Abstract
Streptococcus gordonii is an important member of the oral biofilm. One of its phenotypic traits is the production of hydrogen peroxide (H2O2). H2O2 is an antimicrobial component produced by S. gordonii that is able to antagonize the growth of cariogenic Streptococcus mutans. Strategies that modulate H2O2 production in the oral cavity may be useful as a simple therapeutic mechanism to improve oral health, but little is known about the regulation of H2O2 production. The enzyme responsible for H2O2 production is pyruvate oxidase, encoded by spxB. The functional studies of spxB expression and SpxB abundance presented in this report demonstrate a strong dependence on environmental oxygen tension and carbohydrate availability. Carbon catabolite repression (CCR) modulates spxB expression carbohydrate dependently. Catabolite control protein A (CcpA) represses spxB expression by direct binding to the spxB promoter, as shown by electrophoretic mobility shift assays (EMSA). Promoter mutation studies revealed the requirement of two catabolite-responsive elements (CRE) for CcpA-dependent spxB regulation, as evaluated by spxB expression and phenotypic H2O2 production assays. Thus, molecular mechanisms for the control of S. gordonii spxB expression are presented for the first time, demonstrating the possibility of manipulating H2O2 production for increased competitive fitness.
INTRODUCTION
Oral biofilm development follows distinct spatial-temporal patterns (28). The developmental process is influenced by environmental fluctuations in nutrient availability, oxygen tension, pH, saliva flow, and host innate immune components (29). Individual biofilm members need to adapt to these environmental fluctuations and compete with other species for the available space (20). The most abundant early oral biofilm formers belong to the genus Streptococcus (44). Oral streptococci can initiate biofilm development by expressing surface proteins that specifically interact with components of the salivary pellicle formed on the tooth surface (15, 36). Once the early biofilm has been established, other organisms can join via cell-cell interactions and/or binding to extracellular substances, such as glucans, proteins, and DNA produced or secreted by the early colonizers during biofilm formation (28). Two important pioneer colonizers are the oral commensals Streptococcus sanguinis and Streptococcus gordonii. In addition to their ability to bind to salivary components such as amylase, both species are known to produce bactericidal amounts of hydrogen peroxide (H2O2), possibly as a competitive measure during biofilm development (3, 32, 45). H2O2 originates mostly from the enzymatic activity of the pyruvate oxidase, SpxB (encoded by the spxB gene) (6, 7). SpxB catalyzes the conversion of pyruvate to acetyl phosphate, which is subsequently converted to acetate by acetate kinase (7). Acetyl phosphate is a central metabolite also used in the transfer of phosphoryl groups to two-component response regulators and ADP in order to generate ATP for energy (52). The enzymatic reaction requires oxygen, and the end product H2O2 leaves the cell, probably through diffusion. H2O2 can accumulate to substantial amounts when S. sanguinis and S. gordonii are cultivated as aerobic batch cultures (32). Concentrations between 1 and 10 mM have been reported (45). The ability to produce H2O2 has also been reported for another related species, Streptococcus pneumoniae, and is dependent on SpxB activity (48). Interestingly, interspecies interference between S. pneumoniae and Staphylococcus aureus has been observed in epidemiologic studies (41) and was attributed to the bactericidal activity of H2O2 demonstrated in in vitro experiments (42, 47).
A similar antagonistic relationship between cariogenic species and oral commensal streptococci has been shown in several clinical studies. For example, subjects with a high abundance of S. sanguinis have a lower incidence of caries development, and vice versa (4). Furthermore, initial colonization with S. sanguinis delays colonization with cariogenic Streptococcus mutans (8). This suggests a protective role of oral commensal streptococci against oral disease development. The relevant in vivo mechanisms for this interspecies interference are not known, but several in vitro studies strongly suggest an important role for bacteriocins and H2O2 in this process (30). Interestingly, S. mutans can excrete bacteriocins to engage in interspecies competition with oral commensal streptococci under both aerobic and anaerobic conditions (30, 32). In contrast, S. sanguinis and S. gordonii produce growth-inhibiting amounts of H2O2 only during aerobic growth (32). Further studies with S. gordonii revealed that another environmental component is able to interfere with H2O2 production: when grown with high concentrations of glucose, S. gordonii is no longer able to produce competitive amounts of H2O2 and can no longer inhibit H2O2-susceptible species, such as S. mutans (32). This suggests a carbon catabolite repression (CCR) of H2O2 production. CCR in the Gram-positive model species Bacillus subtilis is regulated mainly by catabolite control protein A (CcpA), belonging to the LacI/GalR transcriptional regulator family (16). The CcpA DNA binding activity is enhanced by the Hpr component of the phosphoenolpyruvate (PEP)-dependent carbohydrate phosphotransferase system (PTS), which is phosphorylated at Ser-46 (Hpr-Ser-P) during growth on glucose or other PTS carbohydrates. The interaction of CcpA and Hpr-Ser-P promotes binding to specific DNA sequences in the promoter region of CCR-controlled genes, designated catabolite-responsive elements (CRE) (10). CCR and CcpA are known to regulate genes involved in carbon and nitrogen utilization as well as virulence genes in several Gram-positive bacteria, including S. gordonii (5, 16, 19).
The regulation of H2O2 production in S. gordonii is of considerable interest because of the potential protective nature of oral commensals against cariogenic species. Increasing the fitness of oral commensal bacteria could be a valuable alternative approach to caries prevention. In this report, the consequences of oxygen and glucose availability for spxB expression and SpxB abundance are further investigated. In addition, evidence is provided that CCR is mediated by several carbohydrate sources and relies on the binding of CcpA to the spxB promoter. This is in contrast to the recently reported observation for S. sanguinis that CcpA-mediated spxB repression is carbohydrate independent (55).
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are listed in Table 1. Strains were routinely grown aerobically (5% CO2) or in an anaerobic chamber (90% N2, 5% CO2, 5% H2) at 37°C on brain heart infusion (BHI) agar plates but in some cases (as stated) were grown in TYE medium (1% tryptone, 0.5% yeast extract, 0.3% K2HPO4). The medium was supplemented with the carbohydrate glucose, galactose, sucrose, maltose, fructose, or lactose from filter-sterilized 20% stock solutions where indicated. For antibiotic selection, cultures were supplemented with the following antibiotics: spectinomycin at 500 μg ml−1, erythromycin at 2 μg ml−1, kanamycin at 300 μg ml−1 for S. gordonii, and ampicillin at 100 μg ml−1 for Escherichia coli.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| DL1 | Wild-type S. gordonii | 39 |
| DL1 spxBp-luc | spxBp luciferase reporter; Spcr | 55 |
| DL1 ΔccpA | ΔccpA; Ermr | This study |
| DL1 compl | DL1 ΔccpA; ccpA complemented; Spcr | This study |
| DL1 Mut CRE 0 | S. gordoniispxB CRE mutagenesis control; Kanr | This study |
| DL1 Mut CRE 1 | S. gordoniispxB CRE 1 mutagenesis; Kanr | This study |
| DL1 Mut CRE 2 | S. gordoniispxB CRE 2 mutagenesis; Kanr | This study |
| DL1 Mut CRE 1 + 2 | S. gordoniispxB CRE 1 + CRE 2 mutagenesis; Kanr | This study |
| DL1 Mut CRE Δ | S. gordoniispxB CRE 1 + CRE 2 deletion; Kanr | This study |
| DH5α | E. coli; cloning strain | 18 |
| BL21(DE3)pLysS | E. coli; protein expression strain | 35 |
| BL21(pET29b+ccpA) | E coli; protein expression strain; pET29b+ccpA; Kanr | This study |
| Plasmids | ||
| pFW5 | Suicide vector; Spcr | 40 |
| pDL278 | E. coli-Streptococcus shuttle vector; Spcr | 13 |
| pLZ2 | pFW5φ(spxBp::luc), luciferase reporter; Spcr | This study |
| pGEM-T Easy | Cloning vector; Ampr | Promega |
| pET-29b(+) | His6 fusion protein expression vector; Kanr | Novagen |
| pDL278+ccpAcompl | ccpA cloned into pDL278; Spcr | This study |
| pET-29b+ccpA | pET29b + ccpA; S. gordoniiccpA overexpression; Kanr | This study |
| pGEM-T-spxB-FLAG | pGEM-T Easy, DL1 spxB-FLAG construction; Ampr | This study |
| pFW5-spxB-flag | pFW5, DL1 spxB-FLAG expression; Spcr | This study |
| pGEM-T-CRE 1 | pGEM + spxBp; CRE 1 mutagenesis; Ampr | This study |
| pGEM-T-CRE 2 | pGEM + spxBp; CRE 2 mutagenesis; Ampr | This study |
| pGEM-T-CRE 1 + 2 | pGEM + spxBp; CRE 1 + CRE 2 mutagenesis; Ampr | This study |
| pGEM-T- CRE Δ | pGEM + spxBp; CRE 1 + CRE 2 deletion; Ampr | This study |
DNA manipulations.
