Abstract
Clostridium perfringens, although a member of the normal gut flora, is also an important cause of intestinal disease in animals and, to a lesser extent, in humans. Disease is associated with the production of one or more toxins, and little is known about environmental influences on the production of these toxins. One of the health-promoting effects of lactic acid bacteria (LAB) is the establishment and maintenance of a low pH in the intestine since an acidic environment inhibits the growth of many potentially harmful bacteria. Here, the effect of the LAB Lactobacillus fermentum on beta2 toxin production by C. perfringens is described. Coculturing of C. perfringens with L. fermentum showed that under in vitro conditions, L. fermentum was capable of silencing beta2 toxin production by C. perfringens without influencing bacterial viability. The reduction in toxin production was shown to be most likely a result of the decline in pH. Quantitative PCR showed that the reduction in beta2 toxin production was due to a decrease in cpb2 mRNA. These results suggest that in the intestine, the production of beta2 toxin by C. perfringens might be regulated by other members of the normal intestinal flora.
INTRODUCTION
The normal intestinal flora comprises several hundred bacterial species and is recognized as being beneficial to its host (16, 28). The species contribute to this phenomenon in various ways. For instance, some species break down undigested or indigestible food whereas others synthesize vitamins or short-chain fatty acids. Additionally, these bacteria prevent the colonization of the gut by potential pathogens (11). Lactic acid bacteria (LAB) such as those belonging to the genera Bifidobacterium, Lactobacillus, Enterococcus, and Streptococcus are members of the normal intestinal flora. They not only prevent colonization by their actual presence, but they also prevent growth of and colonization by pathogens through their production of acid (15). Furthermore, in an attempt to maintain the status quo of their environment, it is known that bacteria may influence intra- and interspecies growth and gene expression through a phenomenon called quorum sensing (14).
Clostridium perfringens, although it is a member of the normal intestinal flora, is regarded as one of the most important causes of intestinal disease in farm animals and wild animals and, to a lesser extent, in humans (20). Strains of C. perfringens are classified into one of five toxin types (A to E) based on the four major toxins they can produce (alpha, beta, epsilon, and iota toxins). Each toxin type is associated with specific diseases of various animal species and of humans (27). In addition to the four major toxins, all types of C. perfringens may carry other toxin-encoding genes, including cpb2, the gene which encodes the beta2 toxin (7). The presence of cpb2-positive C. perfringens strains in the intestine has been associated with intestinal disease in humans (6), ruminants (13), horses (9), and pigs (3, 31). However, cpb2-positive C. perfringens strains have also been reported in animals and humans without any signs of intestinal disease (3, 4, 13).
Most likely, toxin production by C. perfringens and the subsequent induction of disease are initiated by intestinal environmental changes (24, 27). It is known that antibiotic use can lead to such a change by shifting the percentage composition of the intestinal flora species (6). Such a shift is generally thought to favor the proliferation of C. perfringens, with a resultant higher production of toxin(s). Alternatively, it might well be that the shift in the equilibrium of the normal intestinal flora leads to a deregulation of gene expression of the remaining members of the intestinal flora, including C. perfringens.
This study describes the influence of Lactobacillus fermentum, a member of the normal intestinal flora (5, 12, 25), on the production of beta2 toxin by C. perfringens and unravels the underlying mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Cp15, a porcine C. perfringens type A strain harboring cpb2 (originally designated JF2251, kindly provided by J. Frey, Berne, Switzerland) was used as a representative of the beta2 toxin-producing C. perfringens species. L. fermentum strain 104R, isolated from porcine squamous epithelium (8), was used as the representative strain for the L. fermentum species.
Unless otherwise stated, resuspended stationary overnight cultures of C. perfringens strain Cp15 were used in all experiments. After centrifugation (10 min at 4,000 × g) and resuspension of the pelleted bacteria in fresh growth medium to the original volume, the cultures were (metabolically) active, as demonstrated by the production of acid, beta2 toxin, and gyrase mRNA. The use of these active stationary C. perfringens cultures enabled the study of the actual influence of L. fermentum on beta2 toxin production by C. perfringens without the confounding influence of changes in the numbers of C. perfringens bacteria on beta2 toxin production.
All experiments were performed with MRS broth (Oxoid), since this provided the best overall growth environment for both species, as L. fermentum growth was severely impaired in Schaedler broth (Oxoid). Anaerobic conditions were obtained using jars and the Anoxomat gas exchange system (Mart Microbiology). Immediately after the jars were opened, tubes were closed in order to maintain the anaerobic condition as best as possible. Manipulations, such as the removal of supernatants and resuspension of the pellets, were performed under aerobic conditions.
