Abstract
In Gal repressosome assembly, a DNA loop is formed by the interaction of two GalR, bound to two distal operators, and the binding of the histone-like protein, HU, to an architecturally critical position on DNA to facilitate the GalR–GalR interaction. We show that GalR piggybacks HU to the critical position on the DNA through a specific GalR–HU interaction. This is the first example of HU making a specific contact with another protein. The GalR–HU contact that results in cooperative binding of the two proteins to DNA may be transient and absent in the final repressosome structure. A sequence-independent DNA-binding protein being recruited to an architectural site on DNA through a specific association with a regulatory protein may be a common mode for assembly of complex nucleoprotein structures.
Keywords: Transcription repression, DNA looping, protein–protein interaction, HU mutants
Nonspecific DNA-binding proteins are ubiquitous among all organisms. These proteins have wide-ranging effects on DNA conformation, including bending, supercoiling, and compaction (Drlica and Rouviere-Yaniv 1987; Ner et al. 1994; Werner and Burley 1997; Thomas and Travers 2001). The archetype of prokaryotic histone-like protein, HU, binds to DNA with little or no sequence specificity, although it has a preference for DNA containing cruciforms or single-stranded breaks (Bianchi 1994; Nash 1996). HU also bends the DNA to which it binds. The bending induced by HU facilitates optimal protein–protein and/or protein–DNA contacts in an otherwise physically recalcitrant structure, thereby orchestrating the regulated assembly of functional nucleoprotein complexes, such as those involved in DNA replication, recombination, transposition, transcription, and DNA repair (Skarstad et al. 1990; Dri et al. 1992; Haykinson and Johnson 1993; Manna and Gowrishankar 1994; Li and Waters 1998). By playing an architectural role, HU configures and stabilizes DNA conformations required for sustaining a higher-order complex for active DNA metabolic transactions. Consistent with the idea of HU being a passive architectural partner in the nucleoprotein complexes, there is little evidence of HU being involved in any direct contacts with other proteins.
Regulation of transcription initiation in the gal operon of Escherichia coli involves a DNA–multiprotein complex, called repressosome, in which HU plays an essential role. The gal operon is driven by two partially overlapping promoters, P1and P2 (Fig. 1). The binding of two repressor (GalR) dimers to the two spatially separated operators, OE and OI, and of HU to a site (hbs) in between the two operators in negatively supercoiled DNA forms the Gal repressosome, containing a DNA loop. DNA looping, a consequence of interaction between the operator-bound GalR dimers, engenders inhibition of transcription from both promoters (Aki and Adhya 1997; Geanacopoulos et al. 1999; Lewis et al. 1999). The role of HU in the formation of the repressosome is distinguished by the following features (Aki and Adhya 1997): (1) Binding of HU to the hbs occurs at 20-fold lower concentration than its affinity of 10–7M for DNA (Cann et al. 1995). Once bound, HU cannot be competed out of the repressosome by excess unbound HU or heparin, indicating that HU is a stable component of the final nucleoprotein complex, unlike in the HU-containing Mu transposome (Lavoie and Chaconas 1990) or Hin invertosome (Paull et al. 1993) structures. (2) HU is specific in Gal repressosome and cannot be replaced by other bacterial histone-like proteins such as IHF, HNS, or Fis, although it can be replaced by one of the eukaryotic high-mobility group proteins, HMG-17, at a 10-fold higher molar concentration; successful substitution of HU with other bacterial architectural proteins has been shown in hin inversion and Mu DNA transposition (Lavoie and Chaconas, 1994; Paull et al. 1994). Conversely, homologous repressors, like GalS and LacI, when provided with their cognate operators, cannot replace GalR in bringing about HU binding. (3) Finally, there is a tripartite cooperativity between GalR and HU in binding to gal DNA; binding of HU to hbs is absolutely dependent on binding of GalR dimers to both operators and HU binding, in turn, results in increasing the strength of GalR binding. The binding of GalR dimers to OE and OI in the absence of HU is noncooperative (Brenowitz et al. 1990). The synergistic binding of GalR and HU, the strength of HU binding, and the specificity of GalR and HU in the repressosome may originate from a functional interaction between GalR and HU. In this paper, we provide structure-based genetic and biochemical evidence to show a specific and functional GalR–HU interaction, both in vivo and in vitro.
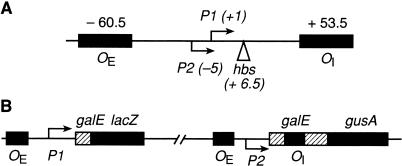
Figure 1.
(A) Map of the gal promoter region showing the location of the P1 and P2 promoters and the HU binding site, hbs. (B) Schematic diagram of the OE+ P2-P1+OI-∼lacZ and OE+P2+P1-OI+∼gusA fusions.
Results
Structure-based genetic analysis of HUα
In E. coli, HU is a heterodimer of two highly homologous subunits, HUα and HUβ. Previous studies showed that the deletion of either of the corresponding genes, hupA or hupB, did not affect the repression of the gal promoters, suggesting that either HU homodimer can substitute functionally for the heterodimer (Aki et al 1996; Lewis et al. 1999). To study any critical GalR–HU interaction, we performed site-directed mutagenesis of the hupA gene to identify any HUα mutants that would be specifically defective in the formation of the repressosome while retaining their ability to bind to DNA with normal affinity. Based on a modeled structure of E. coli HUα homodimer derived from the Bacillus stearothermophilus HU homodimer X-ray and nuclear magnetic resonance structure (Tanaka et al. 1984; Vis et al. 1995), we identified 10 amino acid residues for targeted mutagenesis, following two criteria: (1) the amino acid residues were surface exposed, and (2) they were not on the DNA-binding β-sheet arms or the dimerization interface of the protein (Nash 1996). The amino acid residues at the chosen positions were substituted by residues found at corresponding positions in HU of other bacterial species. The high degree of homology between their primary structures helped us to select amino acids from analogous positions in different species of HU for substitution in HUα, so as to minimize the possibility of destabilization of the mutant proteins (Bordo and Argos 1991). The list of native and substituted amino acid residues in HUα is in Table 1.
