Abstract
Multidrug resistance blaCMY-2 plasmids that confer resistance to expanded-spectrum cephalosporins have been found in multiple bacterial species collected from different hosts worldwide. The widespread distribution of blaCMY-2 plasmids may be driven by antibiotic use that selects for the dissemination and persistence of these plasmids. Alternatively, these plasmids may persist and spread in bacterial populations in the absence of selection pressure if a balance exists among conjugative transfer, segregation loss during cell division, and fitness cost to the host. We conducted a series of experiments (both in vivo and in vitro) to study these mechanisms for three blaCMY-2 plasmids, peH4H, pAR060302, and pAM04528. Results of filter mating experiments showed that the conjugation efficiency of blaCMY-2 plasmids is variable, from <10−7 for pAM04528 and peH4H to ∼10−3 for pAR060302. Neither peH4H nor pAM04528 was transferred from Escherichia coli strain DH10B, but peH4H was apparently mobilized by the coresident trimethoprim resistance-encoding plasmid pTmpR. Competition studies showed that carriage of blaCMY-2 plasmids imposed a measurable fitness cost on the host bacteria both in vitro (0.095 to 0.25) and in vivo (dairy calf model). Long-term passage experiments in the absence of antibiotics demonstrated that plasmids with limited antibiotic resistance phenotypes arose, but eventually drug-sensitive, plasmid-free clones dominated the populations. Given that plasmid decay or loss is inevitable, we infer that some level of selection is required for the long-term persistence of blaCMY-2 plasmids in bacterial populations.
INTRODUCTION
Plasmids found in prokaryotes are typically mobile and variable in size (1.5 to >600 kb) and are generally composed of a “mosaic” of gene sequences that are derived from many sources. This mosaic includes a “plasmid backbone” plus “accessory” genes (36). The backbone includes replication genes plus additional genes for maintenance and transfer capabilities. Accessory genes encode a wide range of phenotypic characteristics that favor survival and reproduction of the host bacterium, such as those encoding catabolic activity, synthetic activity, antimicrobial resistance, and resistance to heavy metals (44, 47). Antibiotic resistance genes encode accessory traits that pose a challenge to human and animal health. One gene of particular interest, blaCMY-2, encodes an AmpC-like β-lactamase that confers resistance to expanded-spectrum cephalosporins (4, 34). blaCMY-2 is often found on large (ca. 140- to 160-kb) IncA/C plasmids (blaCMY-2 plasmids) from a diverse range of bacterial hosts, including Escherichia, Salmonella, Shigella, Klebsiella, Edwardsiella, and Aeromonas (31, 35, 50). These blaCMY-2 plasmid-bearing bacteria have been detected in mammals and fish without a documented or consistent pattern of antibiotic selection pressure (2, 17, 26, 31, 37, 50–52). There is a high prevalence (up to 100%) of blaCMY-2 plasmids reported in commensal and pathogenic bacteria such as Escherichia coli and Salmonella enterica in cattle herds across the United States (1, 15, 30, 42, 51). The blaCMY-2 genes have also been detected in E. coli and Salmonella isolates from ground meat (54). The presence of blaCMY-2 plasmid-bearing Salmonella enterica serovars Newport and Typhimurium presumptively of animal origin has been reported in human clinical cases (17, 18).
The ability of blaCMY-2 plasmids to transfer within and between bacterial populations and the high prevalence of blaCMY-2 plasmids in cattle herds suggest that cattle may act as a reservoir for disseminating blaCMY-2 plasmids or plasmid-bearing bacteria to other animals and human populations. In this context, it is important to note that blaCMY-2 plasmids typically encode multiple antibiotic resistance traits that are already found in terrestrial and aquatic systems (6, 20, 25, 28, 30, 35, 53). Consequently, to invade new populations, blaCMY-2 plasmids should encode a unique trait(s) that allows the plasmid-carrying population to expand where other plasmids cannot convey a similar selective advantage. Resistance to expanded-spectrum cephalosporins is considered the most likely trait, and the use of an expanded-spectrum cephalosporin (ceftiofur) in livestock has been implicated as an important selective pressure responsible for the expansion of blaCMY-2 plasmids in the enteric flora of livestock (40, 43, 48). Nevertheless, when (juvenile and adult) cattle are treated with ceftiofur, the results have not been consistent, suggesting that ceftiofur does not present a strong selective pressure in vivo (9, 24, 43, 48). blaCMY-2 plasmids have also been detected in places where ceftiofur and ceftriaxone are not used, such as in aquaculture farms (Edwardsiella ictaluri isolated from catfish) and in organic cattle farms in the United States (31, 50).
We expect that carriage of plasmids will impose a fitness cost on the host bacterium in the absence of selection (antibiotic) pressure (7, 21, 22, 32). In vitro experiments and mathematical models suggest that the persistence of plasmids over evolutionary time is determined by the combination of (i) the fitness cost to the bacterium due to the plasmid (the cost will be less if the plasmid benefits the host), (ii) the conjugation rate (transfer of the plasmid to new host bacteria), and (iii) plasmid loss due to segregation during cell division (46). While segregation control is needed to retain a plasmid within a given clonal lineage, if the fitness cost due to plasmid carriage is high, then segregation control becomes irrelevant, as the plasmid-bearing bacteria cannot compete with conspecific, plasmid-free bacteria. If plasmids impose a net fitness cost, then plasmid-free segregants will eventually “sweep” through the population and replace the resistant plasmid-bearing strains. Elimination of resistance genes themselves from a plasmid can lower the fitness burden of a plasmid (3, 11), which can also lead to a selective sweep of a less versatile plasmid.
Widespread dissemination and persistence of blaCMY-2 plasmids could be explained by ceftiofur use and/or by other fitness traits that compensate for the burden of plasmid carriage. To begin testing these alternatives, we conducted a series of studies to explore the roles of fitness cost, conjugation efficiency, and the segregation control system in the persistence of multidrug resistance blaCMY-2 IncA/C plasmids peH4H, pAR060302, and pAM04528. Our studies show that in the absence of selection pressure, these blaCMY-2 plasmids confer a measurable fitness cost (in vitro and in vivo) and have variable conjugation efficiencies. Long-term passage without antibiotic selection leads to decay and loss of plasmids. Therefore, for long-term persistence in a bacterial population, blaCMY-2 plasmids clearly require some level of selection pressure.
