt) poloxamer 407 (BASF, Germany). For each flank, the inoculum (5 × 104 CFU in 250 μl) was gently rubbed onto the skin with a sterile pipette tip.
(ii) Clinical follow-up.
Animal infection was monitored twice weekly for 7 weeks by two independent examiners using clinical criteria, as previously described (1, 3). The examiners remained the same for the duration of the experiment. Briefly, both investigators were blinded to the status of the guinea pig flank (infected by the control strain or a mutant strain). A global clinical score was calculated for infected flanks in each guinea pig by adding subjective scores (from 0 to 3) attributed to four clinical criteria (alopecia, erythema, squamosis, and crusts). A mean global clinical score was then calculated for each group (3). For this in vivo experiment, several groups were considered for further analysis. The group of flanks infected with strain AbenKU70M1A (n = 3) was compared with the group of opposite flanks from the same animals infected with the control parental strain (n = 3). Both groups of flanks infected with strain AbenACUEM1A (n = 3) or AbenACUEM1B (n = 3) were compared between themselves and with the group of opposite flanks from the same animals but infected with the control parental strain (n = 6).
(iii) Histology.
Skin biopsy specimens were collected under anesthesia from the guinea pigs 14 days postinoculation (p.i.) with both mutant and control strains. Samples were fixed in 10% neutralized buffered formalin and paraffin embedded for routine processing. Five-micrometer-thick sections were stained with hematoxylin-eosin or periodic acid-Schiff (PAS) to assess the invasion of keratinized skin structures by the various A. benhamiae strains.
(iv) Ethics.
Animal experiments were approved by the local ethic committee of the University of Liège (file no. 750 from 18 March 2008).
(v) Statistical evaluation.
For the in vivo experiments, significant differences between groups were assessed using a mixed linear model, with significance defined as a P value of <0.05. This procedure takes into account repeated measurements on the same subject throughout the experiment. The correlation between the two independent examiners has been verified using the Pearson and Spearman correlation coefficients. Both were positive: 0.791 and 0.762 (P < 0.001), respectively.
Infection of RHE.
The reconstituted human epidermis (RHE), which was used for the analysis of epidermal fungal invasion, was supplied by SkinEthic Laboratories (Lyon, France). According to the instructions of the supplier, the RHE was incubated for 24 h in 1 ml of SkinEthic growth medium at 37°C and in 5% CO2, with saturated humidity. Thereafter, the culture medium was removed, and fresh medium was added. The RHE was infected with 1 × 103 A. benhamiae microconidia, which were derived from MAT agar plates, and resuspended in Dulbecco's phosphate-buffered saline (DPBS; Lonza) at a density of 2 × 105 cells per ml. After 4 days of incubation at 37°C and in 5% CO2 with saturated humidity, invasion of the RHE by the fungal cells was analyzed by microscopy. Infected RHEs as well as noninfected controls were fixed with Karnovsky's fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.05 M cacodylate buffer) and embedded in Technovit 7100 (Heraeus Kulzer). Eight-micrometer-thick sections were stained with PAS reagent and counterstained with hemalum solution.
RESULTS
Targeted gene deletion in the A. benhamiae wild type.
As a first step, the A. benhamiae gene AbenACUE, encoding the putative glyoxylate cycle enzyme malate synthase, was deleted in the wild-type strain Lau2354-2. For this purpose, protoplasts prepared from A. benhamiae germinating microconidia were transformed with a linear deletion cassette in which the AbenACUE coding region had been replaced by the Aspergillus nidulans hygromycin resistance marker (Fig. 1A). Transformation was carried out following a modified protocol of polyethylene glycol-mediated transformation of filamentous fungi (for details, see Materials and Methods) (34). Transformants were selected by their ability to grow on hygromycin-containing Sabouraud agar plates. In order to verify the correct insertion of the deletion cassette at the AbenACUE locus, Southern analysis was performed using upstream and downstream AbenACUE sequences as probes (Fig. 1C). For additional control, ectopic integration of the hygromycin resistance marker in the genome of these transformants was excluded by probing with sequences of the hph resistance gene (data not shown). Two transformation experiments were performed, in which three out of nine and one out of two inspected transformants were proven to be correct. Two independent hygromycin-resistant ΔAbenacuE deletion mutants, AbenACUEM1A and AbenACUEM1B, were used for further investigation.
The frequency of site-specific integration is enhanced in an A. benhamiae Δku70 mutant background.