C. perfringens and L. fermentum coculture experiments.
L. fermentum strain 104R anaerobically cultured in MRS broth (Oxoid) set to pH 7 with NaOH at 37°C for 24 h (stationary phase) was divided into two equal parts. One half was centrifuged, and then the pellet was resuspended in MRS broth (pH 7) and incubated for another hour (L1). The other half was reincubated for another hour without further treatment and centrifuged, and the pellet was resuspended in fresh MRS broth at pH 7 (L). An overnight culture of Cp15 was equally divided among 12 tubes and centrifuged, with the resulting pellets being resuspended in MRS broth at pH 7 (M), in the L. fermentum suspension in fresh MRS broth (pH 7) (L), or in the L. fermentum culture that was incubated for 1 h in fresh MRS broth (pH 7) (L1). For each combination, a tube was incubated for 1, 2, 3, or 4 h. At each time point, the numbers of CFU of the various cultures were determined and the pH was measured. The cultures were centrifuged, and the supernatants were analyzed by Western blot assay for the presence of beta2 toxin.
Establishment of beta2 toxin degradation by L. fermentum.
An overnight culture of L. fermentum was divided into two equal parts. After centrifugation (10 min, 4,000 × g), pellets were resuspended either in a 2-h Cp15 culture in MRS broth (pH 7) or in the supernatant of a 2-h Cp15 culture and incubated anaerobically at 37°C for 2 h. The suspensions were then centrifuged (10 min, 4,000 × g), and the supernatants were analyzed by Western blot assay for the presence of beta2 toxin.
Beta2 toxin production at various pHs.
A stationary (16-h) culture of Cp15 in Schaedler broth was equally divided among 12 tubes and centrifuged for 10 min at 4,000 × g at 37°C. Pellets were resuspended in MRS broth set at pH 5, pH 6, or pH 7 using either HCl or NaOH and incubated anaerobically at 37°C for 1, 2, 3, or 4 h. After each time period, the numbers of CFU of the various cultures were determined and the pH of each culture was measured. The cultures were again centrifuged for 10 min at 4,000 × g, and the supernatants were analyzed for the presence of beta2 toxin by Western blot assay. Pellets were used for RNA isolation and subsequent quantitative PCR (Q-PCR) analysis.
Coculture experiments with Caco-2 cells.
Human-derived, enterocyte-like Caco-2 cells (21) were seeded into 12-well plates (Greiner) at 80,000 cells/cm2 and cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 0.03% l-glutamine (Invitrogen), 1% nonessential amino acids (Flow Laboratories), 10 mM sodium bicarbonate, 25 mM HEPES (Flow Laboratories), 50 μg/ml gentamicin (Gibco), and 20% fetal bovine serum (Lonza) in a humid atmosphere of 5% CO2 at 37°C. At day 5, the medium was removed and the monolayers were washed twice with 0.01 M phosphate-buffered saline (PBS), pH 7.3, and incubated with plain DMEM. After 1 h, the DMEM was removed and MRS broth (pH 7), with or without bacteria, was added to the cells.
A 24 h, MRS (pH 7) broth culture of L. fermentum was divided into two equal parts. One half was centrifuged, and the pellet was resuspended in MRS broth (pH 7) and put on the cells (L1) in a number of wells, while the DMEM in the remaining wells was replaced with MRS broth. Plates were incubated anaerobically for 1 h at 37°C. The other half of the L. fermentum culture was reincubated for another hour without further treatment. Equal parts of an overnight culture of Cp15 were centrifuged and resuspended in overlying MRS broth without lactobacilli (M), in overlying MRS broth already containing L. fermentum for 1 h (L1), or together with the pellet of the reincubated L. fermentum culture resuspended in the pH 7 MRS broth which had overlain the Caco-2 cells for 1 h (L). The plates were then incubated anaerobically for 1, 2, 3, or 4 h. Anaerobic conditions were obtained using jars and the Anoxomat gas exchange system (Mart Microbiology). After each time period, the numbers of CFU of C. perfringens and L. fermentum were determined and the pH was measured. Finally, the cultures were centrifuged (10 min, 4,000 × g) and supernatants were analyzed by Western blot assay for the presence of beta2 toxin.
RNA isolation and cDNA synthesis.