MATERIALS AND METHODS
We used three sequenced blaCMY-2-positive IncA/C plasmids (5) for our studies, including peH4H (∼148 kb, isolated from dairy cow E. coli strain H4H by the Field Disease Investigation Unit, College of Veterinary Medicine, Washington State University, Pullman, WA), pAM04528 (∼155 kb, isolated from human Salmonella enterica serovar Newport strain AM04528, provided by the Centers for Disease Control and Prevention, Atlanta, GA, and referred to here as pAM), and pAR060302 (∼166 kb, isolated from dairy cow E. coli strain AR060302 by R. Singer, University of Minnesota, Minneapolis, MN, and referred to here as pAR). The H4H strain also harbors an unsequenced plasmid, pTmpR (120 kb), that confers resistance to trimethoprim (5). The following transformants were obtained by introducing these plasmids into laboratory strain E. coli DH10B by electroporation (41): DH10B(peH4H), DH10B(pTmpR), DH10B(pTmpR/peH4H), DH10B(pAM), and DH10B(pAR).
Unless otherwise noted, the culture conditions were 37°C with shaking (200 rpm) and the incubation time varied as indicated. Antibiotic concentrations were 40 μg/ml trimethoprim (Sigma, St. Louis, MO), 20 μg/ml ampicillin (Fisher Biotech, Fair Lawn, NJ), 30 μg/ml chloramphenicol (MP Biomedicals, Solon, OH), 50 μg/ml florfenicol (LKT laboratories, Inc., St. Paul, MN), 8 μg/ml ceftiofur (Sigma-Aldrich, St. Louis, MO), 30 μg/ml kanamycin (Fisher Scientific, Pittsburgh, PA), 20 μg/ml nalidixic acid (MP Biomedicals, Solon, OH), 50 μg/ml rifampin (Sigma-Aldrich, St. Louis, MO), and 30 μg/ml tetracycline (GTS, San Diego, CA).
Assessing the conjugation efficiency of blaCMY-2 plasmids.
To determine the conjugation efficiency of plasmids peH4H, pAR, pAM, and pTmpR, we used filter mating experiments. Briefly, equal quantities of overnight cultures of plasmid-bearing donor strains [DH10B(peH4H), DH10B(pTmpR), DH10B(pTmpR/peH4H), DH10B(pAR), and DH10B(pAM)] and plasmid-free recipient strain DH5α (nalidixic acid resistant [Nalr]) were mixed and added to a nitrocellulose membrane overlaid on LB (Luria-Bertani medium; Fisher Biotech, Fair Lawn, NJ) agar plates without antibiotics. After overnight incubation (37°C), the culture was diluted with sterile phosphate-buffered saline (PBS, pH 7.0, 500 μl) and spread onto LB agar plates containing nalidixic acid, ceftiofur, and/or trimethoprim. Putative transconjugants were enumerated, and the conjugation efficiency of plasmids per donor cell was calculated by dividing the number of CFU of transconjugants by the number of CFU of donors. Each experiment was replicated independently three times. The presence of plasmids in the transconjugants was confirmed for a subset of isolates using plasmid profiles and antibiotic resistance phenotypes (8, 38).
Measurement of the relative fitness cost of blaCMY-2 plasmids.
We employed both in vitro and in vivo competition studies to assess the relative fitness cost of blaCMY-2 plasmids. For in vitro studies, equal numbers of CFU (∼109) of blaCMY-2 plasmid-free E. coli DH10B and each blaCMY-2 plasmid-bearing strain [DH10B(peH4H), DH10B(pTmpR), DH10B(pTmpR/peH4H), DH10B(pAR), or DH10B(pAM)] were added into 5 ml LB broth. The different combinations of cultures were passaged (every 24 h, we transferred 5 μl of cultures into a fresh 5 ml of LB broth) for 8 days. On day 8, the plasmid-bearing and plasmid-free cells were enumerated by serial dilution on antibiotic-containing LB agar plates (27). The fitness cost of blaCMY-2 plasmids was determined as the difference in the number of cell doublings between plasmid-free and plasmid-bearing cells, relative to the number of cell doublings of plasmid-free cells (23), as follows: Cost = [log2 (Ntf/N0f) − log2 (Ntp/N0p)]/[log2 (Ntf/N0f)], where N0f and Ntf are the numbers of plasmid-free cells on days 0 and 8 and N0p and Ntp are the numbers of plasmid-bearing cells on days 0 and 8, respectively. A fitness cost above 0 indicates a possible fitness disadvantage that is associated with the carriage of blaCMY-2 plasmids.
For the in vivo experiments, we needed to compare isogenic clones with and without peH4H. Efforts to cure H4H of peH4H have produced only a single plasmid-free strain that exhibited growth defects (D. R. Call, unpublished data). Therefore, in this experiment, we used peH4H-bearing and peH4H-free E. coli H4H strains; the latter arose spontaneously during long-term passage experiments. Both strains were taken from a passage equivalent to ∼ 2,360 generations (H4H2360). In vivo experiments included neonatal calves (2 to 7 days old; n = 5). Both strains retained trimethoprim resistance (i.e., pTmpR was present), and the peH4H-free strain was selected for nalidixic acid resistance (Nalr) (9). Calves (fecal samples) were prescreened for the presence of trimethoprim- and nalidixic acid- or trimethoprim- and ceftiofur-resistant E. coli strains and found to be apparently free of bacteria with these combinations of resistance traits. Equal numbers of CFU (1010) of both peH4H-free and peH4H-bearing E. coli H4H2360 (in 20 ml sterile PBS) were orally administrated to calves using a sterile 12-ml syringe. Fresh fecal material (∼20 g) was collected from the rectum aseptically, and the numbers of CFU of the two E. coli H4H2360 strains were calculated using MacConkey agar plates containing selective antibiotics. Briefly, 1 g of freshly collected fecal sample was diluted in peptone-buffered saline and 10-fold dilutions were plated on selective agar plates. Competitive-index (CI) values within animals across time points were calculated (27). We used pulsed-field gel electrophoresis (PFGE) with XbaI and the PulseNet protocol (10) to verify the strain identities of H4H2360 at the start and end of the experiments. During this experiment, calves were housed individually in vivarium rooms (one calf per room) and fed fresh bulk milk from the Washington State University Dairy. Alfalfa hay, calf pellet rations, and water were provided ad libitum. There were no antibiotic exposures. Animal inoculation studies were approved by the Washington State University Institutional Animal Care and Use Committee.
