Abstract
The Nbs1 complex is an evolutionarily conserved multisubunit nuclease composed of the Mre11, Rad50, and Nbs1 proteins. Hypomorphic mutations in the NBS1 or MRE11 genes in humans result in conditions characterized by DNA damage sensitivity, cell cycle checkpoint deficiency, and high cancer incidence. The equivalent complex in the yeast Saccharomyces cerevisiae (Xrs2p complex) has been implicated in DNA double-strand break repair and in telomere length regulation. Here, we find that xrs2Δ, mre11Δ, and rad50Δ mutants are markedly defective in the initiation of the intra-S phase checkpoint in response to DNA damage. Furthermore, the absence of a functional Xrs2p complex leads to sensitivity to deoxynucleotide depletion and to an inability to efficiently slow down cell cycle progression in response to hydroxyurea. The checkpoint appears to require the nuclease activity of Mre11p and its defect is associated with the abrogation of the Tel1p/Mec1p signaling pathway. Notably, DNA damage induces phosphorylation of both Xrs2p and Mre11p in a Tel1p-dependent manner. These results indicate that the Tel1p/ATM signaling pathway is conserved from yeast to humans and suggest that the Xrs2p/Nbs1 complexes act as signal modifiers.
Keywords: Xrs2p complex, checkpoint, Nijmegen breakage syndrome, Mre11, ATM
Eukaryotic cells have evolved surveillance mechanisms called checkpoints to monitor the integrity of their genome. The DNA damage checkpoint consists of a network of signaling proteins that acts to detect DNA lesions and signal their presence to cell cycle regulators (Michelson and Weinert 2000). This results in the slowing down of the cell cycle, which is thought to provide time for DNA repair factors to prevent the damage becoming fixed as a permanent genetic change (for review, see Foiani et al. 2000).
The mechanisms leading to the detection of DNA lesions by checkpoint factors and the identity of the proteins responsible for the initiation of the checkpoint response have yet to be clarified. However, recent evidence indicates that in many cases the formation of DNA lesions per se may be insufficient to initiate the transduction cascade leading to cell cycle arrest in damaged cells. Indeed, in Saccharomyces cerevisiae rad14 or rad2 mutants, irreparable UV damage remains undetected until the early stages of S phase, whereas nucleotide excision repair (NER)-proficient cells detect this type of damage readily in G1 (Siede et al. 1994; Neecke et al. 1999). Similarly, activation of the p53 response in UV-treated human NER-deficient (XPA−) cells is not observed in the absence of DNA replication (Nelson and Kastan 1994). Together, these observations indicate that UV lesions have to be processed to a signaling-competent state (secondary lesion) before checkpoint responses can be elicited. Accordingly, the existence of a checkpoint-activating nuclease (signal-modifier) has been proposed and recent studies indicate that single-stranded DNA (ssDNA) may be one of the secondary lesions that activates the DNA damage checkpoint (Garvik et al. 1995; Lydall and Weinert 1995; Lee et al. 1998).
The increased ability to detect irreparable UV damage in S phase cells (compared to G1 cells in the examples described above) is not restricted to eukaryotes, as the induction of the SOS response in Escherichia coli also depends on the initiation of DNA replication (Sassanfar and Roberts 1990). This raises the possibility that some nuclease(s) act specifically during S phase to ensure that checkpoint mechanisms are induced efficiently and that no lesions are transformed into stable mutations during the replication process. The critical importance of the intra-S phase checkpoint in preventing the fixation of DNA lesions is particularly relevant in the prevention of cancer. Indeed, individuals afflicted with genetic disorders characterized by a loss of the intra-S phase checkpoint, such as ataxia telangiectasia (AT) and Nijmegen breakage syndrome (NBS), are among those who suffer the most severe predisposition to cancer (for reviews, see Jeggo et al. 1998; Petrini 2000). Cells isolated from AT or NBS patients do not show the typical slowing down of DNA replication seen in the presence of DNA-damaging agents such as bleomycin or ionizing radiation (IR; Painter and Young 1980; Taalman et al. 1983). Instead, they carry on DNA synthesis and cell cycle progression continues unperturbed, a phenomenon known as radio-resistant DNA synthesis (RDS; Painter and Young 1980).
The IR sensitivity and RDS phenotype of AT and NBS cells suggest that the products of the genes mutated in these cells—ATM and Nbs1, respectively—are normally involved in the early steps of the detection and signaling of DNA damage (Petrini 2000). However, whether these proteins are DNA damage sensors, signal-modifiers, or transducers is unclear. Interestingly, the Nbs1 protein has several properties consistent with a role as signal-modifier in the checkpoint transduction cascade. First, despite being IR-sensitive and showing chromosome instability, NBS cells have no gross defects in their abilities to repair DNA damage (Jeggo et al. 1998; Petrini 2000). Second, NBS cells exhibit defects in cell cycle control in S phase (Taalman et al. 1983). Third, Nbs1 is a member of a multisubunit complex that includes the human Rad50 (hRad50) and hMre11 proteins (Carney et al. 1998). Hypomorphic mutations in hMRE11 have been shown to cause an ataxia telangiectasia-like disease (ATLD) which is similar to the checkpoint-deficient AT disorder (Stewart et al. 1999). Furthermore, Mre11 has nuclease activity that can generate extensive regions of ssDNA, which has been shown to activate checkpoints strongly (Garvik et al. 1995; Lydall and Weinert 1995; Lee et al. 1998; Usui et al. 1998). More recently, it has been shown that Nbs1 is phosphorylated by ATM in response to DNA damage and that this is required to mediate an S phase arrest in the presence of DNA damage (for reviews, see Michelson and Weinert 2000; Rhind and Russell 2000). Nevertheless, it has been difficult to identify the precise molecular role(s) of the Nbs1 complex during DNA damage signaling in higher eukaryotes because the genes encoding Mre11, Rad50, and Nbs1 are required for cellular viability (Xiao and Weaver 1997; Luo et al. 1999; Zhu et al. 2001).
The Nbs1 complex is conserved evolutionarily in eukaryotes. hMre11 and hRad50 were originally identified because of their homology with Mre11p and Rad50p, two members of the Xrs2p complex in S. cerevisiae (Alani et al. 1989; Johzuka and Ogawa 1995). Deletion of the genes encoding the members of the Xrs2p complex in yeast result in pleiotropic effects including DNA damage sensitivity, DNA repair deficiency, hyper-recombination, telomere shortening, and impaired meiotic progression (for review, see Haber 1998). Surprisingly, however, no clear checkpoint defects have so far been reported for yeast with mutations in the Xrs2p complex (Kironmai and Muniyappa 1997). The evolutionary conservation of the checkpoint functions of the Xrs2p and Nbs1 complexes has been further put in doubt by the lack of clear sequence homology between Xrs2p and Nbs1 (Carney et al. 1998; Varon et al. 1998). Nevertheless, we show here that the S. cerevisiae Xrs2p complex has a critical role in the initiation of the intra-S phase checkpoint. We discuss these findings in regard to the evolutionary conservation of the ATM signaling pathway and the functions of the Xrs2p/Nbs1 complexes in these events.
Results
Yeast lacking a functional Xrs2p complex are hypersensitive to replicative stress
To gain insight into the molecular basis for the S phase checkpoint defect associated with NBS and ATLD, we decided to study the potential role of the S. cerevisiae Xrs2p complex in S phase regulation. To this end, we examined the sensitivity of various DNA DSB repair and checkpoint mutants to replicative stress induced by the drug hydroxyurea (HU). HU is an inhibitor of ribonucleotide reductase, the rate-limiting enzyme in deoxyribonucleotide (dNTP) biosynthesis. Depletion of dNTPs activates the DNA replication checkpoint, which slows progression through S phase (Desany et al. 1998). Furthermore, initiation of DNA replication in the presence of high levels of HU causes DNA DSBs (Merrill and Holm 1999, and references therein).