Table 1.
Amino acid substitutions in HUα
| Position
|
Native amino acid
|
Substituted amino acid
|
|---|---|---|
| 4 | threonine | alanine |
| 5 | glutamine | aspartate |
| 8 | aspartate | serine |
| 12 | glutamate | phenylalanine |
| 17 | serine | proline |
| 18 | lysine | alanine |
| 19 | threonine | aspartate |
| 22 | lysine | glycine |
| 30 | alanine | glutamate |
| 38 | glutamate | lysine |
The substituted amino acids were chosen from amino acids at the corresponding positions in HU from other species (see text).
HUα mutants defective in repression of gal promoters
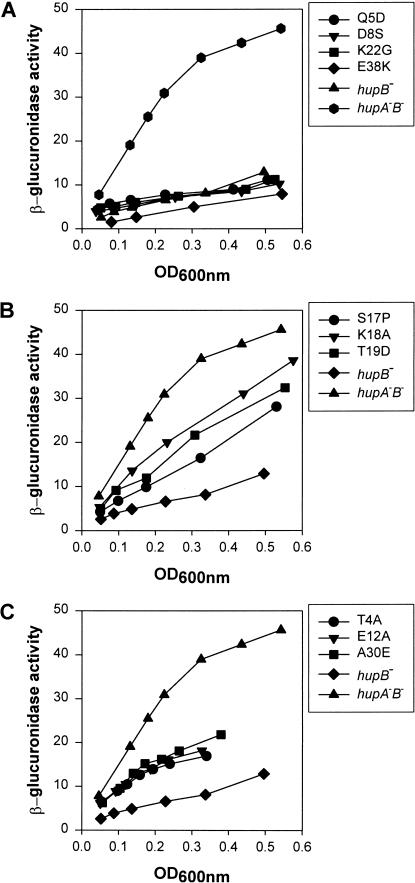
The ability of the different HUα mutants to repress transcription of the gal operon by the formation of the DNA loop was determined in vivo in the reporter strain SK22, which carries a chromosomal fusion of the P2 promoter of gal to the reporter gene gusA. The P1 promoter was inactivated by mutation. In the presence of GalR and HUα, β-glucuronidase synthesis is repressed from the P2∼gus fusion (Lewis et al. 1999). The level of expression of β-glucuronidase from P2 provides a measure of the efficiency of DNA loop formation in this strain. The mutant hupA genes, described in Table 1, were transferred to the chromosome of strain SK22, which is deleted for hupB, to study their effects on P2 repression. The results of β-glucuronidase assays in these strains are shown in Figure 2. Based on these results, we divided the HUα mutants into three groups: (1) Q5D, D8S, and K22G substitutions were as efficient as the wild type in repression of the P2 promoter, whereas E38K repressed P2 better than the wild type (Fig. 2A). (2) S17P, K18A, and T19D substitutions caused derepression of the P2 promoter, presumably because of defects in the formation of the repression loop. Compared with the wild type, these three mutants showed a 2.5- to 4-fold higher rate of β-glucuronidase synthesis (Fig. 2B). (3) T4A, E12A, and A30E substitutions showed partial derepression of the P2 promoter (Fig. 2C). Because T4A, E12A, and A30E showed significantly longer cell doubling times than the wild-type strain at 37°C and the hupA–hupB double null mutant was considerably defective in its growth rate (data not shown), the HU mutations in this class most probably resulted in nonfunctional proteins, leading to impaired growth. The hupA mutants in the first two groups showed no appreciable changes in growth characteristics. The three mutants in the second group, S17P, K18A, and T19D, which were defective in galP2 repression in vivo, were studied further.
Figure 2.
Effect of hupA mutations on DNA looping-dependent repression of the galP2 promoter. Strains containing wild-type or mutant hupA genes were grown in minimal media at 37°C and assayed for expression of the P2∼gusA fusion gene. β-Glucuronidase activities are expressed as change in optical density at 405 nm versus cell density at 600 nm during enzyme assay.
Characterization of the HUα mutants: S17P, K18A, and T19D
We measured the cellular levels of the HUα variants to verify that the results obtained in the P2∼gus reporter gene assays were not due to a difference in their levels in comparison to that of the wild type. Analysis of cell extracts of the three mutants using immunoblotting with antiserum specific to HU showed no apparent deviation in the expressed HUα levels in strains carrying S17P, K18A, or T19D mutation when compared with that in the wild type (data not shown).
Furthermore, we tested whether these HUα mutants that were impaired in P2 repression of gal were compromised in their other cellular functions. Mini-P1 plasmids are not stably maintained in hupA–hupB double null mutants because of the deficiency of ori2-dependent mini-P1 DNA replication in the absence of HU protein (Ogura et al. 1990). The transformation efficiency of mini-P1 plasmids on the three HUα mutants showed no difference when compared with that of the wild-type strain (Table 2, column 1). Bacteriophage Mu is also unable to replicate in hupA–hupB double null mutants (Kano et al. 1989). Growth of phage Mu was tested by measuring the efficiency of plating on lawns of wild type and the P2 repression-defective HU mutants. The plaque-forming efficiency of phage Mu was virtually the same on the wild type and the three HU mutants (Table 2, column 2).
