Abstract
Protozoa are important components of microbial food webs, but protozoan feeding preferences and their effects in the context of bacterial biofilms are not well understood. The feeding interactions of two contrasting ciliates, the free-swimming filter feeder Tetrahymena sp. and the surface-associated predator Chilodonella sp., were investigated using biofilm-forming bacteria genetically modified to express fluorescent proteins. According to microscopy, both ciliates readily consumed cells from both Pseudomonas costantinii and Serratia plymuthica biofilms. When offered a choice between spatially separated biofilms, each ciliate showed a preference for P. costantinii biofilms. Experiments with bacterial cell extracts indicated that both ciliates used dissolved chemical cues to locate biofilms. Chilodonella sp. evidently used bacterial chemical cues as a basis for preferential feeding decisions, but it was unclear whether Tetrahymena sp. did also. Confocal microscopy of live biofilms revealed that Tetrahymena sp. had a major impact on biofilm morphology, forming holes and channels throughout S. plymuthica biofilms and reducing P. costantinii biofilms to isolated, grazing-resistant microcolonies. Grazing by Chilodonella sp. resulted in the development of less-defined trails through S. plymuthica biofilms and caused P. costantinii biofilms to become homogeneous scatterings of cells. It was not clear whether the observed feeding preferences for spatially separated P. costantinii biofilms over S. plymuthica biofilms resulted in selective targeting of P. costantinii cells in mixed biofilms. Grazing of mixed biofilms resulted in the depletion of both types of bacteria, with Tetrahymena sp. having a larger impact than Chilodonella sp., and effects similar to those seen in grazed single-species biofilms.
INTRODUCTION
Protozoa are important microbial predators in aquatic systems, contributing to carbon transfer and nutrient cycling processes and impacting bacterial communities through top-down predation pressures (22, 27, 49, 59). A key aspect of protozoan ecology is the selective nature of protozoan feeding. Protozoan predation may cause shifts in bacterial morphology, physiology, and community composition (22, 27, 39), and selective feeding must contribute to such effects. Protozoan selective feeding may be influenced by prey cell size and shape (10, 50), motility (38), cell surface characteristics (41), and biochemical composition (21, 57), as well as predator feeding history (4). The mechanisms by which protozoa identify and selectively feed on different prey organisms are not well understood, however (42). Surface-bound and dissolved chemical cues are important mediators of microbial interactions, and chemosensory recognition of dissolved signals may be used by predators to locate prey (18, 34, 36, 68) or may deter predation (63, 64). However, elucidating the role of chemical cues in protozoan selective feeding behaviors remains a challenge (23, 42).
Many protozoan feeding studies have focused on flagellates as important consumers of bacteria (7), but ciliates, encompassing considerable diversity in morphology, motility, and feeding strategies, are also important predators of bacteria (24, 58, 69). The feeding preferences shown by ciliates may contrast with those shown by flagellates (26). Recent studies have demonstrated selective ciliate feeding preferences for different diatoms (23), ciliates (54), and flagellates (28) and certain bacteria (4, 21, 66). Dissolved chemical cues may be used by ciliates to discriminate between diatom and ciliate prey species (23, 54), but there is currently a lack of clear evidence for the use of these cues as a basis for ciliate selective feeding on bacteria.
The bacteria and associated ecological processes in streams occur predominantly in biofilms—complex, surface-attached assemblages of microbes, extracellular molecules, and debris (5, 11, 44, 62). However, investigations of protozoan feeding have usually considered planktonic bacteria in pelagic systems (e.g., see references 6, 58, and 60). Clearly, biofilms present a feeding environment for protozoa that is markedly different from that offered by suspensions of planktonic cells. Protozoan grazing may affect biofilm thickness, biomass, and morphology (8, 25, 32, 48), and biofilms may respond to grazing by developing grazing-resistant structures or producing antiprotozoan factors (37, 51, 71). However, there is scarce information on protozoan selective feeding in biofilms and no prior studies have considered protozoan feeding preferences for different species of attached bacteria.
In this study, we investigated the feeding interactions of two contrasting ciliates with bacterial biofilms. The study organisms were all isolated from stream biofilm samples and thus represent ecologically relevant components of a natural microbial food web. The bacterial prey species Pseudomonas costantinii and Serratia plymuthica occur frequently in stream biofilms and are of acknowledged importance in biofilm formation (70). These bacteria were genetically modified to express fluorescent proteins, allowing visualization of cells and biofilm structures. Of the two ciliates used, Tetrahymena sp. is a well-characterized free-swimming filter feeder expected to consume mostly suspended cells (15, 69). In contrast, the common but little-studied protist Chilodonella sp. is a flattened, substrate-crawling raptorial or gulper feeder which preys upon attached cells (8, 55). We hypothesized that these ciliates would show different interactions with bacterial biofilms which could be related to their contrasting motility and feeding modes. We first investigated whether the ciliates showed any differences in the ability to feed upon bacterial cells in suspension or in biofilms. Using novel microcosms allowing spatial separation of bacterial biofilms, we then investigated whether the ciliates showed feeding preferences for biofilms composed of one or the other of the different bacteria and whether dissolved chemical signals played a role in any observed preferences. We then investigated the effects of ciliate grazing on biofilm morphology and composition in light of any detected preferential feeding behaviors.
MATERIALS AND METHODS
Bacterial biofilms and ciliate cultures.
Two bacterial isolates cultured from stream biofilm samples during 2004 were used in experiments. The isolates were identified as Pseudomonas costantinii 8A (from Opanuku Stream, Auckland, New Zealand; 36°53′42″S, 174°35′44″E) and Serratia plymuthica CB007 (from Farm Stream, Canterbury, New Zealand; 43°00′26″S, 171°48′08″E). To allow discrimination of these bacteria by fluorescence microscopy, each culture was genetically modified using the mini-Tn7 transposon system (29, 31). P. costantinii was thus tagged with green fluorescent protein (GFP) and gentamicin resistance (Gmr) markers, while S. plymuthica was tagged with red fluorescent protein (RFP; DsRed-Express) and Gmr markers. Bacterial biofilms were established by filling 12-well plates with R2A broth (Lab M, Lancashire, United Kingdom) to which bacteria were added (∼2 × 106 cells ml−1) and incubating them for 24 h at 28°C. Glass coverslips (16-mm diameter; Marienfeld Superior, Lauda-Königshofen, Germany), glass beads (4-mm diameter; Propper Manufacturing Co., Long Island City, NY), and squares of acetate (16 mm2) were placed in these wells when biofilm establishment on these substrates was required. Prior to experimental use, liquid was removed from wells by pipette and biofilms were gently washed with sterile R2A medium to remove unattached bacteria. Cultures of suspended bacteria were established by incubating R2A medium to which bacteria had been added (∼2 × 106 cells ml−1) on a shaker for 24 h at 28°C.