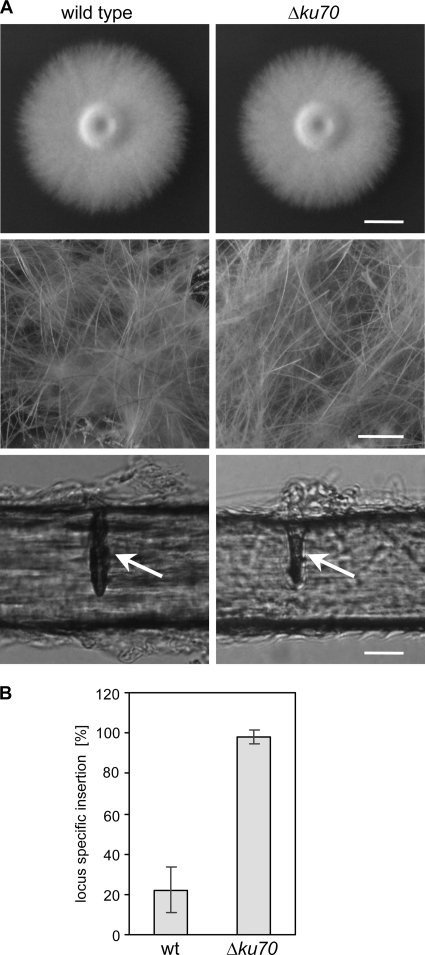
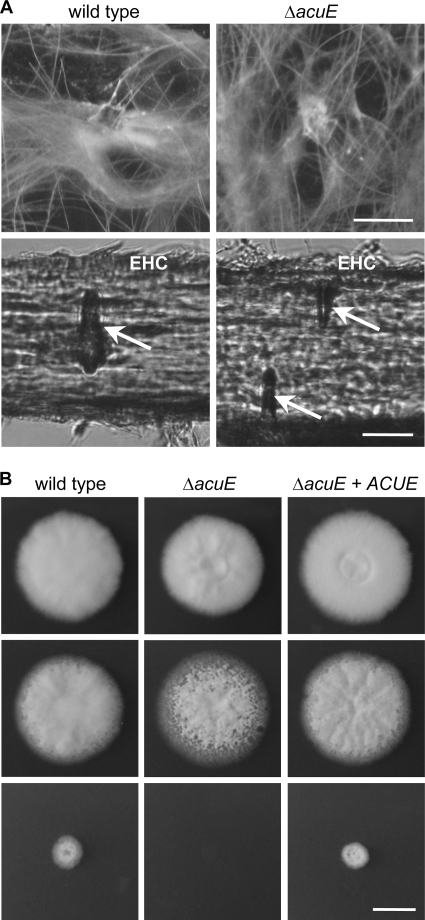
We raised the question of whether the frequency of site-directed integration of linear DNA cassettes by homologous recombination is increased in an A. benhamiae mutant lacking Ku70 recombinase activity. Since locus-specific genetic manipulation is known to be difficult in dermatophyte species (32, 34), by this approach we intended to provide an A. benhamiae strain which supports enhanced homologous insertion. For the specific deletion of the A. benhamiae KU70 gene in the wild-type strain, a DNA cassette was constructed by the use of a second dominant resistance gene, i.e., the neomycin marker (NEOR) (Fig. 2A). This strategy preserves the possibility of using the hygromycin selection marker for subsequent gene targeting experiments in this derivative. Transformation of A. benhamiae wild-type cells with the KU70 deletion construct was performed as described above, except that transformants were selected on G418-containing Sabouraud agar plates. A first transformation experiment resulted in 11 neomycin-resistant clones, 8 of which showed the desired genotype. The correct insertion of the deletion cassette at the AbenKU70 locus was proven by Southern analysis with upstream and downstream AbenKU70 sequences (Fig. 2B). An additional insertion of the selection marker was ruled out by the use of neo sequences as a probe (data not shown). Two independent ΔAbenku70 deletion mutants, AbenKU70M1A and AbenKU70M1B, were stored. The A. benhamiae wild type and ΔAbenku70 mutants were compared in their growth ability and phenotypic appearance on different substrates in vitro (Fig. 3A) and during guinea pig infection (see next paragraph). Growth of the mutants on routine media such as potato dextrose agar was comparable to that of the wild type (the colony size of the ΔAbenku70 mutant may have appeared slightly smaller in diameter as shown in Fig. 3A), and no difference in their ability to grow on or perforate human hair was observed (Fig. 3A). For the genetic manipulation in the Δku70 mutant background, only AbenKU70M1A was used as a parental strain in further experiments. In order to test whether absent Ku70 activity in A. benhamiae allows an increased frequency of targeted insertion, we compared the frequencies of site-specific integration of the AbenACUE deletion cassette at the AbenACUE locus (Fig. 1) in the wild type and the Δku70 mutant. For each strain, three transformations were carried out, and 18 randomly picked hygromycin-resistant transformants per experiment were analyzed by Southern hybridization as described above (data not shown). Twelve out of 54 and 53 out of 54 transformants of the wild type and the Δku70 mutant, respectively, showed the desired genotype, revealing a significant increase (P < 0.05) of the frequency of the locus-specific insertion in the Δku70 mutant background (Fig. 3B). Hence, the A. benhamiae Δku70 mutant represents an additional powerful tool for genetic manipulation in this dermatophyte species. Two independent ΔAbenacuE mutants obtained by transformation of AbenKU70M1A, AbenACUEM2A and AbenACUEM2B, were used for further investigation (Fig. 1D).
Fig. 3.
