Abstract
Nonsense-mediated mRNA decay (NMD) is a conserved surveillance mechanism that eliminates imperfect mRNAs that contain premature translation termination codons (PTCs) and code for nonfunctional or potentially harmful polypeptides. We show that a novel phosphatidylinositol 3-kinase-related protein kinase, hSMG-1, is a human ortholog of a product of Caenorhabditis elegans smg-1, one of seven smg genes involved in NMD. hSMG-1 phosphorylates hUPF1/SMG-2 in vivo and in vitro at specific serine residues in SQ motifs. hSMG-1 can associate with hUPF1/SMG-2 and other components of the surveillance complex. In particular, overexpression of a kinase-deficient point mutant of hSMG-1, hSMG-1-DA, results in a marked suppression of the PTC-dependent β-globin mRNA degradation; whereas that of wild-type hSMG-1 enhances it. We also show that inhibitors of hSMG-1 induce the accumulation of truncated p53 proteins in human cancer cell lines with p53 PTC mutation. Taken together, we conclude that hSMG-1 plays a critical role in NMD through the direct phosphorylation of hUPF1/SMG-2 in the evolutionally conserved mRNA surveillance complex.
Keywords: NMD, PIK-related-protein kinase, smg, upf, PTC, p53
Gene mutation or a failure in mRNA processing can generate imperfect mRNAs containing premature translation termination codons (PTCs) and coding for nonfunctional or potentially harmful truncated polypeptides. Studies on genetic disorders in yeast, Caenorhabditis elegans, and humans have revealed a conserved surveillance mechanism that eliminates aberrant mRNAs containing PTCs. This has been termed nonsense-mediated mRNA decay (NMD) or RNA surveillance (for reviews, see Jacobson and Peltz 1996; Li and Wilkinson 1998; Culbertson 1999; Frischmeyer and Dietz 1999; Hentze and Kulozik 1999; Hilleren and Parker 1999; Maquat and Carmichael 2001).
NMD is a modifier of the phenotypic consequence of gene mutations. One clear example is β-thalassemia. Most mutations in this disease lead to the generation of a PTC in the first two exons of the β-globin gene and are recessive. In individuals carrying only one affected allele, the mutant β-globin mRNA is absent or its level is very low due to NMD, resulting in a clinically asymptomatic phenotype. A subset of individuals heterozygous for β-globin mutations possesses a PTC in the third (last) exon of the β-globin gene. In these cases, the mutated β-globin mRNA is decay-resistant, its levels are not decreased, and it produces truncated dominant-negative β-globin chains, resulting in characteristic inclusions and a clinical phenotype in the heterozygote (Thein et al. 1990; Hall and Thein 1994). In addition to being a modifier of gene mutation, NMD is of particular importance in the physiology of lymphocytes, in which immunoglobulin and T-cell receptor genes undergo somatic recombination and mutation to generate versatility in immune response. This process generates abundant by-products, truncated open reading frames (ORFs), but the resulting mRNAs with PTCs are strongly down-regulated (Carter et al. 1995; Li and Wilkinson 1998).
In mammals, a termination codon is recognized as premature if located upstream of an exon–exon junction and with a distance of at least 50 nucleotides (Carter et al. 1996; Thermann et al. 1998; Zhang et al. 1998a,b). Consistently, introns are rare in the 3′ UTR of mammalian genes. They locate nearly always within 50 nucleotides at the end of the ORF, ensuring a normal termination codon from being not recognized as premature (Nagy and Maquat 1998). Moreover, both general and mRNA-specific inhibitions of translation suppress NMD (Carter et al. 1995; Thermann et al. 1998). However, the molecular processes of NMD have just started to be revealed.
Studies on the molecular processes of NMD have been largely based on yeast genetics. An mRNA containing a PTC is subjected to deadenylation-independent decapping by Dcp1p, followed by 5′ to 3′ decay by exonuclease Xrn1p (Beelman et al. 1996). Recognition of a termination codon as premature requires a cis-acting element, the downstream sequence element (DSE; Peltz et al. 1993; Hagan et al. 1995; Zhang et al. 1995), and Hrp1p interacts with DSE to provide a mark for PTC (Gonzalez et al. 2000). Three additional trans-acting factors are involved in NMD in yeast: Upf1p, Upf2p, and Upf3p.
Genetic studies on C. elegans have also provided the molecular aspects of NMD in higher eukaryotes. In C. elegans, seven smg genes are involved in NMD (Hodgkin et al. 1989; Pulak and Anderson 1993; Cali et al. 1999). In smg mutants, nonsense mutations that are normally recessive often confer a dominant-negative phenotype. For example, when nonsense mutations in unc-54 (myosin heavy chain) are placed in an smg− background, muscle assembly is disturbed by the excess accumulation of truncated unc-54 proteins, resulting in a paralyzed phenotype (Cali and Anderson 1998). Some of the seven smg genes have been cloned and the encoded proteins identified. In particular, SMG-2 is a homolog of Upf1p (Page et al. 1999 ).