Cultures of two ciliates with contrasting swimming and feeding modes, Tetrahymena sp. and Chilodonella sp., were isolated from Opanuku Stream and maintained in modified NAS (Neff's amoeba saline; 45) as previously described (14). Cultures were concentrated by centrifugation (1,000 × g, 5 min) for use in experiments.
For experiments investigating the effects of ciliate feeding on biofilm morphology and composition, ciliate cultures were treated with a mixture of ampicillin and streptomycin (each at 50 μg ml−1) for 12 h to remove extraneous bacteria, which could otherwise affect biofilm development. Nongenetically modified P. costantinii and S. plymuthica cells were then added to ciliate cultures, which were subsequently maintained as previously described (14). Ciliate cultures were further treated with the same antibiotic mixture for several hours immediately before use in experiments to minimize the inclusion of extraneous bacteria.
Ciliate feeding on fluorescent bacteria.
Initial trials investigated the ability of ciliates to ingest fluorescent protein-expressing P. costantinii and S. plymuthica cells from biofilm cultures and bacterial suspensions. Biofilms of each bacterium were established on 4-mm glass beads, which were then washed by transfer into 0.5 ml of fresh R2A medium to remove unattached cells and placed into a further 0.5 ml of fresh R2A. Additionally, 0.5-ml suspensions of each bacterium in R2A medium were prepared such that the number of suspended cells present was approximately equivalent to the number of cells in biofilms grown on glass beads. Approximately 500 Chilodonella sp. or Tetrahymena sp. cells in 0.5 ml NAS were added to bacterial cultures. After 45 min, 1 ml of 10% formalin was added to each experiment and gently mixed to fix the ciliate cells for microscopic examination. Glass beads were removed before the fixed experiments were centrifuged (10,000 × g, 6 min), and pellets were deposited on microscope slides. Fixed ciliate cells were examined for evidence of ingestion of P. costantinii or S. plymuthica by fluorescence microscopy. Green P. costantinii and red S. plymuthica fluorescence was, respectively, visualized using Endow GFP 41017 and tetramethyl rhodamine isocyanate 41002c band-pass filter sets (Chroma Technology, Bellows Falls, VT). The amount of fluorescence per ciliate cell was determined by digital image analysis using Daime (12) to assess the amount of bacteria ingested.
Ciliate feeding preferences for different bacterial biofilms.
For investigation of ciliate feeding preferences for different bacterial biofilms, microcosms allowing spatial separation of P. costantinii and S. plymuthica biofilms were developed. Each microcosm was constructed by pouring a 4-mm-thick layer of agarose (1%) around a three-dimensional mold placed in a petri dish. Molds were removed after the agarose had cooled, resulting in three-dimensional enclosures which were subsequently filled with liquid R2A medium.
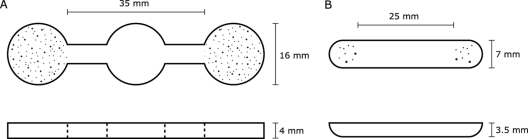
Two different microcosm designs were used. The first microcosm design (type 1) consisted of three connected chambers (Fig. 1 A). Biofilms on glass coverslips (16-mm diameter) of either P. costantinii or S. plymuthica were placed in the two outermost chambers on opposite sides of the microcosm, and ciliates in NAS medium were added to the central chamber, giving them free access to two different biofilms in opposite directions. The numbers of ciliates present on each biofilm and in the central chamber were assessed at regular intervals over several hours. At each interval, ciliate counts were made at nine locations within each biofilm chamber and within the central chamber of the microcosm. Enumeration was carried out using an inverted light microscope (×40 to ×100 magnification), in combination with video obtained using a Spot Insight Firewire digital camera (Diagnostic Instruments Inc., Sterling Heights, MI).
Fig. 1.
Top and side views of microcosms used in ciliate preferential-feeding experiments. In microcosm type 1 (A), biofilms on glass coverslips (16-mm diameter) or blocks of bacterial cell extracts were placed in the outermost chambers, separated by 35 mm, and ciliates were added to the middle chamber. In microcosm type 2 (B), biofilms on squares of acetate (16 mm2) or cell extracts were placed at each end, separated by 25 mm, and ciliates were added to the center.
Microcosm type 1 was successfully used for Tetrahymena sp. experiments, but Chilodonella sp. showed no response to biofilms in this microcosm, possibly due to the microcosm scale and layout in combination with the relatively slow crawling motility of this ciliate. A second microcosm design (type 2) was therefore developed for Chilodonella sp. experiments. Type 2 microcosms were smaller and simpler than type 1 microcosms, consisting of a semicylinder with quarter-spherical ends (Fig. 1B). In type 2 microcosms, biofilms on squares of acetate (16 mm2) were placed at opposite ends of the microcosms, and Chilodonella sp. cells were added at the center. Due to the smaller scale of type 2 microcosms, Chilodonella sp. cells were counted over the entire area of each biofilm and in an equivalent area at the center of the microcosm at regular intervals in each of three replicate microcosms using the same enumeration methods as described for microcosm type 1 experiments.
All microcosm experiments were carried out three times to confirm observed results. The orientation of the microcosms and biofilms was reversed in each repeated experiment. Repeated-measures analysis of variance (ANOVA) was used to test for differences between the numbers of ciliates detected on each biofilm and in the center of the microcosms throughout the experiments. Statistically significant ANOVA results were followed by Tukey honestly significant difference (HSD) post hoc tests to determine whether ciliate counts differed between P. costantinii biofilm and S. plymuthica biofilm or between either biofilm and the microcosm center. These analyses were conducted in PASW Statistics 18 (SPSS Inc., Chicago, IL).
Ciliate responses to bacterial cell extracts.
To investigate whether responses of ciliates to bacterial biofilms could be attributed to chemical cues produced by bacteria, cell-free bacterial extracts were produced by passing up to 5 ml of bacterial cultures in liquid R2A medium through Minisart syringe filters with a pore size of 0.2 μm (Sartorius, Goettingen, Germany). Before filtration, the cell concentrations in bacterial suspensions were assessed by spectrophotometric absorbance measurements (A = 600 nm) and adjusted by addition of R2A medium to give equivalent absorbance for each culture. Agarose powder (1%) was added to each filtered extract sample, which was then gently heated until the agarose dissolved and cooled until solid. Blocks of the resulting solidified extracts were placed in microcosms in the place of bacterial biofilms. Control blocks of bacterial extract-free R2A medium solidified in the same manner were placed in the center of the microcosms. Experiments were then carried out and analyzed as for the biofilm feeding experiments described above.
