Abstract
Chronic wasting disease (CWD) and scrapie can be transmitted through indirect environmental routes, possibly via soil, and a practical decontamination strategy for prion-contaminated soil is currently unavailable. In the laboratory, an enzymatic treatment under environmentally relevant conditions (22°C, pH 7.4) can degrade soil-bound PrPSc below the limits of Western blot detection. We developed and used a quantitative serial protein misfolding cyclic amplification (PMCA) protocol to characterize the amplification efficiency of treated soil samples relative to controls of known infectious titer. Our results suggest large (104- to >106-fold) decreases in soil-bound prion infectivity following enzyme treatment, demonstrating that a mild enzymatic treatment could effectively reduce the risk of prion disease transmission via soil or other environmental surfaces.
INTRODUCTION
Prion diseases, or transmissible spongiform encephalopathies (TSEs), are fatal neurodegenerative diseases that include bovine spongiform encephalopathy (BSE) (“mad cow” disease), scrapie in sheep and goats, chronic wasting disease (CWD) of deer, elk, and moose, and Creutzfeldt-Jakob disease (CJD) in humans (9, 10). The infectious agent of prion diseases is PrPSc, an abnormally folded isoform of a normal cellular protein, PrPc (5, 9, 19). Infectious CWD and scrapie prions are shed from living hosts and present in carcasses and can remain infectious after years in the environment (10, 18). The environment can serve as a long-term reservoir of prion infectivity, which likely facilitates a sustained incidence of CWD in free-ranging cervid populations and complicates efforts to eliminate CWD and scrapie in captive herds (10). PrPSc has recently been detected on gates and fencing at a farm where scrapie is endemic (8), indicating that environmental surfaces can become contaminated with prions. Since prions are known to sorb (i.e., bind) to a wide range of soils and soil minerals and remain infectious (10–12, 15), soil may play an important role in environmental transmission of prion diseases (10).
It is desirable to develop a practical, in situ method for decontaminating soil and other environmental surfaces exposed to prions, since current best practices for prion decontamination, such as incineration, autoclaving, or harsh chemical treatments, are not practical for most environmental applications. We have previously characterized an enzyme-based treatment that is effective at degrading soil-bound CWD PrPSc beyond the limits of Western blot detection at pH 7.4 and 22°C (14). Such a treatment has the potential to lower or eliminate infectivity at identified or presumed CWD- and scrapie-contaminated surfaces in captive and wild settings. However, a number of studies have documented high levels of prion infectivity in the absence of detectable PrPSc (2, 16). Thus, the potential effectiveness of such an enzyme treatment at decreasing the risk of prion transmission remains unclear.
The protein misfolding cyclic amplification (PMCA) method (3, 17) has been used previously to detect low levels of prion infectivity. For serial PMCA (sPMCA), brain homogenate, soil, or other material containing an infectious prion seed is added to uninfected brain tissue (containing PrPc) homogenized in a conversion buffer. The sample is then subjected to repeated cycles of sonication followed by incubation at 37°C. After a specified number of cycles (designated one PMCA round), the samples are diluted in fresh uninfected brain homogenate and subjected to additional cycles of sonication and incubation. In this manner, very small amounts of PrPSc are amplified through conversion of PrPc to PrPSc that can be detected by standard Western blotting (3, 17). Moreover, PMCA can also be used to compare prion replication efficiencies (i.e., the ability to convert PrPc to PrPSc) (1, 15), and a strong correlation between infectious prion titer and PMCA replication efficiency has been observed after one PMCA round (15). Here we developed a serial (multiround) PMCA protocol for correlating PMCA efficiency and infectious prion titer. The objective of this study was to determine any changes in the replication efficiency of soil-bound prions following enzymatic treatment. Prions were adsorbed to a range of soil minerals and whole soils, exposed to a bacterially derived subtilisin enzyme solution, and then subjected to serial PMCA to compare the replication efficiencies of treated and untreated soil-bound prions.