In mammals, human homologs of the three UPF proteins, hUPF1/SMG-2, hUPF2, and hUPF3s, have been identified and characterized (Sun et al. 1998; Bhattacharya et al. 2000; Lykke-Anderson et al. 2000; Mendell et al. 2000; Serin et al. 2001). The RNA helicase activity of hUPF1/SMG-2 is essential for NMD (Sun et al. 1998). Furthermore, hUPF1/SMG-2, hUPF2, and hUPF3, when bound downstream of a termination codon, have been shown to target wild-type β-globin mRNAs to cause NMD (Lykke-Anderson et al. 2000). These results suggest a parallel mechanism of NMD in yeast and mammals.
However, there is a clear difference between yeast, C. elegans and probably mammals. Recent studies on C. elegans have identified essential components of NMD that were not identified in yeast genetics, including smg-1, which encodes a phosphatidylinositol 3-kinase (PIK)-related protein kinase (PIKK; P. Anderson, pers. comm.). Taken together with the fact that SMG-2 is a phosphoprotein and that the other six smg genes can be classified into two groups based on their effect on the phosphorylation of SMG-2 (Page et al. 1999), the identification of SMG-1 as a putative protein kinase raises the possibility that phosphorylation plays a critical role in NMD in C. elegans. In particular, the homolog of SMG-1 is not encoded in the yeast genome (P. Anderson, pers. comm.), suggesting a molecular aspect of the difference between yeast and C. elegans NMD. However, the biochemical and functional importance of protein phosphorylation by SMG-1 remains to be established. Furthermore, the involvement of protein phosphorylation in mammalian NMD and the existence of a mammalian counterpart of SMG-1 remain to be clarified.
PIKKs belong to a unique family of serine/threonine kinases that are conserved from yeast to mammals. In mammals, there are four PIKKs: FRAP (FKBP12-rapamycin-associated protein kinase, also called mTOR/RAFT/RAPT), DNA-PK (DNA-dependent protein kinase), ATM (ataxia telangiectasia mutated), and ATR (ATM and Rad3-related, also called FRP1), all of which are distinguished by their large size and C-terminal protein kinase domain related to those of lipid kinases. FRAP controls cell growth in response to nutrients and growth factors through the regulation of translation and transcription (Thomas and Hall 1997; Hara et al. 1998; Schmelzle and Hall 2000). DNA-PK, ATM, and ATR are considered to participate in response to nuclear cues that activate DNA rearrangement, cell cycle arrest, or apoptosis (Hoekstra 1997; Shiloh 2001).
Here, we have identified cDNAs encoding a novel PIKK member and termed it human SMG-1 based on its overall similarity to C. elegans SMG-1. We show that hSMG-1 can associate with components of the evolutionally conserved mRNA surveillance complex containing hUPF1/SMG-2, hUPF2, and hUPF3, phosphorylates serine residues in SQ motifs of hUPF1/SMG-2 and is critically involved in NMD. These results not only provide direct evidence for the importance of the phosphorylation of the critical component of the surveillance complex but also reveal a novel aspect of the regulatory mechanism of gene expression by NMD through protein phosphorylation.
Results
Identification of cDNA clones encoding a novel protein kinase of the PIKK family
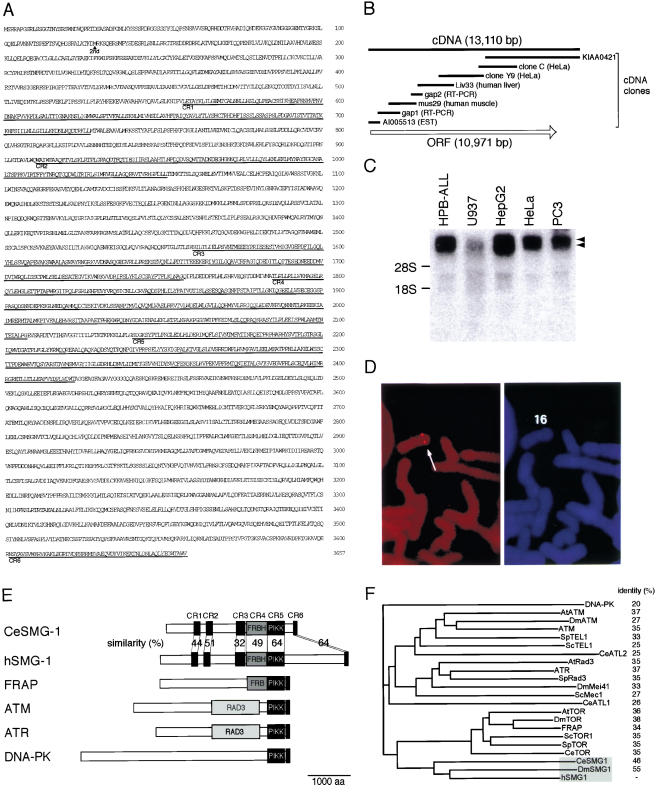
We found that the C-terminal 56-amino-acid sequence encoded by a cDNA clone, KIAA0421, shows close similarity to the C-terminal sequence of the members of the PIKK family (see Fig. 2B, below). Successive backscreenings finally enabled us to isolate overlapping cDNA clones that span 13,110 bp and encode an ORF with 3657 amino acid residues of 409,800 daltons (Fig. 1A,B). Northern blot analysis showed that the corresponding mRNA is more than 10 kb long and is expressed in various human cell lines (Fig. 1C). In situ hybridization revealed that the gene for hSMG-1 locates on 16p12 in human chromosomes (Fig. 1D).