Effects of ciliate feeding upon bacterial biofilm morphology and composition.
For confocal microscopy analysis of ciliate grazing effects on biofilms, single-species and mixed biofilms of fluorescent P. costantinii and S. plymuthica were established in 35-mm-diameter Fluorodish glass bottom cell culture dishes (World Precision Instruments, Sarasota, FL). Trial experiments indicated that a ratio of approximately 15:1 of P. costantinii to S. plymuthica cells was necessary for the establishment of both bacteria in similar abundances in mixed biofilms, and this ratio was used to guide the numbers of bacterial cells added to Fluorodishes (approximately 3 × 107 P. costantinii cells and/or 2 × 106 S. plymuthica cells added to 2 ml R2A medium in each dish). Biofilms were incubated at 28°C for 24 h and subsequently washed once with sterile R2A medium. Approximately 100 Tetrahymena sp. or 500 Chilodonella sp. cells in 0.5 ml NAS plus 0.5 ml R2A medium were then added to biofilms (a higher number of Chilodonella sp. cells was used to compensate for the slower population growth rate observed in this ciliate). After ciliates were added, incubation was done at room temperature for up to 48 h. Biofilms were examined at random locations at a magnification of ×400 using an Andor Revolution XD spinning disc confocal microscope system (Andor Technology, Belfast, United Kingdom) with a Nikon Ti-E inverted microscope. Green P. costantinii fluorescence and red S. plymuthica fluorescence were detected using blue (wavelength, 488 nm) and yellow/green (wavelength, 561 nm) lasers, respectively, in combination with Semrock (Rochester, NY) Brightline multiband band-pass filters with center wavelengths of 512 nm (>90% transmission over 23 nm) and 630 nm (>90% transmission over 91 nm). Resulting images and z stacks were examined using ImageJ (1). All confocal investigations of ciliate feeding upon biofilm morphology were repeated to confirm observations.
RESULTS
Ciliate feeding on fluorescent bacteria.
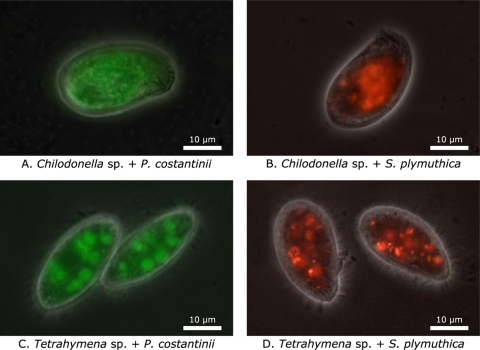
Fluorescence microscopy showed that both Chilodonella sp. and Tetrahymena sp. readily ingested both P. costantinii and S. plymuthica cells when presented with one or the other, both as biofilms and in suspension (Fig. 2). The amount of fluorescence evident in fixed ciliates is likely to reflect the quantity of bacteria ingested and varied widely between individual ciliate cells in all experiments (data not shown). There was little clear evidence of differences between the amounts of biofilm bacteria compared to suspended bacteria ingested by either ciliate.
Fig. 2.
Representative Chilodonella sp. (A and B) and Tetrahymena sp. (C and D) cells after feeding on GFP-expressing P. costantinii (A and C) or RFP-expressing S. plymuthica (B and D) biofilms. Ciliates were added to bacterial biofilms 45 min before being fixed with formalin. Composite images constructed from phase-contrast microscopy images overlaid with fluorescence microscopy images are shown.
Ciliate feeding preferences.
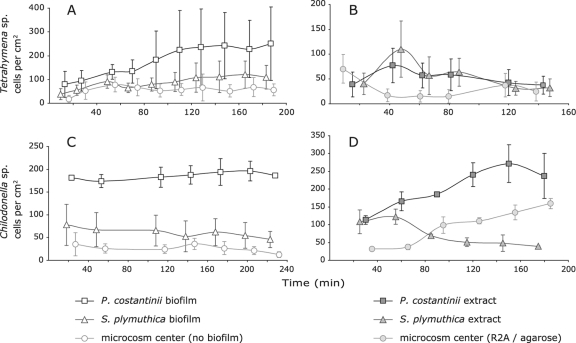
When Tetrahymena sp. cells were introduced into microcosms holding spatially separated P. costantinii and S. plymuthica biofilms (microcosm type 1, Fig. 1A), significant differences in the numbers of ciliate cells were detected at different locations in the microcosm (ANOVA; P = 0.001). Post hoc Tukey HSD tests showed that significantly more Tetrahymena sp. cells were observed on P. costantinii biofilm than on S. plymuthica biofilm or at the center of the microcosm (P ≤ 0.005), suggesting a preference for feeding on P. costantinii (Fig. 3 A). It was noted that the distribution of Tetrahymena sp. cells on biofilms was very uneven, contributing to the relatively large margins of error shown in Fig. 3A.
Fig. 3.
Distribution of Tetrahymena sp. cells in microcosms (type 1) containing spatially separated P. costantinii and S. plymuthica biofilms (A) or bacterial cell extracts (B) and distribution of Chilodonella sp. cells in microcosms (type 2) containing biofilms (C) or bacterial cell extracts (D). Cell counts were made by direct microscopic observations and video recordings. Counts of Chilodonella sp. cells were not made until after sufficient time had elapsed to allow cells added in suspension to settle. Each data point is the mean number of counts at nine different locations on each biofilm (for Tetrahymena sp. experiments) or three separate replicates (for Chilodonella sp. experiments). Error bars represent ± one standard deviation.
When biofilms were replaced with cell-free bacterial extracts, significant differences in the numbers of Tetrahymena sp. ciliate cells in different locations were again detected (ANOVA; P = 0.001). The number of ciliate cells detected in chambers containing bacterial cell extracts was higher than that in control chambers (Tukey HSD; P ≤ 0.005), suggesting that Tetrahymena sp. cells responded positively to bacterium-derived chemical cues present in the extracts (Fig. 3B). A preference for either bacterial extract was not detected, however, and the response to extracts appeared to decrease over time.