MATERIALS AND METHODS
Prion sources and tissue preparation.
Tissue was collected from uninfected hamsters or hamsters infected with the hyper (HY) strain of transmissible mink encephalopathy (TME) agent, as previously described (13). Prion-infected brain tissue was homogenized to 10% (wt/vol) in Dulbecco's phosphate-buffered saline (DPBS) without Ca2+ or Mg2+ (Mediatech, Herndon, VA) using strain-dedicated Tenbroeck tissue grinders (Kontes, Vineland, NJ). For the PMCA substrate, uninfected hamster brain tissue was homogenized to 10% (wt/vol) in ice-cold conversion buffer (DPBS [pH 7.4] containing 5 mM EDTA, 1% [vol/vol] Triton X-100, and a complete protease inhibitor tablet [Roche Diagnostics, Mannheim, Germany]) and centrifuged at 500 × g for 30 s. The supernatant was collected and stored at −80°C.
Prion soil adsorption.
Prion soil adsorption was performed as described previously (14). Brain homogenate from hamsters infected with the HY TME agent was combined with gamma-irradiated fine white sand, Rinda silty clay loam soil (SCL soil), sodium bentonite clay, silicon dioxide powder (SiO2), or humic acid-coated silica gel particles (SiO2-HA), all of which have been previously characterized (12, 14). The sandy loam soil used previously (14) was not used in this study as it is not compatible with the PMCA method (15). Soil-homogenate mixtures (in 1× DPBS) were gently rotated at 24 rpm at 22°C. The incubation time and soil, buffer, and brain homogenate amounts were selected based on previous studies (11, 12, 14, 15) and are detailed in Table 1. PrP adsorptions to silty clay loam, bentonite clay, SiO2 powder, and SiO2-humic acid were conducted in a total volume of 1 ml in 1.5-ml polypropylene tubes (Fisher Scientific). Due to its large particle size, quartz sand could not be processed in bulk and aliquoted into smaller samples; thus, individual sand samples (10 mg) were incubated with brain homogenate in a total volume of 0.2 ml in polypropylene PCR tubes (Fisher Scientific). Samples were removed after incubation and centrifuged at 100 × g for 5 min. The supernatant was removed, and the pellets were washed 5 times with DPBS.
Table 1.
Parameters used to generate soil-bound prions
| Soil or minerala | Adsorption incubation time (h) | Soil concnb (mg/ml) | Brain homogenate (%) | Amt imaged for Fig. 1 (mg) | PMCA spike (1:100) (mg) |
|---|---|---|---|---|---|
| Rinda SCL soil | 24 | 5 | 0.5 | 0.5 | 0.1 |
| Bentonite clay | 24 | 5 | 0.5 | 0.5 | 0.1 |
| SiO2 powder | 24 | 50 | 0.25 | 5 | 2 |
| SiO2-HA | 168 | 10 | 0.5 | 1 | 1 |
| Fine quartz sand | 168 | 50 | 0.5 | 10 | 10 |
SCL, silty clay loam; SiO2, silicon dioxide; HA, humic acid.
The total adsorption solution volume was 1 ml (SCL soil, bentonite clay, SiO2, and SiO2-HA) or 200 μl (sand).
Subtilisin enzyme treatment.
For enzymatic treatment, soil pellets (amounts are shown in Table 1) were exposed to an aqueous enzyme solution known as Prionzyme M (Genencor/Danisco, Rochester, NY). The “Prionzyme” is a proprietary serine protease derived from a Bacillus subtilis strain (6), referred to here as the “subtilisin.” Soil pellets were combined with a 10% Prionzyme dose (the stock solution consisted of 100 Prionzyme units/gram in an aqueous solution containing propylene glycol and sodium formate), and incubated undisturbed for 7 days at pH 7.4 and 22°C as described previously (14). All experiments were performed in triplicate.
sPMCA.