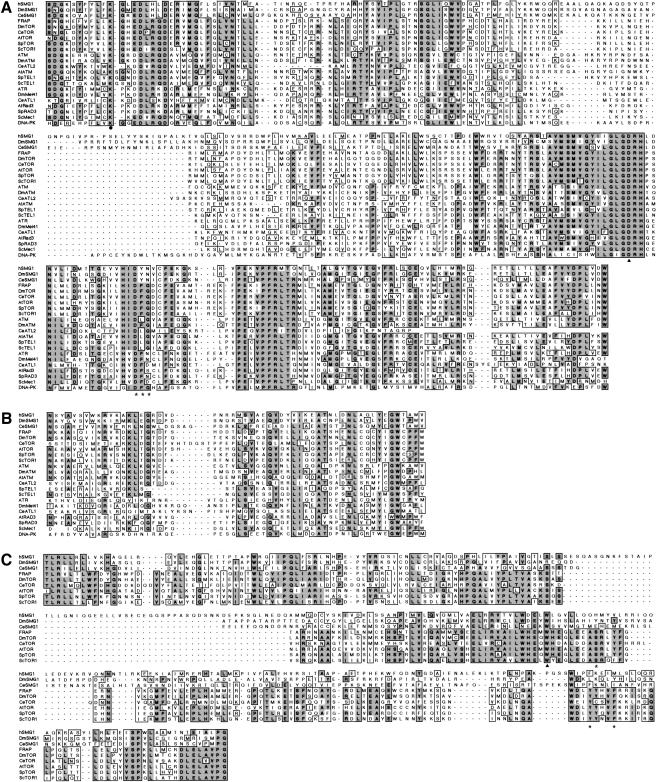
Figure 2.
Sequence comparison of human SMG-1 with other PIK-related protein kinase family members. (A) Alignment of catalytic core domains of PIKKs. Completely and partially conserved amino acids are boxed in dark and light gray, respectively. The amino acid sequence, DFG (asterisks), seen in several members of the PIKK family, is highly conserved between kinases. The solid triangle shows the aspartic acid residue completely conserved in essentially all protein kinases. (B) Alignment of the C-terminal homology region of PIKK. (C) Alignment of the FRBH domain. The tryptophan residue (solid triangle) shows a conserved residue essential for both kinase activity and binding to the FKBP12–rapamycin complex. The asterisks show residues essential for binding to the FKBP12–rapamycin complex.
Figure 1.
Structure of hSMG-1. (A) The predicted translation product of the human SMG-1 (hSMG-1) cDNA clones is shown. CR1–CR6 show regions conserved between CeSMG-1 and hSMG-1. (B) Schematic representation of the hSMG-1 ORF (open arrow) and cDNA clones used to analyze the complete structure. (C) Northern blotting analysis of hSMG-1 mRNA using total RNA from the indicated human cell lines. Arrowheads show the positions of two major signals. Positions for the 18S and 28S ribosomal RNAs are also shown. (D) FISH mapping of hSMG-1. Human mitotic chromosomes with the FISH signal (left panel). The arrow shows the position of the specific signal. In the right panel, the PI (propidium iodide) staining pattern of the same viewing field is shown to observe chromosome morphology. (E) Schematic representation of the members of mammalian PIK-related kinase family in comparison with CeSMG-1. The putative PIKK domain is shown as a dark gray box. The FRBH region is shown as medium gray and the RAD3 homology region is shown as light gray. GenBank accession nos. are as follows: hSMG-1,AB061371; FRAP, L34075; ATM, U33841; ATR, U76308; DNA-PK, U34994. (F) Molecular phylogenetic tree for putative catalytic domains of human PIKK family members.