Bacterial pellets were washed with distilled water and resuspended in STET buffer (0.1 M NaCl, 10 mM Tris/HCl [pH 8.0], 1 mM EDTA [pH 8.0], 5% Triton X-100) containing 20 mg/ml lysozyme (Merck) and 100 μg/ml proteinase K (Merck) and incubated for 4 h at 37°C with continual agitation. Next, the suspensions were centrifuged for 5 min at 16,000 × g and the resultant pellets were used for RNA isolation using Trizol (Invitrogen) according to the manufacturer's protocol, except that after the addition of isopropanol, mixtures were kept at −20°C for 15 min. RNA concentration was measured with a Nanodrop spectrophotometer (Isogen). Residual DNA was removed with RNase-free DNase I (Fermentas). One microgram of RNA was used for cDNA synthesis using the Transcriptor First Strand cDNA synthesis kit (Roche).
Q-PCR analysis.
The amount of cpb2 mRNA in the samples was measured by Q-PCR. The Q-PCR mixture consisted of 5 μl of cDNA, 12.5 μl of SYBR green mix (Bio-Rad Laboratories), 10 μM forward primer (5′-CAAGCAATTGGGGGAGTTTA-3′),10 μM reverse primer (5′-GCAGAATCAGGATTTTGACCA-3′), and 6.5 μl of distilled water. For normalization, the amount of gyrase mRNA was determined simultaneously by using 10 μM forward primer (5′-AGATATAGAAGACTTAATACAAG-3′) and 10 μM reverse primer (5′-AAAGAATAATAAGTTGAGTGTG-3′). The Q-PCR program consisted of 40 cycles of 30 s of denaturation at 95°C, 30 s of annealing at 57°C, and 15 s of extension at 72°C. Results were analyzed using the IQ5 software (Bio-Rad Laboratories) and expressed as CT (cycle threshold) cpb2 − CT gyrA.
Western blot analysis.
Ten microliters of supernatant was tested for the presence of beta2 toxin by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis with a monospecific polyclonal anti-beta2 toxin antibody (kindly provided by J. Frey, Berne, Switzerland), followed by a swine secondary antiserum coupled to peroxidase (Dako). The reaction was visualized by a chemiluminescence detection system (ECL Plus Western blotting detection reagents; GE Healthcare). Protein bands were quantified by densitometry using a Bio-Rad GS-700 densitometer (Bio-Rad Laboratories).
All experiments, with the exception of the degradation experiment (n = 2), were performed three times. Q-PCR of each cDNA sample was performed twice in triplicate.
Statistics.
Due to a low intraclass correlation coefficient in both the experiments with lactobacilli and pH experiments, a general linear model (GLM) was used instead of a linear model, with random effects to account for the correlation between observations within a repetition. For the lactobacillus experiment, the measured outcome was toxin production, which was square root transformed, with the fixed factors being lactobacilli (L, L1, and M), time (1 h, 2 h, 3 h, and 4 h), repetition (1, 2, and 3), and the interaction between lactobacilli and time. For the pH experiment, pH (pH 5, pH 6, or pH 7), time (1 h, 2 h, 3 h, or 4 h), and the number of repetitions (1, 2, or 3) were used as block factors and the interaction between pH and time was used as an explanatory factor. Due to nonconsistency of variance, the best GLM for the experiment with lactobacilli and the pH experiment was again fitted by an iterated reweighted least-squares method (29) using the median absolute difference for weighting the measurements. To analyze the data from the experiment on the production of beta2 toxin in the presence of lactobacilli and Caco-2 cells, a mixed model was used with repetition as a random effect (J. Pinheiro, D. Bates, S. DebRoy, D. Sarkar, and the R Development Core Team, 2009; nlme, Linear and Nonlinear Mixed-Effects Models, R package version 3.1-96). The outcome was square root transformed, and the same explanatory variables were used as for the experiment with lactobacilli. Model selection was based on the Akaikes information criterion. The residuals of the final models were used to check for normality and consistency of variance. For all analyses, the statistical package R version 2.11.1 (23) was used.
RESULTS
Beta2 toxin production by C. perfringens cultured in the presence of L. fermentum.