Table 2.
Mini-Pl plasmid replication and Mu bacteriophage development in looping-defective HU mutants
| Strain
|
No. of transformants/μg of plasmid DNA
|
No. of plaques/mL of phage
|
|---|---|---|
| hupA+B+ | 2.85 × 104 | 3.14 × 109 |
| hupA−B− | 0 | <1 × 103 |
| SKS17P | 3.18 × 104 | 2.78 × 109 |
| SKK18A | 2.96 × 104 | 3.03 × 109 |
| SKT19D | 3.15 × 104 | 2.82 × 109 |
The numbers are averages of three independent sets of experiments.
Effect of HUα mutant proteins on gal transcription in vitro
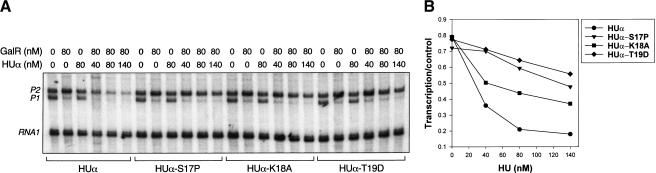
Wild-type and mutant HUα homodimers (S17P, K18A, and T19D) with a hexa-histidine tag at the N-termini were expressed in E. coli and purified as described in Materials and Methods. The purified proteins were used to study their effects as modulators of repression of gal transcription in vitro (Fig. 3A). In the absence of GalR and HU, transcription from wild-type gal template generated two transcripts, a 125-nucleotides long P1 RNA and a 130-nucleotides long P2 RNA (Choy and Adhya 1993). The presence of GalR alone caused repression of P1 with a concurrent activation of P2, as expected. Presence of both GalR and wild-type HUα caused simultaneous repression of both P1 and P2. Wild-type HUα repressed P2 with a concentration-dependent profile, attaining >80% repression at 80 nM HUα (Fig. 3B). However, in the case of the three HUα mutants, S17P, K18A, and T19D, when used at similar concentrations, repression of P2 was significantly reduced for all of them. The level of P2 transcription in the presence of T19D, for example, was similar to that with GalR alone.
Figure 3.
(A) Transcription of galP1 and galP2 promoters in the presence of GalR or wild-type HUα and HUα mutants, S17P, K18A, or T19D. Concentration of GalR was 80 nM; concentrations of HU were 40 nM, 80 nM, and 140 nM, as shown at the top. The 80-bp RNA1 served as an internal control. Details are in the text. (B) Graphical representation of the repression data for galP1 promoter. Transcription is expressed as the percentage of that which was obtained in the absence of GalR.
DNA binding of the HUα mutants
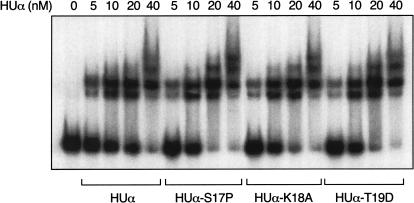
We performed an electrophoretic mobility-shift assay to compare the mutant HUα proteins with the wild-type HUα for their ability to bind to linear DNA. Binding of successive molecules of HU dimers generated nested complexes that were progressively more retarded in electrophoretic mobilities (Fig. 4). Wild-type HUα and the three HUα mutants, at equal concentrations, gave rise to identical retarded species, indicating that within the range of sensitivity and resolution of this assay, the mutants were as proficient as the wild type in DNA-binding.
Figure 4.
Electrophoretic mobility-shift assay of wild-type HUα and HUα mutants with a 266-bp gal DNA fragment, as described in Materials and Methods. Complexes were formed with 0.1 nM DNA and 5 nM, 10 nM, 20 nM, and 40 nM of HUα (lanes 2–4), S17P (lanes 6–8), K18A (lanes 10–12), and T19D (lanes 14–16).
Coimmunoprecipitation of GalR and wild-type HU
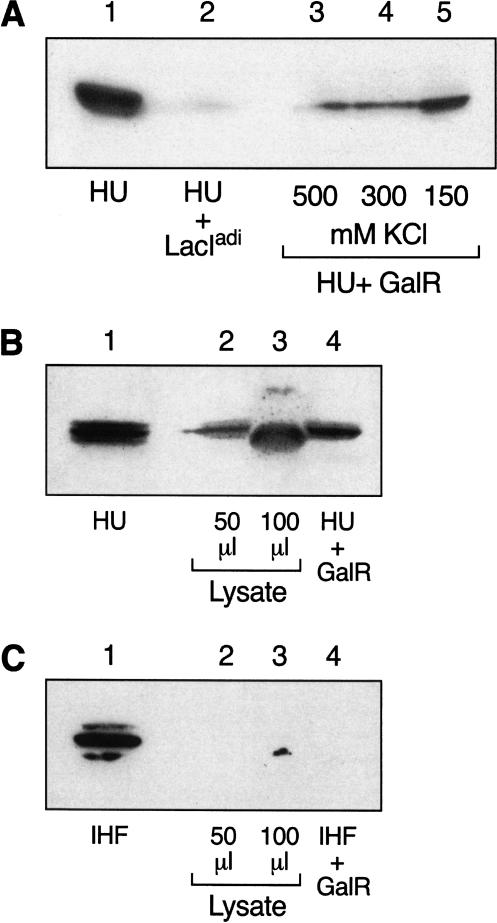
We investigated the potentiality of a physical interaction between GalR and HU biochemically by coimmunoprecipitation reactions. Coimmunoprecipitation using purified GalR and HU with antiserum specific to GalR consistently revealed the presence of HU in these immunocomplexes, as ascertained by Western blotting with HU-specific antiserum. Complex formation was detected even at high salt concentration, indicating a specificity of GalR–HU interaction (Fig. 5A; lanes 3–5). As a control, we used a homologous protein, LacIadi (Brenowitz et al. 1991), instead of GalR, with LacIadi antiserum to pull down the complexes. As shown in Figure 5A, lane 2, LacIadi and HU complex formation was barely detectable under the lowest salt concentration that was used in reactions involving GalR. To test whether GalR forms a specific complex with HU in vivo, we transformed wild-type E. coli cells with an expression vector plasmid encoding GalR with a hexa-histidine tag at the C-terminal end. This was done to make the in vivo concentrations of GalR and HU comparable for easy detection of GalR–HU interaction by immunoprecipitation: The normal in vivo concentration of GalR is 40 dimers per cell (Tokeson 1989) whereas that of HU is 30,000 dimers per cell (Rouviere-Yaniv and Kjeldgaard 1979). After induction of GalR, the cellular extracts were analyzed for GalR–HU complexes by immunoprecipitating with anti-His antibody and immunoblotting with antiserum specific for HU. The histidine-tagged GalR coimmunoprecipitated with HU (Fig. 5B, lanes 2,3). The same extracts were also immunoblotted with anti-IHF antibody to check whether GalR coimmunoprecipitated with IHF, a protein that is highly homologous with HU (Oberto et al. 1994). GalR did not form any complexes with IHF either in crude extracts or with purified proteins (Fig. 5C, lanes 2–4).
