Abstract
We previously showed a pivotal role of the polysaccharide (PS) moiety in the cell wall of the Lactobacillus casei strain Shirota (YIT 9029) as a possible immune modulator (E. Yasuda M. Serata, and T. Sako, Appl. Environ. Microbiol. 74:4746-4755, 2008). To distinguish PS structures on the bacterial cell surface of individual strains in relation to their activities, it would be useful to have a rapid and high-throughput methodology. Recently, a new technique called lectin microarray was developed for rapid profiling of glycosylation in eukaryotic polymers and cell surfaces. Here, we report on the development of a simple and sensitive method based on this technology for direct analysis of intact bacterial cell surface glycomes. The method involves labeling bacterial cells with SYTOX Orange before incubation with the lectin microarray. After washing, bound cells are directly detected using an evanescent-field fluorescence scanner in a liquid phase. Using this method, we compared the cell surface glycomes from 16 different strains of L. casei. The patterns of lectin-binding affinity of most strains were found to be unique. There appears to be two types of lectin-binding profiles: the first is characterized by a few lectins, and the other is characterized by multiple lectins with different specificities. We also showed a dramatic change in the lectin-binding profile of a YIT 9029 derivative with a mutation in the cps1C gene, encoding a putative glycosyltransferase. In conclusion, the developed technique provided a novel strategy for rapid profiling and, more importantly, differentiating numerous bacterial strains with relevance to the biological functions of PS.
INTRODUCTION
It is well documented that bacterial cell surface components and structures are critical factors for pathogenesis, host-microbe interaction, immune modulation, and symbiosis. Toll-like receptors (TLRs) expressed on mammalian epithelial and immune cells act as pattern recognition receptors, which are individually responsible for a variety of different bacterial components, such as lipopolysaccharides (LPSs) from Gram-negative bacteria, peptidoglycan (PG), lipoteichoic acid (LTA), and wall teichoic acid (WTA) from Gram-positive bacteria, flagella, lipoproteins, and nucleic acids. TLRs transfer signals to the innate as well as the acquired immune system (1). Bacterial cell surface components are also recognized by other mammalian signaling molecules, such as nucleo-tide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2, which are intracellular proteins functioning as cytosolic sensors in the regulation of inflammatory responses (17). The human gut is inhabited by an enormous amount of microbes involved in maturation of the host immune system, establishment of commensalism, or induction of the inflammatory response through interaction with host cells. While the mechanism of a typical host-microbe interaction through TLRs and NODs is documented, there are a number of ambiguous interactions between host and microorganism in which bacterial surface components are involved. It has recently been revealed that polysaccharide (PS) A from Bacteroides fragilis stimulates T-cell activation (22), while type III PS from group B streptococci is known to be an immune stimulant, in addition to being a virulence factor (23).
Many strains of lactic acid bacteria (LAB) are used as probiotics, which are defined as microbes that exert a beneficial effect on the host. Various factors produced by LAB have been proposed to actively interact with mammalian host cells (12). For example, soluble proteins produced by probiotic bacteria are known to regulate survival and growth of intestinal epithelial cells (11, 18, 30). Although the cell surface components of LAB such as S-layer proteins, LTA, WTA, and PSs are proposed to be immune modulators (21), the active components directly involved in immune modulation and the molecular nature of these recognition processes are largely unknown.
The probiotic activity of the Lactobacillus casei strain Shirota (YIT 9029) has been extensively analyzed. While the bacterium is known to be a strong Th1-type cytokine inducer (32), of which the active component was proposed to be LTA (12), it exerts anti-inflammatory activity against diabetes mellitus (26) and inflammatory bowel disease (24, 25) in animal models. It was shown that the cell wall preparation containing PS and PG could be the active component of the anti-inflammatory activity (25). We have focused on the role of cell wall PSs in the immune modulation activities of YIT 9029 and have found that high-molecular-mass cell wall PSs (PS-1) are a prerequisite for anti-inflammatory activity (43). In this analysis, we identified a cluster of genes essential for the biosynthesis of the high-molecular-mass PS-1 moiety, which are designated cps1A, cps1B, cps1C, cps1D, cps1E, cps1F, and cps1J.
In the light of recent research on the role of bacterial cell surface structures/components in probiotic or symbiotic action (21), PSs from each bacterial cell wall could have individual and unique roles which are more important than has ever been assumed (4, 19, 22, 46). As described in some reports concerning Bacteroides fragilis polysaccharides (7, 8, 27), bacterial cells have different impacts on host cells, even when various mutants having defects in different predictive polysaccharide biosynthesis genes or deletions showed different immune-modulating effects on cultured splenocytes and T cells. However, the kind of PS structure important for their activity and how these molecules exert their activities on host cells are still open questions. In addition, it has been shown that extracellular PSs of Streptococcus thermophilus (39) and group B Streptococcus capsular PSs are highly diverse (6). It is well-known that pathogenic bacteria, such as hemolytic streptococci, often change their outer surface glycan profile to escape the host immune defense mechanism (5). To investigate the dynamism and diversity of bacterial outer surface structures, especially bacterial glycomes, in relation to their functional characterization, a powerful methodology with high throughput and versatility is needed.
A novel technique called lectin microarray was developed to profile the complex features of glycans expressed in various forms (2, 14, 20, 37, 38). We adopted a unique evanescent-field-activated fluorescence detection principle to detect highly sensitive and reproducible lectin-glycoconjugate interactions on a glass side (20, 38). Using this detection principle, Tateno et al. (2007) developed an application method enabling detection of direct interaction between lectins and whole mammalian cells (37). Similarly, Hsu et al. (14-16) applied lectin microarray technology to targeting of bacterial cells, though they utilized a confocal detection principle.