Long-term passage of blaCMY-2 plasmids.
To evaluate the impact of alternative mechanisms on the persistence of blaCMY-2 plasmids, we passaged the blaCMY-2 plasmid-bearing (wild-type and transformant) bacterial strains, H4H, AM04528, and AR060302 [DH10B(peH4H), DH10B(pTmpR/peH4H), DH10B(pAR), and DH10B(pAM)], in LB broth without antibiotic selection pressure by using the method of De Gelder et al. (14). Briefly a single colony from a freshly streaked freezer stock was inoculated into 5 ml LB broth and incubated at 37°C. After 24 h of incubation, 5 μl of culture was transferred into a fresh 5 ml of LB broth and the passage was continued until 4,000 generations for H4H and DH10B (peH4H- and pTmpR-carrying strains) and up to 2,600 generations for the other strains. Using CFU counts, we estimated a priori that each passage in this system resulted in approximately 10 generations in 24 h (the number of CFU increased from ∼106 to ∼109 in 24 h, which is equivalent to ∼10 doublings). During long-term passage, approximately every 200 generations (20 days), we selected 48 or 96 colonies and inoculated them onto LB agar plates with and without antibiotics (ampicillin, cefoxitin-ceftiofur, florfenicol-chloramphenicol, tetracycline, kanamycin, and trimethoprim) by using a 96-pin replicator. The percentage of antibiotic resistance phenotypes in each population was subsequently calculated, and it was assumed that the assay provided an analytic sensitivity of 1 to 2 orders of magnitude (102). Antibiotic-resistant and -sensitive colonies were tested periodically for the presence or absence of plasmids by plasmid profile determination, followed by Southern blot hybridization using a blaCMY-2-specific probe and a commercial kit (Roche PCR DIG probe synthesis kit and DIG Easy Hyb; Roche Diagnostics GmbH, Penzberg, Germany) (8).
RESULTS
Conjugation efficiency varies.
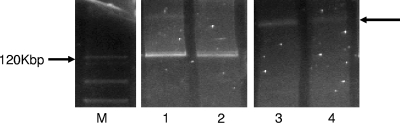
In the absence of antibiotic selection, the conjugation efficiency varied between plasmids. In wild-type strains, we observed low conjugation efficiencies (2.8 × 10−7 to 1.3 × 10−7 for peH4H and 1.2 × 10−8 to 1.2 × 10−7 for pAM) to moderate efficiency for pAR (9.2 × 10−4 to 2.1 × 10−3) and pTmpR (5.6 × 10−3 to 8.0 × 10−3). When E. coli DH10B was used as the donor strain, neither pH4H nor pAM transconjugants were detected while pAR and pTmpR had conjugation efficiencies similar to those observed with the wild-type strains (1.8 × 10−4 to 1.3 × 10−3 and 5.6 × 10−3 to 8.0 × 10−3, respectively). Interestingly, while peH4H conjugation from DH10B was not observed, when we combined peH4H and pTmpR in the DH10B host, we found transconjugants (2.4 × 10−8 to 7.0 × 10−8) that harbored either both peH4H and pTmpR or a larger plasmid that presumably represented a chimera of these two plasmids (∼2/3 of the transconjugant population) (Fig. 1), consistent with pTmpR contributing to the mobilization of peH4H.
Fig. 1.
Plasmid profile of H4H::DH5α transconjugants obtained from conjugation experiments. Lanes: M, BAC-Tracker supercoiled DNA ladder; 1, plasmids peH4H and pTmpR from wild-type strain H4H; 2, plasmids peH4H and pTmpR from H4H::DHα transconjugants; 3 and 4, chimera plasmids (see arrow on right) from H4H::DH5α transconjugants.
Measurable fitness cost involved with carriage of blaCMY-2 IncA/C plasmids.
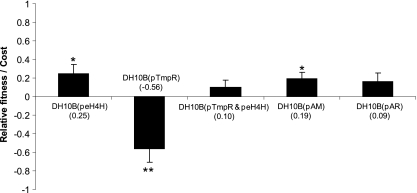
In vitro direct competition studies between plasmid-free and plasmid-bearing E. coli strains showed that there is a measurable fitness cost associated with the carriage of blaCMY-2 plasmids. Plasmid peH4H showed a higher cost (0.25) than plasmids pAR (0.09) and pAM (0.19). In contrast, plasmid pTmpR conferred a clear fitness advantage (−0.56) on the host and appeared to mitigate the fitness cost of peH4H (0.10) when these two plasmids were coresident (Fig. 2). The apparent advantage of pTmpR could be explained in part by “invasion” of plasmid-free competing bacteria by conjugation during the competition assay. To determine if conjugative transfer confounded the assay by creating more plasmid-bearing strains and producing an overestimated fitness advantage for pTmpR, we repeated the experiment using a recipient DH10B strain that could be distinguished from the plasmid-bearing host due to its resistance to rifampin. After 4 days of passage, we found no DH10B(pTmpR) Rifr transconjugants in the population, indicating that conjugation did not significantly affect the original fitness results. Therefore, we conclude that in the absence of antibiotic selection, the three blaCMY-2 plasmids imposed a fitness cost on the E. coli host while pTmpR provided a fitness benefit (Fig. 2).
Fig. 2.