Comparison of A. benhamiae wild type and Δku70 mutant. (A) A. benhamiae wild type and mutants in AbenKU70 display comparable phenotypes during growth on rich potato dextrose agar or human hair. A. benhamiae wild type and Δku70 mutant strains AbenKU70M1A and AbenKU70M1B were grown on PDA (5 days at 30°C) and human hair (50 days at 25°C). A. benhamiae colonies on PDA plates (top; scale bar represents 1 cm); the mycelial, woolen growth of A. benhamiae on human hair is demonstrated by macroscopic inspection (middle; scale bar represents 0.5 cm). Microscopy of single hairs reveals the characteristic hair perforations (arrows) caused by the fungus (bottom; scale bar represents 20 μm). The two independently constructed mutant strains behaved identically, and only one of them is shown. (B) Increased frequency of targeted insertion in the A. benhamiae Δku70 mutant. Wild type and AbenKU70M1A (Δku70 mutant) were transformed with the AbenACUE deletion cassette, the specific integration of which at the AbenAcuE locus was analyzed by Southern analysis for 18 randomly picked, hygromycin-resistant transformants. The results are the means ± standard deviation (SD) of three independent experiments.
The A. benhamiae wild type and Δku70 mutant display comparable virulence in the guinea pig infection model.
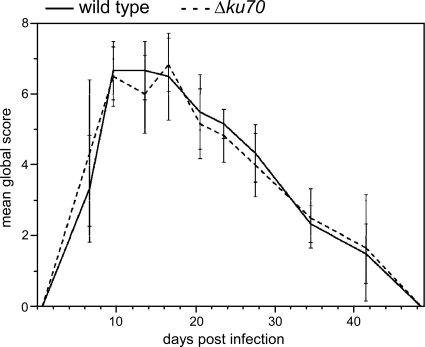
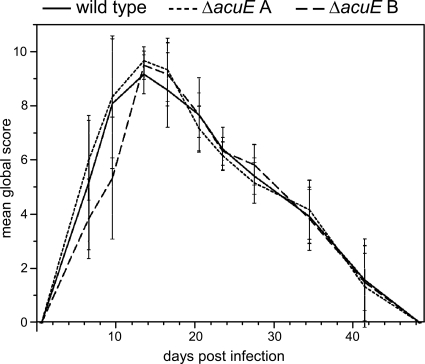
A guinea pig skin infection model was previously developed for A. benhamiae (25). In the present study, this model was used to compare the pathogenic potential of the A. benhamiae strain AbenKU70M1A with that of the parental wild-type strain, an important issue for the future analysis of A. benhamiae derivatives generated by the use of the Δku70-deficient transformant. As a positive finding, the Δku70 mutant behaved like the wild type in the guinea pig model. Both strains, the wild type and AbenKU70M1A, induced highly inflammatory dermatophytosis as previously reported (25). The clinical time course assessed by the two independent investigators was undistinguishable between the two strains, as outlined by the mean global clinical scores that did not significantly differ at any time during infection (P < 0.05), i.e., between day 0 and day 49 p.i. (Fig. 4). Both strains induced similar skin lesions, including erythema, alopecia, scaling, and crusting, which were most severe 14 days p.i. (Fig. 5A and B), and then healed progressively (Fig. 4). Histopathological examination of the skin sections of the injured areas was undistinguishable for both tested strains and revealed inflammatory epidermal, dermal, and follicular lesions similar to those previously described for the A. benhamiae guinea pig model (25). PAS staining allowed the visualization of the fungus in both epidermal and hair follicle keratin by staining cell walls of hyphae or conidia in both control (Fig. 5C and E) and AbenKU70M1A- (Fig. 5D and F) experimentally infected guinea pigs. This finding indicates that the Δku70 mutant cells are able to colonize the keratinized host structures in a way similar to that of the wild-type cells. In conclusion, the A. benhamiae Δku70 mutant strain behaves as virulently as the wild type in our in vivo guinea pig model, which renders this genetically modified A. benhamiae strain suitable for further functional gene analyses under these conditions.
Fig. 4.
A. benhamiae wild-type and Δku70 mutant cells induce comparable symptoms of dermatophytosis in guinea pigs. Kinetics of the mean global score (± SD) reflect the severity of dermatophytic lesions in guinea pig flanks after infection with A. benhamiae strain AbenKU70M1A or the control parental strain Lau2354-2. Guinea pig flanks were infected as described in Materials and Methods (n = 3 in both groups). A global score was calculated for infected flanks in each guinea pig by adding clinical scores (from 0 to 3) attributed blindly by two independent examiners according to four clinical criteria (alopecia, erythema, squamosis, and crusts). A mean global score was then calculated for each group of flanks.
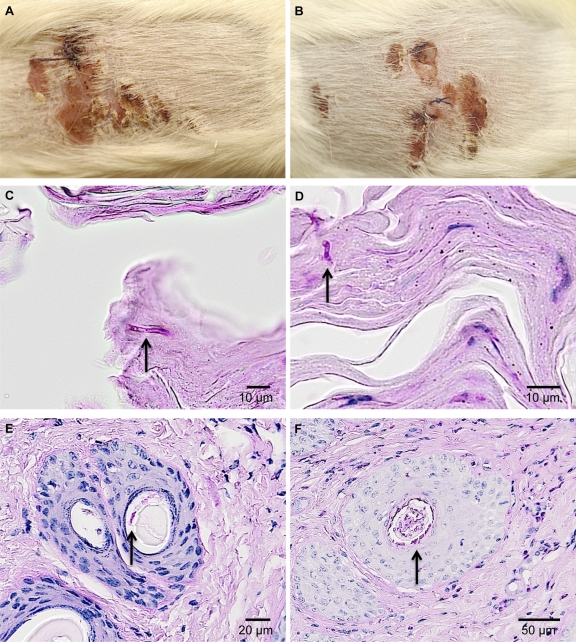
Fig. 5.