Figure 5.
Immunoprecipitation of GalR and HU. (A) Purified GalR and HU. Equimolar amounts of GalR and HU were mixed together in buffer containing varying concentrations of salt. (Lane 3) 150 mM KCl; (lane 4) 300 mM KCl; (lane 5) 500 mM KCl. The proteins were immunoprecipitated with anti-GalR antibody and protein A-agarose. (Lane 2) Equimolar amounts of LacIadi and HU were mixed together in 150 mM KCl and precipitated with anti-LacIadi antibody. (Lane 1) HU. Samples were separated by SDS-PAGE and immunoblotted with anti-HU antibody. (B) GalR and HU in crude cell extracts. BL21(DE3) cells were transformed with plasmid pAP2 containing histidine-tagged GalR coding region under the T7 promoter. Following IPTG induction, cell extracts were made and immunoprecipitated with anti-His monoclonal antibody and protein A-agarose. Samples were divided in two, separated by SDS-PAGE, and immunoblotted either with anti-HU antibody (B) or anti-IHF antibody (C). (Lane 1) HU or IHF; (lane 2) 20 μL cell extract; (lane 3) 40 μL cell extract; (lane 4) Purified GalR with either HU or IHF.
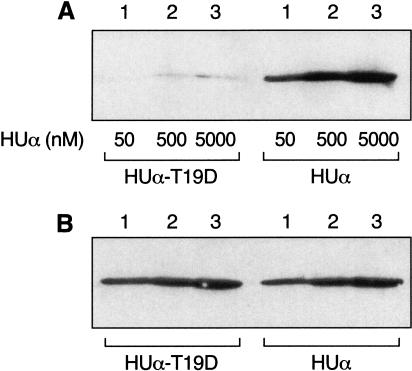
Coimmunoprecipitation of HUα mutants with GalR
We determined whether the repression-defective HUα mutants were impaired in their ability to interact with GalR in immunoprecipitation reactions. Equal amounts of GalR were mixed with different concentrations of wild-type and mutant HUα and precipitated with anti-GalR antibody. Figure 6A shows that, compared with wild type, the HUα mutant T19D was nearly completely defective in interacting with GalR. Figure 6B, which shows the amount of HU protein added to each reaction, confirmed that the differences in the formation of immunocomplexes between the wild-type and mutant HUα were not due to variations in protein concentrations or inadequate antibody recognition by the mutant HUα. HUα mutants S17P and K18A also showed weak interaction with GalR by immunoprecipitation (data not shown).
Figure 6.
Immunoprecipitation of HUα and T19D with GalR. (A) A fixed concentration of GalR (5 nM) was mixed with increasing concentrations of wild-type and mutant HUα, as indicated in the figure. The proteins were immunoprecipitated with anti-GalR antibody followed by immunoblotting with anti-HU antibody. (B) Equivalent amount of HU added to each immunoprecipitation reaction was run on SDS gel and immunoblotted with anti-HU antibody.
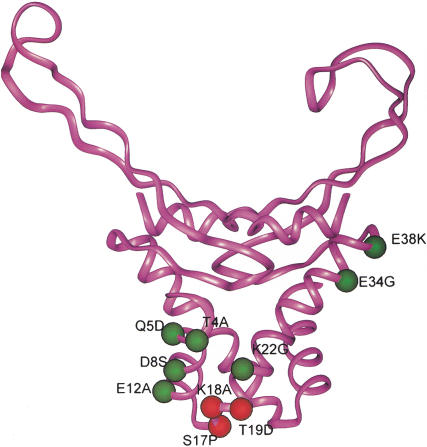
Location of S17, K18, and T19 in the modeled HUα structure
The structure of HU heterodimer in E. coli has not yet been determined experimentally. We used the crystal structure of the HU homodimer (HBs) from B. stearothermophilus to model the E. coli HU. Because the HUα of E. coli and HBs from B. stearothermophilus share a 59% sequence homology (Drlica and Rouviere-Yaniv 1987), the actual three-dimensional structure of HBs was used as a prototype for HUα. The amino acid residues S17, K18, and T19 in HUα, which when substituted showed defect in repression of gal transcription and in interaction of GalR with HU, are located contiguously in a small turn between the first and second alpha helices (Fig. 7). This region lies on the opposite face of the DNA-binding surface, in a prominently accessible portion of HU. All three amino acids have solvent-exposed side chains and are likely candidates to contact GalR.