Serial protein misfolding cyclic amplification (sPMCA) was performed as described previously (15). Briefly, an initial prion seed was combined with PMCA substrate (10% [wt/vol] uninfected brain homogenate in conversion buffer [see above]) for a final volume of 100 μl in 0.2-ml PCR tubes (Fisher Scientific). The amount of soil used as an initial seed is shown in Table 1 for each soil type. These amounts correspond approximately to a 1:100 initial dilution (infectious seed/uninfectious substrate) (15). Samples were then subjected to alternating cycles of sonication and incubation at 37°C. Sonication was performed with a Misonix 4000 sonicator (Misonix, Farmingdale, NY) with amplitude set to level 75, generating an average output of 160 W during each sonication cycle. After 24 h (one PMCA round), an aliquot of the samples was added to fresh PMCA substrate and subjected to a second round of PMCA. Two protocols were used for sonication length and interround dilution. Protocol 1 used 25 s of sonication followed by 9 min 35 s of incubation at 37°C, and samples were diluted 1:1 between rounds. Protocol 2 used 5 s of sonication followed by 10 min of incubation at 37°C, and samples were diluted 1:20 between rounds. Following PMCA, all samples were analyzed by Western blotting for the presence of proteinase K (PK)-resistant PrP (PrPSc). Three replicates containing only uninfected brain homogenate were always run concurrently as negative controls.
Immunoblotting.
SDS-PAGE/Western blot detection of soil-bound PrP and PMCA samples was performed as described previously (14) without modification. Briefly, proteins were desorbed from soil (if soil bound) and denatured by boiling samples in SDS-PAGE buffer (Laemmli buffer containing 2% SDS) at 100°C for 10 min. After electrophoresis using 12.5% acrylamide gels, transfer to polyvinylidene difluoride (PVDF) membranes, and blocking in Blotto, blots were probed with monoclonal antibody (MAb) 3F4 (Chemicon, Temecula, CA) (1:10,000 dilution) and developed with Pierce SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific, Rockford, IL). For proteinase K digestion, samples were incubated at 37°C under constant agitation for 30 min with 25 μg PK (Roche Diagnostics) per ml of sample. PK digestion was terminated by boiling in SDS-PAGE buffer.
RESULTS AND DISCUSSION
Enzymatic digestion of soil-bound PrPSc.
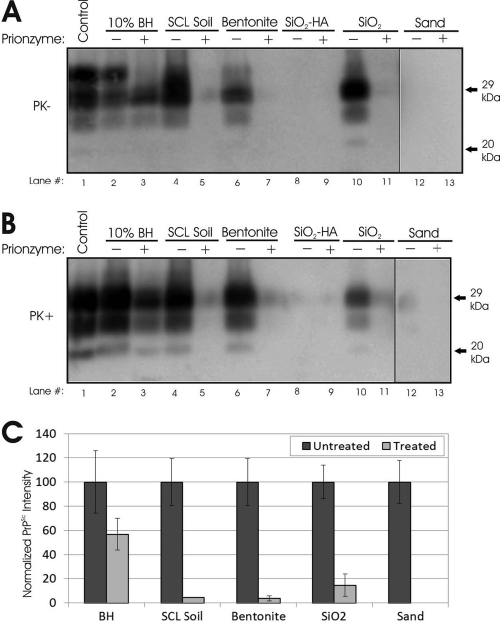
The subtilisin enzyme treatment yielded complete or near-complete loss of soil-bound HY PrPSc (Fig. 1), which agrees with previous results (14). A weak PrPSc signal did remain for SCL soil, bentonite, and SiO2 powder following subtilisin digestion and was most easily detected following PK digestion (4 to 14% compared with untreated controls) (Fig. 1B and C). The unbound PrPSc signal was decreased but remained strong following digestion (57%) (Fig. 1A and B, lanes 3). After the 7-day incubation, both treated and untreated humic acid (SiO2-HA) samples were very faint or undetectable (Fig. 1A and B, lanes 8 and 9), as were all sand samples except for the untreated, PK-digested replicates (Fig. 1B, lanes 12 and 13, and C). Overall, these results indicate large decreases in soil-bound PrPSc following subtilisin enzyme digestion and suggest significant reductions in the infectivity of treated soil samples.