Standard recombinant DNA techniques were used as described previously (55). Restriction enzymes and DNA ligase were obtained from New England Biolabs (Beverly, MA) or Promega (Madison, WI) and were used as specified by the manufacturer. PCR products were cloned into the pGEM-T Easy kit from Promega. All plasmids were extracted and purified from E. coli using a Qiagen (Valencia, CA) miniprep kit. DNA extracted from agarose gels (1%) was purified with a Qiagen QIAquick gel extraction kit. PCR was performed with a G-Storm GS1 thermocycler (Gene Technologies, Essex, United Kingdom) according to the manufacturer's protocol. GoTaq DNA polymerase was obtained from Promega, and Phusion high-fidelity DNA polymerase was obtained from New England Biolabs. The primer sequences (Table 2) were designed using sequence data obtained from the Los Alamos National Laboratory Oral Pathogens Sequence Database (http://www.oralgen.lanl.gov) and were synthesized by Integrated DNA Technologies (IDT; Coralville, IA).
Table 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Purpose |
|---|---|---|
| 16S RNA RT F | AAGCAACGCGAAGAACCTTA | RT-PCR |
| 16S RNA RT R | GTCTCGCTAGAGTGCCCAAC | RT-PCR |
| Sg. spxB RT F | GGATGCTTTGGCTGAAGAC | RT-PCR |
| Sg. spxB RT R | GGACCACCTGAACCTACTG | RT-PCR |
| Sg. spxB F | AGCGGTCGACGGACTGGGATTGACCCTC | spxB reporter |
| Sg. spxB R | AGCGGGATCCTGCTGCAGATGCAGTAAT | spxB reporter |
| Sg. spxB-flag F | AGCGCTCGAGCGTGATTACATGAACAAACTCG | spxB-FLAG tag |
| Sg. spxB-flag R | CGCTAGATCTTTACTTGTCATCATCGTCTTTGTAATCTTTAATTGCGCGTGATTGCA | spxB-FLAG tag |
| Sg CcpA-his tag F | AGCGCATATGAACACAGACGATACAGTAAC | CcpA-His tag |
| Sg CcpA-his tag F | CGCTCTCGAGTTTTCTAGTTGAGTTCCGTTC | CcpA-His tag |
| Sg ccpA Up F | CTTGCTTGATGGCAAAGTCA | ccpA knockout |
| Sg ccpA Up R | GACAACCACCCGACTTTGAA CCATAGAAACCCCTGCTTCA | ccpA knockout |
| ermF | TTCAAAGTCGGGTGGTTGTC | ccpA knockout |
| ermR | GAAGCTGTCAGTAGTATACC | ccpA knockout |
| Sg ccpA Dn F | GGTATACTACTGACAGCTTC GGAAGAGTTGGAAGAACGTGA | ccpA knockout |
| Sg ccpA Dn R | TGGCATCCTTCTAGCCACTT | ccpA knockout |
| Sg ccpA compl F | GATGGATCCTGAAACCTTCTTTCTATGCCTCT | ccpA complementation |
| Sg ccpA compl R | GATGAATTCTGGCAAAAGCAAGTCTGTCT | ccpA complementation |
| Kan F | AGG TGA TAG GTA AGA TTA TAC CG | CRE mutagenesis |
| Kan R | CCC TAT CTA GCG AAC TTT TAG A | CRE mutagenesis |
| Sg spxB cre mut up F | CAGCTCAAAAGAAGCGATCA | CRE mutagenesis |
| Sg spxB cre mut up R | GGTATAATCTTACCTATCACCTGAGGCTGGGACAAAAGTCCT | CRE mutagenesis |
| Sg spxB cre mut Dn F | TAAAAGTTCGCTAGATAGGG CAGCTACAAGTCTTAGAGGTGCAT | CRE mutagenesis |
| Sg spxB cre mut Dn R | TTTCAACAACAGCTGGACCTT | CRE mutagenesis |
| mut Cre1 R | GAACGGAGATACCTCTGGAAACTTTACTATATCAATTTACCA | CRE 1 mutagenesis |
| mut Cre1 F | CCAGAGGTATCTCCGTTCACAA ATGGAAATGTTTTCAA | CRE 1 mutagenesis |
| mut Cre2 R | TAGCAGATATGTCCGTTTGTGAATGAAAACGCTTCAAG | CRE 2 mutagenesis |
| mut Cre2 F | AAACGGACATATCTGCTATAACTGAAAATTTATTTTATTGAAG | CRE 2 mutagenesis |
| mut Cre3 (both) F | CCAGAGGTATCTCCGTTCACAA ACGGACATATCTGCTA | CRE 1 + CRE 2 mutagenesis |
| delete both Up R | CAATAAAATAAATTTTCAGTAAACTTTACTATATCAATTTACCA | CRE 1 + CRE 2 deletion |
| delete both Dn F | ACTGAAAATTTATTTTATTGAAGGAGA | CRE 1 + CRE 2 deletion |
| 5′ RACE PCR adapter | PHOS-TTT AGT GAG GGT TAA TAA GCG GCC GCG TCG TGA CTG GGAGCG C | 5′ RACE PCR |
| 5′ RACE PCR adapter primer | GCG GCC GCT TAT TAA CCC TCA CTA AA | 5′ RACE PCR |
| 5′ RACE PCR spxB gene-specific primer | GTATGAACCTGAACCGTAGTATGAGTTT | 5′ RACE PCR |
| EMSA F primer | CAGCTACAAGTCTTAGAGGTGCAT | EMSA |
| EMSA R primer | Biotin-TGC AGA TGC AGT AAT TTT TCC TTG AG | EMSA |
Construction of a ΔccpA mutant.