In this study, we provide a practical methodology for profiling bacterial cell surface glycomes and show substantial differences in glycan profiles between strains of the same Lactobacillus species.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. Lactobacillus casei YIT 9029 is a commercial strain used in the production of fermented milk. A mutant of L. casei YIT 9029 of which the cps1C gene was knocked out, YIT 9029 Δcps1C (43), was analyzed. YIT 0180 is the neotype strain of L. casei (10). Cells were cultured in MRS medium (Becton Dickinson and Company) for 22 h at 37°C under aerobic conditions.
Table 1.
Lactobacillus casei strains used in this study
| YIT strain no.a | Identification no.b | Source, designation, or description | Reference(s) |
|---|---|---|---|
| YIT 0001 | ATCC 27139 | S-1 (A Murata) | 33 |
| YIT 0003 | IAM 1045 (JCM 20024) | Cheese | |
| YIT 0005 | ATCC 25302 | Saliva | 35 |
| YIT 0006 | ATCC 25303 | Saliva | 35 |
| YIT 0007 | JCM 1109 | Human intestine | |
| YIT 0009 | NIRD C-9 | Human | 46 |
| YIT 0015 | JCM 1053 | T. Mitsuoka S2-5 | |
| YIT 0047 | NIRD A-121 | Human | 45,46 |
| YIT 0091 | IPOD 1766 | YPS-1c | |
| YIT 0123 | ATCC 27216 | Saliva of childd | |
| YIT 0128 | ATCC 4646 | Dental caries | |
| YIT 0226 | PHLS A357/84 | Human blood | |
| YIT 0289 | PHLS A22/73 | Endocarditis | |
| YIT 0290 | PHLS A198/89 | Endocarditis | |
| YIT 9029 | Strain Shirota | Original collection of Yakult | 43 |
| YIT 0180Te | ATCC 334 | Emmental cheese | 10 |
| Mutant of YIT 9029 | Δcps1C | 43 |
YIT, registration number of the culture collections preserved in Yakult Central Institute for Microbiological Research, Tokyo, Japan.
Strains were purchased from the American Type Culture Collection (ATCC; Manassas, VA), Institute of Molecular and Cellular Biosciences (IAM; University of Tokyo, Tokyo, Japan), Japan Collection of Microorganisms (JCM; Wako, Japan), National Institute for Research in Dairying Collection (NIDR; India), International Patent Organism Depositary (IPOD; Tsukuba, Japan), and the Public Health Laboratory Service (PHLS; United Kingdom).
From the collection of the Yakult Central Institute for Microbiological Research.
Type of L. casei subsp. alactosus.
Neotype strain.
Fluorescent staining of Lactobacillus casei cells.
Cells cultured in 4 ml MRS medium (1 × 109 to 2 × 1010) were harvested by centrifugation (4,000 × g for 5 min at 4°C) and washed three times with 10 mM phosphate-buffered saline (PBS; pH 7.0). The cells were resuspended in 4 ml of 70% ethanol and agitated (100 rpm for 30 min at room temperature [RT]) using a Personal-11 shaker (Taitec Co., Ltd.). The cells were harvested by centrifugation (4,000 × g for 5 min at 4°C) and washed three times with PBS. The cells were then resuspended in 4 ml of PBS, before incubation with 1 to 50 μM SYTOX Orange nucleic acid stain (Molecular Probes Co., Ltd.) for 5 min at RT (41, 42). Cells labeled with SYTOX Orange were washed three times with PBS and finally resuspended in 360 μl of PBS containing 1% bovine serum albumin (PBS-BSA). The fluorescence intensity of 2 × 108 labeled cells was measured using an ARVO X3 apparatus (PerkinElmer) within 1 h of labeling.
To visualize the cells labeled with SYTOX Orange, they were embedded in Vectorshield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Glass slides were examined using a Leica Q550FW system, and fluorescent images were analyzed using Image-Pro Plus software (Media Cybernetics, Silver Spring, MD) (36).
Lectin microarray preparation and lectin specificities.
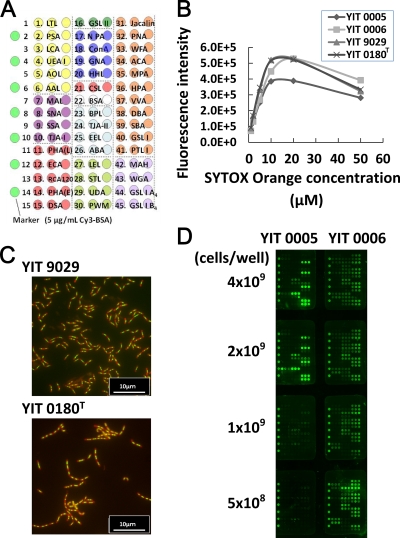
The lectin microarray was prepared as previously described (21-23). Briefly, 44 lectins were dissolved at a concentration of 0.5 mg/ml in a spotting solution (Matsunami Glass) and spotted onto epoxysilane-coated glass slides (Schott) in triplicate using a noncontact microarray printing robot (MicroSys4000; Genomic Solutions). The glass slides were then incubated at 25°C overnight to allow lectin immobilization. The lectin-immobilized glass slides were then washed with probing buffer (25 mM Tris-HCl, pH 7.5, 140 mM NaCl [TBS] containing 2.7 mM KCl, 1 mM CaCl2, 1 mM MnCl2, and 1% Triton X-100) and incubated with blocking reagent N102 (NOF Co.) at 20°C for 1 h. Finally, the lectin-immobilized glass slides were washed with TBS containing 0.02% NaN2 and stored at 4°C until use. The glycan-binding specificities of the lectins are listed in Table S1 in the supplemental material. In addition, CSL, a rhamnose (Rha)-binding lectin isolated from chum salmon (Oncorhynchus keta) eggs (34, 40), was used in this study (see Fig. 2A).