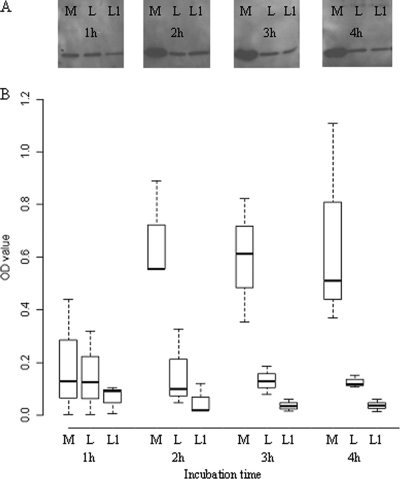
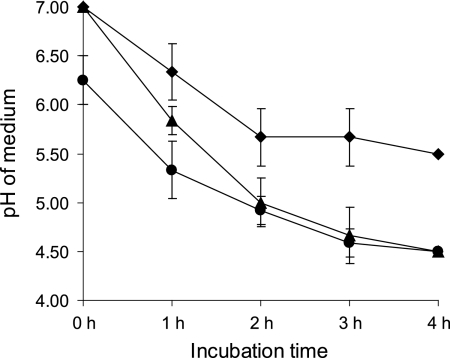
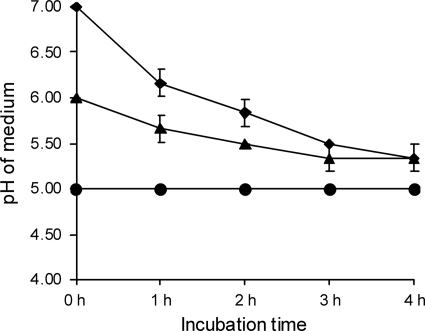
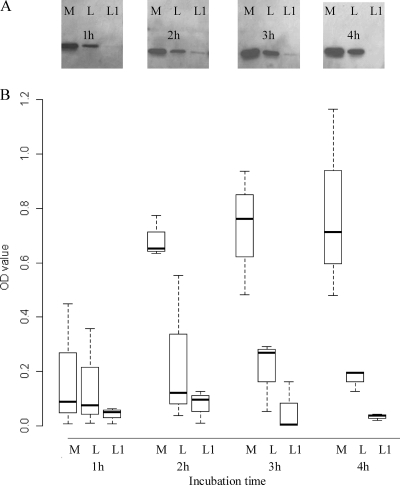
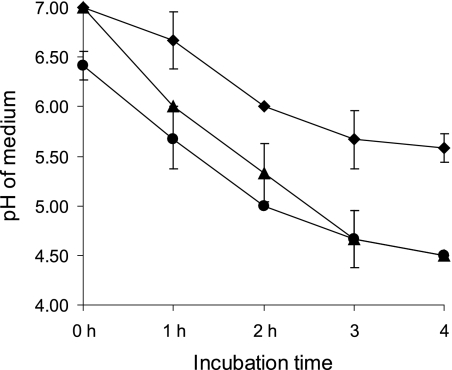
To study the effect of L. fermentum on beta2 toxin production by C. perfringens, Cp15 was cocultured with L. fermentum. The amount of beta2 toxin produced by Cp15 in this coculture (Fig. 1, L) was smaller than the amount of beta2 toxin produced by Cp15 in the absence of L. fermentum (Fig. 1, M). This effect was enhanced when the L. fermentum strain was grown in fresh MRS broth, pH 7, for 1 h before the addition of Cp15 to this culture (Fig. 1, L1). Coculturing of C. perfringens with L. fermentum lowered the pH of the coculture from 7 to 4.5 after 4 h, while the C. perfringens culture without L. fermentum reached a final pH of 5.5 after 4 h (Fig. 2). The pH of culture L1 had already decreased to pH 6.3 by the time Cp15 was added to the culture. After 4 h of coculturing, a final pH of 4.5 was reached (Fig. 2). Coculturing of Cp15 with L. fermentum did not influence the viability of either bacterial species, as the numbers of CFU of the various cultures remained stable for each species during the entire experiment (data not shown).
Fig. 1.
Differences in beta2 toxin production by C. perfringens strain Cp15 alone (M), in the presence of L. fermentum (L), or with a 1-h L. fermentum preincubation step (L1) after 1, 2, 3, and 4 h of incubation. (A) Western blot assay of a single experiment. (B) Mean optical density (OD) values of the various bands of three independent experiments as quantified by densitometry. The P value for the difference between the mean toxin levels of M and L is <0.0001, that for the difference between the mean toxin levels of M and L1 is < 0.0001, and that for the difference between the mean toxin levels of L and L1 is < 0.019.
Fig. 2.
Decline in medium pH due to the growth of C. perfringens strain Cp15 alone (⧫), in the presence of L. fermentum (▴), or with a 1-h L. fermentum preincubation step (•). The start of the experiment is at 0 h, immediately after the C. perfringens cultures were added. Means of 3 individual experiments and standard deviations are shown.
