Abstract
A significant percentage of the human population is exposed to high levels of naturally occurring airborne dusts. Although the link between airborne particulate inhalation and a variety of respiratory diseases has long been established, little is known about the pathogenic role of the microbial component of the dust. In this study, we applied highly multiplexed PCR and a high-density resequencing microarray (RPM-TEI version 1.0) to screen samples of fine topsoil particles and airborne dust collected in 19 locations in Iraq and Kuwait for the presence of a broad range of human pathogens. The results indicated the presence of potential human pathogens, including Mycobacterium, Brucella, Coxiella burnetii, Clostridium perfringens, and Bacillus. The presence of Coxiella burnetii, a highly infectious potential biowarfare agent, was confirmed and detected in additional samples by use of a more sensitive technique (real-time PCR), indicating a high prevalence of this organism in the analyzed samples. The detection of potentially viable pathogens in breathable dusts from arid regions of Iraq and Kuwait underscores the importance of further study of these environments.
INTRODUCTION
It is a well-established fact that the exposure of the human respiratory system to high concentrations of airborne particulate matter (dust) can have significant adverse health effects (15). Desert storm activity is the most significant source of nonoccupational dust exposure in arid and semiarid regions of the world. It is estimated that 0.5 to 5 billion tons of desert topsoil is made airborne every year during high-wind desert storm events globally (50). Exposure to desert dust has been linked to many conditions, including silicosis and asthma (9, 45, 48, 54). Arid areas of Iraq and Kuwait are among the largest sources of airborne dust on the Earth (61) due to the very high content of fine, easily suspendible particles in their topsoils (1). Many respiratory diseases that are linked or suspected to be linked with the inhalation of desert dust, such as desert lung syndrome (24, 25, 46, 51), desert storm pneumonitis (Al Eskan disease) (14, 31, 32), severe acute pneumonitis (12), and acute eosinophilic pneumonia (57), have been reported to occur in this region.
The pathogenic effect of dust inhalation on respiratory tissues is attributed mostly to direct physical action of the dust particles on the epithelium of the human airways and may be exacerbated by toxic effects of the trace elements (including arsenic, mercury, cadmium, and iron) and of biologically active compounds that the dust particles may carry (15). Not much is known, however, about the health impact of the microbiological component of the dust (bacteria, fungi, and viruses) on exposed populations. A few studies linked exposure to African desert dust with the increased risk of bacterial meningitis caused by Neisseria meningitidis in the “meningitis belt” of North Africa, but the role of the dust is not clear (8, 44, 58).
Previous research has shown that although desert dust particles are subjected to extreme physical conditions, such as UV irradiation, desiccation, lack of nutrients, and high temperatures, they contain surprisingly diverse viable microbes and bacterial and fungal spores (21, 28). Furthermore, while the mobilized desert topsoils are most often transported locally, large-scale high-energy wind events can result in long-range movement of the suspended particles, which in some cases cross oceans and even circumnavigate the globe (18, 19, 22). While some data indicate the possibility of intercontinental spread of infectious pathogens (17, 43, 56), very little is known about the role that long-distance airborne particulate transport plays in this phenomenon.
Screening dust for infectious agents requires a method that provides precise identification and a high sensitivity. A method having broad coverage would also be advantageous. The normal approaches, e.g., bacterial culture, microscopy, or molecular analysis of 16S and 18S rRNA, used to study the microbiological content of airborne desert dust do not meet all of these requirements (21, 27, 62). While microbiological culture allows for accurate identification of the bacteria found in the sample, it is capable of detecting only a small fraction of all of the microorganisms present, since <1% of the bacterial strains present in the environment are currently cultivable (3). Microscopic examination and 16S and 18S rRNA-based methods can give insight into the total biodiversity of the sample, but their resolution is limited to genus-level identification for many taxonomic groups (27, 62). Highly multiplexed detection technologies based on multiple molecular signatures with better overall resolution are necessary for screening environmental samples for human pathogens. High-density resequencing microarray technology, in particular, is an example of technology that was shown to be capable of highly accurate broad-range pathogen detection (6, 7, 36, 38, 59).
The focus of this study was to develop a procedure to screen dust samples for the presence of human pathogens that can provide immediate determination of their presence, which allows the directed use of more sensitive follow-on tests to confirm the presence of such pathogens. To achieve this goal, we used a high-density resequencing microarray designed to detect a broad range of biothreat agents, RPM-TEI version 1.0 (33), to analyze samples of the readily suspendible fraction of the desert topsoil and airborne particulates from 19 distinct locations in Kuwait and Iraq. In one case, detection of DNA of a known human pathogen with bioweapon potential (Coxiella burnetii) was investigated further using a highly sensitive real-time PCR assay.
MATERIALS AND METHODS
Soil and dust sample collection. (i) Time and location.
Samples of soil and airborne particulates (dust) were collected between 23 July and 27 September 2007 from 4 locations in Iraq and 15 locations in Kuwait (Fig. 1 and Table 1), near the areas of deployment of U.S. troops, by the U.S. Navy Field Deployable Preventive Medicine Units (USN-FDPMUs).
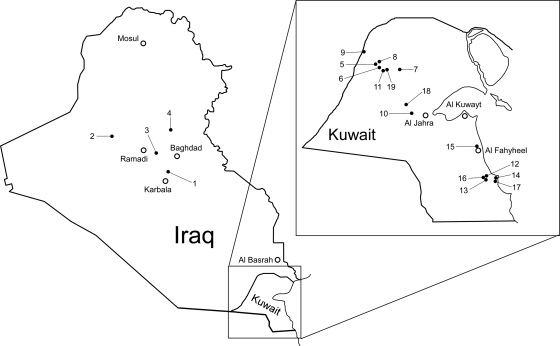
Fig. 1.