Fig. 2.
Optimization of lectin microarray analysis for bacterial cells. (A) Lectin microarray format. Glycan-binding specificities of the lectins used in this article are listed in Table S1 in the supplemental material. The concentration of each lectin was 0.5 mg/ml. (B) Effect of SYTOX Orange concentration on cell fluorescence intensity. Lactobacillus casei YIT 0005, YIT 0006, YIT 9029, and YIT 0180T cells were grown to stationary phase, before labeling with SYTOX Orange (1 to 50 μg/ml, as described in Materials and Methods). The fluorescence intensity of 1 × 108 cells was measured by an ARVO X3 apparatus (PerkinElmer) using a Cy3 filter. The highest fluorescence intensity was 10 μM for each strain. (C) Fluorescent images of L. casei cells labeled with SYTOX Orange and DAPI. L. casei YIT 9029 and L. casei YIT 0180T cells were labeled with 10 μM SYTOX Orange and relabeled with DAPI on a glass slide. Red images were obtained with SYTOX Orange, and green images were obtained with SYTOX Orange and DAPI. The cells were observed to be thin and long using SYTOX Orange and round using DAPI. (D) Dose-dependent fluorescent signals of L. casei cells in the lectin microarray. Various numbers of cells labeled with SYTOX Orange were allowed to bind with the lectin array (0.5 to 4 × 109 cells/well). Bound cells were detected using an evanescent-field fluorescent scanner.
Lectin microarray hybridization.
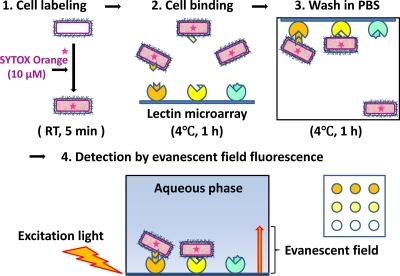
L. casei cells labeled with SYTOX Orange were suspended in PBS-BSA and added to each well of a glass slide containing immobilized lectins (100 μl/0.5 × 109 to 5 × 109 cells/well), followed by incubation at 4°C for 1 h. Unbound cells were mildly removed by immerging the inverted lectin microarray in more than 1 liter of cold PBS at 4°C for 30 min. Bound cells with lectins immobilized on a glass slide were detected with an evanescent-field fluorescence scanner (Fig. 1). Data are shown as the ratio of fluorescence intensities of the 44 lectins relative to the maximal fluorescence intensity on the lectin microarray. The lectin-binding signals for each strain were normalized with the highest signal. Unsupervised hierarchical clusters were generated for the Lactobacillus casei strains, and their glycan profiles were obtained by lectin microarray. The levels of lectin-binding signals are indicated by the color change from green (low binding levels) to red (high binding levels).
Fig. 1.
Profiling system of bacterial cell wall polysaccharides using lectin microarray.
Carbohydrate inhibition assay.
In order to determine the effects of carbohydrates on binding affinity between CSL and YIT 9029 and/or YIT 0047, whose origin is significantly different from that of YIT 9029 (45), d-galactopyranose (Gal), d-glucopyranose (Glc), d-galactosylpyranosyl-(1→4)-d-Glc (Lac), d-mannopyranose (Man), l-rhamnopyranose (Rha), d-fructofuranosyl-(2→1)-d-glucopyranoside (Suc), or (3S,4R,5S,6S)-6-methyltetrahydro-2H-pyran-2,3,4,5-tetraol (Fuc) was added at a concentration of 0.1 μM to 2 × 109 cells labeled with SYTOX Orange in PBS-BSA. The effect of carbohydrate inhibition is shown as a percentage of the control signals with no added carbohydrate.
PS-PG.
Cells grown overnight in 100 ml of MRS medium were harvested by centrifugation (12,000 × g for 10 min at 4°C) and washed three times with distilled water. Cells were resuspended in 4 ml of 5 mM Tris-malate-2 mM MgCl2 (pH 6.4). After boiling for 10 min, 1 mg of N-acetylmuramidase SG (Dainippon Sumitomo Pharma Co., Ltd.), to solubilize polysaccharides by digesting the peptidoglycan network of the cell wall, and 1 mg of benzonate (Merck Japan Ltd., Tokyo, Japan), as an endonuclease, were added to the cell suspension, and the mixture was incubated at 37°C for 18 h. The reaction mix was heated at 100°C for 10 min and then centrifuged at 12,000 × g for 10 min at 4°C. One milligram of pronase (Roche Diagnostics K. K., Tokyo, Japan) in the solution was added to the supernatant to digest all protein components into small fragments, and the reaction mix was incubated at 37°C for an additional 20 h. The resulting solution was thoroughly dialyzed against deionized water using a 3,500-molecular-mass-cutoff dialysis bag and more than 8 exchanges of water for 2 or 3 days at RT. The samples obtained were designated the PS-PG fraction and stored in a refrigerator until further use.
The carbohydrate composition of the PS-PG was analyzed by labeling the sample using an ABEE labeling kit (J-Oil Mills Co. Ltd., Tokyo, Japan), followed by high-performance liquid chromatography analysis using a Honenpak C18 column (75 mm by 4.6 mm [inner diameter]) (44).
RESULTS
Optimization of labeling for Lactobacillus casei.