The amino acid sequence of the ORF shows the closest similarity to that of the C. elegans SMG-1 (CeSMG-1; P. Anderson, pers. comm.) in the overall sequence. The conserved sequences, from CR1 to CR6, are indicated in Figure 1A. Thus, we named the encoded protein human SMG-1 (hSMG-1). hSMG-1, as well as CeSMG-1, shows similarity to the members of the PIKK family in terms of their kinase domain (Figs. 1E and 2A,B). As described above, the putative kinase domain of hSMG-1, as well as that of C. elegans, is separated into two parts by an insert, distinguishing them from other PIKK family members. Interestingly, the CR4 sequence shows similarity to the FKBP12-rapamycin-binding (FRB) region of FRAP (Fig. 2C).
Identification of hSMG-1 isoforms and their intrinsic protein kinase activity
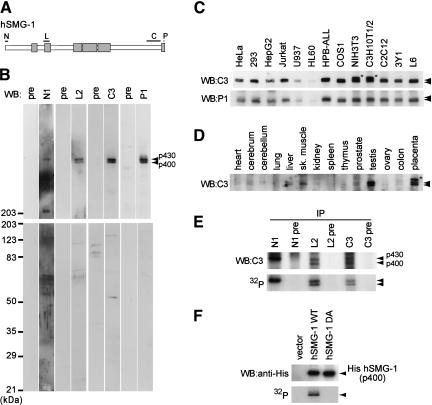
A series of antibodies against four independent regions of the protein, N, L, C, and P, reveal that these antibodies recognize a protein of ∼430 kD (p430) in HeLa cells (Fig. 3A,B). In addition, antibodies L2, C3, and P1 also recognize a protein of ∼400 kD (p400). The nucleotide sequence of the hSMG-1 cDNA reveals that there is a codon, Met129, that fulfills Kozak's criteria of translation initiation. The putative ORF that starts from Met129 encodes a 3529-amino-acid protein of 396,040 daltons. Taken together, p400 is most likely the product of the ORF that starts at the second Met at position 129. The two hSMG-1 proteins, p430 and p400, are expressed in various cell lines of human, monkey, rat, and mouse (Fig. 3C). Moreover, they are expressed in various rat tissues and mouse placenta (Fig. 3D).
Figure 3.
Identification of human SMG-1 and its intrinsic protein kinase activity. (A) The schematic structure of hSMG-1 is shown with the regions (N, L, C, and P) used for antibody generation. CR1–CR6 are shown in boxes. (B) The HeLa cell lysate was analyzed by Western blotting using antisera shown on top and the corresponding pre-immune sera. Two bands, corresponding to 400- and 430-kD proteins (p400 and p430), are indicated by arrowheads. (C) hSMG-1 expression in various cell lines from human, simian, mouse, and rat was examined using C3 or P1 antiserum. The uppermost bands in two cell lines from mouse, NIH3T3 and C3H10T1/2, p460 (asterisk) were not stained with P1 and appeared to be a nonspecific band. (D) hSMG-1 expression in various tissues, mainly from rat, was examined with C3 antiserum. Only the placenta was from mouse, and the asterisk indicates a nonspecific band. (E) hSMG-1 expressed in HeLa cells has intrinsic autophosphorylation activity. The HeLa cell lysate was subjected to immunoprecipitation using N1, L2, or C3 antiserum or their corresponding pre-immune serum. Immunoprecipitated proteins were analyzed by Western blotting using C3 antiserum (top). In vitro kinase reaction with the immuno complex was performed without using any exogenous substrate. This was followed by 5.5% SDS-PAGE and autoradiography. (F) His-tagged hSMG-1 overexpressed in 293T cells has intrinsic autophosphorylation activity. 293T cells were transfected with SR6H (vector), SR6H hSMG-1, or SR6H hSMG-1-DA. Transfected cells were harvested and lysed, followed by immunoprecipitation using an anti-His-tag antibody and in vitro kinase assay. The autoradiogram is shown (bottom). Western blotting results of the immunoprecipitated His-tagged hSMG-1s are also shown (top).
p430 could be immunoprecipitated from HeLa cell lysates using three independent antibodies, N1, L2, and C3, and p400 using the antibody L2 or C3 (Fig. 3E), in agreement with the results of Western blot analysis. When these immunoprecipitates were incubated with ATP in a kinase reaction mixture, both p430 and p400 were phosphorylated (Fig. 3E), supporting the notion that p430 and p400 recognized by anti-hSMG-1 antibodies are protein kinases.