In order to rule out the possibility that the decrease in beta2 toxin was due to degradation by L. fermentum, L. fermentum was added both to a 2-h Cp15 culture and to the supernatant of such a culture and reincubated. No degradation or decrease in the total amount of beta2 toxin in either combination was observed by Western blot assay compared to the beta2 toxin produced by a plain Cp15 culture (Fig. 3). These experiments also demonstrate that pH did not affect the stability of the toxin, since L. fermentum, when metabolically active, lowers the pH over time.
Fig. 3.
Stability of beta2 toxin in a culture of C. perfringens strain Cp15 alone (C), in a Cp15 culture to which L. fermentum was added (CL), in the supernatant of a culture of Cp15 alone (S), or in the supernatant of a CP15 culture to which L. fermentum was added (SL). The 0-h time point is the start of the experiment, immediately after the L. fermentum cultures were added. The 2-h time point is after 2 h of incubation starting at the moment the L. fermentum cultures were added.
Beta2 toxin production by C. perfringens cultured at different pH values.
From the above data, it could be hypothesized that beta2 toxin production by C. perfringens is regulated by the pH of the environment. In order to study this in more detail, Cp15 was grown in MRS broth at pH 5, pH 6, or pH 7 for several time periods. Western blot analysis revealed that at every time point, the amount of beta2 toxin produced was smaller when Cp15 was grown at a lower pH than when it was grown at pH 7 (Fig. 4). The total amount of beta2 toxin produced in broths that had an initial pH of 6 or 7 increased less than expected after 3 and 4 h of incubation (Fig. 4). This was most likely caused by a decrease in the pH of the medium, since the pH of both broths decreased during prolonged incubation. The broth set at pH 7 reached a pH of 5.3 after 4 h, whereas the broth set at pH 6 already reached the (final) pH of 5.3 after 3 h. The pH of the broth set at pH 5 remained the same during the entire experiment (Fig. 5). No influence of the various pHs on the viability of Cp15 was observed, as the numbers of CFU of the various cultures remained stable and no differences in the numbers of CFU between the cultures grown in MRS with different initial pHs were found (data not shown). Efforts to stabilize the pH of the various media were not successful, since the salt concentrations needed to buffer these media at the desired pH influenced bacterial growth/survival.
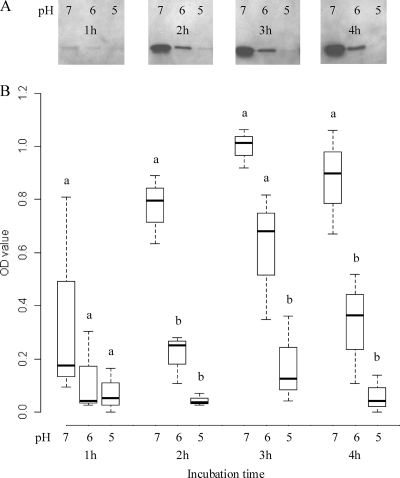
Fig. 4.
Differences in beta2 toxin production by C. perfringens strain Cp15 in MRS broth with an initial pH of 7, 6, or 5 after 1, 2, 3, or 4 h of incubation. (A) Western blot assay of a single experiment. (B) Mean optical density (OD) values of the various bands of three independent experiments as quantified by densitometry. A different letter indicates a significant difference (P < 0.05) in the mean toxin levels at the pH levels and times indicated.
Fig. 5.
Decline in medium pH due to the growth of C. perfringens strain Cp15 in MRS broth with an initial pH of 7 (⧫), 6 (▴), or 5 (•). Means of 3 individual experiments and standard deviations are shown.
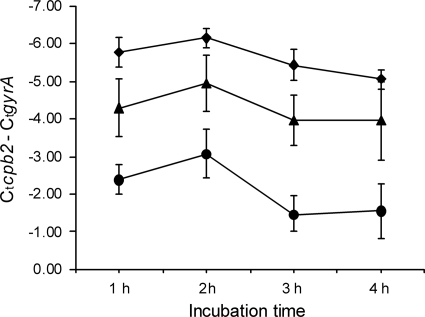
Q-PCRs were performed in order to address the possibility that the smaller amount of beta2 toxin produced in the broth with an initial pH of 5 or 6, compared to the amount produced at an initial pH of 7, was due to a lower level of cpb2 transcription. The gyrase mRNA was used as reference mRNA. In each experiment at a given time point, the CT value of gyrase between the various conditions varied by only approximately 1 CT value, again demonstrating the equal viability of the bacterium under the various conditions. However, in contrast, the amount of cpb2 mRNA was greater in samples grown at a higher pH. There was at least an 8-fold difference between cultures grown at pH 7 and those grown at pH 5, a 2-fold difference between those grown at pH 7 and those grown at pH 6, and a 4-fold difference between those grown at pH 6 and those grown at pH 5 (Fig. 6). The results from the Q-PCR clearly indicated that the pH of the environment influences the transcription of cpb2.