Locations of sample collection sites. The sites are marked with filled circles and numbered. The open circles show the locations of major cities. All locations are approximate. The exact positions were recorded and are available upon request.
Table 1.
Collection site information
| Site no. | Country | Location | Sitea | Typeb | Environmental conditionsc |
||
|---|---|---|---|---|---|---|---|
| Temp (°C) | Humidity (%) | Wind speed (m/s) | |||||
| 1 | Iraq | Iskandariyah FPB | Transformer yard | SA/H | 36.9 | 25.4 | 1.2 |
| 2 | Al-Asad airbase | South of flight line | A/H | 35.6 | 12.5 | 5.0 | |
| 3 | Camp Falluja | Open field | SA/M | 33.7 | 23.9 | 2.6 | |
| 4 | Camp Anaconda | Near firing range | SA/M | 34.1 | 18.1 | 3.3 | |
| 5 | Northern Kuwait | Range 6 | Open field | A/M | 36.0 | 11.0 | 3.3 |
| 6 | Range 7A | Open field | A/M | 29.2 | 15.4 | 0.8 | |
| 7 | Camp Buehring | Staging area | A/M | 54.4 | 6.0 | 4.4 | |
| 8 | FOB Scimitar | Open field | A/M | 36.0 | 11.0 | 3.3 | |
| 9 | K-Crossing | Open field | SA/M | 41.9 | 13.3 | 1.8 | |
| 10 | Life support area | Open field | SA/M | 42.3 | 9.8 | 5.4 | |
| 11 | Range 8 ECP | Open desert | A/M | 31.9 | 6.0 | 2.5 | |
| 12 | Southern Kuwait | Camp Arifjan | Near tent area | E/M | 36.1 | 30.0 | 2.2 |
| 13 | Camp Arifjan | Open field | A/NDd | 42.8 | 7.3 | 0.0 | |
| 14 | Camp Patriot | Commando cell KNB | A/M | 35.8 | 26.5 | 1.3 | |
| 15 | SPOD | Eastern connex yard | E/M | 38.9 | 12.1 | 2.8 | |
| 16 | Camp Arifjan | Zone 6, near ballfield | A/M | 37.1 | 13.0 | 5.8 | |
| 17 | Camp Patriot | Berthing spaud | A/H | 36.8 | 54.0 | 2.2 | |
| 18 | Northern Kuwait | Camp Virginia | Open field | A/M | 40.4 | 10.0 | 3.7 |
| 19 | FOB Lance | Open desert | E/L | 37.6 | 20.7 | 8.0 | |
The approximate geographic locations of collection sites within Iraq and Kuwait are depicted in Fig. 1. The exact GPS coordinates of the sites were collected and are available upon request.
The type of location is defined by the type of vegetation present (code before the slash) and the degree of land transformation (code after the slash). Vegetation codes: SA, semiarid—desert scrublands and grasses; A, arid—diffuse, sparse vegetation; E, extremely arid—no vegetation. Land transformation codes: H, high—farm use, camp use, heavy traffic; M, medium—lightly populated, light traffic; L, low—remote, at least 8 km from populated areas, no traffic.
Environmental condition readings were taken at the time of topsoil collection, which coincided with the starting point of airborne particulate collection, which lasted 24 h.
Degree of land transformation was not recorded for this location. ND, no data.
(ii) Soil sample collection and processing.
The center of the collection area was recorded by taking a global positioning system (GPS) reading (these data may be obtained directly upon request). Soil samples (to a maximum depth of 2 cm) were collected from the center of the collection area and from four additional positions (corresponding to the north, south, east, and west compass points) located 50 m from the center. Approximately 1 liter of surface soil was collected from each site (200 ml from each of the five positions). All soil samples were collected in sterile 50-ml tubes. Sterile working techniques were used for collection and all subsequent handling of the samples.
For each collection site, equal amounts of the soil from all five positions were mixed using sterile techniques to produce a composite sample representative of the area. The composite sample was subjected to a two-stage sieving process using sterilized standardized 3-in.-diameter stainless steel sieves (Gilson Company, Inc., Lewis Center, OH; Endecotts Ltd., London, England; and Arthur S. La Pine & Co., Chicago, IL). The sieves were used to sieve up to 50 g of soil, using 2 sieve stacks. Sieve stack 1 consisted of five sieves and a collection pan. The order of the sieves, from top (coarser) to bottom (finer), was sieve no. 20 (0.840-mm mesh), no. 40 (0.590-mm mesh), no. 60 (0.420-mm mesh), no. 80 (0.177-mm mesh), and no. 100 (0.149-mm mesh). The fraction of soil collected in the collection pan (particles of <0.149 mm) was applied to sieve stack 2, which consisted of four sieves and a collection pan. The order of the sieves from top to bottom was no. 140 (0.106-mm mesh), no. 200 (0.075-mm mesh), no. 270 (0.053-mm mesh), and no. 500 (0.025-mm mesh). The sieving was conducted using a Gilson performer III, model SS-3, sieve shaker (Gilson Company, Inc.) at an amplitude setting of 8 for 30 min. The soil fraction collected in the stack 2 collection pan, containing particles smaller than 0.025 mm (25 μm), was used for nucleic acid extraction. All sieves were cleaned using detergent solutions and sterilized by rinsing with 70% ethanol and irradiating with UV for 15 min before reuse.
(iii) Airborne particulate collection.
