Abstract
Choline is abundant in association with eukaryotes and plays roles in osmoprotection, thermoprotection, and membrane biosynthesis in many bacteria. Aerobic catabolism of choline is widespread among soil proteobacteria, particularly those associated with eukaryotes. Catabolism of choline as a carbon, nitrogen, and/or energy source may play important roles in association with eukaryotes, including pathogenesis, symbioses, and nutrient cycling. We sought to generate choline analogues to study bacterial choline catabolism in vitro and in situ. Here we report the characterization of a choline analogue, propargylcholine, which inhibits choline catabolism at the level of Dgc enzyme-catalyzed dimethylglycine demethylation in Pseudomonas aeruginosa. We used genetic analyses and 13C nuclear magnetic resonance to demonstrate that propargylcholine is catabolized to its inhibitory form, propargylmethylglycine. Chemically synthesized propargylmethylglycine was also an inhibitor of growth on choline. Bioinformatic analysis suggests that there are genes encoding DgcA homologues in a variety of proteobacteria. We examined the broader utility of propargylcholine and propargylmethylglycine by assessing growth of other members of the proteobacteria that are known to grow on choline and possess putative DgcA homologues. Propargylcholine showed utility as a growth inhibitor in P. aeruginosa but did not inhibit growth in other proteobacteria tested. In contrast, propargylmethylglycine was able to inhibit choline-dependent growth in all tested proteobacteria, including Pseudomonas mendocina, Pseudomonas fluorescens, Pseudomonas putida, Burkholderia cepacia, Burkholderia ambifaria, and Sinorhizobium meliloti. We predict that chemical inhibitors of choline catabolism will be useful for studying this pathway in clinical and environmental isolates and could be a useful tool to study proteobacterial choline catabolism in situ.
INTRODUCTION
Catabolism of host-derived molecules is an important link regulating the interactions between eukaryotic hosts and their associated bacterial pathogens, commensals, and symbionts. One host-derived molecule that is important in a number of bacterium-host interactions is the quaternary amine alcohol choline. Choline is present on and around eukaryotes in its free form (10, 23), as a moiety on the soluble molecules choline-O-phosphate and choline-O-sulfate (13, 31), and as the head group moiety on sphingomyelin and phosphatidylcholine (46). Catabolism of host-derived choline is important in many bacteria for generation of the osmoprotectant glycine betaine (GB), which has been linked to enhanced bacterial survival in high-salinity environments (4, 22). For Pseudomonas aeruginosa, catabolism of choline generates GB, which can be further catabolized to dimethylglycine (DMG) (Fig. 1), both of which induce transcription of the hemolytic phospholipase C (plcH) via the GbdR transcription factor (33, 36, 40). P. aeruginosa production of PlcH during infection is important for virulence in a number of animal models (17, 29, 30, 43). Thus, for P. aeruginosa, catabolism of choline directly regulates virulence factor production.
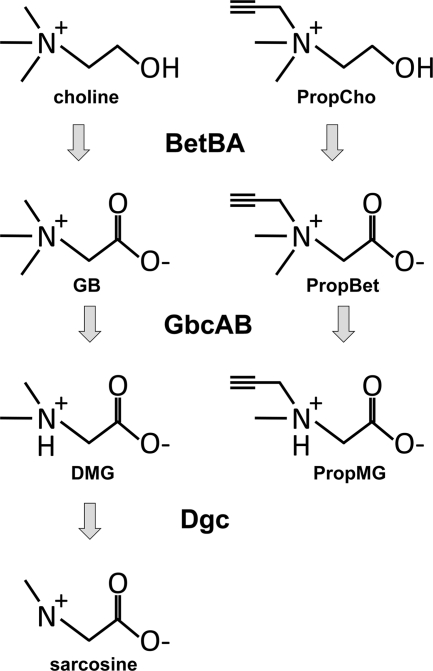
Fig. 1.
Diagram of the choline catabolic pathway in P. aeruginosa. The structures of choline and its catabolic intermediates are shown on the left. PropCho and its predicted catabolic intermediates are shown on the right. Enzymes required for catabolism are noted for each catabolic step.
In order to be catabolized or used as an osmoprotectant, quaternary amines like choline and GB must be transported into the cell. Such transport is typically mediated though ABC family transporters (15, 19) or BCCT family symporters (45, 47). P. aeruginosa possesses the most complex bacterial choline uptake system described to date, which highlights the importance of choline for the organism's physiology (24). Choline transport in P. aeruginosa is accomplished by three major transporters, the ABC transporter, CbcXWV, and two BCCT family symporters, BetT1 and BetT3 (7, 24). These transporters differ in their contributions to growth and osmoprotection in differing salt concentrations, such that CbcXVW and BetT1 contributed most under hypo- and iso-osmolar conditions whereas BetT3 appears to be the primary transporter under hyperosmolar conditions (24). The transcription factor BetI appears to control betT1 and betT3 transcription, while GbdR induces expression of cbcXVW (24). The BetT3 transporter is also regulated by an osmosensory domain, like many BCCT family transporters (6, 24).