A ΔccpA deletion mutant was constructed via double-crossover homologous recombination. To generate the construct, two fragments corresponding to ∼800 bp of the upstream and downstream sequences of ccpA were amplified by PCR using Phusion Hot Start high-fidelity DNA polymerase with the primer pair ccpA Up F/ccpA Up R and the primer pair ccpA Dn F/ccpA Dn R. Each of the primers listed as Up R or Dn F incorporated 25 bases complementary to the erythromycin resistance cassette ermAM (34). The erythromycin resistance cassette ermAM was amplified by PCR using primers ermF and ermR as described previously (55). All three PCR amplicons were purified with the Qiagen PCR purification kit and were mixed in a 1:1:1 ratio. The mixture served as a template for a second round of PCR with the appropriate Up F and Dn R primers. The resulting PCR amplicons were transformed into wild-type (WT) strain DL1 to generate the ΔccpA deletion mutant (DL1 ΔccpA). Deletion was confirmed by PCR. The mutant was complemented with a copy of ccpA expressed in trans from shuttle plasmid pDL278. The complete ccpA open reading frame and 250 bp of the upstream intergenic region encoding the promoter were amplified by PCR with primer pair Sg ccpA compl F/Sg ccpA compl R; the amplicon was digested with BamHI/EcoRI and was ligated into pDL278 digested with the same enzymes to generate the complementation plasmid pDL278+ccpAcompl.
Construction of a pyruvate oxidase carrying a C-terminal FLAG epitope.
For the construction of an SpxB C-terminal FLAG epitope, about 700 bp from the 3′ end of the pyruvate oxidase gene (spxB) was PCR amplified with specific primers incorporating a 6-FLAG epitope sequence (GATTACAAAGACGATGATGACAAG) upstream of the stop codon using primer pair spxB-flag F/spxB-flag R. The PCR product was inserted into the pGEM-T Easy vector to create pGEM-T-spxB-flag. The construct was sequenced to confirm the in-frame presence of the FLAG epitope sequence. The cloned fragment was released with XhoI/BglII and was inserted into pFW5 digested with the same enzymes. Plasmid pFW5 is not able to replicate in S. gordonii and requires integration into the chromosome to confer antibiotic resistance (40). The recombinant plasmid was transformed into wild-type DL1; transformants were selected on BHI plates containing spectinomycin antibiotics and were further analyzed by PCR for integration of the plasmid at the correct chromosomal position.
Western immunoblotting.
Western immunoblotting was performed essentially as described previously (55). Briefly, after cell disruption, cytoplasmic extracts were obtained by centrifugation, and the proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membrane was blocked with a solution of 5% skim milk dissolved in Tris-buffered saline containing 0.1% Tween 20 (TBST) for 1 h. Subsequently, the membrane was incubated in a primary antibody solution (anti-FLAG antibody M2; Stratagene) overnight at 4°C, followed by three wash steps with TBST, and was then incubated with horseradish peroxidase-conjugated secondary antibodies (Thermo Scientific) for 1 h at room temperature. After washing with TBST, the blots were developed using the ECL chemiluminescent detection system (Thermo Scientific). Images of Western immunoblots were obtained by scanning of the respective films. Images were adjusted with GIMP photo processing software, version 2.6.10 (http://www.gimp.org/), applied to all parts of the image.
Determination of transcription start sites.
The FirstChoice RLM random amplification of cDNA ends (RACE) kit (Ambion) was used to determine transcription start sites essentially as described previously (38). RACE-specific primers are listed in Table 2.
RNA isolation, cDNA synthesis, and real-time PCR.
RNA was isolated using the Qiagen RNeasy kit, cDNA was synthesized using StrataScript reverse transcriptase (Stratagene) according to the manufacturer's protocol. Real-time reverse transcriptase PCR (RT-PCR) was performed to detect specific transcripts by the comparative threshold cycle (CT) method using the Bio-Rad (Hercules, CA) MyiQ cycler. Relative changes in gene expression were calculated using the ΔCT method described previously (55). The 16S rRNA gene was used as the housekeeping reference gene. All primers used for real-time RT-PCR are listed in Table 2.
Luciferase assay.
The protocol for the construction of an spxB luciferase reporter strain was described previously (55). Luciferase assays were performed using a method described previously (33). Three parallel cultures were measured at each time point, and the mean value was calculated. Each experiment was repeated at least twice.
Overexpression and purification of CcpA.
PCR amplification of ccpA with the primer pair Sg CcpA-his tag F/Sg CcpA-his tag R was performed using DL1 chromosomal DNA as the template. The ccpA PCR fragment was inserted into the pGEM-T Easy vector, creating plasmid pGEM-T-ccpA. The construct was confirmed by sequencing and was subsequently digested with NdeI/XhoI to release ccpA. The ccpA fragment was ligated into appropriate restriction enzyme sites in pET-29b(+) to generate a CcpA C-terminal His tag. For overexpression, the confirmed plasmid was transformed into E. coli BL21(DE3)pLysS. The overexpressed CcpA His tag protein was purified under nondenaturing conditions using the Ni-nitrilotriacetic acid (NTA) purification system according to the manufacturer's protocol (Invitrogen). The protein concentration was determined using the Bradford protein assay (Bio-Rad protein assay) with bovine serum albumin (BSA) as a standard, and the samples were stored at −80°C for future use.
CRE site mutagenesis.
To study the roles of the two conserved catabolite-responsive elements (CRE) in the S. gordonii spxB promoter, overlap extension PCR was used to introduce mutations into the respective CRE. For the mutation of CRE 1, we used primer pairs cre mut up F/mut Cre1 R and mut Cre1 F/cre mut Dn R to generate two DNA fragments with overlapping ends containing the desired CRE 1 mutation. The two PCR fragments were purified with the Qiagen PCR purification kit, mixed in a 1:1 ratio, and used as a template for a second-round PCR with primers cre mut Dn F/cre mut Dn R. The mutated PCR fragment was purified and ligated into pGEM-T, resulting in plasmid pGEM-T-CRE 1. The same strategy was used to mutate CRE 2 (with primer pairs cre mut up F/mut Cre2 R and mut Cre2 F/cre mut Dn R), to mutate both CRE 1 and CRE 2 [with primer pairs cre mut up F/mut Cre2 R and mut Cre3 (both) F/cre mut Dn R], and to completely delete both CRE (with primer pairs cre mut up F/delete both Up R and delete both Dn F/cre mut Dn R). The plasmids constructed, pGEM-T-CRE 1, pGEM-T-CRE 2, pGEM-T-CRE 1 + 2, and pGEM-T-CRE Δ, were all sequenced to confirm the mutations introduced. The respective CRE mutations were transferred to DL1 using an overlap extension PCR strategy, with subsequent transformation of the PCR fragment by chromosomal integration via homologous recombination. Briefly, ∼700 bp of the upstream sequences of the CRE using wild-type chromosomal DNA as a template was PCR amplified with the primer pair cre mut up F/cre mut up R. The downstream fragments containing the mutated CRE sites were PCR amplified using the respective plasmids with the primer pair cre mut Dn F/cre mut Dn R. As a control, the same region was amplified from DL1 chromosomal DNA. Each of the primers listed as Up R or Dn F incorporated 25 bases complementary to the kanamycin resistance gene aphAIII. The kanamycin resistance gene was amplified by PCR from plasmid pJH1 (50) using primers Kan F and Kan R. All three PCR amplicons (the upstream fragment, kanamycin cassette, and downstream fragment) were purified with the Qiagen PCR purification kit and were mixed in a 1:1:1 ratio. The mixture served as a template for a second-round PCR with the appropriate Up F and Dn R primers. The resulting PCR amplicons were transformed into the wild-type strain DL1 to generate mutagenesis strains with various CRE sites (DL1 Mut CRE 1, DL1 Mut CRE 2, DL1 Mut CRE 1 + 2, DL1 Mut CRE Δ; strain DL1 Mut CRE 0 works as a control to exclude a polar effect from the introduced kanamycin cassette). All strains were confirmed by PCR.
