Abstract
In spite of the wealth of clinical evidence supporting the health benefits of Lactobacillus rhamnosus GG in humans, there is still a lack of understanding of the molecular mechanisms behind its probiosis. Current knowledge suggests that the health-promoting effects of this probiotic strain might be partly dependent on its persistence in the intestine and adhesion to mucosal surfaces. Moreover, L. rhamnosus GG contains mucus-binding pili that might also explain the occupation of its ecological niche as a comparatively less stringent allochthonous intestine-dwelling bacterium. To uncover additional surface proteins involved in mucosal adhesion, we investigated the adherence properties of the only predicted protein (LGG_02337) in L. rhamnosus GG that exhibits homology with a known mucus-binding domain. We cloned a recombinant form of the gene for this putative mucus adhesin and established that the purified protein readily adheres to human intestinal mucus. We also showed that this mucus adhesin is visibly distributed throughout the cell surface and participates in the adhesive interaction between L. rhamnosus GG and mucus, although less prominently than the mucus-binding pili in this strain. Based on primary structural comparisons, we concluded that the current annotation of the LGG_02337 protein likely does not accurately reflect its predicted properties, and we propose that this mucus-specific adhesin be called the mucus-binding factor (MBF). Finally, we interpret our results to mean that L. rhamnosus GG MBF, as an active mucus-specific surface adhesin with a presumed ancillary involvement in pilus-mediated mucosal adhesion, plays a part in the adherent mechanisms during intestinal colonization by this probiotic.
INTRODUCTION
The commensal Gram-positive lactobacilli are one of the first groups of bacteria to inhabit the human gastrointestinal (GI) tract (16) and include some strains (autochthonous) that colonize the intestine stably throughout the lifetime of the host (31). In addition, there are those strains, called allochthonous, that persist only briefly in the intestine, many of these being probiotics (1, 9, 13) and understood to stimulate health-benefiting immune responses in host intestinal cells (for a review, see reference 14) or cause competitive displacement of invading pathogens (for a review, see reference 35). Thus far, the precise molecular mechanisms that differentiate the colonization ability between autochthonous and allochthonous intestinal lactobacilli remain undefined (42), although they are likely to be partly dependent on a diverse range of cell surface adhesion molecule-mediated interactions with the host intestinal mucosa. With that being said, there are a growing number of reports in the literature that indicate that lactobacillar adherence to the intestinal mucosal layer is mediated by surface proteins with a mucus-binding capacity (15, 21, 22, 25, 30, 32, 33, 41). Moreover, homology-driven genome mining in several Lactobacillus spp. (3, 7, 8, 11, 33) has identified the presence of various-sized putative mucus adhesins consisting of one or more copies of an approximately 100- to 200-amino-acid (aa) mucus-binding (MUB) domain repeat, which, given its prevalence in lactic acid bacteria (LAB), can be considered a unique functional feature for promoting host-microbe interplay in the GI tract (7).
In a recent study, we reported that proteinaceous pilus-like structures protrude from the cell surface of a widely used probiotic Lactobacillus strain (21). Earlier studies have primarily characterized Gram-positive pili as virulence factors in pathogen-mediated disease and illness (for a review, see reference 37). However, with the discovery that Lactobacillus rhamnosus GG is a piliated strain (21) and that an intestinal mucus-binding capacity is associated with the corresponding pili (called SpaCBA) (21, 41), new light has been shed on the putative role of piliation as a novel surface-localized feature in mediating intestinal colonization by probiotic lactobacilli. As a typical probiotic, L. rhamnosus GG adopts the characteristic colonization behavior associated with allochthonous bacteria (42) and so persists only transiently in the GI tract (1). However, compared to the genetically similar but non-SpaCBA-piliated L. rhamnosus LC705 strain, piliation might offer a possible explanation for the heightened ability of L. rhamnosus GG to adhere to intestinal mucosal surfaces (39) and to occupy its intestinal niche with greater duration (21). In this context, L. rhamnosus GG might be regarded as a more sustainable allochthonous strain than L. rhamnosus LC705.
Up to now, SpaCBA pilus fibers have been characterized as the predominant surface-localized component for L. rhamnosus GG mucosal adhesion (21, 41). However, there are growing indications that other LPXTG-anchored cell wall proteins, such as MabA (29), also function in the adherence of this probiotic strain to the host intestinal mucosa. As an additional example, the LGG_02337 open reading frame (ORF), which includes the primary structural elements for an N-terminal secretion signal, 4 Pfam-MucBP (mucin-binding protein) domain repeats, and a C-terminal sortase-specific LPXTG cell wall-anchoring domain (21), represents the only predicted surface protein in L. rhamnosus GG that exhibits amino acid identity to a recognized mucus-binding domain (4). Interestingly, a homologous form of this LPXTG-like surface protein (encoded by the LC705_02328 ORF) is found in the less-mucus-adherent L. rhamnosus LC705 strain (39) and, like in L. rhamnosus GG, is the only predicted protein that contains acknowledged mucus-related adhesion domains (21). Typically, the Pfam-MucBP domain repeats are shorter in length (∼50 aa) than the predicted MUB domains present in the large-sized Mub (mucus-binding) proteins produced by various Lactobacillus spp. (7). However, besides being the canonical component of mucus-binding proteins in other LAB, the Pfam-MucBP domain repeat is also a characteristic element of numerous proteins in different Listeria spp. (7). For instance, the Pfam-MucBP domain can be found frequently in the internalin (Inl) family of leucine-rich repeat-containing surface proteins that are exclusive to the food-borne pathogen Listeria monocytogenes (6). Ostensibly due to the shared sequence similarity with the Pfam-MucBP domain repeats found typically in the Inl-like proteins, the ORFs for LGG_02337 in L. rhamnosus GG and LC705_02328 in L. rhamnosus LC705 have been annotated previously as internalins (21).