A diverse array of environmental bacteria and opportunistic pathogens can catabolize choline aerobically (2, 20). Aerobic choline catabolism involves oxidation of choline to GB, followed by progressive demethylation to form DMG, sarcosine, and finally, glycine (5, 11, 20). P. aeruginosa catabolism of choline is summarized in Fig. 1 (left). While the metabolic intermediates of aerobic choline catabolism are conserved, each chemical conversion can be carried out by members of at least two different enzyme families. Different groups of bacteria differ in their complement of enzymes for each conversion step. The oxidation of choline to form GB can be catalyzed by either monomeric choline oxidases or by concerted action of an oxidase and an aldehyde dehydrogenase, which catalyze the stepwise oxidation of choline to betaine aldehyde and betaine aldehyde to GB (16, 22). GB demethylation can proceed through either a GB methyltransferase (GBMT) or a Rieske dioxygenase-dependent demethylase, with cyanobacteria typical of the former and Pseudomonas possessing the latter (1, 37, 42). Both DMG and sarcosine demethylation can proceed through either single-subunit oxidases, including Arthrobacter dimethylglycine oxidase and Bacillus sarcosine oxidase, or via multisubunit dehydrogenases, as in the Pseudomonads (8, 25, 42). Both monomeric and multimeric forms of the DMG and sarcosine demethylases are flavin-containing enzymes in which the flavin moiety plays a direct role in catalysis (38, 44).
Inhibition of flavin-based oxidases, including mammalian sarcosine oxidase, by acetylenic-substituted substrates has been previously reported (3, 21, 39). The inhibitory mechanism has been shown to involve destabilization of the acetylene group in the active site, which reacts to form a covalent bond with the enzyme-linked flavin (3). Inhibitors of this type have been used to understand the enzymatic mechanisms of N-demethylases in mammalian tissues, where propargyl derivatives of dimethylglycine and sarcosine were shown to covalently bind and inhibit dimethylglycine oxidase and sarcosine oxidase, respectively (21). The utility of these propargyl-substituted inhibitors has not been appreciated in microbial systems. Here we report the characterization of the choline analogue propargylcholine and its metabolites for the study of choline catabolism in bacteria, with a focus on Pseudomonas aeruginosa.
MATERIALS AND METHODS
Strains and growth conditions.
Pseudomonas aeruginosa strains PA14 and PAO1, Pseudomonas putida (ATCC 49128), Pseudomonas fluorescens (ATCC 13525), Pseudomonas mendocina ymp, Burkholderia cepacia (ATCC 25416), Burkholderia ambifaria AMMD, Sinorhizobium meliloti RM1021, and Escherichia coli strains were maintained on LB medium. The P. aeruginosa ΔdgcA mutant strain was previously described (42). When necessary, gentamicin was added to final concentrations of 10 μg/ml for E. coli, 50 μg/ml for P. aeruginosa in LB medium, and 25 μg/ml for P. aeruginosa in morpholinepropanesulfonic acid (MOPS) medium. Growth of all species was measured based on the optical density at 600 nm (OD600) after growth for 24 h in MOPS minimal medium with a 20 mM concentration of the sole carbon source, as described elsewhere (42), except for S. meliloti, which was grown in 0.5× MOPS medium with 10 mM carbon source, and growth was measured at 60 h. When inhibitors were added, their concentrations are noted below in Results and the figure legends. P. aeruginosa, B. cepacia, and Burkholderia cenocepacia were grown at 37°C, while P. putida, P. mendocina, P. fluorescens, B. ambifaria, and S. meliloti were grown at 30°C. Except for S. meliloti, all cultures were started at an OD600 of 0.01. Because of poor growth from low-density S. meliloti cultures, these were started at an OD600 of 0.1.
Creation of the ΔgbcA and ΔbetBA deletion strains.
The constructs to generate the gbcA and betBA deletions were made in the pMQ30 plasmid (35), and unmarked deletions in P. aeruginosa PA14 were made by recombination as described previously (34, 42). Briefly, the upstream and downstream regions of the gbcA gene were amplified by PCR from genomic DNA using the following primers: gbcA-GOI-F, 5′-GACGTTGTAAAACGACGGCCAGTGCCAAGCTTGCATGCCTcttcagggtcgaagtcttgc-3′; gbcA-SOE-R, 5′-AAGTACGAAGGCGACTCGACCATGGTGGgcctggtccatactcgaaga-3′; gbcA-SOE-F, 5′-CCATGGTCGAGTCGCCTTCGTACTTgcagcgagaagctgtggt-3′; gbcA-GOI-R, 5′-TAACAATTTCACACAGGAAACAGCTATGACCATGATTACGgtggacgaagaccatgtcg-3′. Similarly, the analogous regions of the betBA locus were amplified using the following primers: betBA-GOI-F, 5′-GGGACAGGGTCTTGTAGTCG-3′; betBA-SOE-R, 5′-AAGTACGAAGGCGACTCGACCATGGctcaatgccaccaccatcat-3′; betBA-SOE-F, 5′-CCATGGTCGAGTCGCCTTCGTACTTgaccgccaatgtagagcttc-3′; betBA-GOI-R, 5′-ACTTGAAGCCCGGATTCTG-3′. Uppercase letters in sequences in the above primers represent regions used for yeast cloning or splice overlap extension. The splice overlap extension PCR product was cloned into pMQ30 using yeast recombination as described previously (35) for the gbcA knockout (gbcA-KO) construct and by E. coli-based cloning for the betBA-KO construct as described previously (41). Donor E. coli S17/λpir carrying the gbcA-KO or betBA-KO constructs was mated with P. aeruginosa PA14, and single-crossover mutants were selected for growth on gentamicin. Recombinants were verified by PCR after selecting for double-crossover events by growth on 5% sucrose LB plates with no NaCl.
Creation of the dgc operon complementation plasmid.