Before using SYTOX Orange, we examined the applicability of Cy3 labeling, which is usually used in fluorescent labeling of glycoproteins and cell surface proteins for microarray analysis. The fluorescein isothiocyanate-labeling method (32) with 10 μg/ml Cy3 successfully labeled 1 × 109 to 2 × 109 cells. However, nonspecific binding between Cy3-labeled cells and the glass microarray slides was detected. This was probably due to increased hydrophobicity of the cells. Labeling with Cell Tracker, another labeling reagent for whole-mammalian-cell staining (37), was also unsuccessful, since the fluorescence intensity of L. casei cells (1 × 1010 to 2 × 1010) labeled with Cell Tracker (50 μg/ml) was too low to be analyzed by this technology.
We then examined the suitability of SYTOX Orange (41, 42), whose maximum excitation wavelength is similar to that of Cy3 (32). We first measured the fluorescence intensity of 1 × 108 cells labeled with different concentrations of SYTOX Orange (Fig. 2B ). The highest fluorescence intensity was obtained at 10 μM for all four strains. This concentration is 100-fold higher than the maximum concentration recommended in the standard protocol to label bacterial cells. This may be caused by the lower permeation of the dye across the cell wall and cell membrane in lactobacilli treated with 70% ethanol than in mammalian cells and other bacterial cells. The higher fluorescence intensity of labeled cells was more suitable for lectin microarray analysis. Another advantage of SYTOX Orange is that the fluorescence intensity of cells labeled with SYTOX Orange was stable for 2 days at 4°C. Consequently, the concentration of SYTOX Orange used to label bacterial cells in this study was 10 μM.
Proper labeling of the cells with SYTOX Orange was confirmed by monitoring the labeled cells with fluorescence microscopy. As shown in Fig. 2C, it was possible to distinguish the area of the cells stained by SYTOX Orange from that stained by DAPI after double staining of L. casei cells. The latter stained only the center of the cells, as DAPI binds to AT-specific chromosomal DNA (31); SYTOX Orange stained the whole area of the cell 2 or 3 times more strongly than DAPI. This is probably because the dye binds not only to double-stranded DNA, as described in the supplier's manual, but also to RNAs that are distributed throughout the whole-cell cytoplasm. It was essential to treat bacterial cells with 70% ethanol for 30 min before staining with SYTOX Orange in order to obtain the full fluorescence intensity, and microscopic analysis suggested that this treatment did not damage cell morphology. Hence, we concluded that 10 μM SYTOX Orange was the most suitable labeling dye for lectin microarray analysis.
Differential profiling of 4 Lactobacillus casei strains.
We determined the required number of SYTOX Orange-labeled cells using 2 different strains of L. casei, YIT 0005 and YIT 0006, at 0.5 × 109, 1.0 × 109, 2.0 × 109, or 4.0 × 109 cells/well. As shown in Fig. 2D, only 2.0 × 109 and 4.0 × 109 cells/well gave the full fluorescence intensity and reproducible results in both strains. It was concluded that 2 × 109 to 4 × 109 cells/well was suitable for this assay.
The lectin-binding activity of YIT 0005, YIT 0006, YIT 9029, and YIT 0180T was then analyzed in more detail. As shown in Fig. 3, each strain had a unique binding profile. Similar to previously reported glycoproteins (20) and mammalian cells (37), the bacterial cells had their own molecular patterns of glycosylation on their cell surfaces. It can be also speculated that YIT 0005, YIT 0006, and YIT 0180T have similar outer surface PS structures. In particular, strong signals were commonly detected in these three strains for ECA (asialo complex-type N-glycan binder), BPL and TJA-II (Gal binders), LEL (chitin binder), and O-glycan binders (WFA, MPA, HPA, VVA, SBA, GSL-I, and PTL-I).
Fig. 3.
Differential profiling of 4 Lactobacillus casei strains. Bacterial cells (4 × 109 cells/well) were subjected to glycan profiling by lectin microarray. Data were analyzed as described in Materials and Methods. Each strain shows a strain-specific lectin-binding affinity. Red boxes indicate the signals bound to Rha-binding lectin (CSL). Though YIT 0005, YIT 0006, and YIT 0180T cells bound to a variety of lectins, YIT 9029 cells bound to CSL only. Data are the averages ± standard deviations of triplicate determinations.
However, YIT 9029 bound only to CSL, a rhamnose-specific lectin (32, 33, 37). It is known that PS from YIT 9018, the parental strain of YIT 9029 which was produced by removing bacteriophage φFSW from the YIT 9018 genome (33), contains Rha, Glc, Gal, N-acetylglucosamine (GlcNAc), and N-acetylgalactosamine (GalNAc) (28). We also confirmed the sugar composition of YIT 9029 PS using the ABEE labeling kit (44) and found the presence of Rha, Glc, Gal, and others (data not shown), consistent with the results of Nagaoka et al. (28). Thus, the binding pattern of YIT 9029 does not directly reflect the sugar composition of its cell wall PS. Similarly, to analyze the relationship between lectin-binding profiles and sugar composition, we confirmed the sugar composition of PSs from YIT 0005, YIT 0006, and YIT 0180T. While the presence of Glc, GlcNAc, and GalNAc was confirmed in all strains, Rha was not detected in YIT 0005 and Gal was confirmed to be present only in YIT 9029. The sugar compositions of YIT 0005 and YIT 0006 in our study are consistent with those reported by Šimelyte et al. (35) (YIT 0005 and YIT 0006 correspond to L. casei ATCC 25302 and L. casei ATCC 25303, respectively). On the basis of our results, the binding profiles of the strains analyzed are not just determined by the sugar composition of the cell wall PSs, but they are also partly determined by the specificity of lectin binding.