Fitness cost of plasmids blaCMY-2 and pTmpR measured during in vitro competition studies with plasmid-bearing and plasmid-free E. coli DH10B. The x axis shows plasmid-bearing E. coli DH10B competition against plasmid-free E. coli DH10B, and the y axis shows the relative fitness or cost (see Materials and Methods). Carriage of peH4H, pAM, and pAR imposes a relative fitness disadvantage on E. coli DH10B, and carriage of pTmpR imposes a fitness advantage. *, P < 0.05; **, P < 0.001 (Student's t test).
Plasmid peH4H does not provide an obvious fitness advantage to E. coli strain H4H2360 in calves.
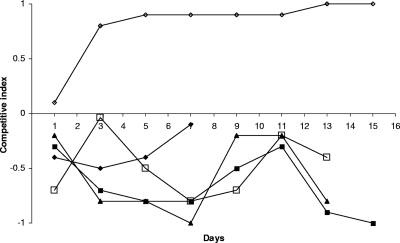
To identify the effect of plasmid peH4H on host fitness under in vivo conditions, we used neonatal calves to conduct competition studies with E. coli H4H bacteria that either had the peH4H plasmid or did not (both strains still harbored pTmpR and were isolated after 2,360 generations of in vitro passage); these bacteria were designated H4H2360). The results showed that in 4 out of 5 calves, the E. coli H4H bacteria that harbored peH4H were outnumbered by the peH4H-free bacteria and did not persist as long as their peH4H-free competitors (Fig. 3). In one calf, the peH4H-bearing bacteria remained dominant in the population. It appears that carriage of peH4H provided a fitness disadvantage from day 1 onward (4/5 calves), and thus, on average, the peH4H-bearing E. coli bacteria lost the competition against the peH4H-free bacteria (Fig. 3). PFGE verified that the H4H2360 bacteria at the start and end of the experiment were identical (data not shown). At 15 days postinoculation, neither type was detected in the calves, suggesting that long-term persistence in calves may be limited in the absence of antibiotic selection pressure.
Fig. 3.
CIs measured during in vivo competition between plasmid peH4H-bearing and peH4H-free E. coli H4H2360. The x axis shows days on which the CIs were calculated, and the y axis shows that the CIs ranged between −1 and +1. The different symbols represent individual animals. A CI value above 0 indicates that plasmid peH4H-bearing strain H4H dominates, while a value below 0 indicates that peH4H-free strain H4H2360 dominates. We did not find strain H4H2360 after day 7 in one animal and after day 15 in two animals.
Emergence of antibiotic-sensitive bacterial phenotypes during long-term passage.
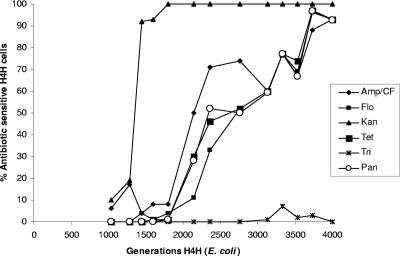
To study the effects of fitness cost, segregation control, and conjugation on plasmid persistence in vitro, we conducted long-term passage experiments with the three blaCMY-2 plasmid-bearing bacterial strains H4H, AM04528, and AR060302. The findings showed that in two trials, different antibiotic sensitivity phenotypes emerged in the three strains at different generations during the long-term passage, as might be expected for a random mutation process (Fig. 4 and 5; Tables 1 and 2). For strain H4H, kanamycin-sensitive (Kans) derivatives were detected (10% of the cells) at around 1,000 generations and swept through the population (∼100% of the cells) by generation 1,600. Ampicillin- and ceftiofur-sensitive (Amps Cefs) cells (∼6% of the cells) were detected at around 1,040 generations and gradually increased their proportion to a majority level (∼93% of the cells) by 4,000 generations. Tetracycline- and florfenicol-sensitive (Tets and Flos) H4H cells were detected at around 1,450 and 1,600 generations, respectively, and these phenotypes gradually increased and dominated the population (93% of the cells) by 4,000 generations. In addition, we found that at 4,000 generations, most (∼93%) of the cells were sensitive to all of the antibiotics tested and we initially detected (∼28% of the cells) this sensitive phenotype at ∼2,140 generations. As expected from competition studies (Fig. 2), most (∼100%) of the H4H cells throughout the experiment were resistant to trimethoprim and retained the pTmpR plasmid up to 4,000 generations, except for a small fluctuation (∼1 to 7% of the cells picked) between 3,100 and 3,700 generations (Fig. 4). A plasmid profile followed by Southern blotting of the antibiotic-sensitive and -resistant H4H strains isolated during the passage experiment showed that the antibiotic-resistant cells had retained and the antibiotic-sensitive cells had lost plasmid peH4H (n = 6 isolates) (data not shown). In addition, identical PFGE profiles confirmed that the chromosomal background of the resistant and sensitive H4H strains was the same at the start and end of passage experiments (n = 6 isolates), indicating that sensitive strains were unlikely to be contaminants (data not shown). When in vitro passage experiments were done with host strain DH10B (with both pTmpR and peH4H), the antibiotic sensitivity phenotypes emerged in a pattern similar to the dynamics of the H4H phenotypes described above (Table 2).
Fig. 4.
Percentages of antibiotic-sensitive E. coli H4H phenotypes (y axis) in different generations (x axis) during long-term passage culture (without antibiotic selection). A gradual increase in antibiotic-sensitive populations was observed after 1,000 generations, and most isolates became pansensitive after 3,000 generations. In contrast, most isolates were resistant to trimethoprim and retained pTmpR even after 4,000 generations. Amp/CF, proportion of isolates sensitive to ampicillin-ceftiofur; Kan, proportion of isolates sensitive to kanamycin; Tet, proportion of isolates sensitive to tetracycline; Flo, proportion of isolates sensitive to florfenicol; Tri, proportion of isolates sensitive to trimethoprim; Pan, proportion of isolates sensitive to Amp-Cef, Kan, Tet, and Flo (Tri sensitivity was excluded from this calculation).