The dgc operon (PA5396, PA5397, dgcA, and dgcB) was amplified from genomic DNA using the primers dgc-F (5′-TTCTCCATACCCGTTTTTTTGGGCTAGCGAATTCGAGCTCGGTACCCGGGGAAGGAGATAcagatgagcccagccgaattgc-3′) and dgc-R (5′-CCAGTGAAAAGTTCTTCTCCTTTACTCATATGTATATCTCCTTCcttgtcgtggctggagagac-3′), giving rise to a 6.3-kbp fragment. Uppercase letters indicate regions utilized in yeast recombination. This fragment was cloned into the pMQ80 vector by using yeast homologous recombination (35). The resulting plasmid, pDgc, was electrotransformed into P. aeruginosa, and transformants were selected for gentamicin resistance. The pDgc plasmid complements the growth defect of the ΔdgcA mutant for growth on choline, GB, and DMG (data not shown). The cloning strategy places the dgc operon under the control of the pBAD promoter on pMQ80. Addition of l-arabinose is required for complementation of the growth phenotype, although there is low expression from the operon in the uninduced state (data not shown).
Reporter fusion assay for plcH induction.
Plasmid preparations of the plcH-lacZ reporter plasmid pMW22 (40) were transformed into P. aeruginosa by electroporation (9), and transformants were selected for growth on gentamicin. After overnight growth in MOPS minimal medium (27) amended with 20 mM sodium pyruvate and 5 mM d-glucose, cells were pelleted and resuspended in MOPS with 20 mM pyruvate and the indicated concentration of choline. Cells were shaken for 4 h under inducing conditions at 37°C unless otherwise stated. The β-galactosidase assays were conducted according to the methods of Miller (26).
LC/MS and NMR methodology.
Liquid chromatography/mass spectrometry (LC/MS) was performed on an Applied Biosystems 4000QTrap Pro Instrument. 1H nuclear magnetic resonance (NMR) spectra were obtained on a Varian Unity Inova 500-MHz high-field NMR spectrometer, and 13C NMR spectra were obtained on a Bruker AXR 500-MHz high-field NMR spectrometer.
Labeled compounds were exposed to P. aeruginosa in a manner similar to that described previously (34). P. aeruginosa was grown overnight on MOPS minimal medium and 20 mM choline, washed twice with Dulbecco's phosphate-buffered saline (PBS), and either exposed to 10 mM 13C-labeled choline and 1 mM propargylcholine (PropCho) or to 10 mM choline and 1 mM 13C-labeled PropCho for 6 h in MOPS medium. After centrifugation, cells were washed twice with Dulbecco's PBS, the labeled compounds were extracted twice with 80% ethanol, and the extracts were dried. The residue remaining after evaporation of the ethanol was resuspended in 100% D2O and analyzed by 13C NMR. NMR was calibrated to 3-(trimethylsilyl)propane-1-sulfonic acid before each run.
Synthesis and validation of choline analogues.
All chemicals were obtained from Sigma-Aldrich at greater than 98% purity unless otherwise noted in the methods.
Propargylcholine (PropCho).
PropCho [IUPAC N-(2-hydroxyethyl)-N,N-dimethylpropan-2-yn-1-aminium] was synthesized using the strategy of Jao et al. (13) by combining dimethylethanolamine and propargylbromide in tetrahydrofuran (THF). The structure was validated by LC/MS ([M]+ 128.1), 1H NMR (500 MHz, dimethyl sulfoxide [DMSO], δ 5.35 (HO, t) 4.94 (CH2, s) 4.08 (HC, s) 3.86 (CH2, q) 3.50 (CH2, t) 3.17 (C2H6, s), and 13C NMR (500 MHz, D2O) δ 81.7(C, t) 70.8 (CH, t) 65.0 (CH2, s) 55.6 (CH2, s) 55.5 (CH2, s) 51.5 (C2H6, s).
[1,2-13C]PropCho and [1,2-13C]choline.
Synthesis of labeled PropCho was identical to that for unlabeled PropCho, but using labeled [1,2-13C]dimethylethanolamine (custom synthesized by Cambridge Isotopes) in place of the natural abundance dimethylethanolamine. Labeled choline was synthesized similarly to labeled PropCho, with iodomethane in place of propargylbromide.
PropMG.
Propargylmethylglycine {PropMG; IUPAC [methyl(prop-2-yn-1- yl)ammonio]acetate} was synthesized using two different methods. The first was a two-step modified version of that described by Kraus et al. (21). Sarcosine ethyl ester HCl was added to diisopropylethylamine and 95% ethanol. While the reaction mixture was being stirred at room temperature, 80% propargylbromide in toluene was added. The mixture quickly turned a deep yellow. The product, propargylmethylglycine ethyl ester, was purified through a cation-exchange resin column (AG 50W-X8) by the pH-dependent release according to the manufacturer's instructions. The ester was then hydrolyzed to the desired acid by adding NaOH to 10%. PropMG was also synthesized by adding N-methylpropargylamine to chloroacetic acid in 95% ethanol and stirring on ice. The product of each method was validated by 1H NMR (500 MHz, DMSO) δ 9.20 (CO2H, s) 4.07 (CH2, s) 3.63 (CH2, d) 3.44 (HC, t) 2.08 (CH3, s) and 13C NMR (500 MHz, D2O) δ 176.2 (CO2H, s) 80.9 (C, s) 75.8 (CH, s) 45.8 (CH2, s) 40.3 (CH2, s) 34.5 (CH3, s).
PropBet.
Propargyl betaine {PropBet; IUPAC [dimethyl(prop-2-yn-1-yl)ammonio]acetate}was synthesized by adding N-dimethylpropargylamine to chloroacetic acid with stirring in cold 95% ethanol. The structure was validated by 1H NMR (500 MHz, DMSO) 3.68 (CH2, d) 3.53 (CH2, s) 3.41 (HC, s) 3.15 (C2H6, s) and 13C NMR (500 MHz, D2O) δ 171.1 (CO2H, s) 84.1 (C, s) 73.9 (CH, s) 65.7 (CH2, s) 56.7 (CH2, s) 45.1 (CH3, s). The acidic proton was poorly resolved in our aqueous solution, given the low predicted pKa of the proton at ∼1.8.