Chilodonella did not demonstrate an obvious response to bacterial biofilms or extracts in microcosm type 1. When Chilodonella sp. cells were added to the smaller and simpler type 2 microcosm (Fig. 1B), there were significant differences in the numbers of Chilodonella sp. cells throughout the microcosm (ANOVA; P ≤ 0.001). Higher numbers of ciliate cells were detected on P. costantinii biofilm than on S. plymuthica biofilm or at the center of the microcosm (Tukey HSD; P ≤ 0.001), suggesting a preference for P. costantinii, much like that of Tetrahymena sp. (Fig. 3C). Similarly, there were significant differences between the numbers of Chilodonella sp. cells at different locations in microcosms containing cell-free bacterial extracts (ANOVA; P = 0.005). Significantly more Chilodonella sp. cells were detected at the microcosm end with P. costantinii extracts than at the end with S. plymuthica extracts or at the center of the microcosm (Tukey HSD; P ≤ 0.01; Fig. 3D), consistent with the response to P. costantinii biofilm and in contrast to the response exhibited by Tetrahymena sp. The number of Chilodonella cells at the S. plymuthica extract end of the microcosm was not significantly different from the number of cells at the center of the microcosm and appeared to decrease throughout the experiment. Unlike the response of Tetrahymena sp. to the bacterial extracts, the response of Chilodonella sp. to P. costantinii extracts was sustained throughout the experiment.
Control experiments in which ciliates were introduced into microcosms containing no biofilms or cell extracts resulted in no significant differences between the numbers of cells observed at any locations within the microcosms (data not shown).
Effects of ciliate grazing on biofilm morphology and composition.
Although Chilodonella sp. and Tetrahymena sp. were added to biofilm grazing experiments at respective rates of 500 and 100 cells ml−1, after 24 h, the number of Tetrahymena sp. cells present on biofilms exceeded the number of Chilodonella sp. cells in equivalent experiments by a factor of about 10 (Table 1). The numbers of ciliates detected on P. costantinii biofilms, S. plymuthica biofilms, or biofilms composed of both bacteria were not significantly different for either Chilodonella sp. or Tetrahymena sp., however.
Table 1.
Abundance of ciliates after 24 h of grazing on bacterial biofilms
| Bacterial biofilm composition | Mean abundance (no. of cells ml−1) of ciliates ± SD a |
|
|---|---|---|
| Childonella sp. | Tetrahymena sp. | |
| P. costantinii | 3,225 ± 526 | 22,590 ± 8,632 |
| S. plymuthica | 2,602 ± 331 | 24,750 ± 2,888 |
| P. costantinii + S. plymuthica | 2,306 ± 548 | 31,320 ± 5,803 |
Either 500 Chilodonella sp. cells ml−1 or 100 Tetrahymena sp. cells ml−1 were initially added to biofilms.
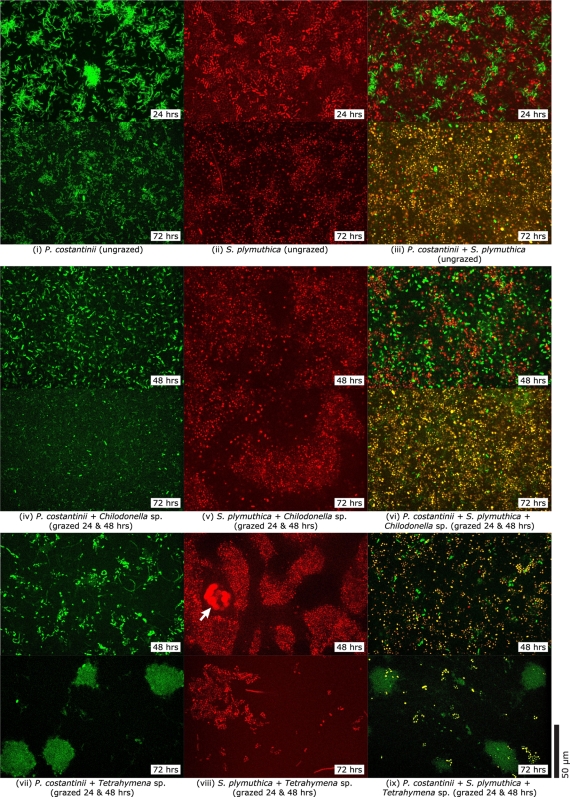
From confocal microscopy observations, ungrazed P. costantinii biofilms (green fluorescence) consisted of patchy scatterings of bacterial cells and clusters up to several cells in height after 24 h of growth (Fig. 4 i). These biofilms showed little further change after 48 and 72 h. In contrast, ungrazed S. plymuthica biofilms (red fluorescence) completely covered the substrate with an irregular layer with a thickness of up to tens of cells after 24 h (Fig. 4 ii). These biofilms expanded into extensive mounds of loosely aggregated cells after 48 and 72 h of growth. Growth of both bacterial species in mixed culture for 24 h resulted in irregular clusters of P. costantinii cells scattered within an extensive S. plymuthica layer some 10 cells thick (Fig. 4 iii). In all experiments which included P. costantinii, the amount of green fluorescence produced by P. costantinii biofilm cells had decreased when examined after 48 and 72 h of biofilm growth, although planktonic P. costantinii cells continued to fluoresce brightly for up to 72 h. To improve the visualization of P. costantinii in mixed biofilms after 48 h and 72 h of growth, the intensity of blue laser illumination was increased but this caused simultaneous fluorescence of S. plymuthica cells. Use of a dual-wavelength filter meant that fluorescence from both bacteria was included in the green image channel, making it difficult to visualize P. costantinii biofilm cells.
Fig. 4.
Representative confocal microscopy images of live bacterial biofilms. Ungrazed biofilms of P. costantinii (i), S. plymuthica (ii), and both bacteria together (iii) after 24 and 72 h of growth. Chilodonella sp.-grazed biofilms of P. costantinii (iv), S. plymuthica (v), and both bacteria (vi) after 48 and 72 h (24 h of biofilm growth followed by 24 h and 48 h of grazing). Tetrahymena sp.-grazed biofilms of P. costantinii (vii), S. plymuthica (viii), and both bacteria (ix) after 48 and 72 h (24 h of biofilm growth followed by 24 and 48 h of grazing). The S. plymuthica-filled vacuoles of a feeding Tetrahymena sp. cell are indicated (arrow). Green fluorescence indicates P. costantinii cells. Red fluorescence indicates S. plymuthica cells. Yellow fluorescence in biofilms composed of both bacteria also represents S. plymuthica cells and is the result of superimposed red and green fluorescence due to the use of high-intensity blue laser illumination causing simultaneous fluorescence of both S. plymuthica and P. costantinii cells in combination with a dual-wavelength filter. The scale bar applies to all images.