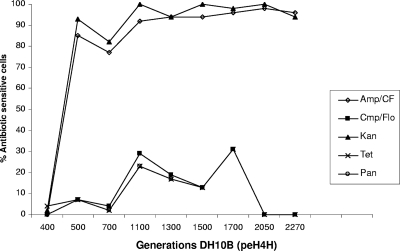
Fig. 5.
Percentages of antibiotic-sensitive E. coli DH10B(peH4H) phenotypes (y axis) in different generations (x axis) during the long-term passage experiment. After 500 generations, there was a rapid increase (up to >80% and >90%) of ampicillin-ceftiofur- and kanamycin-sensitive DH10B(peH4H) isolates, respectively. The florfenicol- and tetracycline-sensitive subpopulations reached >80% of the culture after 1,100 generations. Amp/CF, proportion of isolates sensitive to ampicillin-ceftiofur; Kan, proportion of isolates sensitive to kanamycin; Tet, proportion of isolates sensitive to tetracycline; Flo, proportion of isolates sensitive to florfenicol; Tri, proportion of isolates sensitive to trimethoprim; Pan, proportion of isolates sensitive to Amp-Cef, Kan, Tet, and Flo (Tri sensitivity was excluded from this calculation).
Table 1.
Percentages of bacteria of strains AM04528, AR060302, DH10B(pAM), and DH10B(pAR) with antibiotic sensitivity phenotypes in 2 independent trials during long-term passage experiments
| Trial and no. of generationsa | % of bacteria with antibiotic sensitivity phenotype |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM04528 |
AR060302 |
DH10B(pAM) |
DH10B(pAR) |
|||||||||||||
| Amps Cefsb | Flos | Tets | Pans | Amps Cefs | Flos | Tets | Pans | Amps Cefs | Flos | Tets | Pans | Amps Cefs | Flos | Tets | Pans | |
| Trial 1 | ||||||||||||||||
| 200 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| 400 | 88 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 98 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| 600 | 89 | 0 | 0 | 0 | 73 | 0 | 0 | 0 | 98 | 2 | 2 | 2 | 100 | 0 | 0 | 0 |
| 800 | 98 | 25 | 25 | 25 | 83 | 3 | 0 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| 1,000 | 98 | 8 | 2 | 2 | 99 | 8 | 0 | 0 | 92 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| 1,200 | 90 | 4 | 4 | 4 | 83 | 0 | 0 | 0 | 88 | 0 | 0 | 0 | 96 | 0 | 0 | 0 |
| 1,400 | 50 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 98 | 21 | 21 | 21 |
| Trial 2 | ||||||||||||||||
| 400 | 68 | 0 | 0 | 0 | 84 | 0 | 0 | 0 | 46 | 0 | 0 | 0 | 72 | 0 | 0 | 0 |
| 600 | 86 | 0 | 0 | 0 | 86 | 0 | 0 | 0 | 88 | 0 | 0 | 0 | 86 | 0 | 0 | 0 |
| 800 | 94 | 0 | 0 | 0 | 68 | 0 | 0 | 0 | 92 | 0 | 0 | 0 | 68 | 0 | 0 | 0 |
| 1,000 | 96 | 0 | 0 | 0 | 92 | 0 | 0 | 0 | 92 | 0 | 0 | 0 | 94 | 0 | 0 | 0 |
| 1,200 | 92 | 48 | 48 | 46 | 88 | 12 | 12 | 0 | 86 | 18 | 18 | 18 | 86 | 22 | 18 | 18 |
| 1,400 | 85 | 96 | 96 | 96 | 85 | 0 | 0 | 0 | 50 | 100 | 100 | 50 | 66 | 0 | 0 | 0 |
| 1,600 | 92 | 90 | 80 | 80 | 83 | 90 | 0 | 68 | 83 | 85 | 96 | 83 | 94 | 80 | 94 | 80 |
| 1,800 | 92 | 92 | 86 | 86 | 100 | 88 | 86 | 86 | 88 | 88 | 92 | 88 | 92 | 94 | 100 | 92 |
| 2,000 | 92 | 92 | 88 | 88 | 100 | 85 | 88 | 85 | 86 | 88 | 88 | 86 | 96 | 88 | 88 | 88 |
| 2,200 | 96 | 96 | 92 | 92 | 90 | 85 | 94 | 85 | 92 | 92 | 94 | 92 | 88 | 92 | 92 | 88 |
| 2,400 | 100 | 75 | 92 | 75 | 82 | 100 | 85 | 82 | 96 | 88 | 98 | 82 | 98 | 92 | 94 | 92 |
| 2,600 | 100 | 98 | 100 | 98 | 88 | 88 | 90 | 88 | 88 | 96 | 96 | 88 | 94 | 96 | 96 | 94 |
A generation is binary fission. Based on 1:210 serial dilutions, we calculated that 24 h is equivalent to approximately 10 generations.
Percentages of bacteria with antibiotic sensitivity phenotypes were calculated by selecting a subset of 48 to 96 isolates from the population approximately for every 200 generations and characterizing the antibiotic susceptibility profiles. Amps Cefs, sensitivity to ampicillin-ceftiofur; Tets, sensitivity to tetracycline; Flos, sensitivity to florfenicol; Pans, sensitivity to Amp-Cef, Tet, and Flo.
Table 2.