Fig. 1.
Subtilisin enzyme treatment of soil-bound HY TME PrP. Representative Western blots (n = 3) of treated and untreated samples after 7 days of treatment with a 10% Prionzyme dose at 22°C are shown. Imaged soil amounts are shown in Table 1. (A) No proteinase K (PK) digestion prior to detection. (B) PK digestion prior to detection. (C) Quantification of results shown in panel B. Error bars show ±1 standard error of the mean. BH, brain homogenate. Soil abbreviations: SCL, silty clay loam soil; SiO2HA, humic acid-coated silica beads; SiO2, silicon dioxide powder.
Development of quantitative serial PMCA protocol.
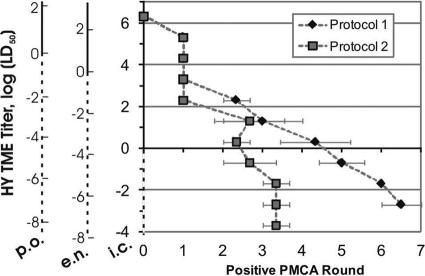
To explore the relationship between PMCA efficiency and sample titer, three replicates of a 10-fold serial dilution of a 10% HY TME agent-infected brain homogenate were subjected to up to seven rounds of serial PMCA using one of two protocols, as described above. Figure 2 reports on the first PMCA round, where a distinguishable positive PrPSc Western blot signal was observed for each dilution. The infectious titer of an HY TME agent-infected brain at terminal disease that we used in this study contains 109.3 intracerebral (i.c.) 50% lethal doses (LD50), 103.7oral (per os) LD50, and 105.5 extranasal (e.n.) LD50 per g of brain by endpoint titration (7). Using protocol 1, the highest dose (a 10−1 homogenate dilution) was readily detected without PMCA, while the next three dilutions were detected after one round. For higher dilutions, an approximately linear relationship existed between spiked titer and initial positive PMCA round. Protocol 2, employing a shorter sonication time and higher interround dilutions, yielded faster amplification, with a 10−11 homogenate dilution detectable after only three PMCA rounds. PrPSc was not detected in the negative controls (n = 3) for either protocol (data not shown).
Fig. 2.
Quantitative serial PMCA of unbound HY TME prions. Results for triplicate 10-fold serial dilutions of HY TME agent-infected brain homogenate using two different PMCA protocols are shown. Error bars show ±1 standard error of the mean. y axes show the oral (p.o.), extranasal (e.n.), and intracerebral (i.c.) infectious titers of the brain homogenate dilutions (7). Values below 0 indicate less than 1 LD50.
These results show a strong, replicable correlation between PMCA efficiency (i.e., the number of rounds required for a positive signal) and the infectious titer of HY TME agent-infected brain homogenate samples. Although protocol 1 required more rounds of PMCA to amplify low levels of infectivity, the larger range of PMCA rounds allowed for better quantification of differences in amplification efficiency. The error of the standard curves shown in Fig. 2 could undoubtedly be reduced by using additional replicates. Although it was beyond the scope of this study to perform an in-depth evaluation of the differences in amplification over a range of PMCA protocols, our results do demonstrate how protocols can be modified to yield maximum amplification or maximum quantification resolution.
Chen et al. recently reported use of a quantitative sPMCA protocol (4). However, their resolving power was ±2 to 4 logs of PrPSc, analogous to that of our protocol 2. While their use of dilutions of enriched PrPSc for the standard curve is potentially useful for quantifying PrPSc levels in fluids and tissues, it may prove to be less helpful in evaluating sample infectivity than using brain dilutions with known titers. Our results suggest that a standard curve of tissue dilutions with known titers generated by sPMCA could be used to quantify sample infectivity, although additional validation work would clearly be required before such an application. It has previously been shown that PMCA efficiency varies with prion strain (1) and species (15); thus, standard curves would need to be developed for each strain and species used.