P. costantinii biofilms that were grazed by Chilodonella sp. for 24 h (i.e., 48 h biofilm growth) had abundant cells scattered homogeneously on the substrate and in the overlying liquid (Fig. 4 iv). After grazing for 48 h (72 h growth), these biofilms showed little further change other than a reduction in the number of brightly fluorescing cells. The effects of grazing by Chilodonella sp. on S. plymuthica biofilms were indistinct after 24 h (48 h growth). Grazing by Chilodonella sp. for 48 h (72 h growth) had clear effects, however, with extensive mounds of S. plymuthica biofilm intersected by irregular channels through which Chilodonella sp. cells were seen crawling (Fig. 4v).
In contrast, the impacts of grazing by Tetrahymena sp. on biofilm morphology were very clear. Grazing by Tetrahymena sp. for 24 h (48 h growth) caused P. costantinii biofilms to form scattered clusters, and after 48 h of grazing (72 h growth), these biofilms formed dense microcolonies with associated extracellular polymers (Fig. 4 vii). Grazing by Tetrahymena sp. for 24 h (48 h growth) on S. plymuthica biofilms caused the appearance of extensive holes and channels (Fig. 4 viii). Biofilm cells were more densely packed together, and fewer cells were suspended in the surrounding liquid than in ungrazed and Chilodonella sp.-grazed S. plymuthica biofilms. After 48 h of grazing (72 h growth), Tetrahymena sp. reduced S. plymuthica biofilms to isolated patches of cells covering little of the substrate.
Mixed biofilms grazed by ciliates showed a combination of the effects observed in single-species biofilms. Mixed biofilms grazed by Chilodonella sp. had abundant brightly fluorescing P. costantinii cells after 24 h of grazing (48 h growth) which largely disappeared after 48 h of grazing (72 h growth; Fig. 4 vi). S. plymuthica in mixed biofilms grazed by Chilodonella sp. formed irregular structures similar to those in equivalent S. plymuthica-only biofilms (Fig. 4 vi). In mixed biofilms grazed by Tetrahymena sp. for 24 h (48 h growth), S. plymuthica cells were less abundant and did not show the clear 3-dimensional structure observed in equivalent S. plymuthica-only biofilms, and few P. costantinii cells were evident (Fig. 4 ix). These biofilms were subsequently reduced to scattered P. costantinii microcolonies interspersed with small patches of S. plymuthica cells. Unfortunately, it was not feasible to quantify changes in P. costantinii abundance in mixed biofilms associated with ciliate grazing due to the faint fluorescence associated with this bacterium after 48 h of biofilm growth.
DISCUSSION
Preferential feeding on bacterial biofilms.
The abilities of protozoa to converge on patches of high prey density are well known, and the role of chemosensory functions in this process has previously been investigated in a variety of studies (35, 36, 68). In this study, both Tetrahymena sp. and Chilodonella sp. evidently used dissolved chemical cues to converge upon bacterial biofilms and cell extracts, although the circumstances under which this occurred differed between the two ciliates. Tetrahymena sp. responded to biofilms in the type 1 microcosm, while Chilodonella sp. responded only to biofilms in the smaller and simpler type 2 microcosm. This suggests that foraging by Chilodonella sp., a slow-moving crawler, is influenced by spatial and topographic factors, in contrast to that by free-swimming Tetrahymena sp. Motility differences may also explain the observed contrasting temporal responses of these ciliates to bacterial cell extracts. Extract-associated chemical signal gradients are likely to have declined as solute molecules diffused throughout the medium, resulting in fast-moving Tetrahymena sp. cells gradually becoming redistributed throughout the microcosm while less-motile Chilodonella sp. cells did not. These results suggest that ciliate feeding is influenced by microhabitat scale and topography and that ciliates with different modes of motility may have grazing effects at different spatial and temporal scales.
Despite their contrasting feeding and motility modes, both Chilodonella sp. and Tetrahymena sp. showed a clear preference for feeding upon P. costantinii biofilm over S. plymuthica biofilm. This suggests that in natural biofilms, bacterivorous ciliates may seek out preferred patches of bacteria to feed upon. Investigations of protozoan feeding in planktonic contexts have shown that selective feeding is influenced by a complex range of factors, including prey cell size (16, 50), motility (38), cell surface properties (41), and biochemical composition (57). Additionally, dissolved chemical cues are of recognized importance in planktonic protist feeding interactions (9, 56). The two bacteria included in this study are Gram-negative rods with similar cell dimensions (approximately 1.5 μm long and 0.5 to 1.0 μm in diameter; 70) and were embedded in biofilms, making it unlikely that either prey dimensions or motility was important in this case. Rather, our investigations with bacterial cell extracts indicated that the observed feeding preference of Chilodonella sp. for P. costantinii was based upon the detection of dissolved chemical cues, although it is unclear whether this was also the case for Tetrahymena sp. It is possible that chemosensory mechanisms for locating and identifying patchily distributed prey in biofilms may be more important for less-motile species such as Chilodonella sp. than for free-swimming ciliates, which can more rapidly forage over a larger area.
The nature of the dissolved cues which caused the feeding preference of Chilodonella sp. for P. costantinii is uncertain. Amino acid, peptide, and lectin molecules have been shown to act as attractants for Tetrahymena cells (2, 30, 34), and attractants for Paramecium include acetate, lactate, folate, cyclic AMP, and NH4+ (67). Conversely, certain amino acids may have inhibitory effects on ciliates (64). Analysis of extracellular metabolites from biofilm cultures of S. plymuthica and a bacterium related to P. costantinii (Pseudomonas syringae) showed that a variety of amino acids and other molecules were present in concentrations which differed significantly between these cultures (70) and may have caused the observed feeding preference of Chilodonella sp. for P. costantinii biofilm.
While Tetrahymena sp. cells clearly responded positively to bacterium-associated dissolved chemical cues, the contribution of these factors to this ciliate's feeding preference for P. costantinii biofilm was less clear. Preferential feeding by Tetrahymena sp. on P. costantinii biofilm may instead have been based upon chemosensory detection of bacterial cell surface properties or biochemical composition upon contact, capture, processing, or ingestion of prey (42). Biofilm bacteria may produce secondary metabolites that have deleterious effects on feeding protists (13, 40). For example, S. plymuthica produces a variety of molecules with antimicrobial effects, including prodigiosin, a red pigment with reported antiprotozoan properties (20, 43, 61, 72). It is possible that such chemical defense factors were detected by Tetrahymena sp. upon contact with S. plymuthica biofilm, making it a less favorable prey option than P. costantinii.
Our results demonstrate that ciliates can differentiate between patches of biofilm separated by modest distances, with identification of favored patches based on the detection of dissolved chemical cues or contact-based detection of bacterial attributes. Chemosensory prey detection in a biofilm may be influenced by the presence of extracellular polymeric material and the mingling of signals from many cells in close proximity. It is not clear whether ciliates can identify and selectively prey upon individual cells, or small clusters of cells, within a closely packed, mixed assemblage of biofilm bacteria. Further investigation is therefore required to determine the spatial limits on preferential feeding in biofilms, along with elucidation of the role of chemical cues within biofilms and the signals which underlie the observed ciliate feeding preferences.