Airborne particulate sampling at each collection site was performed using a deployable particulate sampler (DPS) system (SKC Inc., Eighty Four, PA) equipped with a PM10 size-selective inlet allowing collection of particles of 10 μm or smaller in aerodynamic diameter. The particles were collected for 24 h from the sampled air at a rate of 10 liters per min on 47-mm, 2-μm-pore-size polytetrafluoroethylene (PTFE) filters reinforced with a PMP support ring (SKC Inc.). In each location, a duplicate PTFE filter was taken to the field but not exposed and was used as a negative control (field blank [FB]). The exposed filters were stored short term (approximately 1 month), shipped to the laboratory at ambient temperatures (approximately 25°C), and frozen at −20°C for long-term storage upon arrival at the laboratory. The blank filters were weighed before the exposure to airborne particulates, and filters containing collected particulates were weighed after arriving back at the laboratory. The weight of collected particulates was obtained by subtracting the weight of the empty filter from the combined weight of the filter with particulates (Table 2).
Table 2.
Amounts of airborne particulates collected and DNA contents of collected samplesa
| Site no. | Airborne particulates |
Topsoil DNA contentb (μg/g) | ||
|---|---|---|---|---|
| Total collected (mg) | Total DNA (ng) | DNA content (μg/g) | ||
| 1 | 1.9 | 22.8 | 12.0 | 20.8 |
| 2 | 1.3 | 37.4 | 28.2 | 15.3 |
| 3 | 0.6 | 17.6 | 28.2 | 10.8 |
| 4 | 2.2 | 16.4 | 7.5 | 13.6 |
| 5 | 1.4 | ND | ND | 9.2 |
| 6 | 1.5 | ND | ND | 6.0 |
| 7 | 1.9 | ND | ND | 3.6 |
| 8 | 1.6 | 16.4 | 10.1 | 8.6 |
| 9 | 2.4 | 12.4 | 5.2 | 16.4 |
| 10 | 1.6 | 24.0 | 15.0 | 6.8 |
| 11 | 2.6 | ND | ND | 9.8 |
| 12 | 4.2 | ND | ND | 2.0 |
| 13 | ND | ND | ND | 7.7 |
| 14 | 2.1 | 12.5 | 5.9 | 1.0 |
| 15 | 2.4 | 13.8 | 5.7 | 5.2 |
| 16 | 4.4 | ND | ND | 2.0 |
| 17 | 2.7 | ND | ND | 9.7 |
| 18 | 1.8 | 22.8 | 12.6 | 3.3 |
| 19 | 3.6 | 46.1 | 13.0 | 13.0 |
Data were not obtained for some of the airborne particulate samples. ND, no data.
The topsoil DNA content is based on total DNA extracted from 500 mg of topsoil fraction containing particles smaller than 0.025 mm.
Nucleic acid extraction.
The total nucleic acids were extracted from filter-captured airborne particulates and topsoil samples by use of a FastDNA spin kit for soil (MP Biomedicals, Solon, OH) according to the manufacturer's instructions. For the extraction of nucleic acids from soil samples, 500 mg of the sub-25-μm fraction of composite soil samples was used. The final nucleic acid elution was performed using 50 μl of DNase- and pyrogen-free water. In order to extract nucleic acids from airborne dust particles collected on PTFE filters, the PMP support ring was removed and the whole filter was used for extraction. The filters were folded and inserted into the kit's lysing matrix E tube and subsequently processed in the same way as soil samples. The field blanks were extracted in the same way as the exposed filters. The final nucleic acid elution for airborne particulate samples was performed using 100 μl of DNase- and pyrogen-free water. The DNA concentrations were measured fluorometrically using a Qubit instrument and Qubit dsDNA HS and BR assay reagent kits (Invitrogen, Carlsbad, CA).
RPM-TEI version 1.0 microarray analysis.
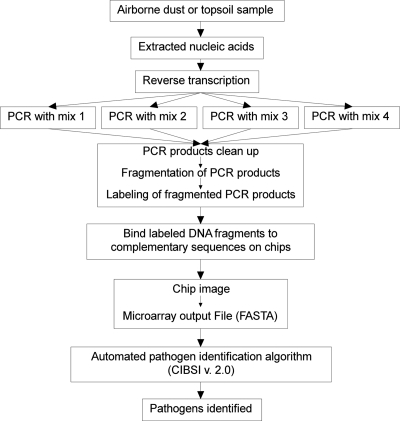
The RPM-TEI version 1.0 (subsequently called RPM-TEI) microarray analysis was conducted as previously described (33) and as summarized in Fig. 2. Briefly, 2 μl of nucleic acid preparation from each soil sample, airborne particulate sample, and field blank was reverse transcribed using random primers to obtain cDNAs for RNA templates potentially present in the sample. The obtained mixture was split into four aliquots of equal volume, and four separate, approximately 50-plex PCRs were conducted using RPM-TEI microarray-specific primer cocktails in order to amplify all of the targets (see reference 33 for detailed information on multiplex PCR mixtures). Subsequently, the PCR amplification products were combined and purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA). Purified PCR products were fragmented using DNase I and labeled with biotinylated ddATP, using terminal transferase. Labeled samples were loaded into preheated RPM-TEI microarray cartridges (TessArae LLC, Potomac Falls, VA) that were previously incubated with prehybridization buffer for at least 10 min, and the hybridization was conducted overnight at 49°C. Next, the sample was removed from the cartridge and the microarray was processed using a GeneChip 450 fluidics station and scanned using a GeneChip 3000 scanner with an autoloader (Affymetrix, Santa Clara, CA). Sequence data based on microarray hybridization patterns were generated by GeneChip 4.0 sequence analysis software (Affymetrix). Pathogen identification was performed by using Computer-Implemented Biological Sequence Identifier (CIBSI) 2.0 software (40), which filters the sequence of base calls from the microarray, and then using the resolved bases as the query for a similarity search of a DNA database (currently NCBI GenBank) to identify the most likely species and variants that matched the base calls from the hybridization observed. Although the microarray was designed with tiles for specific pathogen targets (33), it is capable of detection and correct sequence determination for targets differing by up to 15% from the sequence present on the microarray. This allows for detection of target variants and near-neighbor discrimination. For RPM-TEI, one to six different genome/plasmid sequences per targeted pathogen are present on the microarray. Depending on the concentration and quality of the target, similarity to targeted pathogen, and other factors, the hybridization on the microarray may produce anywhere from nearly complete resequencing of the detected target to short gene sequence fragments to no signal at all. Based on a similarity search of the obtained sequences with GenBank, CIBSI software will provide one of the following four outcomes: (i) no target was detected, (ii) a targeted pathogen was detected, (iii) a near neighbor was detected, and (iv) either a targeted pathogen or a near neighbor was detected, but the microarray sequence data were insufficient to discriminate between them.