PMCA of subtilisin-treated soil samples.
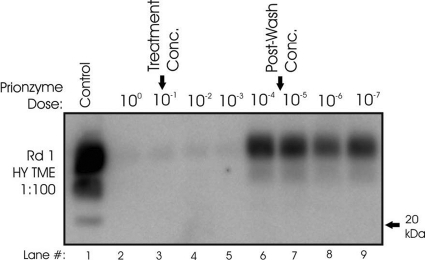
To evaluate any inhibitory effects of the subtilisin solution on the PMCA method, a 10-fold serial dilution of Prionzyme was spiked into identically prepared samples of HY TME agent (1:100 ratio of HY-infected brain homogenate to PMCA substrate), which were then subjected to one round of PMCA (protocol 1). The subtilisin solution completely inhibited HY PrPSc amplification down to a 10−3 dilution (Fig. 3). It is likely that this inhibition was due to subtilisin digestion of PrPc and/or other conversion cofactors in the PMCA substrate. Given this inhibition, we employed a washing step for soil samples (applied to both treated and untreated samples) to lower the subtilisin concentration below inhibitory levels. Soils samples were diluted 1 to 20 in 1× DPBS and then centrifuged (100 × g) to pellet soil particles. The supernatant was removed and this process was repeated twice more, diluting the calculated subtilisin concentration from 10−1 to 10−4.9. Unfortunately, this washing step could not be performed on unbound samples, since this would dilute not only the subtilisin but also the unbound HY TME agent. Thus, the effectiveness of the enzyme treatment on unbound HY TME agent could not be evaluated.
Fig. 3.
Inhibition of PMCA due to the presence of a subtilisin enzyme solution. Representative Western blots (n = 3) of 1:100 HY TME agent samples spiked with a serial dilution of the Prionzyme solution and subjected to one round of PMCA are shown. Arrows indicate the Prionzyme treatment dose (10%) and calculated postwash concentration (10−4.9).
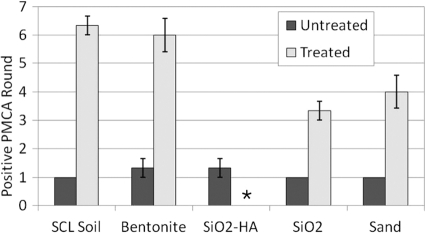
Treated and untreated soil-bound prions were subjected to both PMCA protocol 1 and protocol 2. Soil amounts used for PMCA seeding are shown in Table 1. Figure 4 shows the mean positive round (three replicates) for each soil type. A significant increase between treated and untreated samples was observed for all soil types (P < 0.05 by t test assuming unequal variance [GraphPad Prism, San Diego, CA]). The results using protocol 2 were consistent with protocol 1 results, albeit with smaller differences in mean positive round (data not shown). Note that the presence of soil without PrPSc does not induce PrPSc formation through three PMCA rounds (15).
Fig. 4.
Serial PMCA of treated and untreated soil-bound HY TME prions. Results for triplicate treated and untreated samples using protocol 1 are shown. Error bars show ±1 standard error of the mean. *, treated SiO2-HA samples were not positive through 8 rounds.
The soil type influenced the effectiveness of enzymatic degradation of soil-bound HY TME agent (Table 1). The subtilisin was least effective at decreasing the PMCA efficiency of HY bound to SiO2 and sand samples (2- or 3-round extensions). It was more effective for SCL soil and bentonite (5-round extensions) and was especially effective against humic acid (SiO2-HA)-bound prions, which failed to amplify in any experiment following treatment. Overall, these results further demonstrate that soil-bound prions are not necessarily protected from enzymatic digestion and that the subtilisin appears to be broadly effective against prions bound to a range of soils.