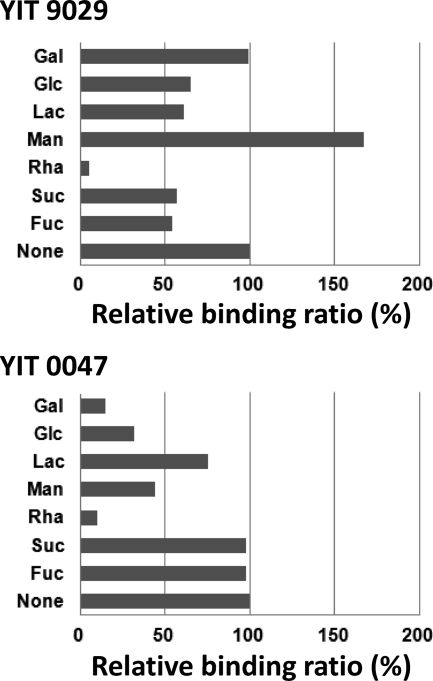
Next, a competitive inhibition assay was performed using monosaccharides and disaccharides as competitors (Gal, Glc, Man, Rha, Suc, Fuc, and Lac) to measure the binding specificity of YIT 9029 and YIT 0047 (NIRD A-121) to CSL. The binding ratios of YIT 9029 and YIT 0047 are shown in Fig. 4. Rha completely inhibited the binding of YIT 9029 and YIT 0047 to CSL, indicating that the binding of both strains to CSL was rhamnose specific. Although the binding of YIT 0047 to CSL was partially inhibited by Gal and Glc, these sugars did not affect the binding of YIT 9029 to CSL, suggesting that Gal and Glc moieties in the cell surface structure of YIT 0047 but not YIT 9029 are present near the binding site of CSL. Hence, the liquid-phase lectin microarray technique is sensitive enough to distinguish similarities and differences in the glycome of bacterial cell surfaces between strains, even in the same species, and is suitable in the identification of individual bacterial strains. At the same time, this technique is very simple and reproducible for characterizing cell wall structures.
Fig. 4.
Carbohydrate inhibition assay. Relative binding ratios of Lactobacillus casei YIT 9029 and YIT 0047 are shown. Gal, Glc, Lac, Man, Rha, Suc, and Fuc were added at 0.1 μM to 2 × 109 YIT 9029 or YIT 0047 cells labeled with SYTOX Orange. None, control assay, without sugars. Data were analyzed as described in Materials and Methods. Rha competes to inhibit CSL in both strains. The other sugars were not as effective in inhibiting CSL.
Profiling 16 L. casei strains.
We examined 16 L. casei strains which are indistinguishable from each other by 16S rRNA sequences using the lectin microarray technique. As shown in Fig. 5, each strain has a unique profile of binding to various lectins, except for YIT 0001, YIT 0091, and YIT 9029 (Fig. 5). YIT 0091 is a clone of YIT 9029 personally stored by a researcher at the Yakult Central Institute for Microbiological Research a long time ago and is thus considered a clone of YIT 9029 (M. Onoue [YIT 0091] and T. Sakurai [YIT 0001], personal communication). YIT 0001 also had the same binding profile as YIT 9029 and will be discussed later.
Fig. 5.
Relative binding of 16 Lactobacillus casei strains with respect to lectin binding. The lectin-binding signals for each strain were normalized with the highest signal. Unsupervised hierarchical clusters were generated for the L. casei strains, and their glycan profiles were obtained by lectin microarray. The levels of lectin-binding signals are indicated by the color change from green (low binding levels) to red (high binding levels). The red box and blue box indicate the lectins that bound to more than five L. casei strains and that did not bind to any L. casei strain, respectively.
Among 16 L. casei/L. paracasei strains, two types of lectin-binding characteristics could be recognized; one group has few lectin responders which bind to only one or two different lectins, and the other group has multiple lectin responders which bind to multiple lectins with different specificities. YIT 0001, YIT 0091, and YIT 9029 bound only to CSL, YIT 0009 bound only to ABA, YIT 0123 bound only to jacalin, and YIT 0226 bound only to BPL and SBA. Similar to YIT 9029, these strains do not necessarily have PSs with simple sugar composition but may contain various sugar compositions. Alternately, there are strains which bind to a number of lectins. For instance, YIT 0003 binds to 23 lectins, YIT 0006 binds to 24 lectins, and YIT 0290 binds to 27 lectins. From all the binding profiles, we could not draw a specific binding profile for L. casei/L. paracasei species but, rather, recognized profiles specific for individual strains. None of the strains bound ACA and AOL, which are specific for Gal-β1-3GalNAc-Thr/Ser and Fuc, respectively.
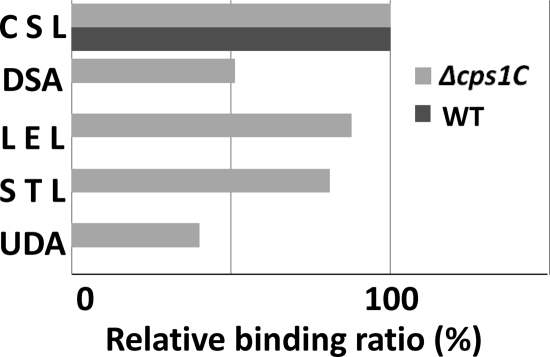
Next, we analyzed the lectin-binding profile of a mutant of YIT 9029 as a typical example of a strain with few lectin binders. As described above, the cell wall PS of YIT 9029 contains several sugar molecules, including Glc, Gal, Rha, GlcNAc, and GalNAc, with different linkages. The mutant of YIT 9029 with the Δcps1C mutation was constructed by site-specific deletion mutagenesis within the cps1C gene encoding a putative glycosyltransferase, which is essential to synthesize PS-1 and most probably lacks a certain glycosyltransferase activity (43). The glycan profile of the strain with the Δcps1C mutation was clearly different from that of YIT 9029: in addition to binding to CSL, the cells of the strain with the Δcps1C mutation bound to an asialo complex-type N-glycan binder, DSA, and chitin binders LEL, STL, and UDA (Fig. 6). Interestingly all these lectins have the binding specificity of the N-acetylglucosamine polymer. No other lectins bound to the mutant. Therefore, the deficiency of the cps1C gene resulted in a partial release of PS structures consisting of at least one GlcNAc moiety in the cell wall PS, in addition to the disappearance of the high-molecular-mass PS-1 (43).