Fig. 2.
Overview of the experimental protocol used with the RPM-TEI version 1.0 microarray. (Adapted from reference 37.)
Real-time PCR.
Real-time PCR for detection of Coxiella burnetii (IS1111 marker) was performed using previously published PCR primers IS1111-F801 (5′-AATTTCATCGTTCCCGGCAG-3′) and IS1111-R901 (5′-GCCGCGTTTACTAATCCCCA-3′) (13). The amplification reactions were conducted using SsoFast EvaGreen reaction mix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. An aliquot of 2 μl of total nucleic acid preparation from each soil, airborne particulate, or field blank sample was used as a template. The reaction was carried out in a 20-μl total volume containing a 500 nM concentration of each primer. Thermal cycling and fluorescent signal detection were performed in a CFX96 real-time PCR detection system (Bio-Rad Laboratories) with the following thermal cycling conditions: initial incubation at 98°C for 2 min, followed by 40 cycles of 98°C for 2 s and 60°C for 5 s. The amplification cycle was followed by melting curve analysis under the following conditions: samples were heated from 75°C to 95°C with 0.2°C increments, and the fluorescence was read after 10 s of incubation at each step. The results were analyzed with CFX Manager v. 1.5.534.0511 software (Bio-Rad Laboratories). A 5-μl aliquot of each C. burnetii-positive sample was analyzed by agarose gel electrophoresis (1.2% FlashGel DNA cassette; Lonza, Walkersville, MD) to confirm that the obtained PCR products had the expected size.
RESULTS
RPM-TEI microarray analysis.
Nucleic acids extracted from all samples (soil, airborne particulate, and field blank samples) were analyzed using a high-density resequencing microarray for detection of tropical and emerging pathogens (RPM-TEI). Microarray analysis indicated that several microorganisms were present in the analyzed samples. The most common bacteria present in these samples were Mycobacterium spp., followed by Brucella spp. and Coxiella burnetii. Clostridium perfringens was found in 3 samples, while Bacillus spp. were found at only one site, in Iraq. The results are summarized in Table 3.
Table 3.
RPM-TEI v. 1.0 microarray identification resultsa
| Country | Site no. | Sample typec | Hybridizing detector tile |
Most likely identificationd | |
|---|---|---|---|---|---|
| Organism | Gene or sequence | ||||
| Iraq | 1 | S | B. anthracis | rpoB | Bacillus cereus group |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | B. anthracis | rpoB | Bacillus cereus group | ||
| A | Brucella melitensis | gyrA | Brucella suis, B. abortus, or B. melitensis | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 2 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | M. tuberculosis | rpoB | Mycobacterium gilvum | ||
| 3 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | Clostridium perfringens | gyrA | Clostridium perfringens | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 4 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | M. tuberculosis | rpoB | Mycobacterium gilvum or M. smegmatis | ||
| Northern Kuwait | 5b | S | Brucella melitensis | gyrA | Brucella suis, B. abortus, or B. melitensis |
| S | Coxiella burnetii | IS1111 | Coxiella burnetii | ||
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 6 | S | Brucella melitensis | gyrA | Brucella suis, B. abortus, or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | No hybridization | ||||
| 7 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | M. tuberculosis | rpoB | Mycobacterium smegmatis | ||
| 8 | S | Coxiella burnetii | IS1111 | Coxiella burnetii | |
| S | M. tuberculosis | rpoB | Mycobacterium gilvum or M. barrassiae | ||
| A | No hybridization | ||||
| 9 | S | Brucella melitensis | gyrA | Brucella suis, B. abortus, or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | M. tuberculosis | rpoB | Mycobacterium gilvum, M. smegmatis, M. barrassiae, or M. vanbaalenii | ||
| 10 | S | Brucella melitensis | gyrA | Brucella abortus or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | M. tuberculosis | rpoB | Mycobacterium gilvum, M. vanbaalenii, or M. lepromatosis | ||
| 11 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | M. tuberculosis | rpoB | Mycobacterium gilvum or M. vanbaalenii | ||
| Southern Kuwait | 12 | S | Brucella melitensis | gyrA | Brucella abortus or B. melitensis |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | Coxiella burnetii | IS1111 | Coxiella burnetii | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae or M. mageritense | ||
| 13 | S | Brucella melitensis | gyrA | Brucella abortus or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | No hybridization | ||||
| 14 | S | No hybridization | |||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae or M. fortuitum | ||
| 15 | S | Brucella melitensis | gyrA | Brucella suis or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 16 | S | M. tuberculosis | rpoB | Mycobacterium barrassiae | |
| A | Clostridium perfringens | gyrA | Clostridium perfringens | ||
| A | Coxiella burnetii | icd | Coxiella burnetii | ||
| A | Coxiella burnetii | IS1111 | Coxiella burnetii | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 17 | S | Clostridium perfringens | colA (kappa toxin) | Clostridium perfringens | |
| S | M. tuberculosis | rpoB | Mycobacterium ulcerans | ||
| A | No hybridization | ||||
| Northern Kuwait | 18 | S | Brucella melitensis | gyrA | Bradyrhizobium |
| S | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
| 19 | S | Brucella melitensis | gyrA | Brucella suis, B. abortus, B. ovis, or B. melitensis | |
| S | M. tuberculosis | rpoB | Mycobacterium ulcerans | ||
| A | M. tuberculosis | rpoB | Mycobacterium barrassiae | ||
All field blanks (FB samples) were negative for all microarray tiles.