Sequence alignment and phylogenetic analysis.
The predicted amino acid sequences for homologues of the dgc genes in a select set of proteobacteria were aligned using MUSCLE v3.8.31 (12) and then subjected to both maximum parsimony and Bayesian phylogenetic analyses, respectively, using the TNT v1.1-64bit (14) and MrBayes v.3.2 (32) programs. For the maximum parsimony analysis, the default settings of the xmult function were used plus 10 cycles of tree drifting. Two trees with a best score of 3,841 steps were retained and combined using a Nelson strict consensus approach. Bootstrap values were then calculated using 100 replications of symmetric resampling and analysis of each data set with a single random addition sequence plus tree-bisection-reconnection. The Bayesian analyses consisted of determining the protein model set using the “prset aamodelpr = mixed” function followed by a tree search using the mcmc command to search 100,000 generations.
The following sequences were used in the analysis: NP_254085.1, YP_350951.1, NP_742477.1, YP_003898529.1, YP_001341572.1, YP_270675.1, YP_573046.1, ZP_03697525.1, YP_440059.1, ZP_02414425.1, YP_105246.1, NP_521619.1, YP_299011.1, ZP_02904657.1, YP_623154.1, YP_001583525.1, YP_003674253.1, YP_998776.1, YP_003070153.1, YP_002422769.1, YP_002498618.1, YP_001524092.1, YP_001234136.1, YP_743894.1, ZP_07298244.1, NP_108417.1, ZP_01013251.1, ZP_01443608, YP_001169396.1, YP_002520848.1, ZP_04683062.1, NP_355484.1, ZP_07475029.1, ZP_07478605.1, YP_765024.1, YP_473024.1, ZP_07592927.1, ZP_05113079.1, ZP_01547668.1, YP_165436.1, ZP_01036476.1, ZP_02147545.1, ZP_00961317.1, ZP_02141049.1, YP_682470.1, and YP_509470.1.
RESULTS
Propargylcholine potently inhibits P. aeruginosa growth on choline, GB, and DMG.
We synthesized the N-propargyl-substituted choline analogue PropCho (Fig. 1), based on the method of Jao and colleagues (18). Growth experiments demonstrated that PropCho was incapable of supporting P. aeruginosa growth as a sole carbon source, and PropCho was not capable of inducing expression from BetI- or GbdR-controlled promoters (data not shown). However, when PropCho was added at 500 μM to MOPS minimal medium with choline (20 mM) as the sole carbon source, we observed complete inhibition of growth (Fig. 2 A). Further growth analyses demonstrated that PropCho inhibited P. aeruginosa growth on GB and DMG, but not on sarcosine (Fig. 2A). The inhibitory effect was specific for choline catabolism, as growth on pyruvate and glucose was unaffected (Fig. 2A). The inhibitory effect of propargylcholine was greatest for growth on DMG, for which the 50% inhibitory concentration was 5.16 ± 0.71 μM (mean ± standard deviation). Data shown are for P. aeruginosa strain PA14, but similar results were seen for PAO1 (data not shown). Based on our previous studies of these catabolic pathways in P. aeruginosa and the knowledge that propargyl-modified analogues can inhibit N-demethylation in flavin-containing demethylases in mammals (21), we predicted that PropCho would inhibit the Dgc enzyme complex (the predicted PropCho catabolic pathway is diagrammed in Fig. 1, right).
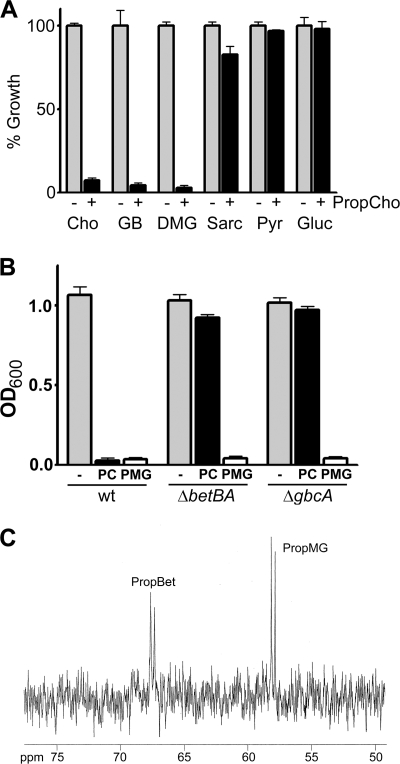
Fig. 2.
Inhibition of P. aeruginosa choline catabolism by propargylcholine and its metabolites. (A) The percent growth of P. aeruginosa PA14 in the absence or presence of 500 μM PropCho, calculated from the OD600 after 24 h of growth in MOPS minimal medium with a 20 mM concentration of the listed carbon source. Growth in the absence of PropCho was set at 100%. (B) Growth of P. aeruginosa wild type (wt), ΔbetBA, and ΔgbcA in the absence and presence of PC and PMG, as measured by absorbance at 600 nm. (C) 13C NMR results of ethanolic extracts from choline-grown cells exposed to [1,2-13C]PropCho. Peaks were validated by comparison to 13C NMR of chemically synthesized PropBet and PropMG. Error bars represent standard deviations of biological triplicates. All data are representative of at least three independent experiments.
Metabolism of propargylcholine to propargylmethylglycine is required for inhibitory activity.