We now present a study performed to determine the contributing role of the LGG_02337-encoded LPXTG-like protein in promoting L. rhamnosus GG adhesion to the intestinal mucosa. The characterization of this putative adhesin involved analyzing its primary structure, assessing its adherence to human intestinal mucus, and demonstrating its cell surface localization and in vivo functionality. Based on our results reported herein, we propose that this LPXTG-like protein be renamed and referred to as the mucus-binding factor (MBF).
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulations.
Escherichia coli strains TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] and BL21(DE3)/pLysS [F− ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS (Camr)] were used for plasmid preparation and protein expression, respectively. E. coli was routinely grown in Luria-Bertani (LB) medium at 37°C with agitation and was supplemented with 50 μg/ml kanamycin when required. The pET28b+ expression vector (Novagen) was used for the cloning and production of recombinant L. rhamnosus GG MBF protein. Lactobacillus rhamnosus GG (ATCC 53103) and L. rhamnosus LC705 (DSM 7061) strains (both kind gifts from Valio Ltd., Finland) were used for antibody-mediated mucus-binding inhibition experiments. Genomic DNA from L. rhamnosus GG was generously provided by Lars Paulin (University of Helsinki, Helsinki, Finland) and was isolated using the procedure described previously (21). Standard DNA protocols were used for molecular cloning and related procedures as described previously (36).
E. coli cloning of the LGG_02337 ORF.
The coding sequence of the L. rhamnosus GG LGG_02337 ORF (21) lacking the DNA region for N-terminal secretion and C-terminal LPXTG-like recognition signals was PCR amplified from genomic DNA using a forward oligoprimer containing an EcoRI restriction site (5′-TAGTCACGTGAATTCGAGCTCGGTGG [EcoRI site is italicized]) and a reverse oligoprimer containing an XhoI restriction site (5′-CTTTTCGTTCTCGAGAGGCAGCCGCCGCTG [XhoI site is italicized]) (Oligomer, Finland). Amplified DNA, cleaved by EcoRI and XhoI restriction endonucleases, was ligated into the pET28b+ expression vector, which was then transformed into chemically competent E. coli TOP10 cells. The resulting plasmid construct (pKTH5323) was subsequently transformed into the E. coli BL21(DE3)/pLysS expression strain for the production of cytosolic C-terminal hexahistidine-tagged MBF protein. The recombinant form of MBF also includes seven residues at the N terminus and two residues preceding the hexahistidine sequence at the C terminus that originated from amino acids encoded by the expression vector.
Production and purification of recombinant MBF protein.
Recombinant MBF protein production and purification was performed using the procedure essentially as described previously (21, 41). Briefly, E. coli cells harboring the pKTH5323 plasmid were grown until mid-log phase at 37°C in kanamycin-supplemented (50 μg/ml) LB medium, and MBF protein production was then induced for 3 h by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). After the cells were harvested by centrifugation, the cell pellet was resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole] and subjected to sonic oscillation in order to disrupt the cells. The lysed cell suspension was centrifuged to remove cellular debris and unbroken cells and was then membrane filtered (0.45-μm pore size) to produce a clarified cell-free protein extract. Immobilized nickel affinity chromatography was used to purify hexahistidine-tagged MBF protein. The cell-free protein extract was loaded onto a nickel nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) column and rinsed with several bed volumes of wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole), and the recombinant MBF protein was then eluted from the column with elution buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 250 mM imidazole). Elution fractions judged to contain purified MBF were pooled, buffer was exchanged with 10 mM Tris-HCl (pH 8.0) by using an EconoPac 10 DG desalting column (Bio-Rad), and protein was concentrated using a 30-kDa Microsep filter (Pall Life Sciences). Protein purity was visualized by SDS-PAGE, and the protein concentration was estimated by A280 measurements. Recombinant L. rhamnosus GG SpaC pilin protein served as a control and was produced and purified according to a method described previously (21).
L. rhamnosus GG MBF antiserum preparation.
Antiserum specific for L. rhamnosus GG MBF protein was produced using the method described previously (21, 41). In brief, rabbits were first injected subcutaneously with a 1-ml volume of 400 μg purified recombinant MBF protein and Freund's complete adjuvant (1:1 mixture), and then 1-ml subcutaneous booster injections of 200 μg MBF protein in Freund's incomplete adjuvant (1:1 mixture) were administered at 3-week intervals over a 9-week period. Blood was collected 14 days after the last booster injection, and the antiserum was prepared as described previously (19).
Construction of L. rhamnosus GG CMPG5351 mutant strain.
The L. rhamnosus GG knockout derivative strain, an inactivated exopolysaccharide (EPS) welE (CMPG5351; ΔwelE::Tcr) mutant, was used for demonstrating MBF cell surface localization by electron microscopic means. Details about its construction have been described previously (23).
Radiolabeling of recombinant MBF protein and L. rhamnosus cells.