No airborne particulate was collected for this location.
S, soil; A, airborne particulate.
“Most likely identification” was a determination made by CIBSI 2.0 software based on matching sequences obtained from the RPM-TEI microarray with sequences in GenBank. See Materials and Methods and reference 35 for details.
Almost all soil samples and the majority of airborne particulate samples (with the exception of a soil sample from site 14 and airborne particulate samples from sites 6, 8, 13, and 17) were found to contain DNA that hybridized to microarray tiles designed for detection of the rpoB gene of Mycobacterium tuberculosis, but other sequences of M. tuberculosis were not positive. The possibility of cross contamination was excluded by inclusion and analysis of negative controls (field blanks), all of which were negative for the presence of Mycobacterium. The sequences read by the microarray indicated, however, that the detected DNAs did not belong to M. tuberculosis. They were most similar to sequences of various Mycobacterium species belonging to the group of rapidly growing mycobacteria (RGM). These species (depending on the particular sample) included Mycobacterium barrassiae, Mycobacterium gilvum, Mycobacterium smegmatis, and Mycobacterium ulcerans. In some cases, determination of a single most likely species was impossible due to insufficient sequence data.
A large number of analyzed samples contained nucleic acids hybridizing with the gyrA tile for Brucella melitensis. Approximately half of the soil samples (from sites 5, 6, 9, 10, 12, 13, 15, and 19) and one airborne particulate sample (from site 1) contained detectable quantities of this DNA. The sequences obtained were found to be most similar to those of Brucella suis, Brucella abortus, or B. melitensis, but the sequence information was not sufficient to make a definitive determination of which of these three species was present in the sample. In one case (soil from site 18), the sequences obtained from this tile were determined to be most similar to GenBank sequences of Bradyrhizobium, although species-level determination was impossible.
The presence of C. burnetii nucleic acids was detected in four samples (soil from sites 5 and 8 in Northern Kuwait and airborne particulates from locations 12 and 16 in Southern Kuwait). In all four cases, strong hybridization with the Coxiella IS1111 tile was observed. The quality of the sequences obtained from this tile allowed for species-level identification. With the airborne particulate sample from site 16, hybridization with microarray tiles containing an additional Coxiella diagnostic sequence, icd, was also detected.
Nucleic acid sequences of another human pathogen, Clostridium perfringens, were found in 3 samples (soil from location 17 and airborne particulates from sites 3 and 16). The species-level identifications in the case of this organism were made based on hybridization with gyrA tiles for samples from locations 3 and 16 and with the colA tile for the airborne particulate sample from site 17.
Sequences hybridizing with Bacillus rpoB tiles were found in both sand and airborne particulates collected in Iraq (site 1). The obtained sequences allowed for determination that detected nucleic acids originated from Bacillus bacteria belonging to the Bacillus cereus group but excluded the possibility that they were from any Bacillus anthracis strain with a sequence submitted to GenBank.
Real-time PCR detection of Coxiella burnetii DNA.
The detection in these samples of Coxiella burnetii, the etiological agent of Q fever and a potential bioweapon, by use of RPM-TEI raised interest and concern that warranted further investigation. Real-time PCR using primers designed by Christensen et al. (13) was conducted to confirm the findings of the microarray and to possibly detect the presence of this organism in additional samples. Similar to the RPM-TEI microarray, this assay takes advantage of the multiple copies of the IS1111 marker present in the C. burnetii genome (30), and IS1111 was previously shown to be very specific and sensitive (13, 26, 39) for C. burnetii detection.
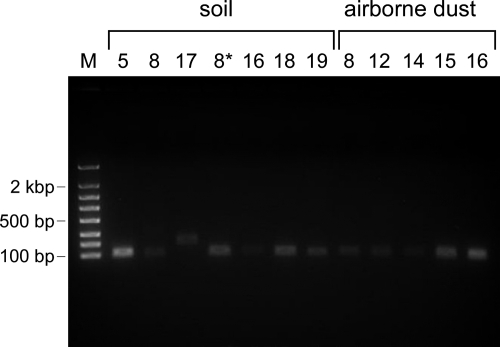
The C. burnetii-specific real-time PCR was run on all collected samples. The field blanks and water were included as negative controls. The PCR analysis confirmed the presence of Coxiella-specific nucleic acids in all samples that tested positive for Coxiella by the RPM-TEI microarray (soil from sites 5 and 8 and airborne dust from sites 12 and 16) but also detected the presence of C. burnetii DNA in six additional samples: soil from sites 16, 18, and 19 and airborne particles from sites 8, 14, and 15. Agarose gel electrophoresis of the amplification products confirmed their correct sizes (Fig. 3). One additional sample (soil from site 17) was found to give a positive amplification; however, analysis of the amplification product (agarose gel electrophoresis, melting curve analysis, and sequencing) indicated that this was a false-positive result. In total, C. burnetii was detected (in either soil or dust) in almost half (8 of 19) of all tested locations. All field blank samples and water resulted in no amplification.
Fig. 3.