Fig. 6.
Differences in lectin-binding affinity between Lactobacillus casei YIT 9029 and its mutant with the Δcps1C mutation, which does not synthesize high-molecular-mass polysaccharide. Data are shown as the ratio of fluorescence intensities of 5 lectins relative to that of CSL, which is the maximal fluorescence intensity.
DISCUSSION
In this study, we have developed a simple and efficient method to profile the bacterial glycome using liquid-phase lectin microarray. We have shown that it is possible to distinguish between strains classified into the Lactobacillus casei/L. paracasei group and identify similarities and differences in the lectin-binding profiles between strains. There are several key processes in this analytical method. First, bacterial cells are pretreated with a 70% ethanol solution for 30 min before labeling. This keeps the integrity of the cell shape and enables the SYTOX Orange dye to penetrate into the bacterial cytosol (Fig. 2C). Second, 10 μM SYTOX Orange was used to label bacterial cells. The concentration of SYTOX Orange dye used to label the bacterial cells was 100-fold higher than that recommended by the supplier (Fig. 2B), but it was the most appropriate concentration in this analysis. The specific application of SYTOX Orange in the analysis of the bacterial glycome differs from the analysis of mammalian cells (37) or glycoproteins (20), for which Cell Tracker and Cy3, respectively, are generally used. An additional advantage of SYTOX Orange is that the dye-stained bacterial cells can be stored for approximately 2 days without a detectable loss of intensity. Lastly, the number of cells used in the binding reaction on the lectin microarray is critical for reproducible and identifiable results. The number of bacterial cells is preferably between 2 × 109 and 4 × 109 cells/well (Fig. 2D). Under these simple conditions, we could successfully analyze the lectin-binding profile of bacterial cells of L. casei/L. paracasei group strains. Since the binding reaction should occur on the outer surface of the cells and since the treatment of bacterial cells in 70% ethanol solution for 30 min is rather mild, we suggest that this technique is also widely applicable to other bacterial genera and species, although it may be necessary to optimize the concentration of SYTOX Orange for individual strains.
During the analysis of L. casei/L. paracasei group strains using the technology developed in this study, we determined some remarkable characteristics of these strains. Although all of the strains are classified into the same species group, L. casei/L. paracasei, the individual strains have very unique lectin-binding profiles. YIT 0091 and YIT 9029 had the same binding profile (Fig. 5). They are known to originate from the same ancestor (M. Onoue, personal communication), and therefore, it could be anticipated that they have the same binding profile. Since YIT 0001 also showed the same binding profile as YIT 0091 and YIT 9029, it is suggested that this strain also shares the same ancestor. Diversity in the outer surface PS structures is often observed among streptococci, including the pathogenic species and other genera. It is believed that there is an active mechanism that enables these strains to frequently alter their outer surface antigenic properties to escape from the host immune mechanism. Numerous reports have shown cell wall structural and genetic diversities among the Lactobacillus species. We confirm this in the L. casei/L. paracasei group strains, using the lectin microarray. There may be a possibility that glycosylated components other than cell wall polysaccharides contribute to the binding of cells to lectins. However, we do not have clear evidence that L. casei/L. paracasei group strains have glycosylated proteins or (lipo)teichoic acids on the cell surface. It is difficult to speculate about the mechanism and frequency of such surface alteration; there may be hot spots of gene conversion or gene exchange in the genomes of these bacteria.
Though none of the lectins binds to every strain used in this study, there are some lectins which bind to a number of L. casei/L. paracasei group strains. Those are CSL (Rha binder), BPL (Gal binder), LEL (chitin binder), ECA, RCA120, and DSA (asialo complex-type N-glycans binders), TJA-II, EEL, and ABA (Gal-type N-glycan binders), and jacalin, WFA, VVA, SBA, GSL-I, and PTL-I (O-glycan binders). In contrast, few strains bound to high mannose-type N-glycan binders (NPA, concanavalin A [ConA], GNA, and HHL) and sialic acid (Sia) binders (MAL, SNA, SSA, and TJA-I); furthermore, none of the strains bound ACA (Gal-β1-3GalNAc-Thr/Ser binder) or AOL (Fuc binder). This suggests that the surface sugar moieties specific for these binders are generally absent among strains of the L. casei/L. paracasei group.
The lectin-binding patterns of lactobacilli were previously reported using 6 different lectins (3). Interestingly, none of the Lactobacillus acidophilus strains bound to any lectins because of autoagglutination. In fact, some strains in this study, for instance, YIT 0005 and YIT 0180T, were found to autoagglutinate in MRS medium; however, agglutination itself did not affect the binding to lectins (Fig. 3). These data may imply that the cell surface glycomes between L. casei/L. paracasei and L. acidophilus would be largely different from each other.