Percentages of strain DH10B(pTmpR/peH4H) bacteria with antibiotic sensitivity phenotypes in 2 independent trials during long-term passage experiments
| Trial and no. of generationsa | % of bacteria with phenotype |
|||||
|---|---|---|---|---|---|---|
| Amps Cefsb | Flos | Kans | Tets | Tris | Pans | |
| Trial 1 | ||||||
| 1,300 | 9 | 0 | 100 | 0 | 0 | 0 |
| 1,420 | 30 | 0 | 100 | 0 | 0 | 0 |
| 1,650 | 67 | 0 | 100 | 0 | 0 | 0 |
| 2,000 | 75 | 0 | 100 | 0 | 0 | 0 |
| 2,370 | 65 | 65 | 100 | 73 | 7 | 65 |
| 2,700 | 91 | 79 | 100 | 95 | 4 | 79 |
| 3,000 | 100 | 92 | 100 | 93 | 0 | 92 |
| 3,270 | 100 | 91 | 100 | 91 | 0 | 91 |
| 3,400 | 97 | 85 | 100 | 92 | 0 | 85 |
| 4,000 | 97 | 85 | 100 | 92 | 0 | 85 |
| Trial 2 | ||||||
| 400 | 0 | 0 | 2 | 4 | 0 | 0 |
| 500 | 85 | 7 | 93 | 7 | 0 | 7 |
| 700 | 77 | 4 | 82 | 2 | 0 | 2 |
| 1,100 | 92 | 29 | 100 | 23 | 0 | 23 |
| 1,300 | 94 | 19 | 94 | 17 | 0 | 17 |
| 1,500 | 94 | 13 | 100 | 13 | 0 | 13 |
| 1,700 | 96 | 31 | 98 | 31 | 0 | 31 |
| 2,050 | 98 | 0 | 100 | 0 | 0 | 0 |
| 2,270 | 96 | 0 | 94 | 0 | 0 | 0 |
A generation is binary fission. Based on 1:210 serial dilutions, we calculated that 24 h is equivalent to approximately 10 generations.
Percentages of bacteria with antibiotic sensitivity phenotypes were calculated by selecting a subset of 48 to 96 isolates from the population approximately for every 200 generations and characterizing the antibiotic susceptibility profiles. Amps Cefs, sensitivity to ampicillin-ceftiofur; Kans, sensitivity to kanamycin; Tets, sensitivity to tetracycline; Flos, sensitivity to florfenicol; Tris, sensitivity to trimethoprim; Pans, sensitivity to Amp-Cef, Kan, Tet, and Flo (Tris was excluded from this category).
Strains AM04528 and AR060302 showed a sharp increase in Amps Cefs phenotypes at around 400 and 600 generations, respectively, in the first long-term passage trial (∼88% of the cells of strain AM04528 and ∼73% of the cells of strain AR060302). These populations were largely replaced by these Amps Cefs AM04528 and AR060302 phenotypes by 800 and 1,000 generations, respectively (Table 1). Interestingly almost 100% of the AM04528 and AR060302 cells were resistant to tetracycline (Tetr) and florfenicol (Flor), except for a minor fluctuation in the Flos phenotype between 800 and 1,250 generations (Table 1). Overall, the peH4H, pAM, and pAR plasmid-bearing E. coli DH10B populations showed similar patterns of the emergence of antibiotic sensitivity phenotypes, but these emerged more quickly than in their wild-type counterparts (Table 1).
DISCUSSION
The most persistent and successful plasmids have a strong segregation control system, a low fitness cost, and a high conjugation rate (45). We studied these traits for the apparently successful multidrug resistance blaCMY-2 IncA/C plasmids peH4H, pAM04528 (pAM), and pAR060302 (pAR). The conjugation efficiency of the three blaCMY-2 plasmids varied from <10−7 to a moderate level (10−3). The latter observation is consistent with the findings of Fricke et al. (19), who also found that the conjugation efficiency of IncA/C plasmids was moderate (10−3) compared to that of more efficient plasmids such as when pB10 (64-kbp IncP-1β plasmid) was transferred to strains of E. coli, Pseudomonas spp., Sinorhizobium meliloti, and Stenotrophomonas maltophilia (>10−1) (16).
Apparent loss of genes that are associated with conjugation and an extensively truncated traC gene may be the reason for the low conjugation efficiency of peH4H and pAM, respectively (5, 26, 39). No transconjugants of peH4H or pAM were detected when these plasmids resided singly in DH10B, and this host strain appeared to be a competent host considering that pAR had no reduction in conjugation efficiency from DH10B. When we added pTmpR with peH4H to the DH10B host, conjugation was successful and this indicates that the coresident plasmid pTmpR probably mobilizes peH4H into other plasmid-free bacteria. Interestingly, for at least 2/3 of these transconjugants, we found a plasmid that is larger (>165 kb) than both plasmids peH4H and pTmpR. The antibiotic resistance pattern and the plasmid profile suggest that these large plasmids are the result of a chimera of plasmids peH4H and pTmpR (Fig. 1). The efficiency of mobilization of peH4H and associated chimeras was 4 orders of magnitude less than that of pTmpR alone. Plasmid pTmpR is coresident with peH4H in wild-type strain H4H. Poole et al. (39) also reported that conjugation-deficient IncA/C plasmids could be mobilized in the presence of compatible conjugative plasmids.
In vitro direct competition studies of blaCMY-2 plasmid-bearing and plasmid-free E. coli DH10B showed a measurable fitness cost (0.09 to 0.25) associated with carriage of the three blaCMY-2 plasmids (peH4H, pAM, and pAR) (Fig. 2). The larger size of blaCMY-2 plasmids and the absence of selection pressure could be the reasons for the significant fitness cost of these blaCMY-2 plasmids. Similar findings were obtained by De Gelder et al., who reported that the cost of plasmid pB10 varies (low to high) for different bacterial hosts. This ranged from a low level (<0.03) for E. coli K-12 and Sinorhizobium meliloti to a moderate level for Pseudomonas spp. (0.037 to 0.15) and a high level for Stenotrophomonas maltophilia (0.59) (13, 23). The reduction in the fitness cost of peH4H upon coresidence with pTmpR may be due to mitigation by the benefits conferred by pTmpR (Fig. 2), although the mechanism underlying the pTmpR fitness advantage has not been determined. In vivo studies using neonatal calves showed a stochastic outcome where blaCMY-2 plasmid-bearing strain H4H2360 was dominated by its peH4H-free counterpart for 4 out of 5 calves (Fig. 3). Overall, the findings of both in vivo and in vitro competition studies indicate a measurable fitness disadvantage associated with the carriage of blaCMY-2 plasmids to the bacterial host. If the in vivo results are representative of events occurring in cattle populations, then the fitness cost associated with blaCMY-2 plasmids should lead to eventual loss of these plasmids from the cattle population if there are no other selection pressures for the maintenance of these traits (Fig. 2).