To further confirm the above finding, we constructed a cDNA expression vector encoding the putative ORF that starts from codon 107 downstream of the His-epitope tag resulting in an ORF encoding a 399,733 dalton protein, as well as a point mutant, hSMG-1-DA, in which codon 2331D is replaced with A. The corresponding aspartic acid in FRAP had been shown to be required for its kinase activity (Brown et al. 1995). Figure 3F shows that His-tagged recombinant hSMG-1 immunoprecipitated using an anti-His antibody from cDNA-transfected 293T cells migrates to ∼400 kD and exhibits protein kinase activity; whereas the hSMG-1-DA mutant does not. These results suggest that p400 and p430 are isoforms of hSMG-1 that possess an intrinsic autophosphorylation activity.
hSMG-1 protein kinase is involved in PTC-dependent degradation of β-globin mRNA
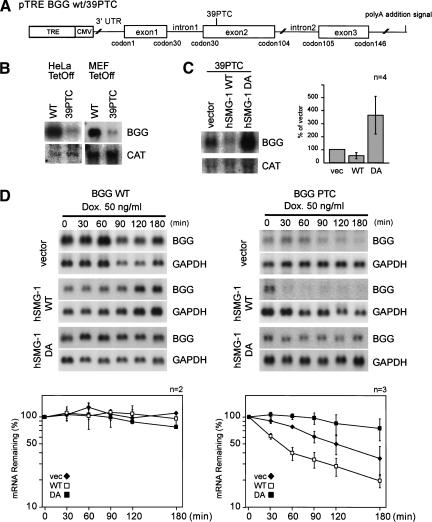
In C. elegans, seven smg genes are involved in NMD. The unexpected finding that a novel member of the PIKK family shows an overall sequence similarity to CeSMG-1 leads us to examine whether hSMG-1 is involved in mammalian NMD. For this purpose, we constructed a reporter gene, in which the genomic sequence with or without PTC at codon 39 of human β-globin (BGG) is placed downstream of the CMV promoter (Fig. 4A).
Figure 4.
SMG-1 is involved in the PTC-dependent degradation of β-globin mRNA. (A) Schematic representation of human β-globin (BGG) gene reporter constructs, BGG-WT and BGG-PTC. The ORF is represented by boxes, and introns and UTRs are represented by lines. (B) The BGG PTC transcript is less abundant than transcripts from the WT reporter. The WT or mutant reporter plasmid was transfected with the CAT plasmid into the HeLa Tet-Off or MEF Tet-Off cell lines. The same amount of total RNA from each cell was analyzed by Northern blotting using a BGG or a CAT probe. (C) Overexpression of hSMG-1 suppresses the accumulation of the BGG 39PTC transcript; whereas that of hSMG-1-DA enhances it. HeLa Tet-Off was transfected with the BGG WT reporter or BGG 39PTC (1.5 μg) with the hSMG-1-WT or hSMG-1-DA (3 μg) and a CAT construct (1.5 μg). Then the accumulated BGG transcripts were evaluated by Northern blotting using CAT as an internal control. The relative amount of transcripts is shown in the bar graph. (D) Overexpression of hSMG-1-WT enhances the decay of the BGG 39PTC transcript; whereas that of hSMG-1-DA suppresses it. The HeLa Tet-Off cell was transfected with the WT (left) or mutant (right) reporter gene, and cells were harvested at an indicated time point after the addition of 50 ng/mL doxycycline to the medium. Total RNA was isolated and analyzed by Northern blotting using the corresponding probe. The relative amount of transcripts is shown in the graph. The BGG RNA levels are normalized to those of GAPDH.
In this construct, the CMV promoter is regulated by the upstream TRE (tetracycline-responsive element) sequence, and when introduced to cell lines with a pTet-Off plasmid, de novo transcription from this reporter gene is specifically and rapidly suppressed in the presence of doxycycline. The reporter plasmid, BGG-WT or BGG-39PTC, was cotransfected with the CAT plasmid in HeLa Tet-Off or MEF Tet-Off cell lines. After incubation in the absence of doxycycline, the accumulation of the BGG mRNA is evaluated. As shown in Figure 4B, BGG-WT mRNA accumulates more abundantly than BGG-39PTC mRNA in either of the cell lines, consistent with the previous observation that BGG-39PTC is degraded through NMD (Thermann et al. 1998; Zhang et al. 1998b). Overexpression of hSMG-1-WT reduces the steady-state amount of BGG-39PTC mRNA; whereas the overexpression of hSMG-1-DA increases it (Fig. 4C).