Amplification products of real-time PCRs for Coxiella burnetii detection. Lane M, FlashGel DNA marker (100 bp to 4 kb). Remaining lanes are labeled with the site number where the sand or airborne particulate was collected. The product of expected size (101 bp) was amplified in all cases except for sample 17, which was determined to be negative based on amplicon sequencing (data not shown). The real-time PCR using the soil sample from location 8 was run in duplicate, using two independently extracted DNA preparations (soil lanes 8 and 8*).
DISCUSSION
In this study, we investigated the potential for sand and airborne dust to contain known human pathogens in samples collected from a number of locations in Iraq and Kuwait. Analysis conducted using an RPM-TEI microarray allowed for detection of nucleic acids of a number of well-known human pathogens, such as Coxiella burnetii and Clostridium perfringens, which may cause disease by inhalation. Nucleic acids were also detected for genera such as Mycobacterium, Bacillus, and Brucella, important potential human pathogens that may also be spread by the respiratory route. Some of them (Coxiella and Brucella) require specialized culture conditions that are not available in most diagnostic laboratories and would not be detected by plating on blood agar, and therefore molecular techniques facilitate their detection. While it was not possible to determine if the detected pathogens were viable based on the microarray results, the detection of their nucleic acids warrants further studies on the health risk associated with breathing fine particles from arid, dusty regions of the Middle East. It should also be noted that there was no significant correlation between the environmental conditions at the collection site and the pathogens detected (Table 1). The analysis also showed that the DNA contents in both airborne dust samples and topsoil were generally within 1 order of magnitude (from micrograms to tens of micrograms per gram of sample) and failed to show any significant connection between the amount of the airborne particulate collected or overall sample DNA content and the pathogen nucleic acids detected (Table 2).
The two most prevalent bacterial targets found that can act as human pathogens were Mycobacterium spp. (11, 60) and Brucella spp. The mycobacteria are commonly found in soil (35, 47), and some RGM might be involved in respiratory disease in humans (2, 49). Brucella spp. include major zoonotic pathogens causing brucellosis in livestock. In addition, some Brucella species (e.g., B. suis in the United States) have been weaponized in the course of offensive biological weapon programs (20). Human infections with Brucella species (mostly with B. melitensis and B. abortus) usually occur from direct or indirect exposure to infected animals (63), but direct infection via inhalation of pathogen-contaminated aerosols is possible (63).
A more serious potential threat to humans detected in a number of analyzed samples was Coxiella burnetii, which causes Q fever and is known to be able to persist for long periods in the environment (41, 52). The identification of this organism by RPM-TEI microarray analysis was based mostly on detection of the IS1111 insertion sequence, which is specific to this species and present in its genome in multiple copies (ranging from 7 to 110) (30). The natural amplification of IS1111 may explain the fact that this marker was detected in all four C. burnetii-positive samples, while the icd marker was detected in only one. A sensitive real-time PCR test confirmed the microarray positive results and revealed the presence of C. burnetii DNA in additional locations, indicating a significant prevalence of this organism in the desert dust in Kuwait. The fact that Q fever was previously observed in this region and the several cases reported for U.S. troops deployed to this part of the world (4, 5, 16, 23, 34) are consistent with our findings. A recent large survey of environmental samples collected from six geographically distinct parts of the United States found that 23.8% of all analyzed samples were Coxiella positive by real-time PCR, suggesting the ubiquity of C. burnetii in the environment, which raises the interesting question of how closely related the organisms are and whether they may be transported intercontinentally or globally in dust particles (29).
Finding nucleic acids for organisms belonging to the genus Bacillus in samples from only one location was surprising, since Bacillus is relatively common in soil. This might have been the result of marker selection geared toward Bacillus anthracis detection (there are 6 markers specific for B. anthracis, including 2 chromosomal and 4 plasmid-borne markers, and only 1 for B. cereus on the microarray). Another possibility is poor nucleic acid recovery from Bacillus spores.
It is worth nothing that all microorganisms for which nucleic acids were detected by microarray analysis, with the possible exception of Mycobacterium, are predominantly zoonotic in nature. This could be the result of sampling in the vicinity of agricultural areas, where infected animals (cattle, sheep, and goats) are likely to contaminate the soil. Also, no viral pathogens were detected, even though the RPM-TEI array is capable of detecting a wide array of viruses. This could be the result of the genuine absence of viral particles from the dust particles due to very harsh environmental conditions affecting the desert dust or an experimental bias introduced by the nucleic acid extraction methods. It should also be noted that although fungi and/or fungal spores may have significant impacts on human health (53, 55), the RPM-TEI array was not designed to detect fungal pathogens. Future development will include potential human fungal pathogens in the microarray design to expand the coverage to this class of pathogens.
In summary, the results of this study indicate that high-density resequencing microarrays have great potential as a tool for broad-spectrum screening of environmental samples for the presence of human pathogens. The RPM-TEI microarray directly detected potential human pathogens and also guided the use of more specific and sensitive follow-up tests. One of the main strengths of the RPM technology, as shown by this study, lies in its ability for simultaneous detection of hundreds of pathogens in a single test. Other sensitive molecular methods, such as real-time PCR, are usually characterized by low multiplexing levels. While these assays are cheaper as standalone tests, running enough of them to provide the same coverage of pathogens with a single sample quickly overtakes RPM microarrays in both cost and logistical complexity of running the analysis. Although other technologies with universal coverage that may be applied to analyze environmental samples exist, they either have low resolution (such as 16S rRNA-based techniques) or are much more complex and expensive (such as high-throughput de novo sequencing).
This study also gives a glimpse at the baseline levels of recognized biothreat agents and their near neighbors that are naturally present in the environment. A number of studies aiming to determine natural background levels have been conducted (10, 42); however, more are needed in order to develop better environmental biothreat sensing technologies and algorithms that are significantly less prone to false-positive results than currently available systems.