Effects of ciliate grazing on bacterial biofilms.
Previous studies have shown that grazing by protists alters biofilm morphology and may cause the development of predation-resistant features such as microcolonies (8, 37, 51, 71). Similarly, in this study, grazing by Tetrahymena sp. induced the formation of dense microcolonies of P. costantinii which appear sparser than but otherwise similar to the patchy microcolonies previously observed in biofilms formed by a toxin-producing Pseudomonas aeruginosa strain grazed by Tetrahymena sp. (71). The bacterium Serratia marcescens may form microcolonies and filamentous structures resistant to flagellate and amoeba grazing (51). In contrast, in this study, S. plymuthica biofilm cells became more closely packed together in response to grazing by Tetrahymena sp. but did not develop any distinct grazing-resistant structures. Biofilm consumption by Tetrahymena sp. is thought to be facilitated by its free-swimming motility and strong feeding currents causing dislodgement of attached cells into suspension and subsequent ingestion (17, 46). The susceptibility of P. costantinii and S. plymuthica biofilms to Tetrahymena sp. grazing is consistent with prior studies in which filter-feeding ciliates have been found to graze efficiently upon biofilms and induce significant changes in biofilm morphology (25, 47, 71).
Grazing by Chilodonella sp. had an effect on P. costantinii biofilms different from that of Tetrahymena sp. The crawling motility of Chilodonella sp. may have caused dislodgement of cells from the substrate, and feeding by this ciliate on surface-associated cells may have promoted the growth of suspended bacteria rather than biofilm cells. Grazing by Chilodonella sp. has previously been shown to stimulate the development of microcolonies in mixed bacterial biofilms (8), in contrast to our observations of effects on P. costantinii biofilms. The mounds and irregular channels observed in S. plymuthica biofilms after 48 h of Chilodonella sp. grazing may, however, indicate the development of similar microcolony forms. Chilodonella sp. had a much lesser impact on biofilms than Tetrahymena sp., which is probably due to the lower abundance of Chilodonella sp. cells and associated reduced levels of predation pressure. This also suggests that surface feeders like Chilodonella sp. may be relatively less successful at feeding upon biofilm cells than suspension feeders. However, comparable ingestion rates have been reported for other surface-dwelling protozoa such as amoebae and Tetrahymena feeding on attached bacteria in mixed-species biofilm (47).
The decrease in green fluorescence observed in P. costantinii biofilm cells was unexpected. The mini-Tn7 transposon used to tag P. costantinii with GFP results in the expression of an unstable GFP molecule, so that sustained green fluorescence requires ongoing GFP gene expression (3, 31). Biofilm formation involves genetic and metabolic changes, and differences in gene expression between biofilm and planktonic bacteria have been detected (33, 44, 52). Our observations suggest that GFP expression in P. costantinii may be affected by changes in genetic regulation associated with biofilm formation. This made detection and quantification of P. costantinii in mixed biofilms difficult, and it is consequently uncertain whether either species of ciliate exercised a selective feeding preference within biofilms composed of both types of bacteria.
The ciliate cultures used in this study were treated with antibiotics in order to limit the inclusion of extraneous bacteria in biofilm grazing experiments, but these bacteria may not have been completely eliminated. Chilodonella sp. did not survive treatment with some agents, constraining the range of antibiotics that could be used. Nonfluorescently tagged P. costantinii and S. plymuthica cells were added to ciliate cultures after antibiotic treatment, however, to promote the development of bacterial populations similar to those in biofilm experiments. Furthermore, biofilms were well established before ciliates were added, suggesting that any residual bacteria in ciliate cultures were insignificant in number compared to the already established biofilm populations and had little effect on our results.
Biofilms play an important ecological role in aquatic environments, providing a wide variety of ecosystem services, including organic matter processing and retention, energy flow, and cycling of nutrients (5). Bacteria, in particular, contribute substantially to these processes, since much of the bacterial biomass, activity, and function can be found in biofilms (19). Protozoan grazing also plays a role in ecosystem functionality, as it is an important factor regulating bacterial productivity and community structure, particularly in high-productivity environments (49, 59, 65). In streams, biofilm-grazing protozoa such as ciliates contribute to carbon and energy transfer from biofilm to higher trophic levels and may stimulate the decomposition of organic material such as leaf litter through grazing pressure on bacteria (53). Bacterial biofilm structure is linked to biogeochemical processes which occur in streams (5). Nutrients and other limiting resources are delivered to the biofilm community via a network of channels (11). Consequently, the increased biofilm porosity and spatial/morphological rearrangements caused by ciliate motility and grazing in this study may enhance nutrient and gaseous transport and exchange within the biofilm and influence biofilm-associated ecological functions.
Conclusion.
The feeding interactions between protozoa and biofilm prey are not well understood. Our results, obtained by investigating interactions of two biofilm-isolated ciliates with contrasting feeding behaviors and motility on bacterial species of likely importance in stream biofilm formation, provide insights into the mechanisms and effects of ciliate grazing and preferential feeding on bacteria in stream biofilms (70). The feeding interactions observed in this study are consequently of likely ecological relevance and indicative of processes occurring in natural stream biofilms. Ciliate feeding behaviors are diverse, however, and different ciliates may vary in their response to, and effect on, biofilm prey. Further studies are therefore needed to extend these investigations to additional ciliate feeding types and their effects in natural biofilm communities, as well as the role of chemical signaling and preferential feeding within biofilms. Our results indicate that grazing on biofilms by ciliates with contrasting morphologies, motilities, and feeding strategies has a range of impacts, contributing to the development of spatial, morphological, and population heterogeneity in stream biofilms.