We next examined the effect of the overexpression of either WT or DA of hSMG-1 on the half-life of either BGG WT or BGG-39PTC mRNA species. In experiments shown in Figure 4D, transcription of either of the BGG reporters was inhibited by the addition of doxycycline, and cells were harvested at the indicated time to measure the BGG mRNA amount. The half-life of BGG WT seems to be very long, as reported previously (Loflin et al. 1999) and is not affected by the overexpression of either hSMG-1-WT or hSMG-1-DA. On the other hand, the half-life of BGG-39PTC (t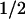 ∼120 min) is decreased by the overexpression of hSMG-1-WT (t
∼120 min) is decreased by the overexpression of hSMG-1-WT (t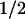 ∼45 min), while it is increased by the overexpression of the hSMG-1-DA mutant (t
∼45 min), while it is increased by the overexpression of the hSMG-1-DA mutant (t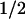 >180 min). These results strongly support the notion that hSMG-1 is involved in the PTC-dependent decay of BGG mRNA.
>180 min). These results strongly support the notion that hSMG-1 is involved in the PTC-dependent decay of BGG mRNA.
hSMG-1 phosphorylates hUPF1/SMG-2 in vitro and in vivo
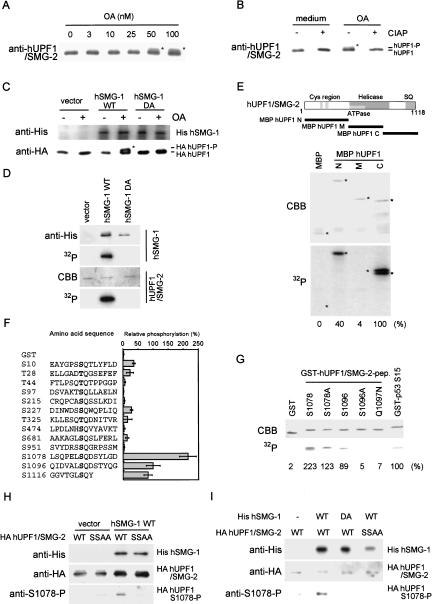
Previous studies have revealed that a human homolog of yeast Upf1p, hUPF1, is involved in mammalian NMD (Sun et al. 1998). One of the seven smg genes (smg 1–7) involved in C. elegans NMD, smg-2, encodes a homolog of Upf1p (Page et al. 1999). SMG-2 is a phosphoprotein and the other six smg genes have been classified into two groups on the basis of the effect of their mutation on the phosphorylation state of SMG-2. Taken together with the identification of SMG-1 as a putative PIKK (P. Anderson, pers. comm.), these observations suggest not only the importance of the phosphorylation of SMG-2, but also the molecular link between SMG-2 and SMG-1 and hence that between hUPF1/SMG-2 and hSMG-1. Consistently, when analyzed by Western blot analysis using an anti-hUPF1/SMG-2 antibody, treatment of HeLa cells with a phosphatase inhibitor, okadaic acid (OA), results in the appearance of an upward-shifted band of hUPF1/SMG-2 (Fig. 5A). This shifted band disappears when the immunoprecipitates were treated with a phosphatase, CIAP (Fig. 5B), indicating that it is the result of phosphorylation. Furthermore, overexpression of hSMG-1, but not its DA mutant, greatly enhances the OA-induced upward-shift of HA-tagged hUPF1/SMG-2, although the OA-induced upward-shift of exogenous HA-tagged hUPF1/SMG-2 is not observed (Fig. 5C). These results provide evidence that hUPF1/SMG-2 is a target molecule of hSMG-1 in vivo.
Figure 5.