A. benhamiae wild-type and Δku70 mutant cells induce comparable symptoms of dermatophytosis and show similar growth in and invasion of guinea pig skin epidermal structures. The development of skin lesion and skin fungal colonization in guinea pigs 14 days postinfection with A. benhamiae strain AbenKU70M1A (B, D, and F) or the control parental strain Lau2354-2 (A, C, and E) is demonstrated. (A and B) Macroscopically visible inflammatory skin lesions consisting of erythema, alopecia, scaling, and crusting are undistinguishable between the lesions induced by control parental strain Lau2354-2 (A) and the mutant AbenKu70M1A (B). (C and D) PAS-stained cross-section microscopy of a scaly and crusty infected area showing similar levels of fungal colonization (arrows) into the keratinized superficial epidermis in flanks infected with the control parental strain Lau2354-2 (C) and the mutant AbenKU70M1A (D). (E and F) PAS-stained cross-section microscopy of an infundibular-isthmal hair follicle showing similar levels of fungal colonization (arrows) of hair keratin in flanks infected with the control parental strain Lau2354-2 (E) and the mutant AbenKU70M1A (F).
The A. benhamiae malate synthase is required for dermatophyte growth on lipids as a sole carbon source.
The observation that the expression of genes encoding key enzymes of the glyoxylate cycle was upregulated in A. benhamiae during experimental infection of guinea pigs encouraged us to investigate the functional role of this metabolic pathway in more detail. The ΔacuE mutants AbenACUEM1A and AbenACUEM1B were analyzed for their ability to grow on potentially pathogenicity-relevant substrates, such as keratin and lipids. No obvious differences between wild-type and mutant strains were detected during growth on human hair as a sole carbon and nitrogen source, i.e., neither mycelium formation nor hair perforation were affected by the deletion of AbenACUE (Fig. 6A). In contrast, the differential growth of wild-type and mutant strains on distinct carbon sources revealed the importance of the A. benhamiae malate synthase for the utilization of acetate, ethanol, and various lipids as sole carbon sources (Fig. 6B; Table 3). Whereas the growth of the A. benhamiae wild type was already comparatively slow on the latter substrates, ΔacuE mutant growth was completely abolished under these conditions; mutant growth on peptone was only slightly slower. Considering that lipids are important skin constituents, different fatty acids which are found in the epidermal stratum corneum, e.g., myristic and palmitic acid, were included in the analysis (13). A. benhamiae ΔacuE mutants failed to grow on these saturated fatty acids, in contrast to the wild type (Table 3). Notably, however, supplementation of lipid-based media with other carbon sources, such as peptone, bypassed this growth defect, suggesting the limited importance of this metabolic pathway if additional carbon sources are provided (Fig. 6B; Table 3). To further prove that the inability of the ΔacuE mutants to grow on lipids is caused by the lack of malate synthase, the AbenACUE gene was reintroduced at its original locus in the two independently constructed ΔacuE mutants, AbenACUEM1A and AbenACUEM1B. Transformation was carried out with a DNA cassette containing the entire coding region of the AbenACUE gene and the neomycin resistance marker (Fig. 1B). Transformants were selected as described above on G418-containing Sabouraud agar plates, and Southern analysis with upstream and downstream AbenACUE sequences was performed to prove the site-specific integration of the complementation cassette (Fig. 1C). Two independent G418-resistant and hygromycin-sensitive transformants, AbenACUEK1A and AbenACUEK1B, were stored and phenotypically characterized. Comparison of the wild type, mutants, and complemented strains on lipid-based media revealed that reinsertion of the AbenACUE gene in the ΔacuE mutants restores the wild-type phenotype (Fig. 6B).
Fig. 6.
The malate synthase AcuE is not required for growth of A. benhamiae on hair but is for growth of the fungus on lipids as the sole carbon source. (A) A. benhamiae wild type and ΔacuE mutants AbenACUEM1A and AbenACUEM1B were grown on human hair for 50 days at 25°C. No differences between wild type and mutants were observed for mycelia formation (top; scale bar represents 0.5 cm) or the ability to form hair perforations (arrows, bottom; EHC, external hair cuticle; scale bar represents 20 μm). (B) A. benhamiae wild type, ΔacuE mutants (AbenACUEM1A and AbenACUEM1B), and ΔacuE::ACUE complemented strains (AbenACUEK1A and AbenACUEK1B) were grown on Aspergillus minimal agar in which peptone (top), olive oil (bottom), or both (middle) were used as a carbon source. Photographs of the colonies were taken after 6 days at 30°C. The scale bar represents 1 cm. The two independently constructed ΔacuE mutants behaved identically, and the complemented strains behaved identically; only one of each is shown.
Table 3.