Purified MBF protein was radiolabeled with 125I as described previously (21, 41) using Pierce iodination reagent according to the manufacturer's instructions (Pierce). Radiolabeled MBF protein was chromatographically separated from residual unbound radiolabel using a D-Salt polyacrylamide desalting column (Pierce). For use as controls in mucus-binding experiments, recombinant SpaC pilin (positive control) and ovalbumin (Sigma) (background control) were radiolabeled using the aforementioned procedure as described previously (21, 41). The L. rhamnosus GG and LC705 strains were metabolically radiolabeled with tritiated thymidine using the procedure described previously (40). Briefly, L. rhamnosus cells were grown overnight in MRS liquid medium containing 10 μl/ml [5′-3H]thymidine (16.7 Ci/nmol) and were then harvested by centrifugation. After two washes in phosphate-buffered saline (PBS) (pH 7.2), the cell pellet was resuspended in the same buffer, and the number of cells was normalized by adjusting the optical density at 600 nm (OD600) to 0.25.
Isolation of mucus from the human intestine.
The source of human intestinal mucus, established previously for use in binding experiments (28), was surgically removed tissue that was provided by patients diagnosed with an operable stage of colorectal cancer and undergoing colonic resection. The ethics committee for the Hospital District of Southwest Finland authorized the isolation and use of the resected human intestinal tissue in this study. In addition, participating patients were fully informed beforehand and had provided their written consent. Noncancerous tissue segments containing intact mucosa were selected, and the mucus gel layer was recovered as described previously (40).
Binding of radiolabeled recombinant MBF to intestinal mucus.
Binding experiments between radiolabeled recombinant MBF protein and human intestinal mucus was carried out using the procedure described previously (21, 41). Briefly, Maxisorp microtiter plate wells (Nunc) containing saturating amounts of intestinal mucus were incubated overnight at 4°C, rinsed several times with PBS to remove unbound mucus, and then incubated in a blocking solution of 0.5% bovine serum albumin (BSA) in PBS for 1 h at room temperature. After the blocking solution was aspirated away, 50 pmol of radiolabeled MBF protein dissolved in blocking solution was added to wells containing immobilized mucus and incubated at 37°C for 1 h. Wells were rinsed three times with PBS, and then the amount of protein-bound 125I was assessed using a Wallac 1480 Wizard 3-inch automatic gamma counter (PerkinElmer). In addition, binding between intestinal mucus and 50 pmol radiolabeled SpaC pilin (positive control) and ovalbumin (background control) was determined.
Antibody-mediated inhibition of L. rhamnosus GG adhesion to intestinal mucus.
The adhesion of L. rhamnosus GG cells to intestinal mucus was inhibited by anti-MBF serum using the procedure described previously (21, 41). Briefly, 3H-labeled wild-type (WT) L. rhamnosus GG cells were first preincubated with anti-SpaC pilin serum (diluted 1:100) and then treated either with or without anti-MBF serum (diluted 1:100). Antibody-untreated WT L. rhamnosus GG served as a positive adhesion control. As an additional control, radiolabeled cells of the L. rhamnosus LC705 strain were treated with or without anti-MBF serum (diluted 1:100). Aliquots (100 μl) of the antiserum-cell suspension were then added to intestinal mucus-coated microtiter plate wells (40) and incubated at 37°C for 1 h. After the wells were rinsed twice with PBS to remove loosely mucus-adhering cells, cells still remaining attached to bound mucus were incubated in lysis solution (1% SDS-0.1 N NaOH) at 60°C for 1 h. The amount of radioactivity in the lysed cell suspensions was then determined by liquid scintillation counting. Adhesion to mucus was expressed as a percentage calculated from the ratio between the amounts of radioactivity measured for the lysed cell suspension and the cell suspension added originally to the wells.
L. rhamnosus cell fractionation and recovery of cell wall proteins.
Cell fractionation and extraction of cell wall-bound proteins from WT L. rhamnosus GG, its EPS (welE) (CMPG5351) knockout mutant derivative, and WT L. rhamnosus LC705 were carried out as described previously (21), with some modifications. In brief, after each of the L. rhamnosus strains were grown overnight, a 1-ml volume of their cells (OD600 of 4.0) was centrifuged, and the pelleted cells were then rinsed once with PBS. Cells were then lysed mechanically three times for 120 s by glass bead disruption (Bühler Vibrogen-Zellmühle) and, after a 0.5-ml volume of PBS was added, low-speed centrifugation (1,000 × g for 1 min) was used to remove the unbroken cells and cellular debris from the disrupted cell suspension. The clarified cell lysate was then fractionated by centrifugation at 16,000 × g for 30 min at 4°C to separate cytosolic and cell wall-associated components. After the removal of the cell-free cytosolic supernatant fraction, cell wall proteins were recovered by resuspension of the pellet fraction in 50 μl of digestion buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 5 mM CaCl2, 10 mg/ml lysozyme, and 150 U/ml mutanolysin) and then incubation at 37°C for 3 h.
Immunoblotting detection of MBF protein in L. rhamnosus cell wall extracts.
Cell wall-associated proteins recovered from WT L. rhamnosus GG, its EPS knockout mutant strain (CMPG5351), and WT L. rhamnosus LC705 were analyzed by SDS-PAGE followed by immunoblotting essentially as described previously (21). All samples were electrophoresed under denaturing conditions on 4 to 15% gradient gels (Bio-Rad) and then electroblotted onto polyvinylidene difluoride (PVDF) Immobilon P membranes (Millipore). Membranes were treated with anti-MBF serum diluted 1:10,000 and then incubated with goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad) diluted 1:100,000. MBF protein was visualized by enhanced chemiluminescence using the Amersham ECL Advance Western blotting detection kit (GE Healthcare Bio-Sciences, Uppsala, Sweden) according to the protocol recommended by the manufacturer.
Electron microscopy and immunogold labeling of L. rhamnosus cells with MBF-specific antiserum.