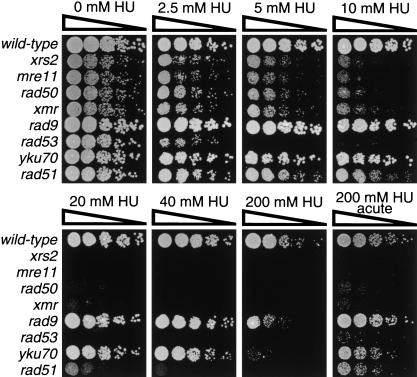
Yeast cells defective in the members of the Xrs2p complex (mre11Δ, rad50Δ, and xrs2Δ) and control mutants defective in DNA repair (rad51Δ, yku70Δ), or defective in the DNA replication (rad53Δ) and DNA damage (rad53Δ, rad9Δ) checkpoints, were serially diluted onto plates containing various concentrations of HU (Fig. 1). High concentrations of HU cause a block to DNA replication and a significant number of DSBs, whereas low concentrations of HU cause only a mild retardation in DNA replication and many fewer DSBs (Merrill and Holm 1999). All mutants tested display significant growth defects on plates containing 200 mM HU (Fig. 1). This lethality is at least partially a consequence of DSB formation because rad9Δ and yku70Δ mutants display little or no sensitivity to growth in the presence of 40 mM HU (Fig. 1). As expected for a DNA replication checkpoint mutant, the rad53Δ strain is sensitive to all the concentrations of HU tested (Fig. 1). Notably, strains with mutations in members of the Xrs2p complex (referred to hereafter as xmr mutants) show marked sensitivity to HU; some sensitivity can be seen with concentrations of HU as low as 5 mM, whereas the xmr mutants become highly sensitive at 10 mM HU and are almost completely dead at 20 mM HU (Fig. 1). The similar responses of the single and triple xmr mutants indicate that null mutations in XRS2, MRE11, and RAD50 are epistatic in their effects on HU sensitivity. Interestingly, the sensitivity of the xmr mutants to 10 mM HU is much greater than that of the rad51Δ mutant (Fig. 1). This is in contrast with the more severe radiosensitivity of rad51 mutants when compared with rad50 mutants (Saeki et al. 1980) and suggests that the HU sensivity of the xmr mutants might not primarily reflect a homologous recombination (HR) defect.
Figure 1.
Hypersensitivity of xrs2Δ, mre11Δ, and rad50Δ mutants to replicative stress. Fivefold serial dilution of yeast cells were plated on YPAD containing various concentrations of HU. For the HU acute hypersensitivity experiment, exponential cultures were grown for 9 h in liquid YPAD containing 200 mM HU before being washed and plated on YPAD plates. Relevant genotypes are shown on the left side of the panels.
To differentiate between a role for the Xrs2p complex in DSB repair or in checkpoint regulation, we examined the ability of the mutant strains to survive a transient period of growth in the presence of high concentrations of HU (HU hypersensitivity assay). This acute treatment causes severe lethality in mutants that are specifically defective in the DNA replication checkpoint but not in mutants affected in DNA repair or in other DNA damage checkpoints (Allen et al. 1994). Thus, it is possible to discriminate between a DNA replication checkpoint defect and a DNA repair defect in a strain sensitive to continuous treatment with HU, on the basis of its additional sensitivity (hypersensitivity) to an acute treatment. Strikingly, whereas the sensitivity of rad51Δ, yku70Δ, and rad9Δ mutant strains to chronic treatment with 200 mM HU is significantly reversed under acute conditions, this is not the case for the xmr mutants (Fig. 1). Indeed, the behavior of the xmr mutants in this assay is similar to that of the rad53Δ mutant, which has a known defect in S phase regulation (Allen et al. 1994). The similarities between the responses of xmr and rad53Δ mutants and the fact that a DNA repair deficiency per se does not result in checkpoint deficiency (Fig. 1; Paulovich et al. 1997) suggest that the HU hypersensitivity of the xmr mutants is due to defective regulation of S phase progression.
The Xrs2p complex is required to slow DNA synthesis in the presence of replicative stress
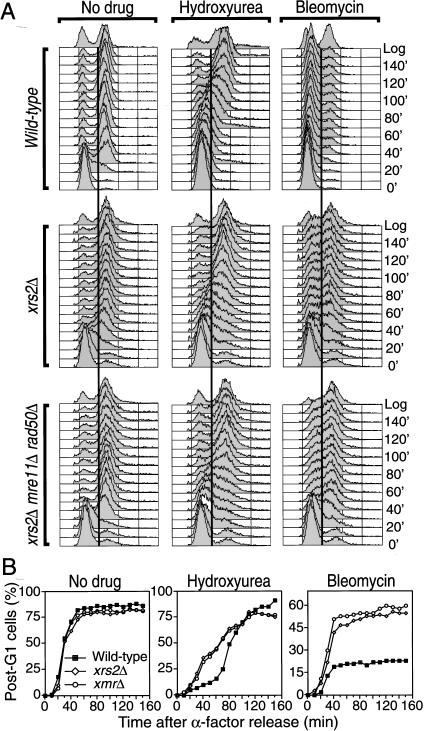
One prediction from the above results is that xmr mutants may be defective in their ability to slow DNA synthesis in the presence of replicative stress. To test this, cells were arrested in G1 with α-factor, released from G1 in the presence of a low concentration (40 mM) of HU, and DNA content was monitored by flow cytometry (FACS) to assess the kinetics of DNA replication (Fig. 2). When released from G1 in the absence of HU, there was no detectable difference in the kinetics of cell cycle progression in the xrs2Δ and xrs2Δ mre11Δ rad50Δ (xmrΔ) mutants compared with the wild-type strain; DNA replication is initiated about 20 min after release from G1 and the bulk of DNA is fully replicated by 50 min (Fig. 2A,B, left panels). We consistently observe, by FACS analysis and immunofluorescence microscopy, that about 5% of the population of cells in xrs2Δ, mre11Δ, rad50Δ, and xmrΔ cultures are irreversibly blocked in a G2-like state (Fig. 2; data not shown) and may represent cells that have failed to fully complete the S phase previous to the G1 synchronization.
Figure 2.
Replicative stress causes premature initiation of DNA synthesis in the absence of a functional Xrs2p complex. (A) α-factor-synchronized cells were released from G1 arrest in medium containing nocodazole and no drug (left panels), 40 mM HU (middle panels), or 35 units/mL bleomycin (right panels), as described in Materials and Methods. Samples were collected at 10-min intervals and analyzed for DNA content by FACS. Yeast strains used were W303-1A (wild-type), DDY004 (xrs2Δ), and DDY022 (xrs2Δ mre11Δ rad50Δ). (B) Quantification of post-G1 cells. Fraction of cells having a DNA content superior to G1 are plotted in function of time below the respective FACS profiles. Numbers are corrected for the presence of cells blocked in G2 at time 0. Typical FACS profiles are shown.