Figure 7.
Three-dimensional model of the HUα dimer showing the location and spatial arrangement of the substituted residues. The amino acid substitutions are shown in green or red in one subunit. The three substitutions, which specifically disrupted looping-mediated repression, are shown in red. Mutations that had no effect on repression are shown in green.
Discussion
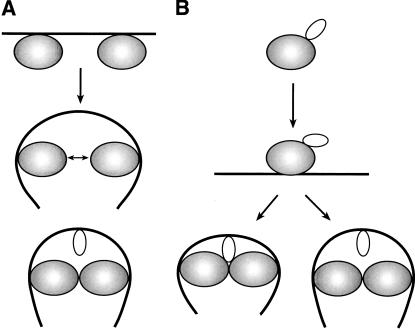
A piggyback model for HU recruitment
In the absence of a stable interaction between the two DNA-bound GalR dimers, the formation of a DNA loop must overcome the energetic barrier imposed by the axial rigidity of the 113-bp DNA segment between the two gal operators. The small nonspecific DNA-binding histone-like protein HU engenders DNA looping and hence, repressosome formation, by binding to and bending the intervening DNA segment by a tripartite cooperativity between itself and the two DNA-bound GalR dimers. The role of HU in the cooperative binding of HU and GalR to DNA can be explained by two models. In model A, the two operator-bound GalR dimers transiently interact and bend the intervening DNA region, facilitating HU binding to the bent region (Fig. 8A). HU binding energetically stabilizes the interaction between the two DNA-bound GalR dimers, resulting in the observed cooperativity. This is the preferred model in the formation of several repressosome-like structures, for example, intasome, transpososome, and enhanceosome. In model B, a direct protein–protein interaction between HU and GalR brings about cooperative binding (Fig. 8B). In this model, HU-bound GalR binds to the gal operators, thereby bringing HU to the vicinity of the gal region and facilitating its DNA binding, which in turn stabilizes DNA-bound GalR tetramerization. We investigated the potentiality of a GalR–HU interaction in the formation of the cooperative complex both genetically and biochemically. By site-directed mutagenesis, we scanned the surface of the HUα homodimer to identify amino acid residues that, when altered, would be defective in DNA looping without the loss of their intrinsic DNA-binding activity. Our experiments discovered three substitutions of HUα— S17P, K18A, and T19D—that were unable to repress the P2 promoter of gal in vivo but retained their ability to support several HU-dependent cellular functions; cells carrying the substitutions showed normal bacterial growth, mini-P1 plasmid replication, and bacteriophage Mu growth. In vitro, the altered HUα proteins showed substantial defect in repression of P2 transcription and exhibited no discernible difference in their DNA-binding ability. We think that the altered function of the mutant proteins originate from a failed interaction with GalR, a specific interaction essential for the assembly of the repressosome structure. Consistently, we demonstrated a complex formation between GalR and wild-type HU by coimmunoprecipitation reactions using both crude extracts and purified proteins. A GalR–HU interaction was also demonstrated by sedimentation ultracentrifugation analysis and fluorescence studies (S. Roy and M. Geanacopoulos, in prep.) and by SELDI protein chip assays (S. Kar and B. Martin, unpubl.). Next, we showed that the repression-defective HUα proteins are defective in forming the immunocomplexes with GalR. In the modeled structure of the HUα dimer, the three residues, S17, K18, and T19, whose replacements resulted in defective transcriptional repression and GalR interaction, are prominently exposed in a peptide turn between two alpha helices on the face of HU near the N-terminal region and farthest from the DNA-binding C-domain and are accessible for interaction with other proteins. From the results described earlier, we conclude that HU specifically interacts with GalR in repressosome formation. The demonstration of an essential GalR–HU contact in the repressosome assembly supports model B described earlier. This is the first instance where a DNA sequence-independent, nucleoid-associated protein has been shown to have a functional interaction with another protein to perform its architectural role in the formation of a higher-order nucleoprotein structure. The formation of complex nucleoprotein structures in which nonspecific DNA-binding proteins are targeted to specific DNA locations by interaction with sequence-specific DNA-binding proteins has been implicated in other cases; for example, the interactions of the HMG1 and HMG2 proteins with p53 (Jayaraman et al. 1998), with steroid hormone receptors (Boonyaratanakornkit et al. 1998), with Hox proteins (Zappavigna et. al. 1996), and with Oct-1 and -2 (Zwilling et al. 1995) in the eukaryotic systems.
Figure 8.
Role of HU and GalR in the formation of Gal repressosome. (A) Two GalR dimers interact transiently to bend the DNA for the HU dimer to bind. (B) HU forms a complex with GalR and is delivered to a specific site on the DNA. The DNA-bound HU could then either maintain or lose contact with GalR in the final repressosome structure. The larger, shaded molecule represents GalR dimer and the smaller molecule represents HU dimer.