The lectin-binding profiles do not always reflect the sugar composition of the bacterial cell wall PSs. In some strains, lectin binding is limited, while in other strains many different types of lectins bind to the cells. We speculate that limited binding is caused by physical hindrance of the binding sites for certain lectins. For instance, YIT 9029 binds only to a Rha-specific lectin CSL in this assay, although the PS of YIT 9029 consists of several sugar components (28; this study). The fact that the binding of CSL to YIT 9029 is inhibited by Rha but not by other sugars (Fig. 4) clearly indicates that this binding occurs in a Rha-specific manner. In one mutant of YIT 9029 with the Δcps1C mutation, which has a defect in a certain sugar transferase gene, we suggest that a high-molecular-mass polysaccharide (PS-1) structure is missing from the cell surface (43). This cps1C mutation has led the mutant to bind multiple lectins (Fig. 6). It is too early to speculate whether PS-1 contains the binding site for CSL and/or whether it would hinder shorter PSs that potentially have the binding sites for CSL and other lectins. Differences in the patterns of inhibition against the binding of different L. casei strains to CSL by various carbohydrates have been indicated. These differences may reflect the structural characteristics of the CSL binding site in these strains. Further biochemical and molecular genetic analyses are required in order to clarify the exact structural features of the cell wall PSs.
The results obtained in this study show the actual interaction between lectins and microbial cells, which is probably via surface PSs. The lectins used in this assay are mostly derived from various plants, except for AOL, which was isolated from Aspergillus oryzae; CSL, which was isolated from salmon eggs (34, 40); and HPA, which was isolated from Helix pomatia; some lectins were isolated from fungi. Recent advances in mammalian cell biology reveal that various lectin-like proteins are expressed on mammalian cell surfaces and may play pivotal roles in cell signaling. In addition, some plant lectins are known to mimic mammalian lectins, but they could easily be applied to the technology described here.
During the host-bacterium interaction in the gut, probiotic bacteria may send their signals through various methods and factors. The data presented in this study clearly show that bacterial PSs can actively interact with lectins. Further analysis on the interaction between bacterial PSs and host cells will enable us to identify novel factors involved in this response.
In conclusion, we have developed a liquid-phase lectin microarray analysis technique to profile the bacterial glycome. The binding profile probably reflects, in part, the content and structure of cell wall PSs. It is a simple and high-throughput system which enables us to distinguish and identify individual bacterial strains within the same species without performing complicated carbohydrate composition and structure analyses. In addition, the interactions between lectins and bacterial cells suggest the presence and importance of this type of interaction in the probiotic actions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoshiko Kubo and Jinko Murakami of the Research Center for Medical Glycoscience at the National Institute of Advanced Industrial Science and Technology for help in preparation of the lectin microarray, Toshihiko Takada of the Yakult Central Institute for Microbiological Research for help with bacterial labeling methods and the electron microscopic photos, and Koich Watanabe for advice on choosing Lactobacillus casei strains. We are deeply indebted to Teruo Yokokura and the late Toshiaki Osawa, who both always encouraged us and gave helpful discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Akira S., Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4: 499–511 [DOI] [PubMed] [Google Scholar]
- 2. Angeloni S., et al. 2005. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology 15: 31–41 [DOI] [PubMed] [Google Scholar]
- 3. Annuk H., Hynes S. O., Hirmo S., Mikelsaar M., Wadström T. 2001. Characterisation and differentiation of lactobacilli by lectin typing. J. Med. Microbiol. 50: 1069–1074 [DOI] [PubMed] [Google Scholar]
- 4. Baik Y. S., Cheong W. J. 2007. Development of SPE for recovery of polysaccharides and its application to the determination of monosaccharide composition of the polysaccharide sample of a lactobacillus KLB 58. J. Sep. Sci. 30: 1509–1515 [DOI] [PubMed] [Google Scholar]
- 5. Bentley S. D., et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cieslewicz M. J., et al. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73: 3096–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cobb B. A., Wang Q., Tzianabos A. O., Kasper D. L. 2004. Polysaccharide processing and presentation by the MHCII pathway. Cell 117: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coyne M. J., et al. 2001. Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 69: 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis G. H. 1955. The classification of lactobacilli from the human mouth. J. Gen. Microbiol. 13: 481–493 [DOI] [PubMed] [Google Scholar]
- 10. Dicks L. M., Plessis E. M., Dellaglio F., Lauer E. 1996. Reclassification of Lactobacillus casei subsp. casei ATCC 393 and Lactobacillus rhamnosus ATCC 15820 as Lactobacillus zeae nom. rev., designation of ATCC 334 as the neotype of L. casei subsp. casei, and rejection of the name Lactobacillus paracasei. Int. J. Syst. Bacteriol. 46: 337–340 [DOI] [PubMed] [Google Scholar]
- 11. Fang Y., et al. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132: 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng Y., Xiao-Min W. 2007. Difference in gene expression of macrophage between normal spleen and portal hypertensive spleen indentified by cDNA microarray. World J. Gastroenterol. 13: 3369–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heng H. H., Tsui L. C. 1993. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 102: 325–332 [DOI] [PubMed] [Google Scholar]
- 14. Hsu K.-L., Mahal L. K. 2006. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat. Protoc. 1: 543–549 [DOI] [PubMed] [Google Scholar]