Immunogold transmission electron microscopy (TEM) of L. rhamnosus GG, its inactivated EPS knockout mutant (CMPG5351), and L. rhamnosus LC705 was performed according to a published procedure (21), but with minor modifications. Briefly, overnight-grown L. rhamnosus cells were recovered by centrifugation and the cell pellets were rinsed once with PBS and then diluted (OD600 of 1.0) in the same buffer. Formvar-carbon-coated copper grids were incubated for 1 h on droplets of the diluted L. rhamnosus cell suspensions, rinsed several times with 0.02 M glycine in PBS, and immersed in a blocking solution of 1% BSA in PBS. The grids were then incubated for 1 h on droplets of blocking solution containing anti-MBF serum (diluted 1:100), rinsed extensively with 0.1% BSA in PBS to wash away unbound antibodies, and treated for 20 min with protein A conjugated to 10-nm-diameter gold particles (pAg) diluted 1:55 in blocking solution. Incubation of the grids with rabbit preimmune serum was also included and served as the negative control. After rinsing four times with PBS, each of the grids was fixed with 1% glutaraldehyde, rinsed eight times with distilled water, and then negatively stained with 1.8% methylcellulose-0.4% uranyl acetate. The grids were examined and the magnified images of L. rhamnosus cells were obtained with a JEOL 1200 EX II transmission electron microscope.
Statistical analysis.
To account for the statistical significance of the mucus-binding data for radiolabeled proteins (125I) or L. rhamnosus cells (3H), pairwise comparisons were performed among the measured samples using Student's t test. Calculated P values of 0.05 or less were regarded as significant.
RESULTS AND DISCUSSION
Domain organization and primary structure comparison of L. rhamnosus GG MBF protein.
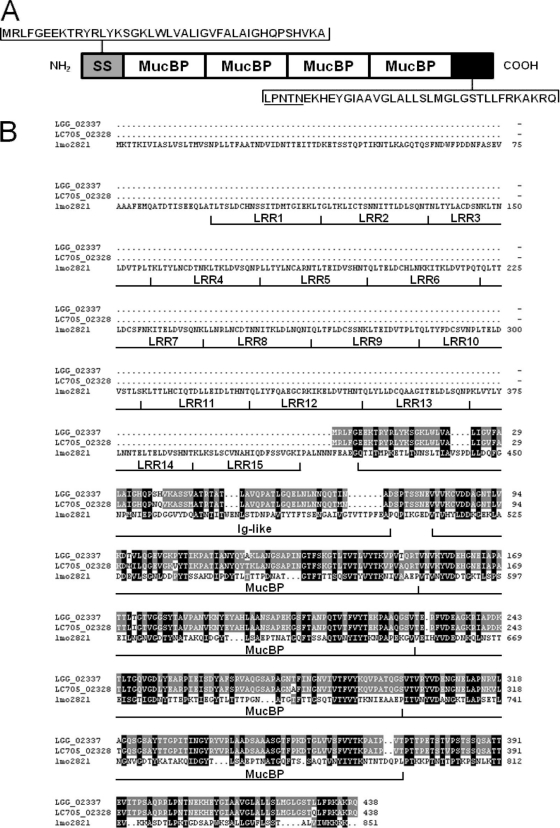
In previous work, the predicted 438-aa product of the LGG_02337 ORF in L. rhamnosus GG was identified as a potential cell wall-associated adhesin (21) which we now call the mucus-binding factor (MBF) protein. Here, in our present analysis of the MBF primary structure, we determined the sequence for a 41-aa N-terminal secretion signal by using the SignalP 3.0 prediction server (5) (http://www.cbs.dtu.dk/services/SignalP/) (Fig. 1A). Moreover, we manually identified a 37-aa C-terminal LPXTG-like anchoring domain beginning with the LPNTN motif, followed by a stretch of predominantly aliphatic amino acids, and ending with a cluster of four positively charged residues (Fig. 1A). Since the presence of both domains is an essential prerequisite for sortase-catalyzed attachment of cell wall proteins, there is little doubt that MBF is a substrate for sortase action, and thus the processed ∼38-kDa protein is likely to be a cell wall-anchored surface component. Given that the deduced amino acid sequence for MBF protein contains four domain repeats with homology to the Pfam-MucBP domain (Fig. 1A) (21), a mucus-binding specificity for this LPXTG-like protein would be a likely possibility. Interestingly, although some homology has been reported to exist between the MucBP repeats and the LAB-associated MUB domains (7), a primary structure alignment between the L. rhamnosus GG MBF protein and the characteristic N-terminal region of the MUB domain (7) did not produce any substantive sequence similarity (data not shown). This seems to suggest that the MBF protein is different from the MUB-containing proteins found in other lactobacilli.
Fig. 1.
Structural organization and amino acid sequence comparison of L. rhamnosus GG MBF protein. (A) A schematic illustration (not to scale) of the domain organization for MBF protein is based on the deduced primary structure of the LGG_02337 ORF from L. rhamnosus GG. Amino acid sequences for the N-terminal secretion signal (SS; gray box) and the C-terminal cell wall-anchoring domain (black box) beginning with the LPNTN motif (underlined) are depicted. Four regions in the primary structure with homology to the Pfam-MucBP domain repeats (MucBP; white box) determined previously (21) are shown. (B) The multiple-amino-acid sequence alignment of L. rhamnosus GG MBF protein (LGG_02337) with the MBF homolog from L. rhamnosus LC705 (LC705_02328 ORF) and the InlJ protein (lmo2821) from L. monocytogenes EGD-e was performed using the MultAlin program (12) (http://multalin.toulouse.inra.fr/multalin/multalin.html) and formatted with the GeneDoc program (27) (http://www.nrbsc.org/gfx/genedoc/index.html). The location of domains for 15 leucine-rich repeats (LRR), one Ig-like interrepeat (Ig-like), and four Pfam-MucBP repeats (MucBP) in L. monocytogenes InlJ protein are shown, and the corresponding sequences are indicated according to results described previously (34). Amino acids that are similar between the sequences for the L. rhamnosus GG and LC705 MBF proteins (gray shading) and for the L. rhamnosus MBF proteins and the L. monocytogenes InlJ protein (black shading) are indicated.