In model B, the delivery of HU by piggyback to its destination on the DNA may be followed by one of the two scenarios (Fig. 8B): (1) After HU binds to the specified position on the DNA, it dissociates from GalR, leaving behind a GalR dimer–dimer contact required for DNA looping; (2) HU binds to DNA while remaining GalR bound. In the proposed models for the assembly of Gal repressosome, the stoichiometry of GalR and HU was not taken into account. Although further studies are needed to distinguish between these two models, we note that energetic considerations favor a model in which HU is not in contact with GalR in the final repressosome structure (Geanacopoulos et al. 2001). Nevertheless, the ability of architectural proteins without any sequence preference to function in specific DNA contexts by virtue of piggyback delivery by a sequence-specific regulatory partner may be a common theme for the assembly of higher-order nucleoprotein structures.
Materials and methods
Bacterial strains and plasmids
DM025 (OE+ P2−P1+OI−∼lacZ and OE+P2+P1−OI+∼gusA, apR, tetR, cmR, Δcya854, ΔhupB) has been described previously (Lewis et al. 1999). KM22, Δ(recC ptr recB recD)::Plac-bet exo kan), was obtained from K.C. Murphy. Strain SK22 was constructed by transduction of DM025 by bacteriophage P1 grown on strain KM22 and selection for kanamycin-resistant colonies. Plasmid pSA509, containing a 288-bp segment of the gal control region (−197 to +91), has been described previously (Choy and Adhya 1993). Maintenance of mini-P1 plasmid and growth of bacteriophage Mu were performed using standard microbiological methods.
Site-directed mutagenesis of HUα
PCR was used to amplify the upstream region (from –1038 to –95) of the hupA gene from wild-type E. coli MG1655 with an EcoRI-containing primer at the 5′ end and a BamHI-containing primer at the 3′ end. The fragment was cloned between the EcoRI–BamHI sites of plasmid pEM7(Zeo) (pSKHU/US). The downstream region of the hupA gene, from +299 (17 bp downstream of the stop codon) to +1005, was amplified using a 5′ end PCR primer containing an HindIII site and a 3′ end PCR primer containing a KasI site. The purified PCR product was subcloned into pSKHU/US at the HindIII–KasI segment, creating the plasmid pSK/USDS. The wild-type hupA and the hupA containing point mutations, generated by PCR, were cloned into pSK/USDS between the upstream and downstream sequences described earlier. The wild-type hupA was amplified from MG1655 using a forward primer, HU/pR3, which contained a BamHI restriction site, and a reverse primer, HU/pR4, which contained a HindIII site. The hupA mutants were generated by a two-step PCR recombination process. The first round of PCR was performed in two reactions, one using the HU/pR3 primer along with one of the mutant downstream primers and the other using the HU/pR4 along with one of the mutant upstream primers. The two PCR products were used in a second round of PCR with the original HU/pR3 and HU/pR4 primers to generate the final mutant hupA fragments. The PCR products containing the wild-type and mutant hupA genes were cleaved with BamHI and HindIII and cloned in pSK/USDS. To clone an antibiotic resistance gene upstream of hupA, we cleaved the plasmids with StuI at a site +248 from the start codon. Plasmid pSE418 (a kind gift from Dr. D. Chattoraj) was used to generate a spectinomycin-resistance cassette. A 2.4-kb BamHI fragment containing the entire spectinomycin resistance gene was cleaved from pSE418, end-filled, and ligated to the StuI site in each of the individual hupA plasmids. The entire cloned DNA region was verified by DNA sequencing in each plasmid.
Chromosomal construction
The mutant hupA genes were transferred from the plasmid into the bacterial chromosome as described by Murphy (1998). A 4.2-kb fragment containing the wild-type or mutant hupA was amplified from the respective plasmid and electroporated into E. coli strain SK22. Spectinomycin-resistant colonies were selected and purified. The chromosomal hupA gene from each spectinomycin-resistant recombinant was sequenced to confirm that the mutations have been transferred to the chromosome.
Assay of β-glucuronidase activity
β-Glucuronidase activities were measured as described previously (Lewis et al. 1999) in log-phase cells growing in M63 supplemented with 0.4% (w/v) d-fructose, 0.1% casamino acids, and 0.0004% (w/v) vitamin B1. The activity of β-glucuronidase was measured by Softmax microplate spectrophotometer system. The rate of β-glucuronide hydrolysis was determined at 405 nm at 37°C.
Purification of HUα
The wild-type hupA and hupA genes containing the mutations S17P, K18A, and T19D, after PCR amplification with primers containing Nde1 and BamH1 sites at the N-terminal and C-terminal ends, respectively, were cloned in expression vector plasmid pET15b (Novagen), which contains an N-terminal hexa-histidine tag. The cloned plasmids were transformed into BL21(DE3) cells (Stratagene). The proteins were induced with 1 mM IPTG for 3 h at an optical density of 0.7. After induction, the cells were harvested by centrifugation and resuspended in ice-cold Buffer A containing 5 mM imidazole, 500 mM NaCl, 10 mM HEPES (pH 7.9), 2 mM PMSF, and 0.1% TritonX-100. The bacteria were lysed by sonication and centrifuged again to remove the cellular debris. The His-tagged proteins were purified by Buffer A equilibrated Ni-NTA agarose (Qiagen). The gel matrix was washed extensively with Buffer B containing 20 mM imidazole, 500 mM NaCl, 10 mM HEPES (pH 7.9), 0.2 mM PMSF, and 0.1% TritonX-100. Proteins were eluted with a step gradient of imidazole ranging from 50 to 250 mM and the fractions were run on 4%–20% Tris-glycine gels. Fractions containing pure HUα proteins were pooled and dialyzed against 2 mM HEPES (pH 7.9).