Electrophoretic mobility shift assays (EMSA).
Biotin-labeled DNA probes were generated by PCR using Phusion Hot Start high-fidelity DNA polymerase with primer pair EMSA F/EMSA R; primer EMSA R was biotin labeled at the 5′ end by IDT. Chromosomal DNA from the wild type or the respective cre site-mutated strains (DL1 Mut CRE 1, DL1 Mut CRE 2, DL1 Mut CRE 1 + 2, and DL1 Mut CRE Δ) served as a template. All five PCR amplicons were purified with the Qiagen PCR purification kit, and biotin-labeled DNA probes (1 ng) were incubated with varying concentrations of purified CcpA in binding buffer [10 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol, 1 mM EDTA, 50 mM KCl, 0.05 μg/μl poly(dI-dC), 1 mM MgCl] for 30 min at room temperature. After incubation, samples were mixed with bromophenol blue loading buffer and were separated by electrophoresis using 6% acrylamide gels in 0.5× Tris-borate-EDTA buffer (pH 8.0) that had been prerun for 30 min prior to sample loading. The samples were electroblotted from the gels onto positively charged Zeta-Probe nylon membranes (Bio-Rad). The membranes were UV cross-linked (Stratagene), and the biotinylated probes were detected according to the manufacturer's protocol for the LightShift chemiluminescent EMSA kit (Thermo Scientific).
Measurement of H2O2 production.
The production of H2O2 during planktonic cultivation was performed essentially as described previously (55). H2O2 indicator plates were prepared as described elsewhere (46).
RESULTS
Growth and expression of spxB in S. gordonii strain DL1.
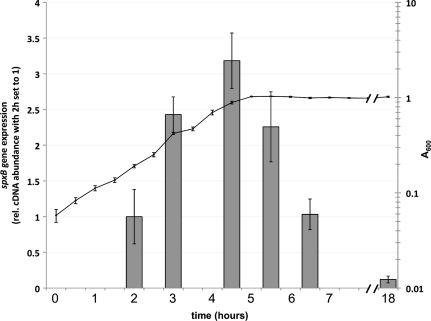
The growth of S. gordonii was tested in BHI and TYE media under conditions of low (0.2%) or high (2%) glucose concentrations. No significant difference in growth was observed between 0.2% and 2% glucose (data not shown). The expression of spxB over time was determined by quantitative real-time PCR. Planktonic cells were grown in TYE medium plus 0.2% glucose, and cDNA abundance was determined at the time points indicated in Fig. 1. The initial cDNA abundance was low but increased steadily during exponential growth. The transition from late-logarithmic phase to early-stationary phase resulted in a decrease of cDNA abundance (Fig. 1). Further overnight incubation gave the lowest cDNA abundance, about 20-fold lower than that in logarithmic phase. We also used an spxB luciferase reporter and confirmed the results from real-time PCR (data not shown). The expression profile of spxB suggests that the spxB promoter is most actively transcribed during the logarithmic growth phase of S. gordonii. Subsequent experiments were therefore performed with cells grown to mid-logarithmic phase. Cells were grown in TYE medium to test for carbohydrate effects; for all other experiments, cells were grown in BHI.
Fig. 1.
Growth phase-dependent regulation of spxB expression. DL1 was grown in TYE medium plus 0.2% glucose. cDNA abundance and optical density were measured at the indicated time points. The 16S rRNA gene was used as the housekeeping reference gene. The means and standard deviations from two independent experiments are presented. rel., relative.
Oxygen influences spxB transcription and SpxB abundance.
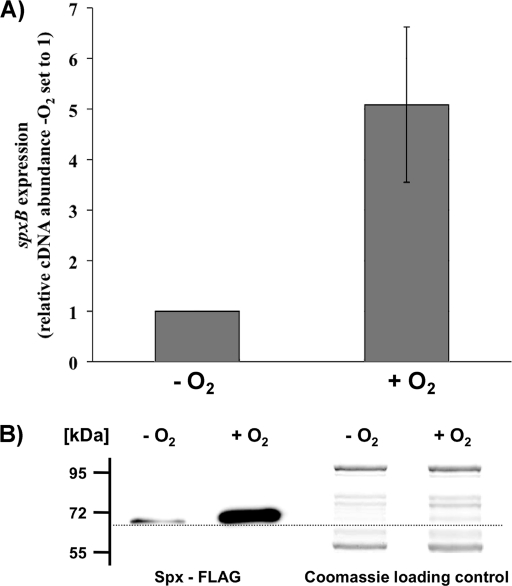
SpxB-dependent H2O2 production requires aerobic conditions (32). Oxygen is one of the SpxB substrates for the conversion of pyruvate into acetyl phosphate (7). The oxygen dependence of spxB expression has a profound effect on the competitiveness of S. gordonii. During anaerobic growth, S. gordonii is not able to produce growth-inhibiting amounts of H2O2 to antagonize H2O2-susceptible species such as S. mutans (32). To test if S. gordonii adjusts spxB gene expression and SpxB abundance according to oxygen availability, expression and protein production were determined under aerobic and anaerobic growth conditions. Cells were grown in BHI with 0.2% glucose. Real-time PCR was used to compare the spxB expression of cells grown with and without oxygen. As shown in Fig. 2 A, spxB expression increased about 5-fold in the presence of oxygen. To correlate gene expression with protein production, a FLAG epitope was engineered onto the C-terminal end of SpxB. This allows for chromosomal spxB-FLAG expression from its own promoter. The FLAG epitope did not influence the production of H2O2 under aerobic conditions, which confirmed the functionality of the protein (data not shown). The band intensity in Western immunoblot analysis appeared to be increased with aerobically grown cells (Fig. 2B). Interestingly, a band was detected for anaerobically grown cells, although no H2O2 is produced under these conditions (32). This suggests that S. gordonii couples spxB expression and protein production but ensures the presence of SpxB even under anaerobic conditions, possibly so as to produce competitive H2O2 instantaneously when oxygen becomes available.
Fig. 2.
Effects of oxygen on spxB transcription and SpxB abundance. (A) spxB transcription was measured by real-time RT-PCR analysis in wild-type DL1. Cells were grown anaerobically or aerobically on BHI agar plates for 7 h at 37°C before RNA isolation. The expression level for spxB from cells grown aerobically is presented relative to that from cells grown anaerobically, which was arbitrarily assigned a value of 1. The 16S rRNA gene was used as the housekeeping reference gene. Averages and standard deviations from three independent experiments are presented. (B) Western blot analysis of SpxB protein abundance. Cells were grown anaerobically or aerobically on BHI agar plates for 7 h at 37°C. Pyruvate oxidase production was detected using Western immunoblot analysis with an antibody against the FLAG epitope. A replicate sodium dodecyl sulfate gel was prepared and stained with Coomassie blue as a loading control. The PageRuler prestained protein ladder (Fermentas) was used as a size reference. Relative protein sizes are given on the left.
Glucose represses spxB expression and SpxB abundance.