Our results (data not shown) obtained from BlastP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches against existing databases for proteins with sequence homology to the predicted LGG_02337 ORF product revealed a high level of similarity with proteins from taxonomically related species of L. rhamnosus GG, including Lactobacillus paracasei, Lactobacillus casei, and other L. rhamnosus strains. Moreover, moderate amounts of similarity were observed with proteins primarily from species of Listeria and, to a lesser extent, Enterococcus. As shown in Fig. 1B, L. rhamnosus GG MBF protein and the predicted homolog from the related L. rhamnosus LC705 strain are nearly identical (97.9%) to each other based on an alignment of their amino acid sequences. Presently, the LGG_02337 ORF in L. rhamnosus GG and the LC705_02328 ORF in L. rhamnosus LC705, each of which encode the MBF protein, are annotated as an internalin J (InlJ) protein. However, despite displaying a reasonable amount of amino acid sequence similarity with the four Pfam-MucBP repeats in the lmo2821 ORF-encoded InlJ from the L. monocytogenes EGD-e strain (18) (Fig. 1B), neither of the two L. rhamnosus MBF ORFs encode a sequence for the internalin domain, an N-terminal canonical structural feature consisting of ∼15 leucine-rich repeat domains and one immunoglobulin (Ig)-like interrepeat domain (6). Because the internalin domain is implicated in the functionality of InlJ for mediating protein-protein interactions during the infection process of L. monocytogenes (10), it seems unlikely that MBF would have a similar role, let alone be a member of the internalin protein family, without the presence of this important functional domain in the primary structure. Consequently, we suggest that the likely primary role of MBF is for cell-mediated mucosal adhesion and that the renaming of this LPXTG-like surface protein and its gene (designated mbf) is appropriate.
L. rhamnosus GG MBF adheres to human intestinal mucus.
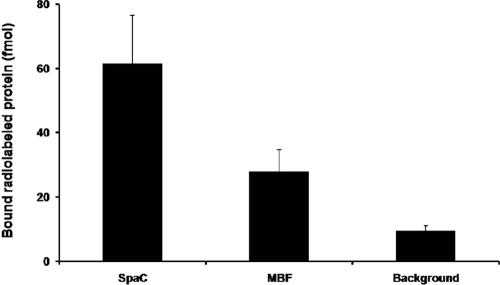
With the aim of providing physical evidence for the predicted role of L. rhamnosus GG MBF as a mucus-binding adhesin, we used purified recombinant MBF protein to assess its adherence to human intestinal mucus by utilizing a microtiter plate-based assay method developed previously for radioiodinated recombinant-produced pilin protein (21, 41). As demonstrated in the mucus-binding profile shown in Fig. 2, recombinant MBF readily adhered to intestinal mucus, although the amount of binding was about 2-fold less than that of the pilus-derived SpaC protein (positive control). Expectedly, mucus binding for recombinant MBF and SpaC both exceeded the measured background (ovalbumin) level by 3- and 6-fold, respectively (Fig. 2). Taken together, these results indicate that the ability of L. rhamnosus GG MBF protein to bind to a mucosal surface is fully consistent with a predicted function based on shared primary structural homology with a known mucus adhesion domain (see previous section).
Fig. 2.
Adherence of L. rhamnosus GG MBF protein to human intestinal mucus. The binding of 50-pmol amounts of 125I-labeled recombinant MBF protein and SpaC pilin to intestinal mucus was carried out according to methods described in Materials and Methods. The mucus adherence of 50 pmol radioiodinated ovalbumin represented the background level of binding to mucus. All binding data represent means ± standard deviations for four to six measurements and the differences between data (P ≤ 0.05) are considered significant. Additional details about the statistical analysis are described in Materials and Methods.
Cell surface localization of MBF in L. rhamnosus GG. Immuno-electron microscopy.
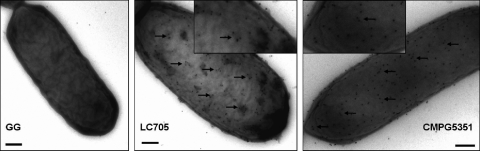
To determine the availability of the MBF for cell-mediated adherence to the intestinal mucosa, we used electron microscopic means for demonstrating its localization as a cell wall-associated surface protein. However, despite several attempts to show the presence of MBF proteins on the cell surface of L. rhamnosus GG, we were unable to do so convincingly with the WT strain by immunogold TEM using antiserum directed against recombinant MBF protein. Our TEM experiments revealed that MBF proteins are not easily visible on the surface of L. rhamnosus GG cells, which was apparent from the absence of observed gold particles (Fig. 3, left). Nonetheless, given the high degree of sequence similarity between MBF and its homolog from L. rhamnosus LC705 and the likelihood for shared recognition of both proteins by L. rhamnosus GG-derived MBF antiserum, we also examined the L. rhamnosus LC705 strain to assess the functionality of the MBF-specific antibodies. In contrast to L. rhamnosus GG or when preimmune serum was used (data not shown), a high number of gold particles was observed on the cell surface of L. rhamnosus LC705 (Fig. 3, center). This not only indicated that the MBF antiserum was functional but also provided visual evidence for the surface localization of the MBF homolog in this related L. rhamnosus strain. Since these two genetically similar L. rhamnosus strains have been reported to differ in the composition of their EPS layers (21, 23), we suspected that the longer galactose-rich EPS repeating units found in L. rhamnosus GG, but not in L. rhamnosus LC705, might be the reason for the observed difference in surface-localized MBF protein. To test this possibility, we examined the uncovered surface of an inactivated EPS knockout mutant of L. rhamnosus GG (CMPG5351), which was used previously for establishing the EPS layer as a potential shield of certain surface adhesins (23). As shown in Fig. 3 (right panel), but not when using preimmune serum (data not shown), gold particles were visibly found throughout the exposed surface of the EPS-deficient mutant cell, providing suggestive evidence that MBF is a frequently occurring surface component of L. rhamnosus GG.