We predicted that PropCho inhibited the Dgc enzymes and, therefore, must be catabolized into its active form, the DMG analogue PropMG (Fig. 1). To test this prediction we compared the ability of the wild type and ΔbetBA and ΔgbcA mutants to grow on DMG in the presence or absence of PropCho. In both mutants, addition of PropCho failed to inhibit growth on DMG (Fig. 2B), supporting our hypothesis that PropCho must be catabolized into an active form by the BetBA and GbcAB enzymes. Our genetic data suggested that PropCho was oxidized by BetBA to form PropBet and demethylated by GbcAB to form PropMG (Fig. 1). To test this hypothesis, we chemically synthesized PropMG. As shown in Fig. 2B, PropMG was able to inhibit growth on DMG without the need to be acted upon by the BetBA or GbcAB enzymes. Although previous work suggested that PropMG inhibition of flavin-containing enzymes would likely be covalent, choline transport rates (7, 24) and levels of DgcA in the cytosol led us to predict that uptake and conversion rates of the inhibitor would provide inhibitor in the cytosol at an abundance that would exceed the prevalence of DgcA catalytic sites. Therefore, we predicted there would be detectable levels of unbound inhibitor remaining in the cytosol. Based on our genetic data, labeled PropCho added to cells growing on choline would be predicted to accumulate as labeled PropMG. We showed that addition of [1,2-13C]PropCho to wild-type cells growing on choline resulted in accumulation of both [1,2-13C]PropMG and [1,2-13C]PropBet (Fig. 2C). Identities of the PropCho metabolites were confirmed by comparison to synthesized standards. Buildup of the 13C-labeled betaine analogue, PropBet, in addition to the labeled DMG analogue, PropMG, was similar to the situation observed with labeled choline in a dgc mutant and is predicted to represent feedback inhibition of gbcAB expression or GbcAB enzyme activity (42).
Evidence that propargylmethylglycine targets the Dgc enzyme complex.
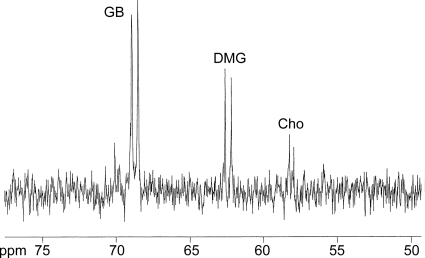
Our genetic evidence and growth data pointed to small-molecule inhibition of the Dgc enzyme complex. This hypothesis was supported by showing that [1,2-13C]choline fed to PropCho-treated cells accumulated as [1,2-13C]DMG and [1,2-13C]GB (Fig. 3). Identities of the choline metabolites were confirmed by comparisons to standards. Similar to the data presented for 13C-labeled PropCho (Fig. 2C), the buildup of 13C-labeled GB in addition to labeled DMG was previously observed in a dgc mutant and is predicted to represent feedback inhibition of gbcAB expression or GbcAB enzyme activity (42). The ability of PropCho to phenocopy a dgc mutation in terms of DMG accumulation strongly supports inhibition of Dgc enzyme activity.
Fig. 3.
NMR analysis for [1,2-13C]choline-exposed cells. Results shown are from 13C NMR of ethanolic extract from P. aeruginosa grown in the presence of 7 mM [1,2-13C]choline and exposed to 500 μM PropCho. Peaks were validated by comparison to carbon NMR of natural abundance GB and DMG as standards. Data are representative of three independent experiments.
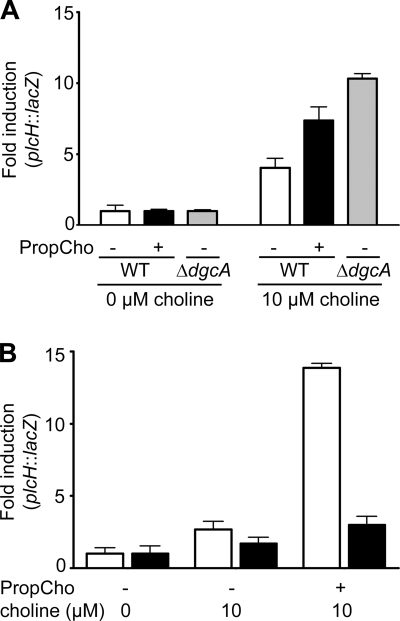
The accumulation of DMG and GB in a ΔdgcA mutant exposed to choline resulted in hyperinduction of GbdR-regulated transcripts, including plcH (Fig. 4 A). Treatment of cells with PropCho phenocopies a dgcA deletion with regard to hyperinduction of plcH reporter activity (Fig. 4A). This further supports the Dgc enzyme complex as the target of PropMG.
Fig. 4.
Evidence that PropMG targets the Dgc enzyme complex in P. aeruginosa. (A and B) Induction (based on changes in Miller units) from a plasmid-based plcH-lacZ reporter fusion (40). (A) plcH reporter activity in the ΔdgcA mutant and wild type (wt) in the absence or presence of 500 μM PropCho. (B) plcH reporter activity in the absence or presence of 500 μM PropCho, with overexpression of the dgc operon from an arabinose-inducible plasmid (pDgc [black bars]) compared to an empty vector control (pMQ80 [white bars]).
The responsiveness of a system to an inhibitor should be dependent on the concentration of the target enzyme. By using hyperinduction of the plcH-lacZ reporter fusion as our readout, we tested whether overexpression of the dgc genes abrogated plcH hyperinduction. As shown in Fig. 4B, expression of the dgc operon from a plasmid (pDgc) resulted in a profound reduction in plcH-lacZ hyperinduction.
Effects of salinity on the inhibitory effects of propargylcholine and propargylmethylglycine.