ACKNOWLEDGMENTS
We acknowledge the members of the Field Deployable Preventive Medicine Units (Amy Mocatelli, Carl Druhl, Richard Ankney, Edward Benchoff, Steward Bullock, Reatha Candler, and Robin Lenon) for conducting the sample collection which enabled this research effort.
The funding for developing the RPM-TEI array was provided by the Office of Naval Research.
The opinions and assertions contained herein are those of the authors, and none are to be construed as those of the Department of Defense, Department of the Navy, or any other military service or government agency at large.
Footnotes
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Abdulla S. A. A., Alrizzo H. M., Cyril M. M. 1988. Particle-size distribution of Iraqi sand and dust storms and their influence on microwave communication-systems. IEEE Trans. Antennas Propag. 36:114–126 [Google Scholar]
- 2. Adekambi T., Raoult D., Drancourt M. 2006. Mycobacterium barrassiae sp. nov., a Mycobacterium moriokaense group species associated with chronic pneumonia. J. Clin. Microbiol. 44:3493–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amann R. I., Ludwig W., Schleifer K. H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson A. D., Smoak B., Shuping E., Ockenhouse C., Petruccelli B. 2005. Q fever and the US military. Emerg. Infect. Dis. 11:1320–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aronson N. E., Sanders J. W., Moran K. A. 2006. In harm's way: infections in deployed American military forces. Clin. Infect. Dis. 43:1045–1051 [DOI] [PubMed] [Google Scholar]
- 6. Berthet N., et al. 2007. Massively parallel pathogen identification using high-density microarrays. Microb. Biotechnol. 1:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berthet N., et al. 2010. High-density resequencing DNA microarrays in public health emergencies. Nat. Biotechnol. 28:25–27 [DOI] [PubMed] [Google Scholar]
- 8. Besancenot J. P., Boko M., Oke P. C. 1997. Weather conditions and cerebrospinal meningitis in Benin (Gulf of Guinea, West Africa). Eur. J. Epidemiol. 13:807–815 [DOI] [PubMed] [Google Scholar]
- 9. Blades E., Naidu R., Mathison G. 1998. The microbiological analysis of Saharan dust and its association with asthma in Barbados. West Indian Med. J. 47:3410368623 [Google Scholar]
- 10. Brodie E. L., et al. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown T. H. 1985. The rapidly growing mycobacteria—Mycobacterium fortuitum and Mycobacterium chelonei. Infect. Control 6:283–288 [DOI] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention 2003. Severe acute pneumonitis among deployed U.S. military personnel—Southwest Asia, March-August 2003. MMBR Morb. Mortal. Wkly. Rep. 52:857–859 [PubMed] [Google Scholar]
- 13. Christensen D. R., et al. 2006. Detection of biological threat agents by real-time PCR: comparison of assay performance on the RAPID, the LightCycler, and the Smart Cycler platforms. Clin. Chem. 52:141–145 [DOI] [PubMed] [Google Scholar]
- 14. Clooman C. C., Tenglin R., Butler F., Leitch R. A. 2000. Six degrees of Kevin Bacon—Al Eskan disease and “dirty dust.” Mil. Med. 165:iv–v [PubMed] [Google Scholar]
- 15. Cook A. G., Weinstein P., Centeno J. A. 2005. Health effects of natural dust—role of trace elements and compounds. Biol. Trace Elem. Res. 103:1–15 [DOI] [PubMed] [Google Scholar]
- 16. Ellis S. B., et al. 2008. Outbreak of sandfly fever in central Iraq, September 2007. Mil. Med. 173:949–953 [DOI] [PubMed] [Google Scholar]
- 17. Garrison V. H., et al. 2006. Saharan dust—a carrier of persistent organic pollutants, metals and microbes to the Caribbean? Rev. Biol. Trop. 54:9–21 [Google Scholar]
- 18. Gillies J. A., Nickling W. G., McTainsh G. H. 1996. Dust concentrations and particle-size characteristics of an intense dust haze event: inland delta region, Mali, West Africa. Atmos. Environ. 30:1081–1090 [Google Scholar]
- 19. Goudie A. S. 2009. Dust storms: recent developments. J. Environ. Manage. 90:89–94 [DOI] [PubMed] [Google Scholar]
- 20. Greenfield R. A., et al. 2002. Bacterial pathogens as biological weapons and agents of bioterrorism. Am. J. Med. Sci. 323:299–315 [DOI] [PubMed] [Google Scholar]
- 21. Griffin D. W. 2007. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin. Microbiol. Rev. 20:459–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Griffin D. W., Kellogg C. A. 2004. Dust storms and their impact on ocean and human health: dust in Earth's atmosphere. EcoHealth 1:284–295 [Google Scholar]
- 23. Hartzell J. D., et al. 2007. Atypical Q fever in US soldiers. Emerg. Infect. Dis. 13:1247–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hawass N. E. 1987. An association between desert lung and cataract—a new syndrome. Br. J. Ophthalmol. 71:694–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirsch M., Barziv J., Lehmann E., Goldberg G. M. 1974. Simple siliceous pneumoconiosis of Bedouin females in Negev-Desert. Clin. Radiol. 25:507–510 [DOI] [PubMed] [Google Scholar]
- 26. Howe G. B., et al. 2009. Real-time PCR for the early detection and quantification of Coxiella burnetii as an alternative to the murine bioassay. Mol. Cell. Probes 23:127–131 [DOI] [PubMed] [Google Scholar]
- 27. Janda J. M., Abbott S. L. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kellogg C. A., Griffin D. W. 2006. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 21:638–644 [DOI] [PubMed] [Google Scholar]