Fig. 3.
Visualization of cell surface-localized MBF in L. rhamnosus GG. Immunogold labeling with anti-MBF serum and electron microscopy analysis of the WT L. rhamnosus GG (GG; left) and LC705 (LC705; center) strains and the L. rhamnosus GG EPS-deficient welE mutant strain (CMPG5351; right) were carried out as described in Materials and Methods. Arrows randomly identifying small dark dots indicate gold particle labeling of MBF protein. The scale bar in each panel represents 0.2 μm. An inset shows an enlarged view of cells from the panels of the L. rhamnosus LC705 and L. rhamnosus GG welE mutant strains.
Immunoblotting.
To confirm whether the L. rhamnosus GG EPS layer is merely shielding the MBF epitopes and that comparable amounts of cell wall-bound MBF protein are produced in L. rhamnosus GG and its EPS-deficient mutant derivative (CMPG5351), mutanolysin-extracted cell wall proteins were recovered from the WT and CMPG5351 strains and then examined for the presence of the MBF by immunoblotting with anti-MBF serum. Cell wall proteins extracted from the L. rhamnosus LC705 strain were included as a control. Mature or fully processed monomeric MBF protein, which lacks the residues of the N-terminal secretion signal and the C-terminal region after the cleavage site in the LPXTG-like motif, and recombinant-produced MBF could be expected to correspond to predicted molecular weights of 38.2 and 39.8 kDa, respectively. As shown in Fig. 4, protein bands of similar intensity migrating just below the 40-kDa marker were detected for the WT (lane 1) and EPS-deficient mutant (lane 2) L. rhamnosus GG strains. As anticipated, similar-sized bands for the L. rhamnosus LC705 strain (Fig. 4, lane 3) and purified recombinant MBF protein (Fig. 4, lane 4) were also apparent on the immunoblot. Unexpectedly, but in part due to an inexact fractionation process, we also observed in these three strains additional bands that likely represent MBF during the different stages of protein processing and peptidoglycan anchoring. For example, based on calculated sizes, we consider the band migrating near the 50-kDa molecular size marker to be completely unprocessed MBF (∼46 kDa) that still contains the N- and C-terminal signaling domains, whereas the faint diffuse band found just above 40 kDa likely represents a partially processed form of MBF (∼42 kDa) with an intact C-terminal membrane-spanning domain.
Fig. 4.

Production of cell wall-associated MBF in L. rhamnosus GG. Immunoblotting with anti-MBF serum of mutanolysin-extracted cell wall proteins from normalized cultures of WT L. rhamnosus GG (lane 1), the L. rhamnosus GG EPS knockout mutant (CMPG5351) (lane 2), and WT L. rhamnosus LC705 (lane 3) was performed as described in Materials and Methods. Purified recombinant MBF protein was included as a control (lane 4). The positions of unprocessed (UP), partially processed (PP), and fully processed (FP) forms of MBF protein are indicated on the right. Molecular weight markers and their positions are indicated on the left.
We interpret the above findings by suggesting that the more extended repeating units of carbohydrate associated with the surrounding L. rhamnosus GG EPS layer, while initially allowing the recognition of cell wall-bound MBF protein by antibody molecules to occur, might later form a physical obstruction that limits the accessibility of the immunoglobulin Fc region to which the protein A-gold particles normally bind. However, even so, our electron microscopic observations, which revealed that MBF is scattered regularly throughout the cell surface of both L. rhamnosus GG and LC705, corroborate the predicted localization of this mucus adhesin as a cell wall-anchored surface protein.
MBF-mediated adhesion of L. rhamnosus GG and LC705 to intestinal mucus.
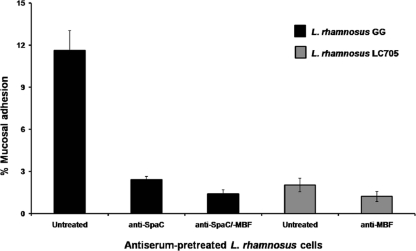
To associate the physical availability of surface-localized MBF proteins with cell-mediated mucosal adhesion, we investigated if this cell wall-associated adhesin contributes to the mucus-binding capacity of L. rhamnosus GG and LC705 cells by performing antibody-mediated inhibition experiments. Using SpaC pilin-specific antiserum to remove the contribution of SpaCBA pili to mucosal adhesion in WT L. rhamnosus GG, we were able to normalize the level of mucus binding for L. rhamnosus GG and LC705, thereby allowing a comparative assessment of MBF-mediated adherence between these two strains. As shown in Fig. 5, WT L. rhamnosus GG cells that were antibody untreated or pretreated with anti-SpaC serum adhered to mucus much in the same way as was reported previously (21, 41), such that SpaC-specific antibody binding to SpaCBA pili blocked a large portion of L. rhamnosus GG-mediated adherence to mucus. Interestingly, when L. rhamnosus GG (anti-SpaC serum-pretreated) and L. rhamnosus LC705 (non-SpaCBA-piliated) cells were treated with anti-MBF serum, we found that the blocking action of the antibodies had caused a relatively modest but statistically significant reduction in mucus binding for each strain (Fig. 5). As these results would imply, MBF proteins are responsible for mediating a low level of mucosal adhesion in L. rhamnosus GG and LC705 cells. Consequently, whereas MBF likely represents one of the key mucus adhesins on the cell surface of the less adhesive L. rhamnosus LC705 strain, this surface-localized protein accounts for only a small fraction of the total mucus-binding capacity in L. rhamnosus GG. For that reason, MBF protein in SpaCBA-piliated L. rhamnosus GG, much like MabA protein described previously (29), will likely have a lesser adhesive role in establishing intimate interactions with intestinal cells.