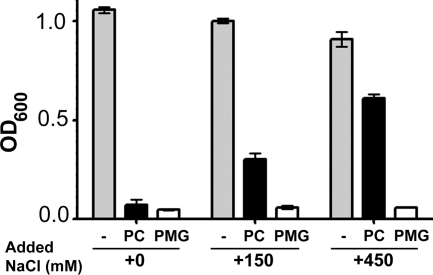
In order to function as an inhibitor, choline analogues need to be transported into the cytosol. The previous experiments were conducted in MOPS medium, which is slightly hypo-osmolar (24). Because regulation and transport specificity change with added salt, we tested the sensitivity of P. aeruginosa to PropCho and PropMG at increasing NaCl concentrations (Fig. 5). Inhibition of P. aeruginosa choline growth by PropMG was not altered under hyperosmolar conditions. However, high salt concentrations decreased the inhibitory effect of PropMG significantly. We predict that this may be due to differing transport specificity between CbcXWV operating under low-osmolarity conditions and BetT3, which appears to be the primary transporter under high-salt conditions (24).
Fig. 5.
Effects of a high salt concentration on choline growth inhibition by PropCho and PropMG. Growth of P. aeruginosa PA14 in the absence and presence of PC and PMG under increasing NaCl concentrations was measured based on the absorbance at 600 nm. Inhibitors were present at 1 mM in these experiments. Error bars represent standard deviations of biological triplicates and are representative of multiple independent experiments.
Propargylcholine and propargylmethylglycine effects on quaternary amine catabolism in other proteobacteria.
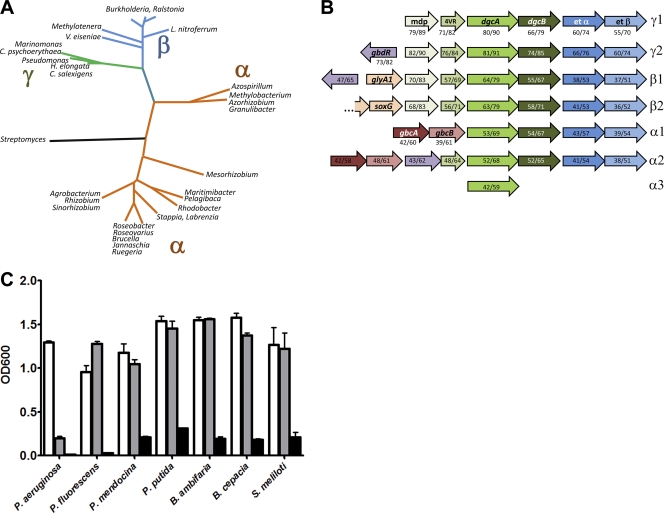
Our data demonstrated that the target of PropMG inhibition is at the level of DMG catabolism. Many of the proteobacteria that grow aerobically on choline have genes for DMG demethylation that are highly homologous to the P. aeruginosa dgcA gene (Fig. 6 A and B). The predicted amino acid sequence for homologues of the dgcA genes in a select set of proteobacteria were aligned and subjected to both maximum parsimony and Bayesian phylogenetic analyses as described in Materials and Methods. All non-Pseudomonas sequences were selected as representatives of their genera in the top 100 BLASTP results by using the predicted DgcA amino acid sequence and excluding the taxon “Pseudomonas.” The resultant summary radial tree (Fig. 6A) is representative of the full tree (see Fig. S1 in the supplemental material, including bootstrap analyses). The phylogenetic analysis revealed a consistent relationship between the predicted DgcA amino acids (as determined from both the maximum likelihood and Bayesian analyses). The predicted dgcA homologues and their surrounding genes are schematically shown in Fig. 6B. We predicted that PropCho and/or PropMG would be capable of inhibiting choline growth in a representative set of choline-catabolizing proteobacteria that carried dgcA homologues. As shown in Fig. 6C, while only P. aeruginosa was susceptible to PropCho, PropMG was capable of inhibiting growth in all of these organisms, although not completely. As controls, PropCho and PMG did not alter S. meliloti growth on succinate, nor did they affect growth on glucose for all other tested species (data not shown).
Fig. 6.
Utility of propargylcholine and propargylmethylglycine in other choline-catabolizing proteobacteria. (A) Radial phylogenetic tree derived from analysis of alignments of predicted amino acid sequences of dgcA homologues. This tree is adapted from the source tree shown in Fig. S1 of the supplemental material. (B) Representation of the dgcA homologues and their genomic surroundings in the major groups depicted in panel A. Only adjacent genes with homology to P. aeruginosa choline catabolism-related genes are shown. The percent similarity over the entire length is noted in parentheses below each homologous gene. γ2′ is the gene arrangement for Chromohalobacter salexigens, and all other gammaproteobacteria fall into the γ1′ arrangement. β2′ is the gene arrangement for Verminephrobacter eiseniae and Methylotenera; all other betaproteobacteria have the β1′ arrangement. The α1′ gene arrangement is found in Methylobacteria spp. and α2′ is found in Granulibacter, Azorhizobium, and Acidophilum species, while all other alphaproteobacteria have the α3′ arrangement. Abbreviations: mdp, membrane dipeptidase homologue; 4VR, 4-vinyl reductase homologue; etα and etβ, α- and β-subunits of an electron transfer protein encoded by PA5400 and PA5401 in P. aeruginosa; dgcA, dgcB, gbcA, gbcB, soxG, glyA1, and gbdR all represent homologues of the corresponding genes in P. aeruginosa. (C) Growth of select proteobacteria in minimal medium with choline (20 mM) as the sole carbon source (10 mM choline for S. meliloti). Each species was grown in choline alone or in choline with the addition of either PropCho or PropMG at 500 μM. Error bars represent standard deviations of biological triplicates. Data are representative of three independent experiments.