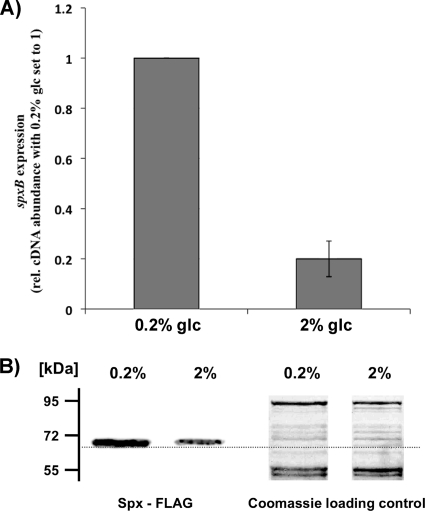
Nutrient availability routinely fluctuates tremendously within the oral cavity. For example, the concentration of glucose can increase as much as 1,000 fold during food intake (1). For S. mutans, it has been shown that growth under conditions of nutrient excess represses the production of bacteriocins. It was suggested that the hostile bacteriocin-producing behavior is not required when carbohydrate abundance allows for optimal cell proliferation (30). To test if S. gordonii spxB expression is influenced by glucose, cells were grown in TYE medium containing 0.2% or 2% glucose under aerobic conditions until the mid-logarithmic growth phase. To exclude any pH-dependent effect on spxB expression, the pHs were measured and found to be similar (pH 6.8 with 0.2% glucose and pH 6.7 with 2% glucose). As shown in Fig. 3 A, spxB expression decreased about 5-fold in the presence of 2% glucose as measured by RT-PCR (RT-PCR). SpxB protein abundance appeared to decrease as well under the same growth conditions (Fig. 3B). This is in agreement with the inability of S. gordonii to inhibit S. mutans in an antagonism assay performed in the presence of 1% glucose (32). As with anaerobic conditions, the immunoblot band intensity appeared reduced for 2% glucose, but the enzyme was still present. The amount of H2O2 produced under these conditions is not high enough to inhibit S. mutans (32). These results strongly suggest that spxB expression is regulated by CCR.
Fig. 3.
Glucose-dependent repression of spxB expression and SpxB abundance. (A) Comparative real-time RT-PCR analysis was used to determine the effect of glucose on spxB transcription. Wild-type DL1 was grown in TYE medium supplied with 0.2% glucose or 2% glucose. The expression level for spxB from cells grown with 2% glucose is presented relative to that from cells grown with 0.2% glucose, which was arbitrarily assigned a value of 1. The 16S rRNA gene was used as the housekeeping reference gene. Averages and standard deviations from three independent experiments are presented. (B) Western blot analysis of SpxB protein abundance in cells grown with 0.2% or 2% glucose. Pyruvate oxidase production was detected using Western immunoblot analysis with an antibody against the FLAG epitope. A replicate sodium dodecyl sulfate gel was prepared and stained with Coomassie blue as a loading control. The PageRuler prestained protein ladder (Fermentas) was used as a size reference. Relative protein sizes are given on the left.
CCR of spxB expression is not restricted to glucose.
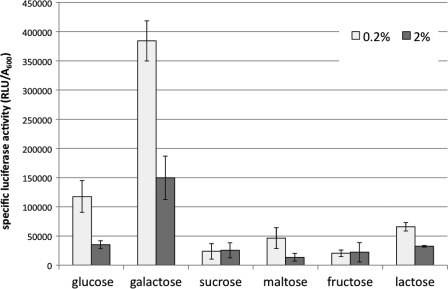
To further characterize the extent of CCR, other carbohydrates were tested for their abilities to repress spxB expression. Cells were grown under aerobic conditions in TYE medium supplemented with either 0.2% or 2% glucose, galactose, sucrose, maltose, fructose, or lactose. No significant difference in growth was observed between low and high carbohydrate concentrations (data not presented). Growth in the presence of 2% glucose confirmed the RT-PCR results, showing 4-fold reduction in luciferase activity (Fig. 4). Growth in the presence of 0.2% galactose resulted in the highest luciferase activity measured. However, at 2% galactose, there was a 3-fold decrease in luciferase activity. This result is consistent with earlier reports of galactose exerting CCR in S. gordonii (49), whereas in S. mutans, galactose is nonrepressing (49). Reductions in luciferase activity were also observed with 2% maltose and 2% lactose, while luciferase activity was not changed with increasing concentrations of sucrose and fructose (Fig. 4). The decrease in luciferase activity suggests that spxB expression is subject to CCR by a broad range of carbohydrate sources. This is in contrast to a recent study with S. sanguinis demonstrating that spxB expression was not significantly repressed by any of the carbohydrates tested, including glucose, maltose, and lactose (55).
Fig. 4.
Carbohydrate-dependent repression of spxB expression. The DL1 spxBp-luc reporter strain was grown in TYE medium supplemented with the indicated carbohydrate at 0.2% or 2%. Luciferase activity and optical density were measured for cells grown to mid-logarithmic phase (A600, ≈0.6). Means and standard deviations from three independent experiments, normalized to optical density, are presented.
CCR of spxB expression is mediated through CcpA.
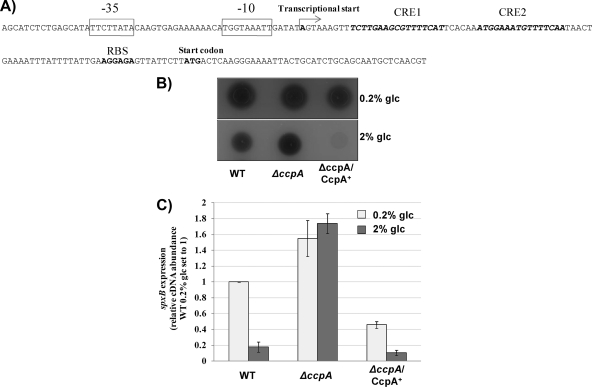
CCR is usually mediated through CcpA (51). CcpA-dependent CCR in S. gordonii has been demonstrated for the gene encoding amylase-binding protein A and the arginine deiminase operon (12, 43). To test if CCR is mediated through CcpA, the promoter sequence was analyzed for the presence of a catabolite-responsive element (CRE), the binding site for CcpA. As shown in Fig. 5 A, two potential CRE, CRE 1 and CRE 2, could be identified upstream of the ribosome binding site. The transcriptional start site for spxB was therefore determined using RACE PCR to confirm a potential overlap with CRE 1 and CRE 2. Transcription starts right after a potential extended −10 region, but before CRE 1 (Fig. 5A).
Fig. 5.
CCR of spxB expression is mediated through CcpA. (A) Organization of the spxB promoter region. Presented is the sequence of the promoter region showing the transcriptional start, the predicted −35 and −10 regions, the ribosome binding site (RBS), and the translation start site of spxB. The putative CRE are shown in italics. The transcriptional start site was mapped by RACE PCR and is indicated by an arrow. (B) The influence of CcpA on H2O2 production was determined with indicator plates. A reaction with H2O2 leads to the formation of a blue precipitate (Prussian blue). H2O2 production was determined for cells grown with 0.2% glucose or 2% glucose. (C) Real-time RT-PCR analysis of the wild type, the ΔccpA mutant, and the complemented strain. Cells were grown on BHI agar plates supplemented with 0.2% glucose or 2% glucose for 7 h aerobically at 37°C, and RNA was subsequently isolated. The data are presented as spxB expression levels relative to that of the wild type grown on BHI agar plates supplemented with 0.2% glucose, which was arbitrarily assigned a value of 1. The 16S rRNA gene was used as the housekeeping reference gene. Averages and standard deviations from three independent experiments are presented.