Fig. 6.
Differences in amounts of beta2 mRNA produced by C. perfringens strain Cp15 in MRS broth with an initial pH of 7 (⧫), 6 (▴), or 5 (•). The amount of beta2 mRNA produced is expressed relative to the amount of gyrase mRNA produced. Means of 2 individual experiments done in triplicate and standard deviations are shown.
Beta2 toxin production by C. perfringens cocultured with L. fermentum on Caco-2 cells.
Finally, we performed coculture experiments in the presence of Caco-2 cells in order to study whether the effect of L. fermentum on the beta2 toxin production of C. perfringens seen in in vitro experiments is also found in a semi-in-vivo setting. The results were that for Cp15 not cocultured with L. fermentum, large amounts of beta2 toxin were produced at all time points, whereas in coculture, little (L) or no toxin (L1) could be detected (Fig. 7). The drop in pH of the various cultures (Fig. 8) was comparable to that in the coculture experiments without Caco-2 cells. No effects of coculturing or the presence of Caco-2 cells on the viability of L. fermentum or C. perfringens were observed, as the numbers of CFU of both species remained stable during the entire experiment (data not shown).
Fig. 7.
Differences in beta2 toxin production by C. perfringens strain Cp15 alone (M), in the presence of L. fermentum (L), or with a 1-h L. fermentum preincubation step (L1) on a monolayer of Caco-2 cells after 1, 2, 3, or 4 h of incubation. (A) Western blot assay of a single experiment. (B) Mean optical density (OD) values of the various bands of three independent experiments as quantified by densitometry. The P value for the difference between the mean toxin levels of M and L is <0.010, that for the difference between the mean toxin levels of M and L1 is < 0.0001, and that for the difference between the mean toxin levels of L and L1 is < 0.067.
Fig. 8.
Decline in medium pH due to the growth of C. perfringens strain Cp15 alone (⧫), in the presence of L. fermentum (▴), or with a 1-h L. fermentum preincubation step (•) on a monolayer of Caco-2 cells after 1, 2, 3, or 4 h of incubation. The 0-h time point is the start of the experiment, immediately after the C. perfringens cultures were added. Means of 3 individual experiments and standard deviations are shown.
DISCUSSION
Although differing in composition, the resident intestinal flora of both animals and humans contributes to the well-being of the host (2, 10). This microflora forms a delicate ecosystem in which the different bacterial species may, to a certain extent, regulate each other's growth and gene expression. Regulation is achieved through the phenomenon of quorum sensing, in which there is inter- and intraspecies communication via released signal molecules (1). It has also been shown that bacteria belonging to the microflora influence each other and other bacteria by their metabolic products (10).
Here we present a study of the possible influence of acid produced by L. fermentum on the production of beta2 toxin by C. perfringens. Results from our initial experiments in which beta2-producing C. perfringens strain Cp15 was cocultured with L. fermentum strain 104R clearly indicate that beta2 toxin production was negatively regulated by L. fermentum. However, this regulation could have been achieved either through the bacterium itself or through one of its products.
It is known that the transcription of cpb2, the gene encoding the beta2 toxin, is under the control of the two-component VirR/VirS regulatory system (18). This VirR/VirS system is demonstrated to be sensitive to the signal molecule encoded by C. perfringens agrBD (agrBDCp), a homologue of Staphylococcus aureus agrBD. In S. aureus, agrBD encodes the propeptide AgrD and its modifying protein AgrB. Once AgrD is processed, the resulting autoinducing peptide (AIP) is secreted into the environment and acts as a quorum-sensing signal molecule (19). This accessory gene regulator (agr) system is conserved within the phylum Firmicutes, which comprises, among others, the orders Clostridiales and Lactobacillales (32). From this, it could be hypothesized that beta2 toxin production in the coculture experiments is regulated via quorum sensing. The production of beta2 toxin might then be influenced by the total amount of AIP produced by C. perfringens and L. fermentum. However, at each experiment, the number of bacteria (either C. perfringens or L. fermentum) was determined and it was concluded that the viability and the total number of CFU of either strain remained stable in all experiments. This finding made regulation of beta2 toxin production via quorum sensing unlikely and indicated a role for products of L. fermentum in the downregulation of beta2 production.