DISCUSSION
We have demonstrated that an alkyne-containing choline analogue, PropCho, is processed by P. aeruginosa catabolic enzymes to an active form, PropMG, which inhibits the Dgc enzyme. PropCho and PropMG are specific for inhibition of the choline catabolic pathway. We have demonstrated that the BetBA and GbcAB enzymes are necessary to metabolize PropCho to the inhibitor PropMG. The presence of homologous enzymes for the catabolism of DMG between P. aeruginosa and a large variety of proteobacteria led us to hypothesize that our inhibitors would block choline catabolism in organisms containing these homologous genes. Here we have demonstrated the broad utility of PropMG to inhibit choline catabolism in members of the Alpha-, Beta-, and Gammaproteobacteria. The failure of PropCho to inhibit growth of organisms other than P. aeruginosa suggests some enhanced selectivity in either transport or choline oxidation in the other studied taxa. We suspect that our inhibitors will be useful for dissecting the importance of choline catabolism in less genetically tractable organisms in these groups. By examining the select organisms represented in the phylogenetic tree of the predicted DgcA amino acid sequence (Fig. 6A), we noted a preponderance of organisms with intimate links to eukaryotes, including symbionts, commensals, and pathogens. We therefore hypothesize that choline, GB, and DMG catabolism play an important role in host-microbe interactions that has been underappreciated.
The alignment of the predicted DgcA homologues shows strong conservation of the dgc operon members in the Gamma- and Betaproteobacteria. These members include a putative dipeptidase (PA5396), a predicted 4-vinyl reductase (PA5397), the predicted catalytic subunit (dgcA), a transmembrane protein predicted to be a 4Fe-4S reductase (dgcB), and the α- and β-subunits of an electron transfer protein (PA5400 and PA5401). In Chromohalobacter salexignes and the Betaproteobacteria, the predicted dgc genes are associated with an AraC family transcription factor with predicted homology to the GB/DMG-sensing transcription factor GbdR from P. aeruginosa (42). This association suggests a detection and transcriptional regulation regimen in these organisms that is similar to that described for P. aeruginosa. The gene encoding the predicted GbdR homologue is also found adjacent to the dgc operon in Granulibacter, Azorhizobium, and Acidophilum, suggesting that this transcriptional regulatory mechanism extends to the Alphaproteobacteria.
Small-molecule analogues that can inhibit specific enzymes are an attractive method for evaluation of catabolic pathways for a variety of reasons. First, they necessitate no genetic changes in the organism, enabling conditional blockage of specific pathways upon application of the inhibitor. Second, small-molecule inhibitors enable examination of the pathway of interest in large numbers of isolates and in organisms containing homologous enzymatic pathways that may not be genetically tractable. Finally, small-molecule inhibitors with effects on many different species are of interest to address the role of the specific metabolic pathway in mixed populations and environmental samples.
Even in genetically tractable organisms, there are instances in which generation of mutant strains is not feasible for one reason or another. One good example in P. aeruginosa is the study of large numbers of clinical isolates from cystic fibrosis patients. While a reasonable portion of these clinical isolates are genetically tractable, generating a specific mutation in many tens or hundreds of isolates is not a reasonable proposition for most labs. We propose that PropCho and its metabolites will allow us to examine the role of choline catabolism in the regulation of phospholipase C (plcH) expression in clinical isolates. This is an important question, as choline-inducible phospholipase production has been linked to decreased lung function in P. aeruginosa-infected cystic fibrosis patients (28). In a more general fashion, to emphasize an idea that has been reiterated many times by others, we predict that inhibitors and other small-molecule probes will complement existing genetic tools, bringing us a step closer to turning our genetic knowledge into therapeutic tools.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeanne Harris (University of Vermont) for the S. meliloti isolate, Ken Hampel (University of Vermont) for useful discussions and critical reading, and Adrian Salic (Harvard University) for helpful suggestions regarding propargylcholine synthesis.