In vitro transcription
Transcription reactions were performed as described by Geanacopoulos et al. (1999). Supercoiled DNA template (2 nM) was preincubated with or without proteins at 37°C in a 45-μL reaction mixture containing 20 mM Tris acetate (pH 7.8), 10 mM magnesium acetate, 100 mM potassium glutamate, 1 mM ATP, 1 mM DTT, and 20 nM RNA polymerase. After incubation for 5 min, transcription was initiated by the addition of 5 μL of NTP mix containing 0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP, and 20 μCi of [α-32P] UTP (3000 Ci/mmole) (ICN). Reactions were terminated after 10 min by addition of equal volume of RNA loading buffer (80% [v/v] deionized formamide, 1× Tris-borate-EDTA [TBE], 0.025% bromophenol blue, 0.025% xylene cyanol). The reactions were analyzed on an 8% polyacrylamide-urea gel followed by autoradiography. The transcription products were quantified with Phosphorimager, using the RNA1 transcript as an internal control for each lane.
Electrophoretic mobility-shift assay
Gel retardation was performed in 10 μL final volume of 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM EDTA, and 0.5 mM DTT. A 266-bp gal DNA fragment was isolated from plasmid pSA509 by cleaving with restriction enzymes EcoR1 and Pst1. The purified DNA fragment was labeled with (α-32P) dATP by end-labeling with Klenow fragment. Labeled DNA fragments (5 ng and 1000 cpm) and proteins (5 nM, 10 nM, 20 nM, and 40nM) were incubated at room temperature for 15 min. The reactions were loaded onto a 5% nondenaturing polyacrylamide gel. After completion of electrophoresis, the gels were dried and autoradiographed.
Coimmunoprecipitation
For immunoprecipitation reactions with purified proteins, 5 μM HU and 5 μM GalR or LacIadi were incubated in 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.2% Tween 20, 0.02% NP-40, 1 mM DTT, and varying concentrations of KCl, as indicated, in a total volume of 20 μL at room temperature for 30 min. The mixtures were diluted 10-fold with buffer containing appropriate concentrations of KCl, and 10 μg of anti-GalR or anti-LacIadi antibody was added to each mixture. Precipitation was performed at 4°C for 4 h. Fifty microliters of buffer-equilibrated Protein-A agarose (Sigma) was added next and incubations continued at room temperature for two more hours on a rotary shaker. The slurries were then poured in small disposable columns and washed extensively with immunoprecipitation buffer, as described earlier. After transferring the slurry to microfuge tubes, the beads were pelleted and suspended in 50 μL SDS sample buffer. Following boiling for 5 min, the reactions were separated on 4%–20% Tris-glycine gels (Invitrogen). The proteins were then transferred to PVDF membranes and probed with anti-HU antibody. Immunoprecipitation of purified HUα and T19D proteins with GalR was done basically as described earlier except a fixed concentration of GalR (5 μM) was mixed with varying concentrations (50 nM, 500 nM, and 5 μM) of either HUα or T19D proteins.
For immunoprecipitations using cell extracts, plasmid pAP2, which carried the galR gene with a hexa-histidine tag at the N-terminal end and situated downstream of a lac promoter, was used. After induction of the galR gene using 0.4 mM IPTG for 3 h, cell extracts were made by sonication and centrifuged to remove the cellular debris. Twenty microliters of monoclonal anti-His antibody coupled to protein A-agarose (Sigma) was added to lysate volumes of 50 and 100 μL. Immunoprecipitation was performed essentially as described earlier for purified proteins. After transfer, the blots were developed with anti-HU or anti-IHF antibodies. Immunoprecipitation of purified HUα and T19D proteins with GalR was done basically as described earlier except a fixed concentration of GalR (5 μM) was mixed with varying concentrations (50 nM, 500 nM, and 5 μM) of either HUα or T19D proteins.
Western blotting
Proteins were transferred to PVDF membranes, which were blocked with 5% nonfat dry milk in TBST (10 mM Tris-HCl at pH 7.5, 150 mM NaCl, and 0.1% Tween 20). The blots were then incubated with primary antibody at 4°C overnight. They were then incubated with horseradish peroxidase (HRP)-conjugated antibody (Sigma) and developed with Supersignal (Pierce). The dilutions of the antibodies were anti-HU antibody (1: 60,000), anti-IHF antibody (1:100,000), HRP conjugated anti-rabbit antibody (1:120,000), and HRP conjugated anti-mouse antibody (1:10,000).
Acknowledgments
We are grateful to George Vasmatzis and Bangalore Sathyanarayana for their help with the computer modeling of HU. We also thank Susan Garges, Dhruba Chattoraj, Dale Lewis, and Mark Geanocopoulos for their helpful suggestions and encouragement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sadhya@helix.nih.gov; FAX (301) 480-7687.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.920301.