The long-term passage of blaCMY-2 plasmid-bearing bacterial strains (H4H, AR060302, and AM04528 and their DH10B counterparts) showed a slow emergence of different antibiotic sensitivity phenotypes in all of the populations tested. Similar findings were also reported in various long-term passage (500 to 1,100 generations) studies conducted with conjugative multidrug resistance plasmids such as pB10, pACYC184, and pBR322 in E. coli and other bacterial species (7, 13, 33, 49). The reduced prevalence of the peH4H plasmid in the population is consistent with the finding of De Gelder et al. (12), who detected the emergence of pB10-free E. coli K-12 at around 500 generations. In the present study, however, plasmid-free (peH4H, pAM, and pAR) strains typically did not emerge until between 800 and 1,200 generations, which is consistent with a relatively high level of plasmid stability.
The long-term passage experiments showed that in the absence of selection pressure, plasmids can shed antibiotic resistance genes, resulting in a presumably less costly plasmid. The arrangements of these antibiotic resistance genes between the mobile elements on the IncA/C backbone may contribute to the differential extrication of these genes from the plasmid backbone (5). For example, early loss of the kanamycin resistance gene may be due to its position between the two direct terminal repeats (insB3 and insB4), leading to instability of the transposable kanamycin resistance elements through homologous intermolecular recombination, as has been shown for Tn1525 (29). Similarly, De Gelder et al. (12) reported the loss of a region containing tet(A) and tetR, which were flanked by long repeat sequences. The blaCMY-2-containing sequence is flanked by inversely oriented insertion sequences (ISEcp1), indicating that it may be a composite transposon that favors the extrication of blaCMY-2 genes from the plasmid backbone (26). The Flor Tetr genes are also flanked by insertion sequences insB1 and insB2. The sequential emergence of Kans, Amps Cefs, and Flos Tets phenotypes probably reflects a combination of the frequency of recombination (loss) and the relative fitness cost of the plasmid segments that encode these resistance traits. To examine this question further, we hybridized four isolates (representing different antibiotic resistance phenotypes) from generation 2,360 to an oligonucleotide microarray that included probes for the open reading frames from peH4H (unpublished data). As expected, insertion sequences with antibiotic resistance genes were lost in sensitive strains that still harbored a peH4H plasmid. For example, a ceftiofur-sensitive isolate lost the blaCMY-2 insert region. A tetracycline- and florfenicol-sensitive strain harbored a peH4H plasmid that was missing the tet(A)-floR region and the blaTEM-1 region, but resistance to ceftiofur and ampicillin was retained because the blaCMY-2 region was retained. Another isolate, which was sensitive to ampicillin, ceftiofur, tetracycline, and florfenicol, showed loss of all associated regions of the plasmid. All of these strains were sensitive to kanamycin, and three of four were missing all or part of the region encompassing aacC. The one exception was a kanamycin-sensitive strain that showed no apparent loss of resistance genes, but our microarray was not complete for this region and this result may reflect an inability of our microarray to detect single-nucleotide changes or small indels that could render a gene nonfunctional. In totality, these findings are consistent with earlier observations that blaCMY-2-positive IncA/C plasmids differ largely by the loss or gain of resistance traits that are encoded by insertion sequences (5, 19).
Although the pattern of emergence of antibiotic-sensitive DH10B was similar in the long-term passage culture, the earlier and rapid emergence and dominance of Kans and Amps Cefs blaCMY-2 plasmid-bearing E. coli DH10B cells compared to their wild-type counterparts is not unexpected because E. coli DH10B is not the natural host of these blaCMY-2 plasmids (Fig. 5; Tables 1 and 2). In fact, it is surprising that the blaCMY-2 plasmids were retained in the populations for several hundred generations. Overall, the long-term passage culture experiments in the absence of selection suggest that the plasmids are very stable but that random mutations accumulate in the plasmid, leading to plasmid decay, and eventually the entire plasmids were lost from the populations (2, 9, 13, 24, 29). We observed some variance in the loss of different resistance phenotypes during the long-term passage trials. We attributed this variance to the timing of losses (a random process) and the different combinations of mutations coupled with the population bottleneck that occurred during every 24-h passage to fresh medium.
While long-term passage experiments indicate that blaCMY-2 plasmids decay or are lost completely, presumptively intact blaCMY-2 plasmids were recovered beyond 800 generations. Studies with strain H4H show that the fitness cost of peH4H can be mitigated in part by another plasmid. Furthermore, while peH4H-free strains dominated in the calf model, this was not the case for one calf. We surmise that even though loss of blaCMY-2 plasmids is a deterministic outcome, in the absence of selection, stochastic events and periodic selection (e.g., ceftiofur use) will most likely prolong persistence and this probably contributes to the successful proliferation of blaCMY-2 plasmids.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Lisa Orfe and Patrick Friel.