When wild-type cells are released from G1 arrest in the presence of 40 mM HU, DNA synthesis is delayed by about 50 min and is completed ∼130 min following release (Fig. 2A,B). In contrast, xrs2Δ and xmrΔ cells growing in the presence of HU show significant DNA synthesis 30 min following release. For example, ∼40% post-G1 cells are observed at 50 min compared to 12% for wild-type (numbers are corrected for the presence of cells blocked in G2 at time 0; Fig. 2B). At 60 min, this increases to almost 50% post-G1 cells in xrs2Δ and xmrΔ cultures compared to 18% for wild-type yeast. Consistent with this, the elongation of the mitotic spindle in xrs2Δ and xmrΔ mutants treated with 40 mM HU precedes by at least 30 min the elongation of the mitotic spindle in wild-type cells (see Supplemental Materials). Eventually, however, DNA synthesis in wild-type cells reaches essentially the same levels as in xrs2Δ and xmrΔ mutants (Fig. 2).
We also examined the contribution of the Xrs2p complex to the DNA damage-induced intra-S phase checkpoint. Cells were synchronized with α-factor as before and then released in fresh medium for 5 min to allow exit from G1. Bleomycin, a radiomimetic drug that produces DNA DSBs (Povirk 1996), was then added and samples were collected at intervals. As seen in Figure 2 (right panels), bleomycin induces a strong arrest in wild-type cells; only a small proportion of the culture (maximum of ∼20% of cells) replicate their DNA whereas the majority of cells remains with a G1 DNA content throughout the experiment. By contrast, both xrs2Δ and xmrΔ yeast initiate significant DNA replication at 30 min, with ∼25%–30%; of cells showing post-G1 DNA content, which then increases to 50%–60% of cells from 80 min until the end of the experiment (Fig. 2B). Surprisingly, DNA synthesis is never completed in the xrs2Δ and xmrΔ mutant treated with bleomycin, suggesting that the intra-S phase checkpoint is not completely lost in these mutants (see below). These results are consistent with those obtained with HU and indicate that the Xrs2p complex is required for the efficient activation of the intra-S phase checkpoint. Essentially identical results were obtained with mre11Δ and rad50Δ single mutants, indicating that null mutations in the members of the Xrs2p complex are fully epistatic for their effects on the intra-S phase checkpoint (Figs. 2,3, and see 5C below).
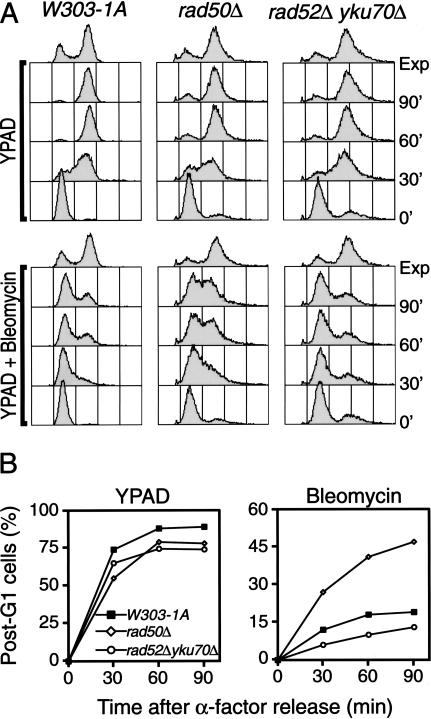
Figure 3.
The intra-S phase checkpoint is functional in cells deficient in DNA DSB repair. (A) α-factor-synchronized cells were released from G1 arrest in medium containing nocodazole and either no drug (top panels) or 35 units/mL bleomycin (bottom panels). Bleomycin was added 10 min after the release from G1 and samples were collected at 30-min intervals and analyzed for DNA content by FACS, as described in Materials and Methods. Yeast strains used were W303-1A (wild-type), DDY008 (rad50Δ), and DDY062 (rad52Δ yku70Δ). (B) Quantification of post-G1 cells. Fraction of cells having a DNA content superior to G1 are plotted in function of time. Numbers are corrected for the presence of cells blocked in G2 at time 0. Typical FACS profiles are shown.
Loss of DNA DSB repair does not impair the intra-S phase checkpoint
Previous studies have shown that defects in DNA repair genes do not cause deficiencies in the intra-S phase checkpoint or in the DNA replication checkpoint (Allen et al. 1994; Paulovich et al. 1997). However, the members of the budding yeast Xrs2p complex are unique in that they are involved in both nonhomologous end-joining (NHEJ) and HR (for review, see Haber 1998). This raised the possibility that the loss of both NHEJ and HR was responsible for the checkpoint defect observed in the xmr mutants. To address this issue, we created a mutant (rad52Δ yku70Δ) that is disabled in both NHEJ and HR and then compared its bleomycin-induced intra-S phase checkpoint with that of a rad50Δ mutant and wild-type cells. As seen in Figure 3, bleomycin induced an arrest in wild-type cells whereas a significant proportion of rad50Δ cells engaged in DNA synthesis by 90 min. In contrast, the rad52Δ yku70Δ double mutant behaved like wild-type yeast and induced a strong cell cycle arrest during the whole experimental time course. (We consistently observe that a small proportion of rad52Δ yku70Δ cells are blocked in a G2-like state even in the presence of α-factor; Fig. 3.) yku70Δ and rad52Δ single mutants behaved similarly to the double mutant and wild-type cells in this assay (data not shown). Together, these results indicate that loss of NHEJ and HR does not in itself cause an intra-S phase checkpoint deficiency in yeast and that the checkpoint function of the Xrs2p complex is a novel and previously uncharacterized function of this complex.
The Xrs2p complex is required for efficient checkpoint signaling
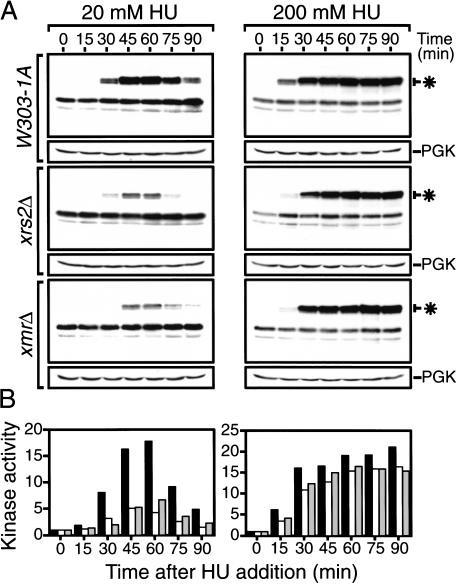
To address the potential role(s) of the Xrs2p complex in S phase checkpoint signaling, we evaluated the activation of Rad53p by an in situ assay (ISA assay; Pellicioli et al. 1999). Rad53p is a protein kinase that is activated by the yeast homologs of human ATR and ATM, Mec1p and Tel1p, respectively (Sanchez et al. 1996). In the presence of replicative stress, Mec1p and Tel1p phosphorylate Rad53p, resulting in activation of Rad53p kinase activity and transduction of the checkpoint signal to downstream effectors (Foiani et al. 2000). Wild-type and mutant cells were synchronized in G1 and Rad53p activity was examined by ISA at different times after G1 release into medium containing 20 mM or 200 mM HU. For wild-type cells treated with 20 mM HU, Rad53p activation is detected 30 min post-release, reaches a peak at about 60 min, and then decreases as cells complete S phase (Fig. 4A). By contrast, xrs2Δ and xrmΔ cells treated with 20 mM HU show a considerable decrease in the extent of Rad53p activation (Fig. 4A). Indeed, quantification of the results reveals that the maximal Rad53p activity is reduced by about 70%–75% in the mutants compared with the controls (Fig. 4B). Essentially identical results were obtained with mre11Δ and rad50Δ single mutants, indicating that null mutations in the XMR genes are epistatic for their effects on the Rad53p response (Fig. 5B; data not shown).