We have previously shown that sand- and SiO2-bound prions (untreated) have higher PMCA efficiencies than SCL soil-, bentonite-, and SiO2-HA-bound prions (15). Moreover, we did not observe consistent variance in enzyme effectiveness against bound PrPSc (i.e., Western blot results) with respect to soil type (Fig. 1) (14). Thus, the variance in enzyme effectiveness observed in this study may be due to variance in the PMCA efficiency of remaining (undigested) soil-bound prions and not due to variance in the ability of the subtilisin to digest prions bound to different soils.
Inhibition of soil-bound prion replication with an enzyme solution.
A key motivation for this work was determining the efficacy of a topically applied enzyme treatment for reducing prion infectivity on environmental surfaces such as soil, thereby potentially reducing the risk of prion disease transmission from an environmental reservoir. Using the data presented in Fig. 2 and Fig. 4 (from protocol 1), it is possible to correlate the mean positive PMCA round for treated and untreated soil samples with a corresponding PMCA efficiency of known HY TME agent-infected brain homogenate titer (Table 2). The observed increases in PMCA rounds required for a positive PrPSc detection following enzymatic treatment correspond to reductions in infectious HY TME titer of 104 (SiO2), 104.5 (sand), 105 (bentonite), 106.5 (SCL soil), and >106.5 (SiO2-HA), suggesting large decreases in sample infectivity. It has previously been shown that the presence of soil can inhibit the PMCA method (and that this inhibition is not soil dependent) (15). Furthermore, taking into account this inhibition, prions bound to SCL soil, bentonite, and SiO2-HA exhibit reduced PMCA efficiency compared to prions bound to SiO2 and sand and to unbound controls. At least in the case of SCL soil, this reduction in PMCA replication corresponds to a reduction in intracerebral infectious titer (15). These previous results were considered when estimating the equivalent titers of untreated samples (Table 2), since a range of titers are readily amplified after one PMCA round.
Table 2.
Estimated reductions in equivalent HY TME agent titer based on PMCA efficiency
| Soil or mineral | Equivalent titer (HY TME agent i.c. log LD50) |
|
|---|---|---|
| Untreateda | Treated | |
| SCL soil | 4 | −2.5 |
| Bentonite clay | 3.5 | −1.5 |
| SiO2-HA | 3.5 | <−3 |
| SiO2 powder | 5 | 1 |
| Sand | 5 | 0.5 |
Untreated equivalent titers are based in part on reference 15.
It is important to note that this work was conducted using HY TME hamster prions, with which we have extensive previous PMCA experience (1, 15, 17). HY TME prion interactions with soil may not necessarily recapitulate that of CWD or other naturally occurring prions (11, 13); nevertheless, we previously observed equal or greater susceptibility to subtilisin digestion for soil-bound CWD PrPSc compared with HY TME PrPSc (14). Thus, it could be reasonably inferred that treated soil-bound CWD prions would exhibit similar decreases in PMCA replication efficiency.
The decisive test for prion infectivity remains animal bioassay, and thus, to demonstrate definitive decreases in infectivity of soil-bound prions due to enzymatic treatment, bioassay will need to be completed. Nevertheless, the subtilisin used (Prionzyme) has a high efficacy for reducing infectivity of unbound prions. A previous study observed a 7-log decrease in 301V mouse prions when using the Prionzyme at pH 12 and 60°C (6). Moreover, our previous work using PMCA with soil has supported its use in correlating sample PMCA efficiency and infectivity (15). Thus, the significant decrease in replication efficiency observed for subtilisin-treated soil samples in the present study is highly indicative of significant decreases in infectious titer. We therefore propose that a mild enzymatic treatment of environmental surfaces, whether using Prionzyme or an alternative enzyme, could potentially reduce the risk of CWD or scrapie transmission in the environment. We suggest that locating environmental “hot spots” of prion infectivity is now the most significant priority for applying a pilot-scale enzyme treatment program to attempt to lower the risk of environmental prion disease transmission.