This project was supported in part by the National Institutes of Health, U.S. Department of Health and Human Services, under contract No1-AI-30055 and by the Agricultural Animal Health Program, College of Veterinary Medicine, and College of Agriculture, Human, and Natural Resource Sciences Agriculture Research Center, Washington State University, Pullman. E. Top was also in part supported by award R01AI084918 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Alcaine S. D., et al. 2005. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob. Agents Chemother. 49:4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barlow M., Hall B. G. 2002. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bergstrom C. T., Lipsitch M., Levin B. R. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155:1505–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bush K., Jacoby G. A., Medeiros A. A. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Call D. R., et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carattoli A., et al. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 12:1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dahlberg C., Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daniels J. B., Call D. R., Besser T. E. 2007. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl. Environ. Microbiol. 73:8005–8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daniels J. B., et al. 2009. Role of ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75:3648–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis M. A., et al. 2009. Multilocus variable-number tandem-repeat method for typing Salmonella enterica serovar Newport. J. Clin. Microbiol. 47:1934–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis M. A., Cloud-Hansen K. A., Carpenter J., Hovde C. J. 2005. Escherichia coli O157:H7 in environments of culture-positive cattle. Appl. Environ. Microbiol. 71:6816–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Gelder L., et al. 2004. Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics 168:1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Gelder L., Ponciano J. M., Joyce P., Top E. M. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153:452–463 [DOI] [PubMed] [Google Scholar]
- 14. De Gelder L., Williams J. J., Ponciano J. M., Sota M., Top E. M. 2008. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178:2179–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donaldson S. C., et al. 2006. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72:3940–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dröge M., Puhler A., Selbitschka W. 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263:471–482 [DOI] [PubMed] [Google Scholar]
- 17. Egorova S., et al. 2008. Ceftriaxone-resistant Salmonella enterica serotype Newport, France. Emerg. Infect. Dis. 14:954–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fey P. D., et al. 2000. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 342:1242–1249 [DOI] [PubMed] [Google Scholar]
- 19. Fricke W. F., et al. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Galimand M., et al. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337:677–680 [DOI] [PubMed] [Google Scholar]
- 21. Godwin D., Slater J. H. 1979. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K12. J. Gen. Microbiol. 111:201–210 [DOI] [PubMed] [Google Scholar]
- 22. Helling R. B., Kinney T., Adams J. 1981. The maintenance of plasmid-containing organisms in populations of Escherichia coli. J. Gen. Microbiol. 123:129–141 [DOI] [PubMed] [Google Scholar]
- 23. Heuer H., Fox R. E., Top E. M. 2007. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 59:738–748 [DOI] [PubMed] [Google Scholar]
- 24. Jiang X., Yang H., Dettman B., Doyle M. P. 2006. Analysis of fecal microbial flora for antibiotic resistance in ceftiofur-treated calves. Foodborne Pathog. Dis. 3:355–365 [DOI] [PubMed] [Google Scholar]
- 25. Kaldhone P., et al. 2008. Characterization of Salmonella enterica serovar Heidelberg from turkey-associated sources. Appl. Environ. Microbiol. 74:5038–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang M. S., Besser T. E., Call D. R. 2006. Variability in the region downstream of the blaCMY-2 beta-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khachatryan A. R., Hancock D. D., Besser T. E., Call D. R. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim M. J., et al. 2008. Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Labigne-Roussel A., Briaux-Gerbaud S., Courvalin P. 1983. Tn1525, a kanamycin R determinant flanked by two direct copies of IS15. Mol. Gen. Genet. 189:90–101 [DOI] [PubMed] [Google Scholar]
- 30. Lindsey R. L., Fedorka-Cray P. J., Frye J. G., Meinersmann R. J. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75:1908–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McIntosh D., et al. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61:1221–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Modi R. I., Wilke C. M., Rosenzweig R. F., Adams J. 1991. Plasmid macro-evolution: selection of deletions during adaptation in a nutrient-limited environment. Genetica 84:195–202 [DOI] [PubMed] [Google Scholar]
- 33. Modi R. I., Adams J. 1991. Coevolution in bacteria-plasmid populations. Evolution 45:656–667 [DOI] [PubMed] [Google Scholar]
- 34. Nordmann P. 1998. Trends in beta-lactam resistance among Enterobacteriaceae. Clin. Infect. Dis. 27(Suppl. 1):S100–S106 [DOI] [PubMed] [Google Scholar]
- 35. O'Brien T. F., et al. 1982. Molecular epidemiology of antibiotic resistance in salmonella from animals and human beings in the United States. N. Engl. J. Med. 307:1–6 [DOI] [PubMed] [Google Scholar]
- 36. Osborn M., et al. 2000. The evolution of bacterial plasmids, p. 301–361 In Thomas C., et al. (ed.), The horizontal gene pool . Harwood Academic Publishers, Amsterdam, The Netherlands [Google Scholar]
- 37. Pan J. C., et al. 2008. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 52:3829–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phornphisutthimas S., Thamchaipenet A., Panijpan B. 2007. Conjugation in Escherichia coli. Biochem. Mol. Biol. Educ. 35:440–445 [DOI] [PubMed] [Google Scholar]
- 39. Poole T. L., et al. 2009. Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack bla(CMY) in the A/C plasmid backbone. Foodborne Pathog. Dis. 6:1185–1194 [DOI] [PubMed] [Google Scholar]
- 40. Raymond M. J., Wohrle R. D., Call D. R. 2006. Assessment and promotion of judicious antibiotic use on dairy farms in Washington State. J. Dairy Sci. 89:3228–3240 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY [Google Scholar]
- 42. Sawant A. A., et al. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 73:156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singer R. S., Patterson S. K., Wallace R. L. 2008. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 74:6956–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smalla K., Osborn A., Wellington E. 2000. Isolation and characterization of plasmids from bacteria, p. 207–248 In Thomas C. (ed.), The horizontal gene pool . Harwood Academic Publishers, Amsterdam, The Netherlands [Google Scholar]
- 45. Sørensen S. J., Bailey M., Hansen L. H., Kroer N., Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3:700–710 [DOI] [PubMed] [Google Scholar]
- 46. Stewart F. M., Levin B. R. 1977. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87:209–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas C. M., Helinski D. R. 1989. Vegetative replication and stable inheritance of IncP plasmids, p. 1–25 In Thomas C. M. (ed.), Promiscuous plasmids of Gram-negative bacteria. Academic Press, London, United Kingdom [Google Scholar]
- 48. Tragesser L. A., Wittum T. E., Funk J. A., Winokur P. L., Rajala-Schultz P. J. 2006. Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67:1696–1700 [DOI] [PubMed] [Google Scholar]
- 49. Turner P. E., Cooper V. S., Lenski R. E. 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52:315–329 [DOI] [PubMed] [Google Scholar]
- 50. Welch T. J., et al. 2009. IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen Edwardsiella ictaluri. Antimicrob. Agents Chemother. 53:845–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Welch T. J., et al. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winokur P. L., et al. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. You Y., et al. 2006. Survival of Salmonella enterica serovar Newport in manure and manure-amended soils. Appl. Environ. Microbiol. 72:5777–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao S., et al. 2001. Identification and expression of cephamycinase bla(CMY) genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]