This research was supported by grants to M.J.W. from the NIH NCRR (P20-RR021905) and the Cystic Fibrosis Foundation (WARGO09F0).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Barra L., et al. 2006. Interrelations between glycine betaine catabolism and methionine biosynthesis in Sinorhizobium meliloti strain 102F34. J. Bacteriol. 188:7195–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bater A. J., Venables W. A. 1977. The characterisation of inducible dehydrogenases specific for the oxidation of D-alanine, allohydroxy-D-proline, choline and sarcosine as peripheral membrane proteins in Pseudomonas aeruginosa. Biochim. Biophys. Acta 468:209–226 [DOI] [PubMed] [Google Scholar]
- 3. Binda C., et al. 2004. Crystal structures of monoamine oxidase B in complex with four inhibitors of the N-propargylaminoindan class. J. Med. Chem. 47:1767–1774 [DOI] [PubMed] [Google Scholar]
- 4. Boch J., Kempf B., Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boncompagni E., Osteras M., Poggi M. C., le Rudulier D. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 65:2072–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C., Beattie G. A. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J. Bacteriol. 190:2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C., Malek A. A., Wargo M. J., Hogan D. A., Beattie G. A. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chlumsky L. J., Zhang L., Jorns M. S. 1995. Sequence analysis of sarcosine oxidase and nearby genes reveals homologies with key enzymes of folate one-carbon metabolism. J. Biol. Chem. 270:18252–18259 [DOI] [PubMed] [Google Scholar]
- 9. Choi K.-H., Kumar A., Schweizer H. P. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397 [DOI] [PubMed] [Google Scholar]
- 10. Csonka L. N., Hanson A. D. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569–606 [DOI] [PubMed] [Google Scholar]
- 11. Diab F., et al. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395–1406 [DOI] [PubMed] [Google Scholar]
- 12. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fitzgerald J. W., Luschinski P. C. 1977. Further studies on the formation of choline sulfate by bacteria. Can. J. Microbiol. 23:483–490 [DOI] [PubMed] [Google Scholar]
- 14. Goloboff P. A., Farris J. S., Nixon K. C. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24:774–786 [Google Scholar]
- 15. Horn C., et al. 2006. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357:592–606 [DOI] [PubMed] [Google Scholar]
- 16. Ikuta S., Imamura S., Misaki H., Horiuti Y. 1977. Purification and characterization of choline oxidase from Arthrobacter globiformis. J. Biochem. 82:1741–1749 [DOI] [PubMed] [Google Scholar]
- 17. Jander G., Rahme L. G., Ausubel F. M. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182:3843–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jao C. Y., Roth M., Welti R., Salic A. 2009. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. U. S. A. 106:15332–15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kempf B., Bremer E. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270:16701–16713 [DOI] [PubMed] [Google Scholar]
- 20. Kortstee G. J. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71:235–244 [PubMed] [Google Scholar]
- 21. Kraus J.-L., Yaouanc J.-J., Sturtz G. 1975. Role of the substituants N-allyl and N-propargyl on the mechanism of flavoprotein enzyme inhibition: N-demethylase and monoamineoxidase. Eur. J. Med. Chem. 10:507–513 [Google Scholar]
- 22. Landfald B., Strom A. R. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165:849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Rudulier D., Strom A. R., Dandekar A. M., Smith L. T., Valentine R. C. 1984. Molecular biology of osmoregulation. Science 224:1064–1068 [DOI] [PubMed] [Google Scholar]
- 24. Malek A. A., Chen C., Wargo M. J., Beattie G. A., Hogan D. A. 8 April 2011. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J. Bacteriol. doi:10.1128/JB.00160-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meskys R., Harris R. J., Casaite V., Basran J., Scrutton N. S. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur. J. Biochem. 268:3390–3398 [DOI] [PubMed] [Google Scholar]
- 26. Miller J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring, NY [Google Scholar]
- 27. Neidhardt F. C., Bloch P. L., Smith D. F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nguyen D., et al. 2007. Clinical response to azithromycin in cystic fibrosis correlates with in vitro effects on Pseudomonas aeruginosa phenotypes. Pediatr. Pulmonol. 42:533–541 [DOI] [PubMed] [Google Scholar]
- 29. Ostroff R. M., Vasil M. L. 1987. Identification of a new phospholipase C activity by analysis of an insertional mutation in the hemolytic phospholipase C structural gene of Pseudomonas aeruginosa. J. Bacteriol. 169:4597–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rahme L. G., et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 31. Rivoal J., Hanson A. D. 1994. Choline-O-sulfate biosynthesis in plants (identification and partial characterization of a salinity-inducible choline sulfotransferase from species of Limonium (Plumbaginaceae). Plant Physiol. 106:1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 33. Sage A. E., Vasil A. I., Vasil M. L. 1997. Molecular characterization of mutants affected in the osmoprotectant-dependent induction of phospholipase C in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 23:43–56 [DOI] [PubMed] [Google Scholar]
- 34. Schweizer H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831–834 [PubMed] [Google Scholar]
- 35. Shanks R. M., Caiazza N. C., Hinsa S. M., Toutain C. M., O'Toole G. A. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shortridge V. D., Lazdunski A., Vasil M. L. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol. 6:863–871 [DOI] [PubMed] [Google Scholar]
- 37. Waditee R., Incharoensakdi A. 2001. Purification and kinetic properties of betaine-homocysteine methyltransferase from Aphanothece halophytica. Curr. Microbiol. 43:107–111 [DOI] [PubMed] [Google Scholar]
- 38. Wagner M. A., Khanna P., Jorns M. S. 1999. Structure of the flavoco- enzyme of two homologous amine oxidases: monomeric sarcosine oxidase and N-methyltryptophan oxidase. Biochemistry 38:5588–5595 [DOI] [PubMed] [Google Scholar]
- 39. Walsh C. T., Schonbrunn A., Lockridge O., Massey V., Abeles R. H. 1972. Inactivation of a flavoprotein, lactate oxidase, by an acetylenic substrate. J. Biol. Chem. 247:6004–6006 [PubMed] [Google Scholar]
- 40. Wargo M. J., Ho T. C., Gross M. J., Whittaker L. A., Hogan D. A. 2009. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect. Immun. 77:1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wargo M. J., Hogan D. A. 2009. Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155:2411–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wargo M. J., Szwergold B. S., Hogan D. A. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J. Bacteriol. 190:2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wiener-Kronish J. P., et al. 1993. Alveolar epithelial injury and pleural empyema in acute P. aeruginosa pneumonia in anesthetized rabbits. J. Appl. Physiol. 75:1661–1669 [DOI] [PubMed] [Google Scholar]
- 44. Willie A., Edmondson D. E., Jorns M. S. 1996. Sarcosine oxidase contains a novel covalently bound FMN. Biochemistry 35:5292–5299 [DOI] [PubMed] [Google Scholar]
- 45. Wood J. M. 2007. Bacterial osmosensing transporters. Methods Enzymol. 428:77–107 [DOI] [PubMed] [Google Scholar]
- 46. Zeisel S. H. 1990. Choline deficiency. J. Nutr. Biochem. 1:332–349 [DOI] [PubMed] [Google Scholar]
- 47. Ziegler C., Bremer E., Kramer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78:13–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.