Fig. 5.
MBF antibody-mediated inhibition of mucus adhesion in L. rhamnosus GG and LC705. Metabolically labeled (3H) cells from normalized cultures of the WT L. rhamnosus GG and LC705 strains were pretreated with the corresponding antiserum, and their capacity for mucus adhesion was determined according to the method described in Materials and Methods. The binding data are means ± standard deviations for at least three measurements, and the differences between data are considered significant (P ≤ 0.05). Further information about the statistical analysis is described in Materials and Methods.
Intriguingly, even with its inherent EPS layer (23), which was recently reported to wield a shielding effect against host protective factors (24), WT L. rhamnosus GG cells are apparently able to mediate mucosal adhesion through the surface-localized MBF. This might reflect the need for a closely interwoven association between the L. rhamnosus GG cell surface and the mucosal layer so that MBF proteins can bind optimally to mucus-related molecules. In keeping with the binding strategies of proposed adhesion models for various other piliated Gram-negative and Gram-positive bacteria, we suggest that only after the elongated mucus-specific pilus fibers make their initial contact and attach L. rhamnosus GG cells to the mucosal surface will MBF proteins be in such close proximity that their binding also becomes a possibility. However, unlike Gram-negative pili, which, due to the structural elasticity of their subunit constituents, would be able to retract reversibly and pull microbes closer to the host cells (17, 20, 26), pili from Gram-positive bacteria appear to consist of inextensible pilin subunits (2), making them more rigid and less apt to function in a similar manner. Consequently, the SpaCBA pilus-mediated binding in L. rhamnosus GG that brings LPXTG-like surface adhesins (e.g., MBF and MabA) closer to the mucus layer could expectedly be less dynamic and more arbitrary, possibly relying on the “zipper-like” mechanistic explanation of adhesion in piliated Gram-positive pathogens described previously (38). On the other hand, collectively, such a multiplicity of adhesive interactions, including those of MBF, will undoubtedly allow L. rhamnosus GG to be better embedded in the intestinal mucosa for what can be regarded as its increased capacity of persistence in the intestine. In comparison, L. rhamnosus LC705, by being demonstratively less adhesive to mucosal surfaces (39), due likely to the absence of mucus-binding pili, is more susceptible to the washout conditions of the intestine and is a less persistent strain (21).
Conclusions.
In our present study, we have now provided evidence that an additional LPXTG-like protein, which we renamed the mucus-binding factor, participates as another surface-exposed component in the interaction between adherent L. rhamnosus GG and the human intestinal mucosa. As an allochthonous probiotic, L. rhamnosus GG is incapable of forming a lifelong association with the intestinal host. However, based on our findings about MBF, together with those from previous studies on the SpaCBA pilus (21) and MabA (29), we put forward the idea that in L. rhamnosus GG, at least these three different surface-localized components are part of a protein-mediated adhesion mechanism, whose collective network of mucus-binding interactions might contribute to the increased capacity of this probiotic strain to survive as a comparatively more persistent and less stringent allochthonous intestinal inhabitant.
ACKNOWLEDGMENTS
This investigation was supported financially by Academy of Finland research grants 118165 and 117877 and was part of the Center of Excellence in Microbial Food Safety Research (MiFoSa) at the University of Helsinki and the Academy of Finland coordinated Research Program on Nutrition, Foods, and Health (ELVIRA). This study was also partly supported by a grant (G.0236.07) from the Fund for Scientific Research Flanders (FWO-Vlaanderen). Willem M. de Vos is a Finland Distinguished Professor and funded by the Finnish Funding Agency for Technology and Innovation (TEKES). Sarah Lebeer is funded as an FWO-Vlaanderen postdoctoral scientist.