Growth of the A. benhamiae wild type, ΔacuE mutants, and complemented strains on various carbon sources
| Carbon source | Wild typea | ΔacuE mutanta | ΔacuE::ACUE complemented straina |
|---|---|---|---|
| 2% glucose | ++ | ++ | ++ |
| 1% peptone | +++ | +++ | +++ |
| 1% peptone/0.5% olive oil | +++ | +++ | +++ |
| 0.5% olive oil | ++ | − | ++ |
| 2% ethanol | + | − | + |
| 100 mM sodium acetate | + | − | + |
| 0.5% myristic acid | ++ | − | ++ |
| 0.5% palmitic acid | ++ | − | ++ |
Growth of A. benhamiae colonies was estimated on Aspergillus minimal medium, supplemented with the indicated substrates as the sole carbon source after 6 days at 30°C as + (weak), ++ (good), +++ (very good), and − (not visible). The two independently constructed ΔAbenacuE mutants (AbenACUEM1A and AbenACUEM1B) behaved identically, and the complemented strains (AbenACUEK1A and AbenACUEK1B) behaved identically.
The A. benhamiae malate synthase AcuE is not required for virulence in the guinea pig infection model.
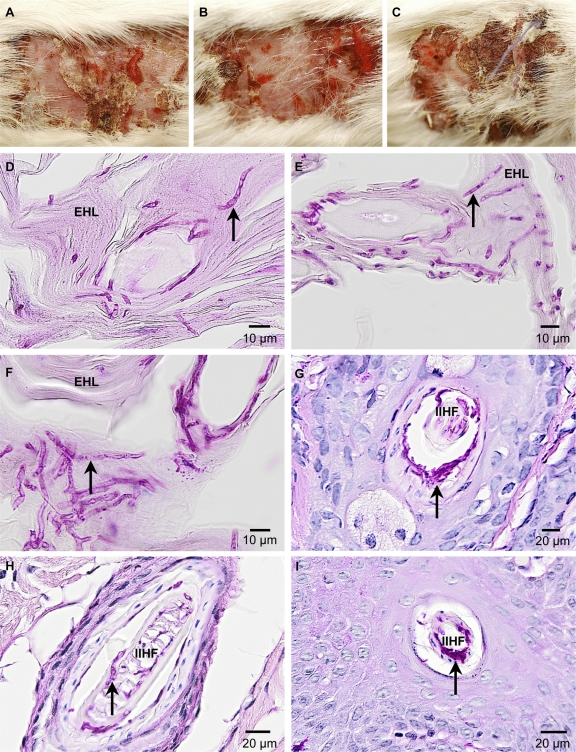
The previously observed upregulation of the AbenACUE gene during guinea pig infection (25) particularly raised the question of whether malate synthase activity is required for the virulence of A. benhamiae in this model. To test this possibility, the two independently constructed malate synthase mutant strains, AbenACUEM1A and AbenACUEM1B, were compared in terms of virulence with each other and the wild-type control. The three strains induced similar levels of highly inflammatory dermatophytosis with undistinguishable clinical time courses, as shown by the mean global clinical scores that did not significantly differ at any time during infection (P < 0.05) (Fig. 7 and 8A to C). These scores were globally higher than those observed with guinea pigs from the previous in vivo experiment which were infected with the A. benhamiae wild type and AbenKU70M1A (Fig. 4). This difference is not surprising since dermatophytes, including A. benhamiae, induce skin lesions which are generally more severe in younger animals (B. Mignon, personal observation; for details on the age of the infected animals, see also Materials and Methods). Histopathological examination of the skin sections of the injured areas was undistinguishable for both the two malate synthase-deficient strains and the control parental strain, showing inflammatory epidermal, dermal, and follicular lesions similar to those previously described for the A. benhamiae guinea pig model (25) (data not shown). PAS staining allowed the visualization of large amounts of fungal elements in both epidermal and hair follicle keratin in guinea pigs infected with the wild type (Fig. 8D and G) and malate synthase-deficient strains (Fig. 8E, F, H, and I), respectively. This result indicates that both ΔacuE mutants were able to colonize the keratinized structures in a way similar to that of the wild-type control. In conclusion, the comparable virulence of the A. benhamiae wild type and malate synthase mutants suggested that AbenACUE is not required for fungal pathogenicity and/or growth during inflammatory dermatophytosis in the tested in vivo model. Thus, carbon sources other than skin lipids likely circumvent the need for malate synthase in the fungal cells during infection of the guinea pig epidermis.
Fig. 7.
A. benhamiae wild-type and ΔacuE mutant cells induce comparable symptoms of dermatophytosis in guinea pigs. Kinetics of the mean global score (± SD) reflect the severity of dermatophytic lesions in guinea pig flanks after infection with A. benhamiae strains AbenACUEM1A and AbenACUEM1B or the control parental strain Lau2354-2. Guinea pigs flanks were infected as described in Materials and Methods (n = 3 in both the AbenAcuEM1A and AbenACUEM1B groups, n = 6 in the Lau2354-2 group). A global score was calculated for infected flanks in each guinea pig by adding clinical scores (from 0 to 3) attributed blindly by two independent examiners according to four clinical criteria (alopecia, erythema, squamosis, and crusts). A mean global score was then calculated for each group of flanks.
Fig. 8.