Figure 4.
The Xrs2p complex is required for efficient checkpoint signaling. G1-synchronized cells were released from α-factor arrest and entered S phase synchronously in the presence either 20 mM (left panels) or 200 mM HU (right panels). Samples were collected at 15-min intervals and protein extracts were prepared. (A) In situ analysis of Rad53p activity. The position of phosphorylated Rad53p is indicated by a star. Western blot analysis of 3-phosphoglycerate kinase (3-PGK) was performed in parallel (shown below Rad53p ISAs) to confirm equal loading in each lane. (B) Quantification of Rad53p autophosphorylation. Bars represent the relative fold-activation of wild-type (black), xrs2Δ (white), and xrs2Δ mre11Δ rad50Δ (grey) strains. The activity of Rad53p was quantified using Fujifilm BAS-2500.
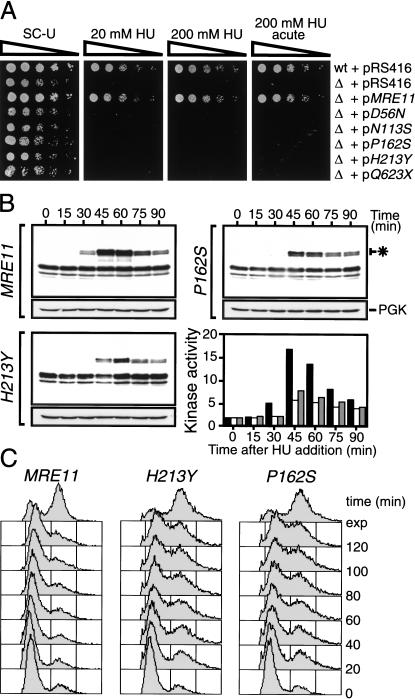
Figure 5.
Hypersensitivity of mre11 nuclease mutants to replicative stress. (A) Fivefold serial dilution of mre11Δ yeast containing single-copy plasmids expressing wild-type Mre11–ProA or various nuclease mutants were plated on SC-uracil containing no drug, 20 mM HU, or 200 mM HU. Exponential cultures were also grown for a further 9 h in liquid SC-uracil containing 200 mM HU before being washed and plated on SC-uracil plates, as described in Materials and Methods. Wild-type (WT) cells carrying an empty plasmid were included for comparison. Δ indicates mre11Δ cells carrying various MRE11 alleles or an empty plasmid. (B) The nuclease activity of Mre11p is required for efficient checkpoint signaling. G1-synchronized mre11Δ cells expressing wild-type Mre11p and nuclease mutants (H213Y and P162S) were released into a synchronous S phase in the presence of 20 mM HU. Rad53p activity was evaluated in situ, as described in Figure 4. The lower right panel is a quantification of Rad53p autophosphorylation; bars represent the relative fold activation of wild-type (black), H213Y mutant (white), and P162S mutant (grey) strains. (C) Mutations in the nuclease domain of Mre11p result in premature initiation of DNA synthesis in the presence of bleomycin. Cells expressing wild-type Mre11p and nuclease mutants (H213Y and P162S) were synchronously released into S phase in the presence of nocodazole and 25 units/mL bleomycin (added 5 min following release), and processed for FACS analysis.
The residual Rad53p activity seen in xmr cells treated with 20 mM HU suggests that the signaling pathway can be activated through alternative routes in these mutants. If this is the case, we reasoned that increasing the intensity of the initial signal might bypass a requirement for the Xrs2p complex. To test this, we performed a Rad53p ISA experiment in the presence of 200 mM HU, a concentration that completely blocks DNA replication (Allen et al. 1994). Figure 4A (right panels) shows that wild-type cells treated with 200 mM HU activate Rad53p at the earliest time point after release from G1 and that activation persists until the end of the experiment. Similarly, Rad53p is strongly activated in xrs2Δ and xmrΔ mutants in the presence of high concentrations of HU (Fig. 4B). The initiation of the Rad53p response was delayed by about 15 min in the mutants treated with 200 mM HU, but a quantification of the results indicates that the maximal Rad53p response in these mutants is only marginally decreased (by 10%–15%) when compared with that of an otherwise isogenic wild-type strain (Fig. 4B, right panel). This result is consistent with the partial restoration of the mitotic spindle elongation delay seen when the xmr mutants are treated with high doses of HU (data not shown). Activation of Rad53p by bleomycin was also significantly curtailed in xmr yeast, indicating that the Xrs2p complex is also required for the efficient signaling of DNA DSB (see Supplemental Materials).
Mre11p nuclease activity is required for efficient initiation of the S phase checkpoint
Because Mre11p is a nuclease, one way that it could potentiate checkpoint-inducing signals in S phase is by generating ssDNA at the sites of stalled replication forks or by otherwise modifying the primary DNA lesion. To test this hypothesis, we evaluated the checkpoint integrity of mre11Δ cells complemented with centromeric plasmids expressing wild-type Mre11p or derivatives mutated in the nuclease domain. Five Mre11p mutants were generated, four affecting evolutionarily conserved nuclease motifs in the enzyme. Two of these mutations (D56N and H213Y) abrogate the exonuclease and endonuclease activities of Mre11p in vitro (Furuse et al. 1998; Usui et al. 1998). The N113S and Q623X mutations correspond to the two hMre11 mutations found in cancer-prone ATLD patients (Stewart et al. 1999). Finally, the P162S mutation affects a proline residue that is conserved in the nuclease domain of all known eukaryotic Mre11 proteins and renders the protein totally inactive at 30°C (not at 23°C, a temperature-sensitive mutant; Johzuka and Ogawa 1995). Figure 5A shows that the ability to grow on HU is severely compromised by mutations in the nuclease domain of Mre11p. Because growth on HU can be a measure of both DNA repair and checkpoint proficiency (Fig. 1), we performed an HU hypersensitivity (acute treatment) experiment to address the integrity of the checkpoint in these mutants (Allen et al. 1994). Notably, inactivating specific nuclease residues in Mre11p completely abrogates survival after transient periods of growth in HU (Fig. 5A).
To characterize further the checkpoint defects associated with the nuclease mutations in Mre11p, we evaluated their effects on Rad53p activation. Because some mutations reduce the stability of Mre11p (D. D'Amours and S.P. Jackson, unpubl.), we selected two mutants (H213Y and P162S) that are stably expressed to wild-type levels for further studies (for expression levels, see Fig. 7B, below). These mutants were released synchronously into S phase in the presence of 20 mM HU and extracts were prepared at 15-min intervals. Figure 5B shows that the Rad53p response of the deletion mutant complemented with wild-type MRE11 is essentially identical to that of the wild-type strain (cf. Fig. 4). However, Rad53p activation is significantly impaired in strains expressing the mutant alleles of MRE11 (Fig. 5B). Indeed, Rad53p activation is reduced by 50%–60% in the absence of nuclease activity when the response is at its maximum (45–60 min), which also results in an apparent reduction in the observable duration of Rad53p activation (Fig. 5B).
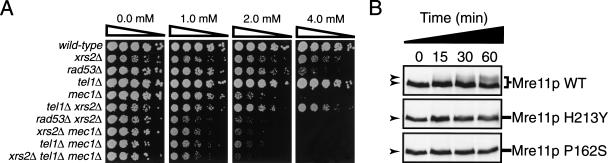
Figure 7.