- 15. Hsu K.-L., Pilobello K. T., Mahal L. K. 2006. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2: 153–157 [DOI] [PubMed] [Google Scholar]
- 16. Hsu K.-L., Gildersleeve J. C., Mahal L. K. 2008. A simple strategy for the creation of a recombinant lectin microarray. Mol. Biosyst. 4: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inohara N., Nuñes G. 2003. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 3: 371–382 [DOI] [PubMed] [Google Scholar]
- 18. Kankainen M., et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106: 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kullberg M. C. 2008. Soothing intestinal sugars. Nature 453: 602–604 [DOI] [PubMed] [Google Scholar]
- 20. Kuno A., et al. 2005. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat. Methods 2: 851–856 [DOI] [PubMed] [Google Scholar]
- 21. Lebeer S., Vanderleyden J., De Keersmaecker S. C. J. 2008. Genes and molecules of Lactobacillus supporting probiotic action. Microbiol. Mol. Biol. Rev. 72: 728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C. H., Lee S. M., VanLare J. M., Kasper D. L., Mazmanian S. K. 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. U. S. A. 105: 3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mancuso G., Tomasello F., von Hunolstein C., Orefici G., Teti G. 1994. Induction of tumor necrosis factor alpha by the group- and type-specific polysaccharides from III group B streptococci. Infect. Immun. 62: 2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto S., et al. 2005. Probiotic Lactobacillus-induced improvement in murine chronic inflammatory bowel diseases is associated with the down-regulation of proinflammatory cytokines in lamina propria mononuclear cells. Clin. Exp. Immunol. 140: 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsumoto S., et al. 2008. A component of polysaccharide peptidoglycan complex on Lactobacillus induced an improvement of murine model of inflammatory bowel disease and colitis-associated cancer. Immunology 128: e170–e180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuzaki T., et al. 1997. Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS 105: 643–649 [DOI] [PubMed] [Google Scholar]
- 27. Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118 [DOI] [PubMed] [Google Scholar]
- 28. Nagaoka M., et al. 1990. Structure of polysaccharide-peptidoglycan complex from the cell wall of Lactobacillus casei YIT 9018. J. Biochem. 108: 568–571 [DOI] [PubMed] [Google Scholar]
- 29. Nitta K., Kawano T., Sugawara S., Hosono M. 2007. Regulation of globotriaosylceramide (Gb3)-mediated signal transduction by Rha-binding lectin. Yakugaku Zasshi 127: 553–561 [DOI] [PubMed] [Google Scholar]
- 30. Pretzer G., et al. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187: 6128–6136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schweizer D. 1981. Counterstain-enhanced chromosome banding. Hum. Genet. 57: 1–14 [PubMed] [Google Scholar]
- 32. Shida K., Kiyoshima-Shibata J., Nagaoka M., Watanabe K., Nanno M. 2006. Induction of interleukin-12 by Lactobacillus strains having a rigid cell wall resistant to intracellular digestion. J. Dairy Sci. 89: 3306–3317 [DOI] [PubMed] [Google Scholar]
- 33. Shimizu-Kadota, et al. 2000. Insertion of bacteriophage phiFSW into the chromosome of Lactobacillus casei Shirota (S-1): characterization of attachment sites and integrase gene. Gene 249: 127–134 [DOI] [PubMed] [Google Scholar]
- 34. Shirai T., Watanabe Y., Lee M., Ogawa T., Muramoto K. 2009. Structure of Rha-binding Lectin CSL3: Unique pseudo-tetrameric architecture of a pattern recognition protein. J. Mol. Biol. 391: 390–403 [DOI] [PubMed] [Google Scholar]
- 35. Šimelyte E., Rimpiläinen M., Lehtonen L., Zhang X., Toivanen P. 2000. Bacterial cell wall-induced arthritis: chemical composition and tissue distribution of four Lactobacillus strains. Infect. Immun. 68: 3535–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takada T., Matsumoto K., Nomoto K. 2004. Development of multi-color FISH method for analysis of seven Bifidobacterium species in human feces. J. Microbiol. Methods 58: 413–421 [DOI] [PubMed] [Google Scholar]
- 37. Tateno H., et al. 2007. A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology 17: 1138–1146 [DOI] [PubMed] [Google Scholar]
- 38. Uchiyama N., et al. 2006. Development of a lectin microarray based on an evanescent-field fluorescence principle: a new strategy for glycan profiling. Methods Enzymol. 415: 341–351 [DOI] [PubMed] [Google Scholar]
- 39. Vaningelgem F., et al. 2004. Biodiversity of exopolysaccharides produced by Streptococcus thermophilus strains is reflected in their production and their molecular and functional characteristics. Appl. Environ. Microbiol. 70: 900–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Watanabe Y., et al. 2009. The function of Rha-binding lectin in innate immunity by restricted binding Gb3. Dev. Comp. Immunol. 33: 187–197 [DOI] [PubMed] [Google Scholar]
- 41. Yan X., et al. 2000. Development of a mechanism-based, DNA staining protocol using SYTOX Orange nucleic acid stain and DNA fragment sizing flow cytometry. Anal. Biochem. 286: 138–148 [DOI] [PubMed] [Google Scholar]
- 42. Yan X., et al. 2005. Probing the kinetics of SYTOX Orange stain binding to double-stranded DNA with implications for DNA analysis. Anal. Chem. 77: 3554–3562 [DOI] [PubMed] [Google Scholar]
- 43. Yasuda E., Serata M., Sako T. 2008. Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides. Appl. Environ. Microbiol. 74: 4746–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yasuno S., Kokubo K., Kamei M. 1999. New method for determining the sugar composition of glycoproteins, glycolipids, and oligosaccharides by high-performance liquid chromatography. Biosci. Biotechnol. Biochem. 63: 1353–1359 [DOI] [PubMed] [Google Scholar]
- 45. Yokokura T., Kodaira S., Ishiwa H., Sakurai T. 1974. Lysogeny in lactobacilli. J. Gen. Microbiol. 84: 277–284 [DOI] [PubMed] [Google Scholar]
- 46. Yuki N., et al. 1999. Survival of a probiotic, Lactobacillus casei strain Shirota, in the gastrointestinal tract: selective isolation from feces and identification using monoclonal antibodies. Int. J. Food Microbiol. 48: 51–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.