A. benhamiae ΔacuE mutants induce symptoms of inflammatory dermatophytosis comparable to those of the wild type. Shown is the development of skin lesions and skin fungal colonization in guinea pigs 14 days postinfection with A. benhamiae strain AbenACUEM1A (B, E, and H) and AbenACUEM1B (C, F, and I) or the control parental strain Lau2354-2 (A, D, and G). (A to C) Macroscopically visible inflammatory skin lesions consisting of erythema, alopecia, moderate scaling, crusting, and severe excoriations due to pruritus are similar between those induced by the control parental strain Lau2354-2 (A) and the mutant strains AbenACUEM1A (B) and AbenACUEM1B (C). (D to F) PAS-stained cross-section microscopy of a scaly and crusty infected area showing similar levels of fungal colonization (arrows) of the keratinized superficial epidermis (epidermal horny layer [EHL]) in flanks infected with the control parental strain Lau2354-2 (D) and the mutant strains AbenACUEM1A (E) and AbenACUEM1B (F). (G to I) PAS-stained cross-section microscopy of an infundibular-isthmal hair follicle (IIHF) showing similar levels of fungal colonization (arrows) of hair keratin in flanks infected with the control parental strain Lau2354-2 (G) and the mutant strains AbenACUEM1A (H) and AbenACUEM1B (I).
A. benhamiae malate synthase is not required for fungal invasion of reconstituted human epidermis.
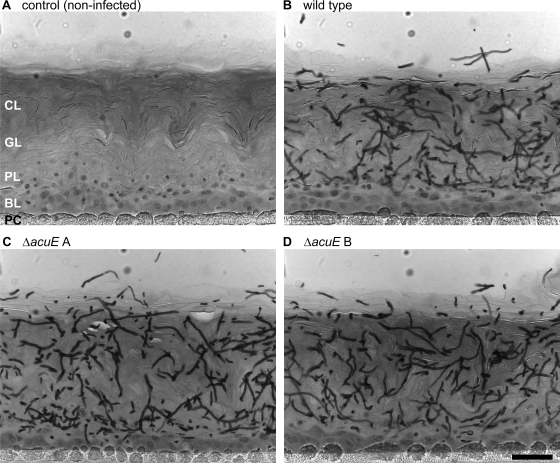
In order to study a potential role of the A. benhamiae malate synthase for fungal invasion of human epidermis, a commercially available model of RHE (SkinEthic Laboratories, Lyon, France) was established for the analysis of A. benhamiae. For this approach, human keratinocytes, cultured on inert polycarbonate filters, were infected with a defined number (1 × 103) of A. benhamiae microconidia and incubated at the air-liquid interface (for details, see Materials and Methods). Epidermal invasion of the cornified cell layers by fungal cells of the wild type and malate synthase mutants AbenACUEM1A and AbenACUEM1B was inspected by microscopy (Fig. 9). Under the tested conditions, massive tissue invasion by fungal filaments of all the tested strains was detected after 2 to 4 days, a time period when in vitro growth of A. benhamiae on lipids or keratin was not yet accomplished. As depicted in Fig. 9, no obvious differences in tissue destruction and/or fungal growth were observed for either mutant. These findings, together with the results from the guinea pig infection, let us conclude that a pathogenicity-related role could not be assigned to the A. benhamiae glyoxylate cycle gene AbenACUE in epidermal invasion.
Fig. 9.
Comparable epidermal invasion of A. benhamiae wild-type and ΔacuE mutant cells in a model of RHE. The RHE was infected with 1 × 103 A. benhamiae microconidia and incubated at 37°C in 5% CO2. Invasion of the RHE was analyzed 4 days after infection by microscopy as described in Materials and Methods. Microscopy of representative PAS-stained sections shows similar degrees of fungal invasion of the RHE by the wild-type strain Lau2354-2 (B) and the mutant strains AbenACUEM1A (C) and AbenACUEM1B (D). An uninfected control sample is shown for comparison (A), and the scale bar represents 50 μm. CL, cornified layer of the RHE; GL, granular layer; PL, prickle cell layer; BL, basal layer; PC, polycarbonate filter.
DISCUSSION
The advent of dermatophyte genome sequencing projects offers fundamental information for genetic research in these major fungal pathogens. At the same time, however, straightforward molecular tools are required to allow for efficient, targeted genetic manipulations in these microorganisms. Whereas in many other fungal pathogens the generation of specific mutants has meanwhile been almost standardized, strain construction in dermatophytes is still challenging. Only recently, advanced protocols for transformation, as well as suitable parental strains, have become available for selected dermatophyte species (32, 34). In the present study, successful transformation and targeted integration of DNA cassettes has been demonstrated for the zoophilic dermatophyte A. benhamiae. Our results revealed that efficient gene targeting was achieved already in the A. benhamiae wild-type background. From previous results for other filamentous fungi and especially from studies of dermatophytes, a less promising rate of specific integrants was initially expected (30, 31, 34). In the present study, targeted gene deletion was accomplished in wild-type cells for two loci, the AbenACUE and the AbenKU70 genes. Yet, it cannot be excluded that other genes are less efficiently targeted during transformation by the present protocol. Therefore, the constructed A. benhamiae Δku70 mutant is offered as an alternative parental strain. Using this mutant, an increased frequency of specific integration was confirmed during mutagenesis of AbenACUE, and efficient gene targeting in the Δku70 parental strain can also be expected for other loci.