Functional and epistatic interactions between the Xrs2p complex and Tel1p. (A) HU sensitivity of various mutants affected in the XRS2, RAD53, TEL1, and MEC1 genes. Fivefold serial dilution of yeast cultures were plated on YPAD medium containing low concentrations of HU. Survival was scored after 2–3 d of growth at 30°C. (We consistently observe that a tel1Δ mutation partially suppresses the slow growth phenotype associated with a xrs2Δ mutation, possibly because Tel1p is inappropriately activated in the absence of Xrs2p.) Yeast genotypes are on the left side of the panels. All the strains tested have a sml1-1 mutation. (B) Nuclease mutants of Mre11p are not phosphorylated in response to DNA damage. Exponential cultures of mre11Δ mutants expressing wild-type or nuclease-defective (H213Y and P162S) Mre11–ProA were damaged with 4-NQO (25 μM) and samples were taken at timed intervals. Protein extracts and Western blot analysis were performed as described in Figure 6.
We also tested the ability of the H213Y and P162S mutants to arrest DNA synthesis in response to bleomycin. Figure 5C shows that the mre11Δ mutant complemented with wild-type MRE11 displays normal S phase arrest in response to bleomycin treatment (cf. wild-type responses in Figs. 2 and 5C). In contrast, a significant level of DNA synthesis can be seen with both point mutants, which appear to be particularly affected in their early responses (Fig. 5C). Furthermore, post-G1 DNA accumulates gradually during the experiment and corresponds to almost half of the total DNA content (for P162S and H213Y mutants, 45% and 42%, respectively) at the end of the time course (Fig. 5C). Conversely, the accumulation of post-G1 DNA with wild-type Mre11p is marginal and remains below 22% of the total DNA content throughout the experiment (Fig. 5C). These results indicate that Mre11p nuclease activity is required to efficiently signal bleomycin-induced damage in S phase and are consistent with a role of the Xrs2p complex as signal modifier. The checkpoint defects of Mre11p point mutants are not as strong as those obtained with the deletion mutants, suggesting that there is some nuclease-independent function of the Xrs2p complex in checkpoint activation (Fig. 5B,C). Nevertheless, the phenotype associated with the nuclease mutations in MRE11 is biologically significant because it results in full lethality in response to acute treatment with HU (Fig. 5A). We have used four mutations each spaced by about 50 residues and covering a region of 150 residues in Mre11p nuclease domain. Although we cannot exclude secondary effects on other aspects of the protein, the above data strongly implicate the nuclease functions of Mre11p in the efficient activation of S phase checkpoints.
Xrs2p and Mre11p are phosphorylated after DNA damage in a Tel1p-dependent manner
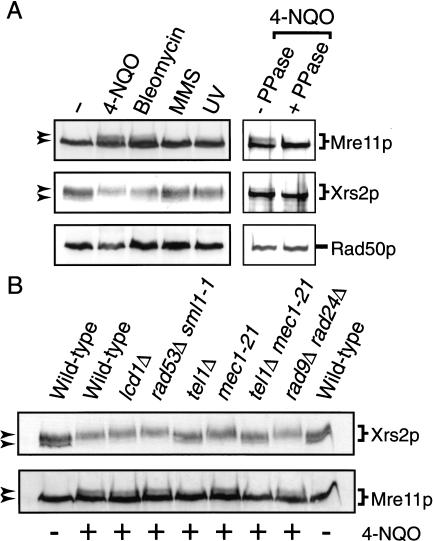
Because checkpoint cascades are regulated by phosphorylation (Foiani et al. 2000), we investigated whether the members of the Xrs2p complex are phosphorylated in response to DNA damage and, if so, which kinase(s) is responsible for this. Protein extracts were prepared from exponentially growing cells expressing epitope-tagged Rad50p, Mre11p, or Xrs2p. (These strains were fully complemented for all the xrm phenotypes tested; D. D'Amours and S.P. Jackson, unpubl.). Extracts from cells treated for 1 h with 4-nitroquinoline-1-oxide (4-NQO), bleomycin, methylmethane sulfonate (MMS), or UV light were resolved by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis. Figure 6A shows that Mre11p and Rad50p migrate as single bands in extracts obtained from untreated cultures, whereas Xrs2p appears as a mixture of heterogeneously-migrating bands, as observed previously (Usui et al. 1998). Upon exposure to 4-NQO or bleomycin, slower migrating forms of Mre11p and Xrs2p are detected (Fig. 6A; for Xrs2p, the faster migrating bands selectively disappear relative to the slower migrating forms). Interestingly, only marginal enrichment of the slower migrating forms of Mre11p and Xrs2p is seen in samples treated with MMS or UV. No significant retardation of the Rad50p band was observed with any of the DNA damaging agents tested (Fig. 6A). Samples treated with phosphatase contained a single band that corresponds to the faster migrating species in the untreated Mre11p and Xrs2p samples (Fig. 6A, right panel). This confirms that the DNA damage-dependent modification of Mre11p and Xrs2p is due to phosphorylation. (Xrs2p was partly dephosphorylated during the immunoprecipitation procedure, but the slow migrating form is still clearly visible in the untreated sample.) In contrast, Rad50p electrophoretic mobility was unchanged by phosphatase treatment (Fig. 6A).
Figure 6.
Xrs2p and Mre11p are phosphorylated in response to DNA damage. (A) Exponentially growing cultures of yeast expressing Mre11–13Myc, Xrs2–13Myc and Rad50–3FLAG were analyzed for protein phosphorylation in response to DNA damage (left panels). Cells were untreated (−) or treated with either 4-NQO (25 μM), bleomycin (350 units/mL), MMS (0.02%), or UV (60 J/m2). Arrows indicate the position of the phosphorylated and unphosphorylated bands (left). For dephosphorylation experiments (right panels), Mre11p, Xrs2p, and Rad50p were immunoprecipitated from soluble extracts of yeast treated with 4-NQO. Half of each immunoprecipitate was dephosphorylated with λ phosphatase; the remaining half was mock treated. (B) Genetic requirements for phosphorylation of Mre11p and Xrs2p. Exponential cultures of various checkpoint mutants expressing Myc-tagged Xrs2p or Mre11p were treated with 4-NQO (25 μM). Untreated (−) and treated (+) wild-type cultures were also included as controls. Protein extracts were prepared 1 h after the addition of 4-NQO and analyzed by Western blotting. Arrows indicate the position of the basal and phosphorylated bands (left). Mutant strains are as follows: tel1Δ (Y662), mec1–21 (Y663), tel1Δ mec1–21 (Y664), rad53Δ sml1–1 (U960–5C), rad9Δ rad24Δ and lcd1Δ. The mec1–21 allele used here is a hypomorphic allele that does not require a second site mutation for the suppression of mec1Δ-associated lethality (Desany et al. 1998). Although it is not known whether this allele is completely defective for checkpoint functions, the similar results obtained with the lcd1Δ/ddc2Δ mutant (which is fully epistatic with MEC1; Rouse and Jackson 2000) and the mec1-21 mutant supports the view that we observe the full effect of the loss of Mec1p in our phosphorylation assay.