To confirm the CcpA-dependent repression of spxB expression, a CcpA mutant was constructed. Initially, the effect of the CcpA mutation on the ability to produce H2O2 was tested phenotypically using a colorimetric plate assay (Fig. 5B). As expected, under low-glucose conditions (0.2% glucose), no obvious difference in H2O2 production was detected between the wild type, the CcpA mutant, and a complemented strain expressing ccpA in trans from a shuttle plasmid. When grown in the presence of 2% glucose, the wild type produced less H2O2 than the CcpA mutant, confirming that CcpA represses spxB expression. The complemented mutant showed very low H2O2 production under the conditions tested (Fig. 5B), possibly due to the increased abundance of CcpA as a result of multicopy plasmid-encoded ccpA.
The expression of spxB was measured to confirm the phenotype. Cells were grown aerobically in TYE medium with low or high glucose concentrations. In agreement with H2O2 production, the most obvious difference was observed in the presence of 2% glucose. spxB expression in the CcpA mutant increased about 10-fold over that in the wild type, but that in the complemented strain was reduced about 2-fold (Fig. 5C). The results suggest a CcpA-dependent repression of spxB expression when the preferred carbohydrate sources are available. To exclude any CcpA-mediated regulation during the switch from aerobic to anaerobic growth, spxB abundance was determined in the CcpA mutant under conditions of anaerobic growth. spxB expression was similarly reduced in anaerobically grown CcpA mutant and wild-type cells. CcpA therefore seems not to be the transcriptional regulator mediating spxB repression under anaerobic conditions (data not shown).
Functional characterization of CcpA regulation.
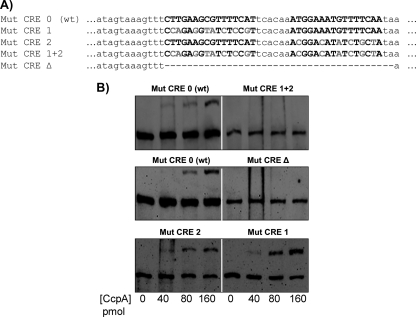
The presence of two or more CRE in CcpA-controlled promoter regions is not unusual (17). It has been demonstrated previously that CcpA binding covers more than the chromosomal region harboring the respective CRE, ranging from 20 to 40 bp (the predicted sizes of CRE 1 and CRE 2 are 16 bp) (17, 25, 26). The predicted CRE 1 and CRE 2 are located in close proximity, separated by 6 bp, suggesting that binding of CcpA to either one could sterically block the binding of an additional CcpA to the free CRE. To determine whether CcpA directly repressed spxB expression, EMSA analysis was performed. CcpA was overexpressed with an engineered C-terminal His tag and was purified using nickel affinity chromatography. To assess the importance of the individual CRE in the binding of CcpA, nucleotide exchanges were introduced into CRE 1 (Mut CRE 1), CRE 2 (Mut CRE 2), or CRE 1 and CRE 2 (Mut CRE 1 + 2). In addition, both CRE 1 and CRE 2 were deleted (Mut CRE Δ) (Fig. 6A). As shown in Fig. 6B, Mut CRE 1 + 2 prevented the binding of CcpA to the spxB promoter sequence used for EMSA; no shift in the DNA band was observed. The wild-type DNA sequence enabled CcpA binding, and a shift was observed, with increasing band intensity concomitant with larger amounts of CcpA. Mut CRE Δ prevented CcpA binding as well. The individual Mut CRE 1 or Mut CRE 2 mutant still exhibited a mobility shift (Fig. 6B). In conclusion, CcpA likely regulates spxB expression in S. gordonii by directly binding to the CRE sites in the spxB promoter, with both CRE 1 and CRE 2 promoting CcpA protein-DNA interactions.
Fig. 6.
EMSA for CcpA DNA binding studies. (A) Sequence of the spxB promoter used for site-directed mutagenesis. The putative CRE are in boldface (wild-type CRE or mutated CRE). Gray letters represent exchanged nucleotides, and dashes indicate deletions. All other bases in the DNA fragments are identical. (B) EMSA analysis was performed using purified recombinant CcpA and a biotin-labeled DNA probe containing wild-type CRE or various mutated CRE. Increasing concentrations of CcpA (40, 80, and 160 pmol) were incubated with 1 ng of the indicated DNA probe before the reactions were run on a nondenaturing polyacrylamide gel.
Consequences of CRE site mutations for H2O2 production.
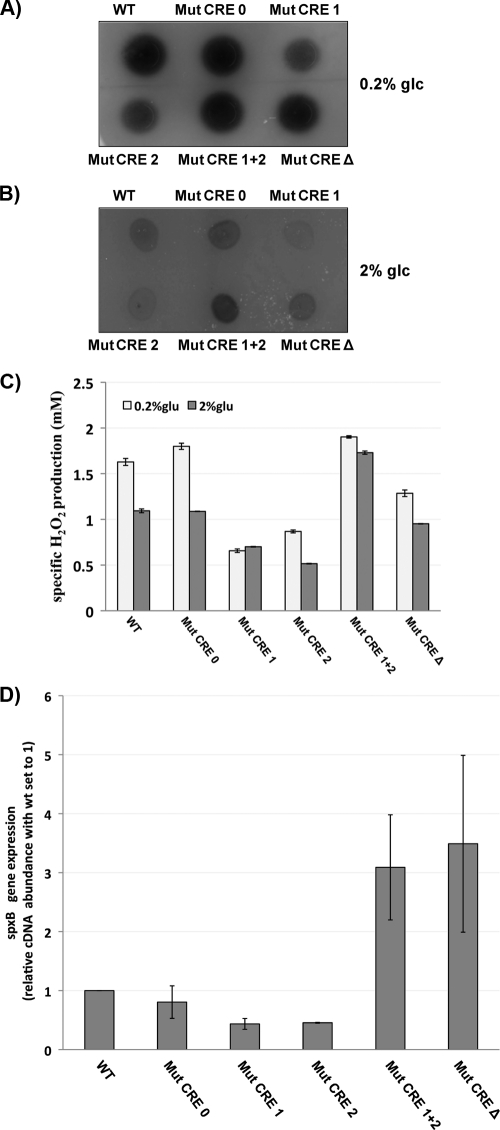
To further characterize the roles of the individual CRE sites in the regulation of spxB expression, the Mut CRE 1 and Mut CRE 2 mutations, as well as the Mut CRE 1 + 2 and Mut CRE Δ muations, were transferred to the chromosome of S. gordonii using double-crossover homologous recombination. This required the introduction of a kanamycin resistance cassette upstream of the spxB promoter for selection. To prove that the kanamycin cassette did not cause any adverse effects on spxB expression, the same strategy for homologous recombination with the mutated promoter sequences was used with the wild-type spxB promoter (Mut CRE 0). Compared to the wild-type CRE, the wild-type CRE carrying the kanamycin cassette (Mut CRE 0) produced equal amounts of H2O2 in the colorimetric plate assay when grown with 0.2% and 2% glucose, respectively. An increase in H2O2 production was observed for the Mut CRE 1 + 2 mutant grown with 2% glucose, confirming the observation from the EMSA showing no binding of CcpA if both sites are mutated. Therefore, no repression occurred. Surprisingly, the individual Mut CRE 1 or Mut CRE 2 mutant seemed to promote CcpA repression. Both mutants showed decreased H2O2 production, even when grown in the presence of 0.2% glucose (Fig. 7A and B).
Fig. 7.