hSMG-1 phosphorylates hUPF1/SMG-2 in vivo and in vitro. (A) Upward-shift of hUPF1/SMG-2 induced by OA treatment. HeLa cells were treated with the indicated concentrations of OA for 4.5 h. The lysate was analyzed by Western blotting using an anti-hUPF1/SMG-2 antibody. Positions of the shifted bands are marked by asterisks. (B) Upward-shift of hUPF1/SMG-2 (marked by an asterisk) is due to phosphorylation. Immunoprecipitate prepared as in A was treated with CIAP (+) or without CIAP (−), followed by Western blotting using an anti-hUPF1/SMG-2 antibody. (C) Overexpression of hSMG-1-WT induces OA-dependent phosphorylation of hUPF1/SMG-2 in 293T cells. Cells were transfected with a WT or a DA mutant hSMG-1 expression plasmid (10 μg) or a vehicle (vector) with an HA-tagged hUPF1/SMG-2 construct (0.5 μg). The upward shift was evaluated as shown in B. The position of the shifted HA- hUPF1/SMG-2 is marked by an asterisk. (D) hSMG-1 directly phosphorylates hUPF1/SMG-2. 293T cells were transfected with the expression plasmid for WT or DA mutant His-tagged hSMG-1 or a vehicle (vector). Immunocomplex using an anti-His antibody was subjected to in vitro kinase assay using purified HA-tagged hUPF1/SMG-2 as an exogenous substrate. An autoradiogram showing hUPF1/SMG-2 phosphorylation is shown (bottom). (E) hSMG-1 directly phosphorylates hUPF1/SMG-2 at its N- and C-terminal regions. In vitro kinase assay was performed using recombinant MBP-fused hUPF1/SMG-2 proteins (MBP hUPF1/SMG-2-N, M and C). The pattern of CBB staining of the gel and its autoradiogram are shown. The extent of phosphorylation relative to MBP-hUPF1/SMG-2-C is shown as percentages at the bottom. Positions of the major bands of each recombinant protein are indicated by asterisks. (F) hSMG-1 phosphorylates GST-fused 14mer SQ- or TQcontaining peptides derived from hUPF1/SMG-2. In vitro kinase assay was performed as in E using GST-fused peptides. Numbers are of the serine or threonine residue adjacent to glutamine. Data in the graph are means ± SD from three to seven independent experiments. (G) Phosphorylation of the GST-hUPF1/SMG-2 peptides with mutations at S1096 was carried out according to a protocol similar to that described in C. (H) Phosphorylation of hUPF1/SMG-2 at S1078 in vitro. 293T cells were transfected with SR6H (vector) or SR6H-hSMG-1-WT. His-SMG-1 was immunoprecipitated from SR6H (vector) or SR6H-hSMG-1-WT-transfected 293T cells using a His-tag antibody. In vitro kinase assay was performed using purified HA-hUPF1/SMG-2-WT or SSAA mutant in the presence of 200 μM ATP. Phosphorylated HA-UPF1/SMG-2 was visualized by Western blotting using anti-S1078-P (bottom) and anti-HA (middle) antibodies. Immunoprecipitated His-hSMG-1 was also probed using an anti-His antibody (top). (I) Overexpression of hSMG-1-WT enhances the phosphorylation of hUPF1/SMG-2 S1078 and S1096; whereas that of hSMG-1-DA suppresses it. 293T cells were transfected with SR6H-SMG-1-WT or SR6H-SMG-1-DA (10 μg) in combination with either SRHA-hUPF1/SMG-2-WT or SRHA-hUPF1/SMG-2-SSAA (0.5 μg). HA-hUPF1/SMG-2 was immunoprecipitated using an anti-HA antibody and analyzed by Western blotting with anti-HA (top) and anti-S1078-P (bottom) antibodies.
We next examined whether hSMG-1 directly phosphorylates hUPF1/SMG-2. We used purified recombinant proteins, HA-tagged hUPF1/SMG-2, His-tagged hSMG-1, and its kinase-deficient point mutant, hSMG-1-DA. As shown in Figure 5D, hSMG-1, but not its DA mutant, phosphorylates HA-hUPF1/SMG-2 in vitro, indicating that hUPF1/SMG-2 serves as a direct substrate of hSMG-1. The PIKK family kinases phosphorylate the serine or threonine residues in SQ or TQ motifs (Kim et al. 1999). Interestingly, hUPF/SMG-2, as well as SMG-2, contains repeats of SQ motifs in its C-terminal region (Page et al. 1999). The C-terminal and the N-terminal fragments of hUPF1/SMG-2 serve as substrates of hSMG-1 in vitro (Fig. 5E). Taken together with the fact that these proteins are not phosphorylated by SMG-1 DA (data not shown), we conclude that hSMG-1 has intrinsic kinase activities to phosphorylate hUPF/SMG-2 fragments in vitro. These data also suggest the importance of SQ motifs in hSMG-1-mediated phosphorylation of hUPF/SMG-2.
To further clarify this point, we next generated a series of GST-fusion proteins in which the hUPF1/SMG-2 sequence with SQ or TQ motifs was fused downstream of GST with corresponding surrounding 12 amino acid residues. A summary of the in vitro phosphorylation experiments using recombinant hSMG-1 is shown in Figure 5F. Typical examples are also shown in Figure 5G. These experiments clearly establish the substrate specificity of hSMG-1. hSMG-1 phosphorylates a control protein containing the SQ motif in p53 (S15). In addition, three peptide constructs, S1078, S1096, and S1116, are efficiently phosphorylated by hSMG-1. The hSMG-1 DA mutant did not phosphorylate these constructs (data not shown). The protein S1078 contains two serine residues in the SQ sequence and is most efficiently phosphorylated. The mutant, S1078A, was phosphorylated but with reduced efficiency (Fig. 5G), suggesting that both S1073 and S1078 are phosphorylated efficiently. On the other hand, neither S1096A nor Q1097N was phosphorylated (Fig. 5G), supporting the importance of Q next to S for the phosphorylation by hSMG-1. Moreover, these results establish the notion that hSMG-1 phosphorylates at least S in the SQ motif and that the sites for phosphorylation of hUPF1/SMG-2 by hSMG-1 contain the serine residues at positions 1073, 1078, 1096, and 1116.