We next investigated the genetic requirements for Mre11p and Xrs2p phosphorylation. To this end, various checkpoint mutants were transformed with centromeric plasmids expressing tagged Mre11p or Xrs2p and their ability to phosphorylate these proteins was evaluated. Perhaps surprisingly, the levels of phosphorylation of both Xrs2p and Mre11p remained unchanged in rad9Δ, rad24Δ, rad53Δ, lcd1Δ/ddc2Δ, or mec1-21 mutant strains (Fig. 6B, see legend for details), which represent the main epistasis groups known to be involved in checkpoint responses (for review, see Foiani et al. 2000). In marked contrast, DNA damage-dependent phosphorylation of Xrs2p was no longer detected in tel1Δ or tel1Δ mec1-21 mutants (Fig. 6B). Similarly, the absence of Tel1p resulted in a significant decrease in the phosphorylation of Mre11p and the residual phosphorylation was eliminated by an additional mutation in MEC1. Mre11p and Xrs2p are the only proteins so far shown to be preferentially phosphorylated in a Tel1p-dependent manner during the cellular response to DNA damage.
TEL1 and XRS2 are in the same epistasis group
A role for Tel1p in checkpoint regulation has been inferred from the ability of overexpressed Tel1p to suppress the DNA damage sensitivity of a Mec1p-deficient strain and from the fact that a tel1 mutation exacerbates the HU and DNA damage sensitivity of a mec1 mutant (Morrow et al. 1995). Accordingly, we have observed that Tel1p-deficient yeast have a weak intra-S phase checkpoint defect and that the loss of the TEL1 gene in a mec1-21 background enhances the checkpoint defect of this strain (see Supplemental Materials). Taken together with the above data, these observations suggest that TEL1 might be in the same epistasis group as the products of the XRM genes. To test this idea, we generated strains deficient in the XRS2, TEL1, MEC1, and RAD53 genes as single or combination mutants and then examined their sensitivity to low doses of HU. Because some combinations of mutation in a mec1Δ background result in a senescence phenotype, we isolated survivor clones of these strains that grow at a rate comparable to that of other mutants (Ritchie and Petes 2000). This allowed us to perform a semi-quantitative analysis of the HU sensitivity of each mutant.
Figure 7A shows the results of the epistatic analysis. The point at which the single mutants become significantly sensitive to HU is as follows: xrs2Δ, 4mM; rad53Δ, 2–4 mM; tel1Δ, not sensitive; mec1Δ, 2 mM (Fig. 7A). In comparison, all the double mutants, except the tel1Δ xrs2Δ strain, became significantly sensitive to HU at a concentration of 1 mM. Interestingly, the sensitivity of the xrs2Δ mec1Δ mutant is not significantly exacerbated by the addition of a tel1Δ mutation (both xrs2Δ mec1Δ and xrs2Δ mec1Δ tel1Δ cells are sensitive to the same extent at 1 mM HU), which indicates that xrs2Δ and tel1Δ mutations are epistatic. Indeed, in this system, combining mec1Δ and tel1Δ mutations (which exacerbates the mec1Δ phenotype) serves as a positive control for a Tel1p-specific effect because Tel1p-deficient mutants have no HU sensitivity on their own (Fig. 7A; Morrow et al. 1995). These results are consistent with xrs2Δ and tel1Δ mutations working epistatically in regard to a chromosome rearrangement phenotype (Myung et al. 2001). Interestingly, we could not obtain any viable mec1Δ xrs2Δ spores in the absence of SML1 mutations (either sml1Δ or sml1-1 alleles; data not shown) during the construction of our double mutants. This result indicates that loss of XRS2 does not suppress the lethality of mec1Δ (by increasing the levels of dNTPs) and makes it unlikely that the faster rates of DNA replication that we see in Figure 2 are due to altered dNTPs pools.
Tel1p-dependent phosphorylation of Mre11p is a feedback response to checkpoint activation
Our results are consistent with a model in which the Xrs2p complex acts with Tel1p at the level of the DNA damage to activate the checkpoint. If the nuclease activity of Mre11p is required to modify the initial damage so that it can activate the checkpoint more efficiently, we would expect that checkpoint kinases, including Tel1p, would not be fully activated in strains expressing nuclease mutants of Mre11p. Alternatively, if the nuclease activity of Mre11p is not required for Tel1p and Mec1p activation, we would expect that the nuclease mutants of Mre11p would be phosphorylated as efficiently as the wild-type enzyme. To address these two possibilities, we tested whether the nuclease activity of Mre11p is required for its Tel1p-dependent phosphorylation. Yeast expressing either wild-type or Mre11p mutants affected in the nuclease domain (H213Y and P162S) were grown to early log-phase, treated with 4-NQO, and extracts were prepared at 15 min-intervals to analyze the kinetics of Mre11p phosphorylation. Figure 7B shows that a weak phosphorylation of wild-type Mre11p can be seen early on during the experiment, which increases significantly at 30 min and reaches a maximal level 60 min after the addition of the DNA-damaging drug. In contrast, no phosphorylation is seen with either Mre11p mutant at any time point during the experiment (Fig. 7B). Taken together with the current literature (Lee et al. 1998; Pellicioli et al. 2001), our results strongly suggest that the nuclease activity of Mre11p is required for the efficient activation of the checkpoint kinases and is consistent with the view that the phosphorylation of Mre11p is a feedback response from the checkpoint signaling pathway.
Discussion
The Xrs2p complex is required for the initiation of S phase checkpoints
Here, we describe a previously uncharacterized role for the Xrs2p complex in checkpoint regulation during S phase. We show that yeast lacking a functional Xrs2p complex are defective in both the HU-induced checkpoint and the bleomycin-induced intra-S phase checkpoint. Specifically, we see that Xrs2p-deficient yeast experiencing replicative stress initiate DNA synthesis earlier than wild-type cells, are unable to inhibit the elongation of their mitotic spindle, and fail to efficiently activate Rad53p, a central checkpoint regulator. The role of the Xrs2p complex in checkpoint regulation does not appear to reflect its role in NHEJ and HR because loss of these DNA repair pathways in a rad52Δ yku70Δ double mutant does not impair the intra-S phase checkpoint. Taken together, our results indicate that the role of the Xrs2p complex in S phase arrest is a novel and previously uncharacterized function of this complex.
It is noteworthy that a specific defect in the initiation of the intra-S phase checkpoint in xmr mutants results in severe lethality in the presence of replicative stress (Fig. 1). This phenotype is consistent with the results of Desany et al. (1998) who have shown that the inability to respond to replicative stress in the rad53 mutant results in an irreparable catastrophe during DNA replication, possibly due to collapsed replication forks. Our results indicate that Rad53p needs to be fully activated very early during replicative stress to ensure survival, and that later activation of the kinase, as seen in xmr mutants, is not sufficient to restore viability. It is likely that the lethal events in xmr and rad53 mutants experiencing replicative stress are the same because xmrΔ cells fail to properly activate Rad53p.
The Tel1p–Xrs2p signal transduction pathway is conserved from yeast to humans
Cells isolated from AT patients show similar cellular phenotypes to NBS and ATLD cells (Petrini 2000). This led to the early suggestion that the products of the ATM and NBS1 genes would be involved in the same processes during the response to DNA damage. This assumption was confirmed recently when it was shown that ATM phosphorylates Nbs1 in response to DNA damage and that this is necessary to enforce the intra-S phase checkpoint (for review, see Michelson and Weinert 2000). Interestingly, previous studies have shown that the yeast homolog of ATM, Tel1p, has only a minor role in the DNA damage response. Indeed, Tel1p seems to be necessary for DNA damage resistance only in the absence of the predominant kinase Mec1p (Morrow et al. 1995). By contrast, we show that Tel1p is required for the effective phosphorylation of Mre11p and Xrs2p in vivo, and that Mec1p only plays an accessory role in these events. This is the first time that a primary dependence on Tel1p has been demonstrated for the phosphorylation of checkpoint proteins and strongly suggests that the fundamental mechanisms of the signaling pathway are conserved from yeast to humans.