It was already shown in 1964 that the production and stability of toxins produced by C. perfringens were influenced by the pH of the growth medium (22). The various toxins had different optimum pHs for their production, and some, for example, the alpha toxin, appeared unstable regardless of the pH, whereas others, like the beta toxin, were sensitive to pH, while others remained stable regardless of the pH. The beta2 toxin was first described in 1997 (7), but there are no data regarding the influence that pH has on its production or stability.
To study the role of the acid produced by L. fermentum on beta2 toxin production, we performed experiments in which beta2 toxin production was measured in the absence of L. fermentum at different pHs (5, 6, and 7) since these values correspond roughly to the normal pH range in piglet intestines (pH 4.6 to 6.8) (26) and human intestines (pH 5.7 to 7.7) (17). From these experiments, it became apparent that the pH of the environment influences the production of beta2 toxin, indicating that, indeed, the acid produced by L. fermentum has a clear effect on beta2 toxin production by C. perfringens upon coculturing. However, the stability of any beta2 toxin that was produced was not affected by the pH. Q-PCRs performed with mRNA isolated from C. perfringens grown at various pHs indicated that the mechanism through which environmental pH influences beta2 toxin production involves effects on gene transcription. However, no conclusion could be drawn from our experiments about a possible influence at the translational level.
Recently, it has been reported that transcription of cpb2 is rapidly upregulated when a cpb2-harboring C. perfringens type C strain of porcine origin comes into close contact with enterocyte-like Caco-2 cells (30). In the intestine, C. perfringens is in close contact with the epithelial layer. This might indicate that the production of beta2 toxin is indeed under the control of an environmental signal since it can be concluded that if this were not the case, C. perfringens would always produce large amounts of (beta2) toxin in the intestine. To address this hypothesis, we performed C. perfringens and L. fermentum coculture experiments in the presence of a monolayer of Caco-2 cells. These experiments clearly demonstrated that lactobacilli are capable of diminishing or even abolishing the promoting effect of cell-to-cell contact between C. perfringens and Caco-2 cells on beta2 toxin production. Based on the results of the former experiments, it is likely that this effect was achieved through changes in pH. The rapid upregulation of toxin production by C. perfringens after contact with Caco-2 cells is mediated via the VirR/VirS system, as demonstrated in experiments using mutants with a nonfunctional VirR/VirS system (30). VirS, the “signaling component” of the VirR/VirS system, is situated in the plasma membrane (30). This makes it unlikely that pH has a direct effect on the binding of the agrBDCp-encoded signaling protein or another molecule(s) involved in upregulation after cell-to-cell contact. It has been suggested that, at least in the case of upregulation of toxin production through cell-to-cell contact, one or more surface factors of C. perfringens might be involved (30), and it is tempting to speculate that binding to these factors is pH dependent.
It is generally accepted that C. perfringens-related intestinal disease is initiated by the use of antibiotics, which allows C. perfringens overgrowth and excess toxin production after the die-off of other bacterial species of the normal intestinal flora. Our findings that beta2 production is regulated by the environmental pH might also explain the onset of disease after the use of antibiotics by hypothesizing that beta2 production is normally silenced by the acids produced by other members of the normal intestinal flora. Removal of these other members by the use of antibiotics would result in a rise in the local pH and subsequent toxin production by C. perfringens.
In conclusion, the results presented here clearly suggest that at least one member of the normal intestinal flora is of great importance in preventing toxin production by C. perfringens in the host. Further research should focus on the roles of other members of the normal intestinal flora in silencing toxin production by C. perfringens. Also, the underlying mechanism should be a subject for further studies since understanding this mechanism might lead to new ways of preventing disease without the use of antibiotics.