The A. benhamiae wild type and Δku70 deletion mutants showed comparable basic growth phenotypes under the tested in vitro conditions, including the ability to destruct and utilize hair keratin. The latter characteristic is assumed to be of major importance for the saprophytic growth and pathogenicity of these keratinophilic fungi. In addition, no visible difference was observed in the virulence of the wild type and the Δku70 mutant during inflammatory skin infection of guinea pigs, neither with regard to cutaneous symptoms nor in terms of epidermal invasion or fungal multiplication. The observation that none of our tested transformants showed an altered virulence further revealed that the presence of either selection marker, i.e., a neomycin or hygromycin resistance cassette, did not impact fungal pathogenesis in our infection model. This result is an important prerequisite for future research on the pathogenicity mechanisms of such genetically manipulated strains in dermatophytes.
For a better understanding of dermatophytosis, the identification of dominant virulence factors in dermatophytes appears to be of particular interest, especially since these fungi also infect immunocompetent individuals. In our recent broad-scale transcriptome analysis of A. benhamiae during inflammatory guinea pig infection, activation of genes encoding the putatively pathogenicity-related, major dermatophyte keratinases had not been detected (25). Instead, genes encoding individual metabolic enzymes, such as isocitrate lyase and malate synthase, had been found to be activated in vivo, an observation which pointed to potential metabolic requirements during adaptation of the fungus to lipid-rich host structures. Our present in vitro results supported this assumption, given that A. benhamiae mutants in the putative malate synthase-encoding gene AbenACUE displayed a growth defect under conditions in which lipids served as the sole carbon source. Supplementation of the tested media with other carbon sources, however, abrogated this growth defect, suggesting a minor role of this metabolic pathway in the presence of alternative carbon sources. These findings are in line with previous observations of A. fumigatus (23) and C. neoformans (21), which also did not assign a pathogenic role to the pathway, in contrast to observations of the yeast C. albicans (15). Notably, no defect in growth and/or virulence was monitored in A. benhamiae ΔacuE mutants in the two infection models analyzed. During infection of guinea pigs, similar skin lesions with erythema, alopecia, scaling, and crusting were produced, and histopathology revealed a comparable fungal load in the infected epidermal layers. However, disease symptoms of inflammatory infections by zoophilic dermatophytes, such as A. benhamiae, are strongly related to host reactions, in contrast to chronic dermatophyte infections, which are associated with long-term fungal persistence, e.g., T. rubrum-induced onychomycosis. Therefore, the possibility remains that other types of dermatophytoses require different metabolic adaptation mechanisms in the fungal cells, and hence, a role of glyoxylate cycle enzymes in dermatophytosis cannot per se be excluded.
In contrast to those in guinea pig infection, tissue invasion and fungal growth in the tested reconstituted epidermis model were accomplished comparatively fast. Therefore, carbon sources other than lipids or keratin likely support the growth of the fungal cells in this model. Such carbon sources can be derived from the tissue itself or from the cell culture medium. An alternative to explain the rapid invasive fungal growth may be the lack of some skin immune components, e.g., leukocytes, under these conditions. In line with this assumption, previous studies of reconstituted human epithelia infected with C. albicans revealed that supplementation of the model with polymorphonuclear leukocytes strongly impaired tissue invasion by this pathogenic yeast (22). In the past, various in vitro models using reconstituted epidermis or skin explants have been established and were also proven successful for the analysis of dermatophytosis (1, 2, 5, 7, 19, 20, 24, 26). Nevertheless, the commercially available RHE model, which was employed in the present study for the analysis of A. benhamiae mutants, may further advance the research of dermatophytosis and the identification of basic virulence attributes of dermatophytes. The development of such in vitro infection models has been proven successful for the identification of pathogenicity-associated factors in other fungi (35), thereby circumventing the use of animal experiments.
In conclusion, the presented genetic system for A. benhamiae, together with the availability of adequate infection models and full genome sequence information for our strain, will further advance the research on dermatophytes, fungal pathogens of major clinical importance.
ACKNOWLEDGMENTS
This work was supported by the DFG-funded excellence graduate school Jena School for Microbial Communication, by the Hans Knoell Institute, and by grant 3.4535.08 from Fonds de la Recherche Scientifique Médicale (FRSM).