It is important to note that there is not a simple causative link between checkpoint defects and DNA damage sensitivity. This is clearly evidenced by the behavior of chk1 mutant in yeast (Sanchez et al. 1999), by the identification of checkpoint-deficient alleles of MEC1 that are not DNA damage sensitive (Weinert et al. 1994), by the general lack of DNA damage sensitivity of mammalian p53−/− cell lines, and by the fact the DNA damage sensitivity and checkpoint defect of AT cells can be dissociated (for review, see Jeggo et al. 1998). In light of this, it is not necessarily surprising that Tel1p could be involved in important checkpoint responses and yet tel1 mutant strains are not markedly hypersensitive to HU or DNA damage.
Interestingly, the phosphorylation of Mre11p by Tel1p appears to be dependent on the nucleolytic activity of the Xrs2p complex, suggesting that Mre11p phosphorylation is a feedback response to Tel1p activation. It is tempting to speculate that this phosphorylation alters the biochemical properties of Mre11p and redirects the Xrs2p complex toward other activities (such as DNA repair) once the intra-S phase checkpoint has been triggered. There is some evidence in humans for a biphasic redistribution of the Nbs1 complex during the DNA damage response, which suggests that the Nbs1 complex has two distinct functions that are temporally separated (Carney et al. 1998). It would perhaps make sense if the Nbs1/Xrs2p complex would need to activate the intra-S phase checkpoint first so that the cell cycle is slowed and time is provided to repair DNA damage. Mre11p phosphorylation might then act as a switch to commit the Xrs2p complex to DNA repair activities following checkpoint activation. Clearly, careful biochemical studies will be required to address these important issues.
The Xrs2p complex as a signal-modifier during S phase
The identity of the DNA damage sensors and the nature of the process(es) leading to DNA damage recognition during checkpoint activation are currently unclear. DNA can be damaged in several ways, and each type of lesion has its own typical structure and requirements for detection (for review, see Friedberg et al. 1995). To address the structural diversity of DNA lesions, the process of DNA repair utilizes several distinct pathways, each of which is concerned with a class of structurally similar lesions. Perhaps surprisingly, however, current data suggest that DNA damage activates a common set of checkpoint proteins independently of the nature of the DNA lesion (Foiani et al. 2000). This indicates either that there is a multitude of DNA damage sensors acting in checkpoint pathways or that it is not the primary lesion itself that is detected but a common intermediate resulting from the processing of the damage. This latter mechanism would seem to be structurally less demanding for the cell and is attractive because the common intermediate could be easily provided by signal modifiers associated with DNA repair or DNA replication processes. Accordingly, it has been shown that irreparable UV lesions are only efficiently detected during S phase in both human and yeast cells, which indicates that the initial damage is processed to a signaling-competent state during DNA replication (Nelson and Kastan 1994; Neecke et al. 1999). This conclusion is also supported by recent results indicating that alkylation and UV-induced lesions are converted to DSBs during S phase (Galli and Schiestl 1999). Whether DSBs can induce the checkpoint response directly or have to be processed first is unknown. However, it is tempting to speculate that DSBs may serve as ideal intermediates for exonucleases to produce ssDNA, a structure that has been shown to be a potent activator of checkpoint responses (Garvik et al. 1995; Lydall and Weinert 1995; Lee et al. 1998).
Mre11p is a strong candidate for this nuclease activity because it has already been shown to be required for the formation of ssDNA regions in vivo at meiotic DSBs and during mating-type switching (Lee et al. 1998; Usui et al. 1998). Furthermore, our finding that the Xrs2p complex is required to signal the presence of low levels of DNA damage to the checkpoint machinery, and that this role can be partly bypassed by increasing the levels of DNA-damaging agent, suggests that the Xrs2p complex potentiates the checkpoint-inducing properties of DNA damage. The simplest model to accommodate these observations is that Mre11p nuclease processes DNA DSBs into checkpoint-activating lesions during S phase. Evidence supporting this interpretation has also been obtained recently from the work of Lee et al. (1998) and Pellicioli et al. (2001) who have shown that cells deficient in Mre11p adapt more readily to the presence of a single HO-induced DSB than wild-type cells. Their interpretation of the data is that Mre11p is normally required to produce the ssDNA required to maintain the checkpoint and that in the absence of this signal, cells abrogate the checkpoint response and exit cell cycle arrest (adaptation). However, it is notable that the nuclease activity of Mre11p in vitro is of the opposite polarity (3′→5′) to the polarity of HO-induced DSB resection in vivo (5′→3′). Because of this discrepency and because MRE11 deletion mutants have been used in previous studies, there is still some debate as to whether Mre11p is directly responsible for the nucleolytic degradation of DNA DSBs or whether it is required to activate a second nuclease that is then responsible for the nucleolytic degradation.
Our analyses of Mre11p nuclease mutants address this issue and strongly suggest that Mre11p nuclease activity is directly responsible for creating the checkpoint-inducing signal. All the exonuclease mutants of Mre11p we have tested so far are highly defective in the intra-S phase checkpoint, are deficient in Rad53p activation, show premature DNA synthesis, and are defective in Tel1p-dependent phosphorylation. The importance of Mre11p nuclease activity for the activation of the intra-S phase checkpoint is reinforced by the fact that a mutation affecting a conserved residue in the nuclease domain of hMre11 causes RDS in ATLD patients (Stewart et al. 1999; see discussion in Rhind and Russell 2000). Although it is difficult to formally exclude potential secondary effects associated with nuclease mutations, the available data strongly suggest that the Xrs2p complex acts as a signal modifier to potentiate the checkpoint-inducing properties of primary DNA lesions during S phase.
Materials and methods
Plasmid and strain construction
Yeast strains with complete deletion of the coding sequence of XRS2, MRE11, RAD50, RAD51, RAD52, or TEL1 genes were constructed according to Baudin et al. (1993) in the original W303-1A background (Table 1). Construction of multiple mutant strains was performed by mating the appropriate single mutants. Genotypes were confirmed by PCR and by complementation with single-copy plasmids (pRS416) carrying the promoter, full-length coding sequence and transcription terminator of the deleted gene. pRS416-based plasmids expressing Xrs2–13Myc, Mre11–13Myc, Mre11–ProA (Protein A), and Rad50–3FLAG were created by subcloning the appropriate tags (Longtine et al. 1998) in frame with the last codon of the genes. All plasmids were shown to be fully functional by complementation of the appropriate mutant strains. Mutagenesis of the nuclease domain of Mre11–ProA was performed with the QuickChange mutagenesis kit (Stratagene).
Table 1.