ACKNOWLEDGMENTS
We thank Ellen van der Wiel, Julie Duval, and Geert de Vrieze for excellent technical assistance. Hélène Verheije and Alan Wolfe are acknowledged for critical reading of the manuscript.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Bassler B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582–587 [DOI] [PubMed] [Google Scholar]
- 2. Berg R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430–435 [DOI] [PubMed] [Google Scholar]
- 3. Bueschel D. M., Jost B. H., Billington S. J., Trinh H. T., Songer J. G. 2003. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet. Microbiol. 94:121–129 [DOI] [PubMed] [Google Scholar]
- 4. Carman R. J., et al. 2008. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe 14:102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dogi C. A., Perdigón G. 2006. Importance of the host specificity in the selection of probiotic bacteria. J. Dairy Res. 73:357–366 [DOI] [PubMed] [Google Scholar]
- 6. Fisher D. J., et al. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56:747–762 [DOI] [PubMed] [Google Scholar]
- 7. Gibert M., Jolivet-Reynaud C., Popoff M. R. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203:65–73 [DOI] [PubMed] [Google Scholar]
- 8. Henriksson A., Szewzyk R., Conway P. L. 1991. Characteristics of the adhesive determinants of Lactobacillus fermentum 104. Appl. Environ. Microbiol. 57:499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herholz C., et al. 1999. Prevalence of β2-toxigenic Clostridium perfringens in horses with intestinal disorder. J. Clin. Microbiol. 37:358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holzapfel W. H., Haberer P., Snel J., Schillinger U., Huis in't Veld J. H. J. 1998. Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85–101 [DOI] [PubMed] [Google Scholar]
- 11. Hooper L. V., Bry L., Falk P. G., Gordon J. I. 1998. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays 20:336–343 [DOI] [PubMed] [Google Scholar]
- 12. Kinoshita H., et al. 2007. Quantitative evaluation of adhesion of lactobacilli isolated from human intestinal tissues to human colonic mucin using surface plasmon resonance (BIACORE) assay. J. Appl. Microbiol. 102:116–123 [DOI] [PubMed] [Google Scholar]
- 13. Lebrun M., et al. 2007. The expression of Clostridium perfringens consensus beta2 toxin is associated with bovine enterotoxaemia syndrome. Vet. Microbiol. 120:151–157 [DOI] [PubMed] [Google Scholar]
- 14. Miller M. B., Bassler B. L. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165–199 [DOI] [PubMed] [Google Scholar]
- 15. Naidu A. S., Bidlack W. R., Clemens R. A. 1999. Probiotic spectra of lactic acid bacteria (LAB). Crit. Rev. Food Sci. Nutr. 39:13–126 [DOI] [PubMed] [Google Scholar]
- 16. Nicholson J. K., Holmes E., Wilson I. D. 2005. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3:431–438 [DOI] [PubMed] [Google Scholar]
- 17. Nugent S., Kumar D., Rampton D. S., Evans D. F. 2001. Intestinal luminal pH in inflammatory bowel disease: possible determinants and implications for therapy with aminosalicylates and other drugs. Gut 48:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohtani K., Kawsar H. I., Okumura K., Hayashi H., Shimizu T. 2003. The VirR/VirS regulatory cascade affects transcription of plasmid-encoded putative virulence genes in Clostridium perfringens strain 13. FEMS Microbiol. Lett. 222:137–141 [DOI] [PubMed] [Google Scholar]
- 19. Ohtani K., et al. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petit L., Gibert M., Popoff M. R. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 21. Pinto M., et al. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323–330 [Google Scholar]
- 22. Pivnick H., Habeeb F. S. A., Grenstein B., Stuart P. F., Hauschild H. W. 1964. Effect of pH on toxinogenesis by Clostridium perfringens type C. Can. J. Microbiol. 10:329–344 [DOI] [PubMed] [Google Scholar]
- 23. R Development Core Team 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 24. Schotte U., Truyen U., Neubauer H. 2004. Significance of β2-toxinogenic Clostridium perfringens infection in animals and their predisposing factors—a review. J. Vet. Med. B Infect. Dis. Vet. Public Health 51:423–426 [DOI] [PubMed] [Google Scholar]
- 25. Shirkey T. W., et al. 2006. Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp. Biol. Med. (Maywood) 231:1333–1345 [DOI] [PubMed] [Google Scholar]
- 26. Snoeck V., Cox E., Verdonck F., Joensuu J. J., Goddeeris B. M. 2004. Influence of porcine intestinal pH and gastric digestion on antigenicity of F4 fimbriae for oral immunisation. Vet. Microbiol. 98:45–53 [DOI] [PubMed] [Google Scholar]
- 27. Songer J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tancrède C. 1992. Role of human microflora in health and disease. Eur. J. Clin. Microbiol. Infect. Dis. 11:1012–1015 [DOI] [PubMed] [Google Scholar]
- 29. Venables W. N., Ripley B. D. 2002. Modern applied statistics with S. Fourth edition. Springer, New York, NY [Google Scholar]
- 30. Vidal J. E., Ohtani K., Shimizu T., McClane B. A. 2009. Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell. Microbiol. 11:1306–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waters M., et al. 2003. Genotyping and phenotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J. Clin. Microbiol. 41:3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Wuster A., Babu M. M. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190:743–746 [DOI] [PMC free article] [PubMed] [Google Scholar]