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Baldo A., et al. 2010. Secreted subtilisin Sub3 from Microsporum canis is required for adherence to but not for invasion of the epidermis. Br. J. Dermatol. 162:990–997 [DOI] [PubMed] [Google Scholar]
- 2. Baldo A., et al. 2008. Secreted subtilisins of Microsporum canis are involved in adherence of arthroconidia to feline corneocytes. J. Med. Microbiol. 57:1152–1156 [DOI] [PubMed] [Google Scholar]
- 3. Brouta F., et al. 2003. Humoral and cellular immune response to a Microsporum canis recombinant keratinolytic metalloprotease (r-MEP3) in experimentally infected guinea pigs. Med. Mycol. 41:495–501 [DOI] [PubMed] [Google Scholar]
- 4. Burmester A., et al. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol. 12:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coquette A., Berna N., Vandenbosch A., Rosdy M., Poumay Y. 1999. Differential expression and release of cytokines by an in vitro reconstructed human epidermis following exposure to skin irritant and sensitizing chemicals. Toxicol. In Vitro 13:867–877 [DOI] [PubMed] [Google Scholar]
- 6. da Silva Ferreira M. E., et al. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duek L., Kaufman G., Ulman Y., Berdicevsky I. 2004. The pathogenesis of dermatophyte infections in human skin sections. J. Infect. 48:175–180 [DOI] [PubMed] [Google Scholar]
- 8. Fachin A. L., Ferreira-Nozawa M. S., Maccheroni W., Jr., Martinez-Rossi N. M. 2006. Role of the ABC transporter TruMDR2 in terbinafine, 4-nitroquinoline N-oxide and ethidium bromide susceptibility in Trichophyton rubrum. J. Med. Microbiol. 55:1093–1099 [DOI] [PubMed] [Google Scholar]
- 9. Ferreira-Nozawa M. S., et al. 2006. The pH signaling transcription factor PacC mediates the growth of Trichophyton rubrum on human nail in vitro. Med. Mycol. 44:641–645 [DOI] [PubMed] [Google Scholar]
- 10. Fumeaux J., et al. 2004. First report of Arthroderma benhamiae in Switzerland. Dermatology 208:244–250 [DOI] [PubMed] [Google Scholar]
- 11. Idnurm A., Howlett B. J. 2002. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot. Cell 1:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krappmann S., Sasse C., Braus G. H. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lampe M. A., et al. 1983. Human stratum corneum lipids: characterization and regional variations. J. Lipid Res. 24:120–130 [PubMed] [Google Scholar]
- 14. Liu O. W., et al. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lorenz M. C., Fink G. R. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86 [DOI] [PubMed] [Google Scholar]
- 16. McKinney J. D., et al. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738 [DOI] [PubMed] [Google Scholar]
- 17. Mignon B., et al. 2008. Immunization and dermatophytes. Curr. Opin. Infect. Dis. 21:134–140 [DOI] [PubMed] [Google Scholar]
- 18. Ninomiya Y., Suzuki K., Ishii C., Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Onyewu C., et al. 2007. Targeting the calcineurin pathway enhances ergosterol biosynthesis inhibitors against Trichophyton mentagrophytes in vitro and in a human skin infection model. Antimicrob. Agents Chemother. 51:3743–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rashid A., Edward M., Richardson M. D. 1995. Activity of terbinafine on Trichophyton mentagrophytes in a human living skin equivalent model. J. Med. Vet. Mycol. 33:229–233 [PubMed] [Google Scholar]
- 21. Rude T. H., Toffaletti D. L., Cox G. M., Perfect J. R. 2002. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect. Immun. 70:5684–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schaller M., et al. 2004. Polymorphonuclear leukocytes (PMNs) induce protective Th1-type cytokine epithelial responses in an in vitro model of oral candidosis. Microbiology 150:2807–2813 [DOI] [PubMed] [Google Scholar]
- 23. Schöbel F., et al. 2007. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect. Immun. 75:1237–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smijs T. G., Bouwstra J. A., Schuitmaker H. J., Talebi M., Pavel S. 2007. A novel ex vivo skin model to study the susceptibility of the dermatophyte Trichophyton rubrum to photodynamic treatment in different growth phases. J. Antimicrob. Chemother. 59:433–440 [DOI] [PubMed] [Google Scholar]
- 25. Staib P., et al. 2010. Differential gene expression in the pathogenic dermatophyte Arthroderma benhamiae in vitro versus during infection. Microbiology 156:884–895 [DOI] [PubMed] [Google Scholar]
- 26. Tabart J., et al. 2007. Reconstructed interfollicular feline epidermis as a model for Microsporum canis dermatophytosis. J. Med. Microbiol. 56:971–975 [DOI] [PubMed] [Google Scholar]
- 27. Takashio M. 1972. Sexual reproduction of some Arthroderma and Nannizzia on diluted Sabouraud agar with or without salts. Mykosen 15:11–17 [DOI] [PubMed] [Google Scholar]
- 28. Tüncher A., Sprote P., Gehrke A., Brakhage A. A. 2005. The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A. nidulans and human. J. Mol. Biol. 352:517–533 [DOI] [PubMed] [Google Scholar]
- 29. Weitzman I., Summerbell R. C. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. White T. C., Oliver B. G., Gräser Y., Henn M. R. 2008. Generating and testing molecular hypotheses in the dermatophytes. Eukaryot. Cell 7:1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamada T., Makimura K., Abe S. 2006. Isolation, characterization, and disruption of dnr1, the areA/nit-2-like nitrogen regulatory gene of the zoophilic dermatophyte, Microsporum canis. Med. Mycol. 44:243–252 [DOI] [PubMed] [Google Scholar]
- 32. Yamada T., et al. 2009. Enhanced gene replacements in Ku80 disruption mutants of the dermatophyte, Trichophyton mentagrophytes. FEMS Microbiol. Lett. 298:208–217 [DOI] [PubMed] [Google Scholar]
- 33. Yamada T., et al. 2009. Agrobacterium tumefaciens-mediated transformation of the dermatophyte, Trichophyton mentagrophytes: an efficient tool for gene transfer. Med. Mycol. 47:485–494 [DOI] [PubMed] [Google Scholar]
- 34. Yamada T., Makimura K., Uchida K., Yamaguchi H. 2005. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes. Med. Mycol. 43:533–544 [DOI] [PubMed] [Google Scholar]
- 35. Zakikhany K., et al. 2007. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell. Microbiol. 9:2938–2954 [DOI] [PubMed] [Google Scholar]