References
- Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;12:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aki T, Choy HE, Adhya S. Histone-like protein HU as a specific transcriptional regulator: Co-factor role in repression of gal transcription by gal repressor. Genes Cells. 1996;2:179–188. doi: 10.1046/j.1365-2443.1996.d01-236.x. [DOI] [PubMed] [Google Scholar]
- Bianchi ME. Prokaryotic HU and eukaryotic HMG-1: A kinked relationship. Mol Microbiol. 1994;14:1–5. doi: 10.1111/j.1365-2958.1994.tb01261.x. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altman M, Ronfani L, Bianchi ME, Taraseviciene L, Nordeen SK, Allegretto EA, Edwards DP. High-mobility chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordo D, Argos P. Suggestions for ‘safe’ residue substitutions in site-directed mutagenesis. J Mol Biol. 1991;217:721–729. doi: 10.1016/0022-2836(91)90528-e. [DOI] [PubMed] [Google Scholar]
- Brenowitz M, Jamison E, Majumdar A, Adhya S. Interaction of the Escherichia coli gal repressor protein with its DNA operators in vitro. Biochemistry. 1990;29:3374–3383. doi: 10.1021/bi00465a033. [DOI] [PubMed] [Google Scholar]
- Brenowitz M, Mandal N, Pickar A, Jamison E, Adhya S. DNA-binding properties of a lac repressor mutant incapable of forming tetramers. J Biol Chem. 1991;266:1281–1288. [PubMed] [Google Scholar]
- Cann JR, Pfenninger O, Pettijohn DE. Theory of the mobility-shift assay of non-specific protein-DNA complexes governed by conditional probabilities: The HU:DNA complex. Electrophoresis. 1995;17:881–887. doi: 10.1002/elps.11501601148. [DOI] [PubMed] [Google Scholar]
- Choy HE, Adhya S. RNA polymerase idling and clearance in gal promoters: Use of a supercoiled minicircle DNA template made in vivo. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dri A, Moreau P, Rouviere-Yaniv J. Role of histone-like proteins OsmZ and HU in homologous recombination. Gene. 1992;120:11–16. doi: 10.1016/0378-1119(92)90003-8. [DOI] [PubMed] [Google Scholar]
- Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geanacopoulos M, Vasmatzis G, Lewis DEA, Roy S, Lee BK, Adhya S. GalR mutants defective in repressosome formation. Genes & Dev. 1999;13:1251–1262. doi: 10.1101/gad.13.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geanacopoulos M, Vasmatzis F, Zhurkin VB, Adhya S. Gal repressosome contains an antiparallel DNA loop. Nat Struct Biol. 2001;8:432–436. doi: 10.1038/87595. [DOI] [PubMed] [Google Scholar]
- Haykinson MJ, Johnson RC. DNA looping and the helical repeat in vitro and in vivo: Effect of HU protein and enhancer location on Hin invertosome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L, Moorthy NC, Murthy KG, Manley JL, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is an unique activator of p53. Genes & Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y, Goshima N, Wada M, Imamoto F. Participation of hup gene product in replicative transposition of Mu phage in Escherichia coli. Gene. 1989;76:353–358. doi: 10.1016/0378-1119(89)90175-3. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Chaconas G. Immunoelectron microscopic analysis of the A, B, and HU protein content of the bacteriophage Mu transposomes. J Biol Chem. 1990;265:1623–1627. [PubMed] [Google Scholar]
- ————— A second high affinity HU binding site in the phage Mu transpososome. J Biol Chem. 1994;269:15571–15576. [PubMed] [Google Scholar]
- Lewis D, Geanacopoulos M, Adhya S. Roles of HU and DNA supercoiling in transcription repression: Specializes nucleoprotein repression complex at gal promoters in Escherichia coli. Mol Microbiol. 1999;31:451–462. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- Li S, Waters R. Escherichia coli strains lacking protein HU are UV sensitive due to a role for HU in homologous recombination. J Bacteriol. 1998;180:3750–3756. doi: 10.1128/jb.180.15.3750-3756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, D. and Gowrishankar, J. 1994. Evidence for involvement of protein HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia colio. J. Bacteriol. 176: . [DOI] [PMC free article] [PubMed]
- Murphy KC. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA. The E. coli HU and IHF proteins: Accessory factors for complex protein-DNA assemblies. In: Lin ECC, Lynch AS, editors. Regulation of gene expression in E. coli. Georgetown, TX: R.G. Landes Co.; 1996. pp. 149–179. [Google Scholar]
- Ner SS, Travers AA, Churchill MEA. Harnessing the writhe: A role for DNA chaperones in nucleoprotein-complex formation. Trends Biochem Sci. 1994;19:185–187. doi: 10.1016/0968-0004(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Oberto J, Drlica K, Rouviere-Yaniv K. Histones, HMG, HU, IHF: Meme combat. Biochimie. 1994;76:901–908. doi: 10.1016/0300-9084(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Ogura T, Niki H, Kano Y, Imamoto F, Hiraga S. Maintenance of plasmids in HU and IHF mutants of Escherichia coli. Mol Gen Genet. 1990;220:197–203. doi: 10.1007/BF00260482. [DOI] [PubMed] [Google Scholar]
- Paull TT, Haykinson MJ, Johnson RC. The non-specific DNA binding and bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes & Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- ————— HU and functional analogs in eukaryotes promote Hin invertosome assembly. Biochimie. 1994;76:992–1004. doi: 10.1016/0300-9084(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Rouviere-Yaniv J, Kjeldgaard NO. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979;106:297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Baker TA, Kornberg A. Strand separation required for initiation of replication at the chromosome origin of E. coli is facilitated by distant RNA-DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka I, Appelt K, Dijk J, White SW, Wilson KS. 3-A0 resolution structure of a protein with histone-like properties in prokaryotes. Nature (London) 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- Tokeson JPE. Ultrainduction of the Escherichia coli galactose operon. Ph.D thesis. Washington, DC.: Howard University; 1989. [Google Scholar]
- Vis H, Mariani M, Vorgias CE, Wilson KS, Kaptein R, Boelens R. Solution structure of the HU protein from Bacillus stearothermophilus. J Mol Biol. 1995;254:692–703. doi: 10.1006/jmbi.1995.0648. [DOI] [PubMed] [Google Scholar]
- Werner MH, Burley SK. Architectural transcription factors: Proteins that remodel DNA. Cell. 1997;88:733–736. doi: 10.1016/s0092-8674(00)81917-0. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Falciola L, Citterich M, Mavilio F, Bianchi ME. HMG-1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]