Phenotypic consequences of CRE site mutations. The effects of CRE site mutations on H2O2 production were detected using H2O2 indicator plates. (A) Growth in the presence of 0.2% glucose. (B) Growth in the presence of 2% glucose. (C) The specific production of H2O2 was determined with planktonic cells grown under conditions of maximal H2O2 production. The detection limit for the assay was 0.125 mM H2O2. Averages and standard deviations from two independent experiments are presented. (D) The expression of spxB in the wild type and mutants grown in TYE medium with 0.2% Glc was determined by real-time PCR. The 16S rRNA gene was used as the housekeeping reference gene. Averages and standard deviations from two independent experiments are presented.
When grown with 2% glucose, the Mut CRE Δ mutant did not show increased H2O2 production over that for the Mut CRE 1 and Mut CRE 2 strains (Fig. 7B). This was unexpected, since no binding of CcpA was observed during EMSA. Therefore, it was expected that spxB was not repressed under any growth condition. The deletion of a substantial spxB promoter part did not abolish the potential of spxB expression, as shown for H2O2 production during growth in the presence of 0.2% glucose (Fig. 7B). One possible explanation for this observation is that the promoter truncation leads to mRNA instability and therefore faster degradation, although spxB expression might be derepressed in a manner similar to that for the Mut CRE 1 + 2 mutant.
To further quantify the differences in H2O2 production caused by the CRE mutations, the strains were grown under planktonic conditions on a horizontal shaker. H2O2 production was shown to be maximal in the wild type under these conditions (31). Confirming the results from the plate assay, H2O2 production was under the control of CCR; production was reduced in the wild type and the Mut CRE 0 strain during growth with 2% glucose. The Mut CRE 1 and Mut CRE 2 mutants showed a 2-fold reduction in H2O2 production, confirming the unexpected repression when only one binding site is present. The Mut CRE 1 + 2 mutant was not responsive to the increased glucose concentration, and therefore CcpA did not repress spxB expression. The Mut CRE Δ mutant did produce less H2O2 than the wild type but was not derepressed like the Mut CRE 1 + 2 mutant (Fig. 7C). We also measured the expression of spxB in the respective mutants. Similar results were obtained, showing that the individual mutants Mut CRE 1 and Mut CRE 2 were lower in spxB expression, while the inability of CcpA to bind showed the expected derepression of spxB expression (Fig. 7D).
DISCUSSION
In the present study, the regulation of spxB expression and subsequent SpxB abundance in S. gordonii were further explored. SpxB-dependent H2O2 production is an important modulator of interspecies competition and oral biofilm ecology (21, 22, 30, 32). S. gordonii is able to inhibit the growth of oral biofilm bacteria, including cariogenic S. mutans and Actinomyces naeslundii, as demonstrated in deferred dual-species antagonism assays (22, 30). Furthermore, S. sanguinis and S. gordonii SpxB mutants are outcompeted by S. mutans in a dual-species biofilm model, whereas an H2O2 overproduction mutant of S. sanguinis increased its inhibiting activity toward S. mutans (32, 55). The production of H2O2 is coupled to the generation of acetyl phosphate. Acetyl phosphate by itself is an important component of central metabolism. It can serve as a phosphoryl donor for two-component response regulators involved in the regulation of diverse cellular processes (52). Furthermore, the conversion of acetyl phosphate to acetate generates ATP. Therefore, acetyl phosphate has been discussed as an important intracellular signal to couple energy metabolism with cellular signaling (52). A legitimate question, therefore, is whether H2O2 is merely a side product of energy metabolism under aerobic conditions. Compared to the ability of S. mutans to produce bacteriocins under aerobic and anaerobic conditions, the limited H2O2 production during anaerobic growth seems to argue against H2O2 production as a competitive antimicrobial component. However, an alternative explanation may be simply that H2O2 production under aerobic conditions is sufficient to function in interspecies competition. For example, an environment of high oxygen availability is most likely to occur under conditions of low biofilm abundance during the earliest stages of biofilm development. As the cells start to produce H2O2, they would naturally favor the growth of the H2O2-tolerant species at the expense of sensitive species, such as S. mutans. Another consequence of H2O2 production is the release of extracellular DNA (eDNA) (31). eDNA has been shown to be an important structural component of biofilms (14), and H2O2 is the sole agent required to induce eDNA release in S. gordonii (A. Itzek et al., unpublished data). Likewise, it has been shown for several species that eDNA is required for biofilm formation (9, 11, 27), and it can promote cell-cell aggregation in S. sanguinis (31). The production of eDNA might therefore promote the initial adhesion to the tooth surface and subsequent biofilm formation.
The initial observation of a glucose-effect on H2O2 production suggested CCR of spxB expression in S. gordonii. CCR in oral streptococci seems to be mediated by two different mechanisms, one that is CcpA dependent and one that is CcpA independent. In a recent report, Tong et al. demonstrated that the EIIABMan domain (ManL) of the PTS controls CCR in S. gordonii (49). Furthermore, CCR in S. mutans is mediated by the general PTS component Hpr-Ser-P, and the carbohydrate-specific EIIABMan, FruI, and EIILev and CcpA seem to play only a minor role (53, 54). Earlier reports for S. gordonii showed CcpA-dependent control of several other genes, including the arginine deiminase operon and the abpA gene, encoding amylase binding protein A, as well as the α-amylase gene amyB (12, 23, 43). The RegPrecise database, a collection of manually curated inferences of regulons in prokaryotic genomes (37), identified 151 genes in 72 operons regulated by CcpA in S. gordonii, while in S. mutans only 92 genes in 41 operons seemed to be regulated by CcpA. Further experiments are needed to identify the extent of CcpA- and EIIABMan-mediated CCR in S. gordonii in order to identify the dominant mechanism of CCR. This will also help to precisely define the differences in the CCR network between S. gordonii and S. mutans. This is important considering that S. gordonii as a commensal is able to compete with S. mutans and is therefore a promising candidate for enhancement of competitiveness toward cariogenic species in general.
The difference in the glucose response between S. sanguinis and S. gordonii initiated the current investigation. CcpA permanently represses spxB expression in S. sanguinis independently of carbohydrate content (55). Interestingly, the RegPrecise database (37) predicted only one CcpA binding site in the promoter region of spxB in S. sanguinis, while two were predicted for S. gordonii. CcpA in vitro bound both predicted CRE sites in S. gordonii. However, it was surprising that the deletion of either CRE 1 or CRE 2 decreased H2O2 production, suggesting that CcpA mediates spxB repression functions only when one CRE is present. This was true even under nonrepressive conditions, such as low glucose. This resembles the observation reported for S. sanguinis, that the presence of only one binding site confers permanent repression (55). One possible explanation for the regulatory role of two CRE sites in spxB expression in S. gordonii could be steric hindrance. Under low-glucose conditions, other coregulatory elements, such as phosphorylated Hpr-Ser-46 (24), would not be available to modulate the binding activity of CcpA. It is possible that these cofactors are required to alleviate the steric hindrance created when both CRE sites are bound. A survey of promoter sequences encoding two CRE sites in S. gordonii showed that the close association in the spxB promoter is unique. Usually, an extended spacer separates the CRE sites in other promoters under CCR. This was also found for S. sanguinis and S. mutans. Further experimental validation is required to confirm this unusual regulation.
In summary, the present study has provided evidence for the environmental control of spxB expression in S. gordonii. Given the potential role of S. gordonii spxB expression in oral biofilm homeostasis, it is possible that oral commensals such as S. gordonii and S. sanguinis could be exploited for their probiotic potential. For example, salivary lactoperoxidase is an important innate immune component that uses inorganic ions and H2O2 as oxidants to generate antimicrobials generally believed to be more effective than H2O2 itself (2). Thus, strategies that modulate SpxB activity in the oral cavity may be useful as a simple therapeutic mechanism to improve oral health.