Footnotes
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- 2. Almagor M., Ron A., Bar-Tana J. 1981. Chemotaxis in Tetrahymena thermophila. Cell Motil. 1: 261–268 [Google Scholar]
- 3. Andersen J. B., et al. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64: 2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayo B., Latatu A., Artolozaga I., Jürgens K., Iriberri J. 2009. Factors affecting preference responses of the freshwater ciliate Uronema nigricans to bacterial prey. J. Eukaryot. Microbiol. 56: 188–193 [DOI] [PubMed] [Google Scholar]
- 5. Battin T. J., Kaplan L. A., Newbold J. D., Hansen C. M. E. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426: 439–442 [DOI] [PubMed] [Google Scholar]
- 6. Berninger U. G., Finlay B. J., Kuuppo-Leinikki P. 1991. Protozoan control of bacterial abundances in freshwater. Limnol. Oceanogr. 36: 139–147 [Google Scholar]
- 7. Boenigk J., Arndt H. 2002. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Van Leeuwenhoek 81: 465–480 [DOI] [PubMed] [Google Scholar]
- 8. Böhme A., Risse-Buhl U., Küsel K. 2009. Protists with different feeding modes change biofilm morphology. FEMS Microbiol. Ecol. 69: 158–169 [DOI] [PubMed] [Google Scholar]
- 9. Breckels M. N., Roberts E. C., Archer S. D., Malin G., Steinke M. 2011. The role of dissolved infochemicals in mediating predator-prey interactions in the heterotrophic dinoflagellate Oxyrrhis marina. J. Plankton Res. 33: 629–639 [Google Scholar]
- 10. Corno G., Jürgens K. 2006. Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl. Environ. Microbiol. 72: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costerton J. W. 2007. The biofilm primer. Springer-Verlag, Berlin, Germany: [Google Scholar]
- 12. Daims H., Lücker S., Wagner M. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8: 200–213 [DOI] [PubMed] [Google Scholar]
- 13. Deines P., Matz C., Jurgens K. 2009. Toxicity of violacein-producing bacteria fed to bacterivorous freshwater plankton. Limnol. Oceanogr. 54: 1343–1352 [Google Scholar]
- 14. Dopheide A., Lear G., Stott R., Lewis G. 2008. Molecular characterization of ciliate diversity in stream biofilms. Appl. Environ. Microbiol. 74: 1740–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eisenmann H., Harms H., Meckenstock R., Meyer E. I., Zehnder A. J. B. 1998. Grazing of a Tetrahymena sp. on adhered bacteria in percolated columns monitored by in situ hybridization with fluorescent oligonucleotide probes. Appl. Environ. Microbiol. 64: 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epstein S. S., Shiaris M. P. 1992. Size-selective grazing of coastal bacterioplankton by natural assemblages of pigmented flagellates, colorless flagellates, and ciliates. Microb. Ecol. 23: 211–225 [DOI] [PubMed] [Google Scholar]
- 17. Fenchel T. 1987. Ecology of protozoa: the biology of free-living phagotrophic protists. Springer-Verlag, Berlin, Germany: [Google Scholar]
- 18. Fenchel T., Blackburn N. 1999. Motile chemosensory behaviour of phagotrophic protists: mechanisms for and efficiency in congregating at food patches. Protist 150: 325–336 [DOI] [PubMed] [Google Scholar]
- 19. Fischer H., Pusch M. 2001. Comparison of bacterial production in sediments, epiphyton and the pelagic zone of a lowland river. Freshwater Biol. 46: 1335–1348 [Google Scholar]
- 20. Grimont F., Grimont P. A. D. 2005. Genus XXXIV. Serratia, p. 799–811 In Brenner D. J., Krieg N. R., Staley J. T. (ed.) Bergey's manual of systematic bacteriology, vol. 2 Springer, New York, NY: [Google Scholar]
- 21. Gruber D. F., Tuorto S., Taghon G. L. 2009. Growth phase and elemental stoichiometry of bacterial prey influences ciliate grazing selectivity. J. Eukaryot. Microbiol. 56: 466–471 [DOI] [PubMed] [Google Scholar]
- 22. Hahn M. W., Höfle M. G. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35: 113–121 [DOI] [PubMed] [Google Scholar]
- 23. Hamels I., Mussche H., Sabbe K., Muylaert K., Vyverman W. 2004. Evidence for constant and highly specific active food selection by benthic ciliates in mixed diatoms assemblages. Limnol. Oceanogr. 49: 58–68 [Google Scholar]
- 24. Hausmann K. 2002. Food acquisition, food ingestion and food digestion by protists. Jpn. J. Protozool. 35: 85–95 [Google Scholar]
- 25. Huws S. A., McBain A. J., Gilbert P. 2005. Protozoan grazing and its impact upon population dynamics in biofilm communities. J. Appl. Microbiol. 98: 238–244 [DOI] [PubMed] [Google Scholar]
- 26. Jezbera J., Horňák K., Šimek K. 2005. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol. Ecol. 52: 351–363 [DOI] [PubMed] [Google Scholar]
- 27. Jürgens K., Matz C. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81: 413–434 [DOI] [PubMed] [Google Scholar]
- 28. Kamiyama T., Arima S. 2001. Feeding characteristics of two tintinnid ciliate species on phytoplankton including harmful species: effects of prey size on ingestion rates and selectivity. J. Exp. Mar. Biol. Ecol. 257: 281–296 [DOI] [PubMed] [Google Scholar]
- 29. Koch B., Jensen L. E., Nybroe O. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45: 187–195 [DOI] [PubMed] [Google Scholar]
- 30. Kóhidai L., Csaba G. 1996. Different and selective chemotactic responses of Tetrahymena pyriformis to two families of signal molecules: lectins and peptide hormones. Acta Microbiol. Immunol. Hung. 43: 83–91 [PubMed] [Google Scholar]
- 31. Lambertsen L., Sternberg C., Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6: 726–732 [DOI] [PubMed] [Google Scholar]
- 32. Lawrence J. R., Snyder R. A. 1998. Feeding behaviour and grazing impacts of a Euplotes sp. on attached bacteria. Can. J. Microbiol. 44: 623–629 [Google Scholar]
- 33. Lazazzera B. A. 2005. Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr. Opin. Microbiol. 8: 222–227 [DOI] [PubMed] [Google Scholar]
- 34. Leick V., Lindemose S. 2007. Chemokinesis by Tetrahymena in response to bacterial oligopeptides. J. Eukaryot. Microbiol. 54: 271–274 [DOI] [PubMed] [Google Scholar]
- 35. Levandowsky M., Hauser D. C. R. 1978. Chemosensory responses of swimming algae and protozoa. Int. Rev. Cytol. 53: 145–210 [DOI] [PubMed] [Google Scholar]
- 36. Martel C. M. 2006. Prey location, recognition and ingestion by the phagotrophic marine dinoflagellate Oxyrrhis marina. J. Exp. Mar. Biol. Ecol. 335: 210–220 [Google Scholar]
- 37. Matz C., Bergfeld T., Rice S. A., Kjelleberg S. 2004. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6: 218–226 [DOI] [PubMed] [Google Scholar]