ACKNOWLEDGMENTS
We thank Ronald Shikiya and Qi Yuan for technical assistance and Scott Hygnstrom for helpful discussions. Humic acid-coated silica gel particles were kindly provided by Robert Bulman. Thanks go to Michael Beard at Becton Dickinson for gamma irradiation of soils.
This research was supported in part by the USDA/APHIS/Wildlife Services/National Wildlife Research Center, the UNL Research Council, the UNL Othmer and Milton Mohr Fellowships, and the National Center for Research Resources (P20 RR0115635-6 and C06 RR17417-01).
Footnotes
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Ayers J. I., et al. 2011. The strain-encoded relationship between PrPSc replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog. 7:e1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barron R. M., et al. 2007. High titers of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vivo. J. Biol. Chem. 282:35878–35886 [DOI] [PubMed] [Google Scholar]
- 3. Castilla J., Saa P., Hetz C., Soto C. 2005. In vitro generation of infectious scrapie prions. Cell 121:195–206 [DOI] [PubMed] [Google Scholar]
- 4. Chen B., Morales R., Barria M., Soto C. 2010. Estimating prion concentration in fluids and tissues by quantitative PMCA. Nat. Methods 7:519–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deleault N. R., Harris B. T., Rees J. R., Supattapone S. 2007. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U. S. A. 104:9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dickinson J., et al. 2009. Decontamination of prion protein (BSE301V) using a genetically engineered protease. J. Hosp. Infect. 72:65–70 [DOI] [PubMed] [Google Scholar]
- 7. Kincaid A. E., Bartz J. C. 2007. The nasal cavity is a route for prion infection in hamsters. J. Virol. 81:4482–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maddison B. C., et al. 2010. Environmental sources of scrapie prions. J. Virol. 84:11560–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prusiner S. B. 2004. An introduction to prion biology and diseases, p. 1–89In Prusiner S. B. (ed.), Prion biology and diseases, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 10. Saunders S. E., Bartelt-Hunt S. L., Bartz J. C. 2008. Prions in the environment: occurrence, fate, and mitigation. Prion 2:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saunders S. E., Bartz J. C., Bartelt-Hunt S. L. 2009. Influence of prion strain on prion protein adsorption to soil in a competitive matrix. Environ. Sci. Technol. 43:5242–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saunders S. E., Bartz J. C., Bartelt-Hunt S. L. 2009. Prion protein adsorption to soil in a competitive matrix is slow and reduced. Environ. Sci. Technol. 43:7728–7733 [DOI] [PubMed] [Google Scholar]
- 13. Saunders S. E., Bartz J. C., Telling G. C., Bartelt-Hunt S. L. 2008. Environmentally-relevant forms of the prion protein. Environ. Sci. Technol. 42:6573–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saunders S. E., Bartz J. C., VerCauteren K. C., Bartelt-Hunt S. L. 2010. Enzymatic digestion of chronic wasting disease prions bound to soil. Environ. Sci. Technol. 44:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saunders S. E., Shikiya R. A., Langenfeld K. A., Bartelt-Hunt S. L., Bartz J. C. 2011. Replication efficiency of soil-bound prions varies with soil type. J. Virol. 85:5476–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scherbel C., et al. 2007. Infectivity of scrapie prion protein PrPSc following in vitro digestion with bovine gastrointestinal microbiota. Zool. Public Health 54:185–190 [DOI] [PubMed] [Google Scholar]
- 17. Shikiya R. A., Ayers J. I., Schutt C. R., Kincaid A. E., Bartz J. C. 2010. Coinfecting prion strains compete for a limiting cellular resource. J. Virol. 84:5706–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamgüney G., et al. 2009. Asymptomatic deer excrete infectious prions in faeces. Nature 461:529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang F., Wang X., Yuan C. G., Ma J. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]