Yeast strains used in this study
| Strain
|
Genotype
|
Constructed by
|
|---|---|---|
| W303-1A | Mata ade2-1 can1-100 his3-11,15 leu2-3,112 rad5-535 trp1-1 ura3-1 | R. Rothstein |
| U960-5C | W303-1A, sml1-1 rad53::HIS3 | R. Rothstein |
| Y661 | Mata ade2-1 can1-100 his3-11,15 leu2-3,112 rad5-535 trp1-1 ura3-1 | S. Elledge |
| Y662 | Y661, tel1::HIS3 | S. Elledge |
| Y663 | Y661, mec1-21 | S. Elledge |
| Y664 | Y661, tel1::HIS3, mec1-21 | S. Elledge |
| lcd1Δ | W303-1A, lcd1::LEU2 | J. Rouse |
| rad9Δ rad24Δ | W303-1A, rad9::HIS3 rad24::TRP1 | D. Durocher |
| hdf1 | W303-1A, yku70::LEU2 | H. Feldmann |
| DDY004 | W303-1A, xrs2::LEU2 | This study |
| DDY006 | W303-1A, mre11::HIS3 | This study |
| DDY008 | W303-1A, rad50:TRP1 | This study |
| DDY022 | W303-1A, xrs2::LEU2 mre11::HIS3 rad50::TRP1 | This study |
| DDY029 | W303-1A, rad9::URA3 | This study |
| DDY035 | W303-1A, sml1-1 xrs2::LEU2 rad53::HIS3 | This study |
| DDY042 | W303-1A, rad51::HIS3 | This study |
| DDY050 | W303-1A, sml1-1 | This study |
| DDY051 | W303-1A, sml1-1 xrs2::LEU2 | This study |
| DDY052 | W303-1A, sml1-1 tel1::HIS3 | This study |
| DDY053 | W303-1A, sml1-1 mec1::TRP1 | This study |
| DDY056 | W303-1A, sml1-1 xrs2::LEU2 tel1::HIS3 | This study |
| DDY062 | W303-1A, yku70::LEU2 rad52::TRP1 | This study |
| DDY063 | W303-1A, sml1-1 mec1::TRP1 xrs2::LEU2 | This study |
| DDY064 | W303-1A, sml1-1 mec1::TRP1 tel1::HIS3 | This study |
| DDY065 | W303-1A, sml1-1 mec1::TRP1 xrs2::LEU2 tel1::HIS3 | This study |
HU and bleomycin sensitivity assays
For growth on solid medium containing HU or bleomycin, saturated cultures were diluted to A600 of 0.3 and fivefold dilution series were spotted on plates and grown at 30°C for 2 d (>40 mM HU or bleomycin) or 4 d (200 mM HU). Stock solutions of bleomycin (Bleo-Kyowa) were freshly prepared before each experiment at a concentration of 15,000 international units/mL in water. To test for HU hypersensitivity (acute treatment), saturated cultures were diluted to A600 of 0.3 in liquid YPAD (or in SC-uracil for mre11 nuclease mutants) and grown for 1.5 h at 30°C. HU was then added to 200 mM and cultures were incubated for 9 h with shaking at 30°C. Cells were washed with water, diluted to an A600 of 0.3, and fivefold dilution series were spotted on YPAD or SC-uracil plates. Survival was scored after 2–3 d.
Analysis of the S-phase checkpoint
FACS analysis and Rad53p activation assays were performed with synchronized cells as follows. Early log cultures were synchronized in G1 with 5 μg/mL α-factor for 1.5 h. After confirming the efficiency of the arrest by light microscopy (>95%), cells were extensively washed to eliminate α-factor and then released in liquid YPAD. For HU experiments, the drug was added at the onset of the release at a final concentration of 20 mM, 40 mM, or 200 mM, as described in figure legends. For bleomycin, the drug was added after 5–10 min of growth to allow the cells to enter S phase. Samples were taken at various time points and processed for the following assays. S phase progression was monitored by FACS on a Becton Dickinson FACSort cytometer equipped with the CELLQuest software, as previously described (Paulovich et al. 1997), except that nocodazole was added to 15 μg/mL when releasing cells to prevent progression to G1. In situ Rad53p kinase assays (ISA) were performed according to Pellicioli et al. (1999). In addition, extracts were probed with anti-PGK (22C5-D8; Molecular Probes) monoclonal antibody to confirm equal loading.
Phosphorylation analysis
Exponentially growing cells expressing tagged proteins were treated with DNA damaging agents in liquid SC-uracil medium. Cells were treated with 25μM 4-NQO, 350 units/mL bleomycin, or 0.02% MMS for 1 h. For UV treatment, cells were exposed to 60 J/m2 UV and left to recover for 1 h. Trichloroacetic acid extracts were prepared as described by Pellicioli et al. (1999) and proteins were resolved by 8% SDS-PAGE in gels containing a 150:1 ratio of acrylamide to bis-acrylamide. Western blots were performed with anti-Myc-9E10 and anti-FLAG-M2 monoclonal antibodies, as described (Pellicioli et al. 1999). Primary antibodies and Mre11–ProA were detected with HRP-conjugated rabbit anti-mouse IgG antibody (Dako). Soluble extract preparation and dephosphorylation were performed as described by Pellicioli et al. (1999).
Acknowledgments
We thank all members of the Jackson laboratory, especially John Rouse and Daniel Durocher, for helpful discussions. We are also grateful to S. Elledge, R. Rothstein, and H. Feldmann for providing strains and to A. Bardin for instructions on the spindle immunofluorescence procedure. D.D. is supported by scholarships from the Conseil de Recherche en Science Naturelle et en Génie (CRSNG) du Canada, from the Fond pour la Formation de Chercheurs et l'Aide à la Recherche du Québec (Fonds FCAR), and from the Cancer Research Campaign. This work was funded by the Cancer Research Campaign.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL spj13@mole.bio.cam.ac.uk; FAX 1223-334089.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.208701.
References
- Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The Sad1/Rad53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes & Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, LeBeau M, Yates JR, Hays L, Morgan WF, Petrini JHJ. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: ASM Press; 1995. [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, Schiestl RH. Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat Res. 1999;429:13–26. doi: 10.1016/s0027-5107(99)00097-4. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- Jeggo PA, Carr AM, Lehmann AR. Splitting the ATM: Distinct repair and checkpoint defects in ataxia telangiectasia. Trends Genet. 1998;14:312–316. doi: 10.1016/s0168-9525(98)01511-x. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K. Alteration of telo meric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo GB, Yao MS, Bender CF, Mills M, Bladl AR, Bradley A, Petrini JHJ. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Holm C. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics. 1999;153:595–605. doi: 10.1093/genetics/153.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson RJ, Weinert T. Closing the gaps among a web of DNA repair disorders. Bioessays. 2000;22:966–969. doi: 10.1002/1521-1878(200011)22:11<966::AID-BIES2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Morrow M, Tagle DA, Shiloh Y, Collins FS, Hieter P. Tel1, an S. cerevisiae homolog of the human gene mutated in Ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Neecke H, Lucchini G, Longhese MP. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Kastan MB. DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: A new explanation. Proc Natl Acad Sci. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Margulies RU, Garvik BM, Hartwell LH. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, DiFiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Petrini JH. The Mre11 complex and ATM: Collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355:71–89. doi: 10.1016/0027-5107(96)00023-1. [DOI] [PubMed] [Google Scholar]
- Rhind N, Russell P. Checkpoints: It takes more than time to heal some wounds. Curr Biol. 2000;10:R908–R911. doi: 10.1016/s0960-9822(00)00849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Petes TD. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki T, Machida I, Nakai S. Genetic control of diploid recovery after gamma-irradiation in the yeast Saccharomyces cerevisiae. Mutat Res. 1980;73:251–265. doi: 10.1016/0027-5107(80)90192-x. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu QH, Wang B, Elledge SJ. Regulation of Rad53 by the Atm-like kinases Mec1 and Tel1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu FH, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Siede W, Friedberg AS, Dianova I, Friedberg EC. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics. 1994;138:271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NGJ, Raams A, Byrd PJ, Petrini JHJ, Taylor AMR. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Taalman RD, Jaspers NG, Scheres JM, de Wit J, Hustinx TW. Hypersensitivity to ionizing radiation, in vitro, in a new chromosomal breakage disorder, the Nijmegen Breakage Syndrome. Mutat Res. 1983;112:23–32. doi: 10.1016/0167-8817(83)90021-4. [DOI] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Xiao YH, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]