ACKNOWLEDGMENTS
This study was supported by NIH grant 4R00DE018400 to J.K.
We thank Justin Merritt (University of Oklahoma Health Sciences Center) for helpful discussions.
Footnotes
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Abbe K., Carlsson J., Takahashi-Abbe S., Yamada T. 1991. Oxygen and the sugar metabolism in oral streptococci. Proc. Finn. Dent. Soc. 87:477–487 [PubMed] [Google Scholar]
- 2. Ashby M. T., Kreth J., Soundarajan M., Sivuilu L. S. 2009. Influence of a model human defensive peroxidase system on oral streptococcal antagonism. Microbiology 155:3691–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnard J. P., Stinson M. W. 1999. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect. Immun. 67:6558–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becker M. R., et al. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bizzini A., Entenza J. M., Moreillon P. 2007. Loss of penicillin tolerance by inactivating the carbon catabolite repression determinant CcpA in Streptococcus gordonii. J. Antimicrob. Chemother. 59:607–615 [DOI] [PubMed] [Google Scholar]
- 6. Carlsson J., Edlund M. B. 1987. Pyruvate oxidase in Streptococcus sanguis under various growth conditions. Oral Microbiol. Immunol. 2:10–14 [DOI] [PubMed] [Google Scholar]
- 7. Carlsson J., Edlund M. B., Lundmark S. K. 1987. Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiol. Immunol. 2:15–20 [DOI] [PubMed] [Google Scholar]
- 8. Caufield P. W., et al. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das T., Sharma P. K., Busscher H. J., van der Mei H. C., Krom B. P. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76:3405–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deutscher J., Kuster E., Bergstedt U., Charrier V., Hillen W. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049–1053 [DOI] [PubMed] [Google Scholar]
- 11. Dominiak D. M., Nielsen J. L., Nielsen P. H. 2011. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13:710–721 [DOI] [PubMed] [Google Scholar]
- 12. Dong Y., Chen Y. Y., Burne R. A. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunny G. M., Lee L. N., LeBlanc D. J. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flemming H. C., Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 15. Gong K., Mailloux L., Herzberg M. C. 2000. Salivary film expresses a complex, macromolecular binding site for Streptococcus sanguis. J. Biol. Chem. 275:8970–8974 [DOI] [PubMed] [Google Scholar]
- 16. Görke B., Stulke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 17. Gösseringer R., Kuster E., Galinier A., Deutscher J., Hillen W. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266:665–676 [DOI] [PubMed] [Google Scholar]
- 18. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 19. Iyer R., Baliga N. S., Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jakubovics N. S. 2010. Talk of the town: interspecies communication in oral biofilms. Mol. Oral Microbiol. 25:4–14 [DOI] [PubMed] [Google Scholar]
- 21. Jakubovics N. S., Gill S. R., Iobst S. E., Vickerman M. M., Kolenbrander P. E. 2008. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J. Bacteriol. 190:3646–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jakubovics N. S., Gill S. R., Vickerman M. M., Kolenbrander P. E. 2008. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol. Ecol. 66:637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson B. P., et al. 2009. Interspecies signaling between Veillonella atypica and Streptococcus gordonii requires the transcription factor CcpA. J. Bacteriol. 191:5563–5565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones B. E., et al. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272:26530–26535 [DOI] [PubMed] [Google Scholar]
- 25. Kim J. H., Guvener Z. T., Cho J. Y., Chung K. C., Chambliss G. H. 1995. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J. Bacteriol. 177:5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. H., Yang Y. K., Chambliss G. H. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56:155–162 [DOI] [PubMed] [Google Scholar]
- 27. Klein M. I., et al. 2010. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One 5:e13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kolenbrander P. E., et al. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47–79 [DOI] [PubMed] [Google Scholar]
- 29. Kreth J., Merritt J., Qi F. 2009. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 28:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kreth J., Merritt J., Shi W., Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kreth J., Vu H., Zhang Y., Herzberg M. C. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191:6281–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kreth J., Zhang Y., Herzberg M. C. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190:4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loimaranta V., Tenovuo J., Koivisto L., Karp M. 1998. Generation of bioluminescent Streptococcus mutans and its usage in rapid analysis of the efficacy of antimicrobial compounds. Antimicrob. Agents Chemother. 42:1906–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin B., Alloing G., Mejean V., Claverys J. P. 1987. Constitutive expression of erythromycin resistance mediated by the ermAM determinant of plasmid pAMβ1 results from deletion of 5′ leader peptide sequences. Plasmid 18:250–253 [DOI] [PubMed] [Google Scholar]
- 35. Moffatt B. A., Studier F. W. 1987. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell 49:221–227 [DOI] [PubMed] [Google Scholar]
- 36. Nobbs A. H., Lamont R. J., Jenkinson H. F. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Novichkov P. S., et al. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okinaga T., Xie Z., Niu G., Qi F., Merritt J. 2010. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25:165–177 [DOI] [PubMed] [Google Scholar]
- 39. Pakula R., Walczak W. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31:125–133 [DOI] [PubMed] [Google Scholar]
- 40. Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lutticken R. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137–147 [DOI] [PubMed] [Google Scholar]
- 41. Regev-Yochay G., et al. 2004. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. JAMA 292:716–720 [DOI] [PubMed] [Google Scholar]
- 42. Regev-Yochay G., Trzcinski K., Thompson C. M., Malley R., Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. Bacteriol. 188:4996–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rogers J. D., Scannapieco F. A. 2001. RegG, a CcpA homolog, participates in regulation of amylase-binding protein A gene (abpA) expression in Streptococcus gordonii. J. Bacteriol. 183:3521–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rosan B., Lamont R. J. 2000. Dental plaque formation. Microbes Infect. 2:1599–1607 [DOI] [PubMed] [Google Scholar]
- 45. Ryan C. S., Kleinberg I. 1995. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch. Oral Biol. 40:753–763 [DOI] [PubMed] [Google Scholar]
- 46. Saito M., Seki M., Iida K., Nakayama H., Yoshida S. 2007. A novel agar medium to detect hydrogen peroxide-producing bacteria based on the Prussian blue-forming reaction. Microbiol. Immunol. 51:889–892 [DOI] [PubMed] [Google Scholar]
- 47. Selva L., et al. 2009. Killing niche competitors by remote-control bacteriophage induction. Proc. Natl. Acad. Sci. U. S. A. 106:1234–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spellerberg B., et al. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803–813 [DOI] [PubMed] [Google Scholar]
- 49. Tong H., Zeng L., Burne R. A. 2011. The EIIABMan phosphotransferase system permease regulates carbohydrate catabolite repression in Streptococcus gordonii. Appl. Environ. Microbiol. 77:1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trieu-Cuot P., Courvalin P. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5″-aminoglycoside phosphotransferase type III. Gene 23:331–341 [DOI] [PubMed] [Google Scholar]
- 51. Warner J. B., Lolkema J. S. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolfe A. J. 2010. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr. Opin. Microbiol. 13:204–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zeng L., Burne R. A. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol. Microbiol. 75:1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeng L., Das S., Burne R. A. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J. Bacteriol. 192:2434–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zheng L., Chen Z., Itzek A., Ashby M., Kreth J. 2011. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J. Bacteriol. 193:516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]