- 38. Matz C., Jürgens K. 2005. High motility reduces grazing mortality of planktonic bacteria. Appl. Environ. Microbiol. 71: 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matz C., Jürgens K. 2003. Interaction of nutrient limitation and protozoan grazing determines the phenotypic structure of a bacterial community. Microb. Ecol. 45: 384–398 [DOI] [PubMed] [Google Scholar]
- 40. Matz C., et al. 2008. Marine biofilm bacteria evade eukaryotic predation by targeted chemical defense. PLoS One 3(7): e2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Monger B. C., Landry M. R., Brown S. L. 1999. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol. Oceanogr. 44: 1917–1927 [Google Scholar]
- 42. Montagnes D. J. S., et al. 2008. Selective feeding behaviour of key free-living protists: avenues for continued study. Aquat. Microb. Ecol. 53: 83–98 [Google Scholar]
- 43. Moons P., et al. 2006. Role of quorum sensing and antimicrobial component production by Serratia plymuthica in formation of biofilms, including mixed biofilms with Escherichia coli. Appl. Environ. Microbiol. 72: 7294–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Toole G., Kaplan H. B., Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54: 49–79 [DOI] [PubMed] [Google Scholar]
- 45. Page F. C. 1988. A new key to freshwater and soil Gymnamoebae. Freshwater Biological Association, Ambleside, England: [Google Scholar]
- 46. Parry J. D. 2004. Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54: 167–196 [DOI] [PubMed] [Google Scholar]
- 47. Parry J. D., Holmes A. K., Unwin M. E., Laybourn-Parry J. 2007. The use of ultrasonic imaging to evaluate the effect of protozoan grazing and movement on the topography of bacterial biofilms. Lett. Appl. Microbiol. 45: 364–370 [DOI] [PubMed] [Google Scholar]
- 48. Pederson K. 1990. Biofilm development on stainless steel and PVC surfaces in drinking water. Water Res. 24: 239–243 [Google Scholar]
- 49. Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3: 537–546 [DOI] [PubMed] [Google Scholar]
- 50. Posch T., et al. 2001. Size selective feeding in Cyclidium glaucoma (Ciliophora, Scuticociliatida) and its effects on bacterial community structure: a study from a continuous cultivation system. Microb. Ecol. 42: 217–227 [DOI] [PubMed] [Google Scholar]
- 51. Queck S. Y., Weitere M., Moreno A. M., Rice S. A., Kjelleberg S. 2006. The role of quorum sensing mediated developmental traits in the resistance of Serratia marcescens biofilms against protozoan grazing. Environ. Microbiol. 8: 1017–1025 [DOI] [PubMed] [Google Scholar]
- 52. Resch A., Rosenstein R., Nerz C., Götz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71: 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ribblett S. G., Palmer M. A., Coats D. W. 2005. The importance of bacterivorous protists in the decomposition of stream leaf litter. Freshwater Biol. 50: 516–526 [Google Scholar]
- 54. Ricci N., Morelli A., Verni F. 1996. The predation of Litonotus on Euplotes: a two step cell-cell recognition process. Acta Protozool. 35: 201–208 [Google Scholar]
- 55. Risse-Buhl U., et al. 2009. Detachment and motility of surface-associated ciliates at increased flow velocities. Aquat. Microb. Ecol. 55: 209–218 [Google Scholar]
- 56. Roberts E. C., Legrand C., Steinke M., Wootton E. C. 19 February 2011, posting date. Mechanisms underlying chemical interactions between predatory planktonic protists and their prey. J. Plankton Res. [Epub ahead of print] doi:10.1093/plankt/fbr005 [Google Scholar]
- 57. Shannon S. P., Chrzanowski T. H., Grover J. P. 2007. Prey food quality affects flagellate ingestion rates. Microb. Ecol. 53: 66–73 [DOI] [PubMed] [Google Scholar]
- 58. Sherr E. B., Sherr B. F. 1987. High rates of consumption of bacteria by pelagic ciliates. Nature 325: 710–711 [Google Scholar]
- 59. Sherr E. B., Sherr B. F. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81: 293–308 [DOI] [PubMed] [Google Scholar]
- 60. Sime-Ngando T., Demers S., Juniper S. K. 1999. Protozoan bacterivory in the ice and the water column of a cold temperate lagoon. Microb. Ecol. 37: 95–106 [DOI] [PubMed] [Google Scholar]
- 61. Singh B. N. 1942. Toxic effects of certain bacterial metabolic products on soil Protozoa. Nature 149: 168 [Google Scholar]
- 62. Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56: 187–209 [DOI] [PubMed] [Google Scholar]
- 63. Strom S., Wolfe G., Slajer A., Lambert S., Clough J. 2003. Chemical defense in the microplankton II: inhibition of protist feeding by beta-dimethylsulfoniopropionate (DMSP). Limnol. Oceanogr. 48: 230–237 [Google Scholar]
- 64. Strom S. L., Wolfe G. V., Bright K. J. 2007. Responses of marine planktonic protists to amino acids: feeding inhibition and swimming behavior in the ciliate Favella sp. Aquat. Microb. Ecol. 47: 107–121 [Google Scholar]
- 65. Thelaus J., Haecky P., Forsman M., Andersson A. 2008. Predation pressure on bacteria increases along aquatic productivity gradients. Aquat. Microb. Ecol. 52: 45–55 [Google Scholar]
- 66. Thurman J., Parry J. D., Hill P. J., Laybourn-Parry J. 2010. The filter-feeding ciliates Colpidium striatum and Tetrahymena pyriformis display selective feeding behaviours in the presence of mixed, equally-sized, bacterial prey. Protist 161: 577–588 [DOI] [PubMed] [Google Scholar]
- 67. Van Houten J. 1992. Chemosensory transduction in eukaryotic microorganisms. Annu. Rev. Physiol. 54: 639–663 [DOI] [PubMed] [Google Scholar]
- 68. Verity P. G. 1988. Chemosensory behavior in marine planktonic ciliates. Bull. Mar. Sci. 43: 772–782 [Google Scholar]
- 69. Verni F., Gualtieri P. 1997. Feeding behaviour in ciliated protists. Micron 28: 487–504 [Google Scholar]
- 70. Washington V. 2010. Interactions between bacteria obtained from stream biofilms. Ph.D. thesis University of Auckland, Auckland, New Zealand: [Google Scholar]
- 71. Weitere M., Bergfeld T., Rice S. A., Matz G., Kjelleberg S. 2005. Grazing resistance of Pseudomonas aeruginosa biofilms depends on type of protective mechanism, developmental stage and protozoan feeding mode. Environ. Microbiol. 7: 1593–1601 [DOI] [PubMed] [Google Scholar]
- 72. Williamson N. R., Fineran P. C., Leeper F. J., Salmond G. P. C. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4: 887–899 [DOI] [PubMed] [Google Scholar]