We thank Ilkka Palva (University of Helsinki) for insightful discussions and comments. Seppo Salminen (Functional Foods Forum, University of Turku), Riitta Korpela, Tuomas Salusjärvi, Riina Kekkonen, and Soile Tynkkynen (Valio Ltd., Research and Development, Helsinki, Finland) are also thanked for their support throughout this project. Esa Pohjolainen, Outi Lyytinen, Katariina Kojo, and Marko Sutinen are kindly acknowledged (University of Helsinki) for their excellent technical assistance during protein purification. We also thank Heikki Huhtinen (Department of Surgery, Turku University Central Hospital) for supplying surgically resected specimens of human intestinal tissue and Satu Vesterlund (Functional Foods Forum, University of Turku) for providing extracted intestinal mucus. The Electron Microscopy Unit (Institute of Biotechnology, University of Helsinki) is acknowledged for providing laboratory facilities.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Alander M., et al. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alegre-Cebollada J., Badilla C. L., Fernández J. M. 2010. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J. Biol. Chem. 285:11235–11242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azcarate-Peril M. A., et al. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bateman A., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 6. Bierne H., Sabet C., Personnic N., Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156–1166 [DOI] [PubMed] [Google Scholar]
- 7. Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. 2006. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152:273–280 [DOI] [PubMed] [Google Scholar]
- 8. Boekhorst J., Wels M., Kleerebezem M., Siezen R. J. 2006. The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with the environment. Microbiology 152:3175–3183 [DOI] [PubMed] [Google Scholar]
- 9. Bonetti A., Morelli L., Campominosi E. 2002. Assessment of the persistence in the human intestinal tract of two probiotic lactobacilli Lactobacillus salivarius I 1794 and Lactobacillus paracasei I 1688. Microb. Ecol. Health Dis. 14:228–232 [Google Scholar]
- 10. Bublitz M., et al. 2008. Crystal structure and standardized geometric analysis of InlJ, a listerial virulence factor and leucine-rich repeat protein with a novel cysteine ladder. J. Mol. Biol. 378:87–96 [DOI] [PubMed] [Google Scholar]
- 11. Buck B. L., Altermann E., Svingerud T., Klaenhammer T. R. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:8344–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Champs C., Maroncle N., Balestrino D., Rich C., Forestier C. 2003. Persistence of colonization of intestinal mucosa by a probiotic strain, Lactobacillus casei subsp. rhamnosus Lcr35, after oral consumption. J. Clin. Microbiol. 41:1270–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dicks L. M. T., Botes M. 2010. Probiotic lactic acid bacteria in the gastro-intestinal tract: health benefits, safety and mode of action. Beneficial Microbes 1:11–29 [DOI] [PubMed] [Google Scholar]
- 15. Du L., Liu F., Ju X., Huo G. 2010. Adhesion capability of first two domains at N terminus of NP_785232 protein and their interaction with a UV-absorbing component from human mucus. Lett. Appl. Microbiol. 51:400–405 [DOI] [PubMed] [Google Scholar]
- 16. Edwards C. A., Parrett A. M. 2002. Intestinal flora during the first months of life: new perspectives. Br. J. Nutr. 88(Suppl. 1):S11–S18 [DOI] [PubMed] [Google Scholar]
- 17. Fällman E., Schedin S., Jass J., Uhlin B. E., Axner O. 2005. The unfolding of the P pili quaternary structure by stretching is reversible, not plastic. EMBO Rep. 6:52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 19. Harlow E., Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20. Jass J., et al. 2004. Physical properties of Escherichia coli P pili measured by optical tweezers. Biophys. J. 87:4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kankainen M., et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinoshita H., et al. 2008. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde 3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104:1667–1674 [DOI] [PubMed] [Google Scholar]
- 23. Lebeer S., et al. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl. Environ. Microbiol. 75:3554–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lebeer S., Claes I. J. J., Verhoeven T. L. A., Vanderleyden J., De Keersmaecker S. C. J. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb. Biotechnol. 4:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macías-Rodríguez M. E., Zagorec M., Ascencio F., Vázquez-Juárez R., Rojas M. 2009. Lactobacillus fermentum BCS87 expresses mucus- and mucin-binding proteins on the cell surface. J. Appl. Microbiol. 107:1866–1874 [DOI] [PubMed] [Google Scholar]
- 26. Miller E., Garcia T., Hultgren S., Oberhauser A. F. 2006. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 91:3848–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicholas K. B., Nicholas H. B., Jr., Deerfield D. W. I. I. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNET.NEWS 4:14 http://www.embnet.org/files/shared/EMBnetNews/embnet_news_4_2.pdf [Google Scholar]
- 28. Ouwehand A. C., et al. 2002. Resected human colonic tissue: new model for characterizing adhesion of lactic acid bacteria. Clin. Diagn. Lab. Immunol. 9:184–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perea Vélez M., et al. 2010. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol. Med. Microbiol. 59:386–398 [DOI] [PubMed] [Google Scholar]
- 30. Pretzer G., et al. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187:6128–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reuter G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43–53 [PubMed] [Google Scholar]
- 32. Rojas M., Ascencio F., Conway P. L. 2002. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68:2330–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roos S., Jonsson H. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433–442 [DOI] [PubMed] [Google Scholar]
- 34. Sabet C., Lecuit M., Cabanes D., Cossart P., Bierne H. 2005. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 73:6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salminen S., et al. 2010. Interaction of probiotics and pathogens—benefits to human health? Curr. Opin. Biotechnol. 21:157–167 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 37. Soriani M., Telford J. L. 2010. Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol. 5:735–747 [DOI] [PubMed] [Google Scholar]
- 38. Telford J. L., Barocchi M. A., Margarit I., Rappuoli R., Grandi G. 2006. Pili in Gram-positive pathogens. Nat. Rev. Microbiol. 4:509–519 [DOI] [PubMed] [Google Scholar]
- 39. Tuomola E. M., Ouwehand A. C., Salminen S. J. 2000. Chemical, physical and enzymatic pre-treatments of probiotic lactobacilli alter their adhesion to human intestinal mucus glycoproteins. Int. J. Food Microbiol. 60:75–81 [DOI] [PubMed] [Google Scholar]
- 40. Vesterlund S., Paltta J., Karp M., Ouwehand A. C. 2005. Measurement of bacterial adhesion—in vitro evaluation of different methods. J. Microbiol. Methods 60:225–233 [DOI] [PubMed] [Google Scholar]
- 41. von Ossowski I., et al. 2010. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 76:2049–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74:4985–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]