- 29. Kersh G. J., et al. 2010. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl. Environ. Microbiol. 76:4469–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klee S. R., et al. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korenyiboth A. L., Korenyiboth A. L., Molnar A. C., Fidelusgort R. 1992. Al Eskan disease—desert storm pneumonitis. Mil. Med. 157:452–462 [PubMed] [Google Scholar]
- 32. Korenyiboth A. L., Molnar A. C., Korenyiboth A. L., Fidelusgort R. 1993. Al Eskan disease. Mil. Med. 158:A6. [PubMed] [Google Scholar]
- 33. Leski T. A., et al. 2009. Testing and validation of high density resequencing microarray for broad range biothreat agents detection. PLoS One 4:e6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leung-Shea C., Danaher P. J. 2006. Q fever in members of the United States armed forces returning from Iraq. Clin. Infect. Dis. 43:e77–e82 [DOI] [PubMed] [Google Scholar]
- 35. Leys N. M., et al. 2005. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51:375–388 [DOI] [PubMed] [Google Scholar]
- 36. Lin B., et al. 2007. Application of broad-spectrum, sequence-based pathogen identification in an urban population. PLoS One 2:e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin B. C., et al. 2007. Using a resequencing microarray as a multiple respiratory pathogen detection assay. J. Clin. Microbiol. 45:443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin B. C., et al. 2006. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 16:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loftis A. D., et al. 2006. Rickettsial agents in Egyptian ticks collected from domestic animals. Exp. Appl. Acarol. 40:67–81 [DOI] [PubMed] [Google Scholar]
- 40. Malanoski A. P., Lin B., Wang Z., Schnur J. M., Stenger D. A. 2006. Automated identification of multiple micro-organisms from resequencing DNA microarrays. Nucleic Acids Res. 34:5300–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maurin M., Raoult D. 1999. Q fever. Clin. Microbiol. Rev. 12:518–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Merrill L., Dunbar J., Richardson J., Kuske C. R. 2006. Composition of bacillus species in aerosols from 11 U.S. cities. J. Forensic Sci. 51:559–565 [DOI] [PubMed] [Google Scholar]
- 43. Mims S. A., Mims F. M., III 2004. Fungal spores are transported long distances in smoke from biomass fires. Atmos. Environ. 38:651–655 [Google Scholar]
- 44. Molesworth A. M., Cuevas L. E., Morse A. P., Herman J. R., Thomson M. C. 2002. Dust clouds and spread of infection. Lancet 359:81–82 [DOI] [PubMed] [Google Scholar]
- 45. Norboo T., et al. 1991. Silicosis in a Himalayan village population—role of environmental dust. Thorax 46:341–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nouh M. S. 1989. Is the desert lung syndrome (nonoccupational dust pneumoconiosis) a variant of pulmonary alveolar microlithiasis—report of 4 cases with review of the literature. Respiration 55:122–126 [DOI] [PubMed] [Google Scholar]
- 47. Parashar D., Das R., Sharma V. D., Chauhan D. S., Katoch V. M. 2007. Pathogenic rapidly growing Mycobacterium manitobense in the environment of Agra, north India. Indian J. Med. Res. 126:230–232 [PubMed] [Google Scholar]
- 48. Park J. W., et al. 2005. Effects of ambient particulate matter on peak expiratory flow rates and respiratory symptoms of asthmatics during Asian dust periods in Korea. Respirology 10:470–476 [DOI] [PubMed] [Google Scholar]
- 49. Park S., et al. 2008. Clinical significance of Mycobacterium fortuitum isolated from respiratory specimens. Respir. Med. 102:437–442 [DOI] [PubMed] [Google Scholar]
- 50. Perkins S. 2001. Dust, the thermostat. Sci. News 160:200–201 [Google Scholar]
- 51. Policard A., Collet A. 1952. Deposition of siliceous dust in the lungs of the inhabitants of the Saharan regions. AMA Arch. Ind. Hyg. Occup. Med. 5:527–534 [PubMed] [Google Scholar]
- 52. Reimer L. G. 1993. Q fever. Clin. Microbiol. Rev. 6:193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Revankar S. G., Sutton D. A. 2010. Melanized fungi in human disease. Clin. Microbiol. Rev. 23:884–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saiyed H. N., et al. 1991. Nonoccupational pneumoconiosis at high-altitude villages in Central Ladakh. Br. J. Ind. Med. 48:825–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sharpton T. J., et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shinn E. A., et al. 2000. African dust and the demise of Caribbean coral reefs. Geophys. Res. Lett. 27:3029–3032 [Google Scholar]
- 57. Shorr A. F., et al. 2004. Acute eosinophilic pneumonia among US military personnel deployed in or near Iraq. JAMA 292:2997–3005 [DOI] [PubMed] [Google Scholar]
- 58. Sultan B., Labadi K., Guegan J. F., Janicot S. 2005. Climate drives the meningitis epidemics onset in West Africa. PLoS Med. 2:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taitt C. R., et al. 2008. Discrimination between biothreat agents and ‘near neighbor’ species using a resequencing array. FEMS Immunol. Med. Microbiol. 54:356–364 [DOI] [PubMed] [Google Scholar]
- 60. Tortoli E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Washington R., Todd M., Middleton N. J., Goudie A. S. 2003. Dust-storm source areas determined by the total ozone monitoring spectrometer and surface observations. Ann. Assoc. Am. Geogr. 93:297–313 [Google Scholar]
- 62. Woo P. C. Y., Lau S. K. P., Teng J. L. L., Tse H., Yuen K. Y. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 63. Young E. J. 2005. Brucella species, p. 2669–2674 In Mandell G. L., Bennett J. E., Dolin R. (ed.), Principles and practice of infectious diseases. Elsevier, Philadelphia, PA [Google Scholar]