To analyze the phosphorylation of hUPF1/SMG-2 in vivo, we generated a phospho-specific antibody against S1078. The antibody specifically recognizes the GST-hUPF1/SMG-2 S1078 and S1096 peptides only when phosphorylated by recombinant hSMG-1 (data not shown). Consistently, the HA-hUFP1/SMG-2 phosphorylated by recombinant hSMG-1 in vitro was recognized by the antibody; whereas the point mutant SSAA, in which both serine residues 1078 and 1096 are substituted with alanine, was not (Fig. 5H). Thus, the antibody specifically recognizes hUPF1/SMG-2 phosphorylated at S1078 and S1096.
Next, we examined whether hUPF1/SMG-2 is phosphorylated at S1078 and S1096 in vivo in an hSMG-1-kinase-activity-dependent manner. As shown in Figure 5I, the phosphorylation of HA-hUPF1/SMG-2 at S1078 and S1096 is strongly enhanced by the overexpression of hSMG-1-WT. In contrast, the phosphorylation is suppressed by that of hSMG-1-DA. Consistent with the results in Figure 5H, no phosphospecific signal is observed when hSMG-1-WT and hUPF1/SMG-2-SSAA are coexpressed. These results support the finding that at least S1078 and S1096 of hUPF1/SMG-2 are phosphorylated in vivo by hSMG-1.
hSMG-1 associates with hUPF1/SMG-2, hUPF2, and hUPF3
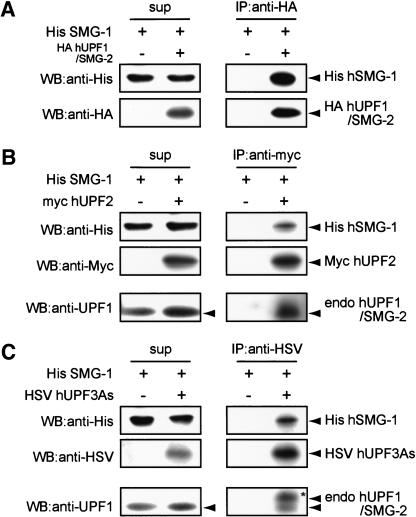
Previous studies on mammalian homologs of yeast, Upf1p, Upf2p, and Upf3p, have revealed that the three proteins form a protein complex, a surveillance complex (Lykke-Anderson et al. 2000; Serin et al. 2001). The next question we addressed is whether hSMG-1 physically interacts with this complex. For this purpose, we performed coexpression and immunoprecipitation experiments using 293T cells. As shown in Figure 6A, the immunoprecipitate of HA-hUPF1/SMG-2 contains His-hSMG-1, indicating a physical interaction between hSMG-1 and hUPF1/SMG-2. Similarly, His-hSMG-1 is also contained in the immunoprecipitate of Myc-hUPF2 or HSV-hUPF3A (Fig. 6B or C). In particular, both of the immunoprecipitates contain endogenous hUPF1/SMG2. These results suggest that hSMG-1 physically interacts with the evolutionally conserved surveillance complex. Interestingly, upward-shifted hUPF1/SMG-2 (hyperphosphorylated form) is preferentially coimmunoprecipitated with HSV-hUPF3A (Fig. 6C, asterisk), whereas no upward-shifted hUPF1/SMG-2 is coimmunoprecipitated with overexpressed hUPF2. This indicates that hyperphosphorylated hUPF1/SMG-2 is preferentially associated with a protein complex containing HSV-hUPF3A. This implies that hUPF3 plays a positive role in the phosphorylation of hUPF1/SMG-2 by hSMG-1.
Figure 6.
Association of hSMG-1 with other NMD components. His-hSMG-1 (10 μg) was coexpressed with either HA-hUPF1/SMG-2 (5 μg), myc-hUPF2 (5 μg), or HSV-hUPF3As (5 μg) in 293T cells. HA-hUPF1/SMG-2 (A), myc-hUPF2 (B), or HSV-hUPF3As (C) was immunoprecipitated using the corresponding anti-tag antibody and the resulting immunoprecipitate was probed using an anti-His antibody to detect hSMG-1. Coimmunoprecipitated endogenous hUPF1/SMG-2 was also probed in B and C. In C, upward-shifted hUPF1/SMG-2 is indicated by an asterisk.
Inhibitors of SMG-1 stabilize endogenous p53 mRNA species with PTC and accumulate their truncated protein products
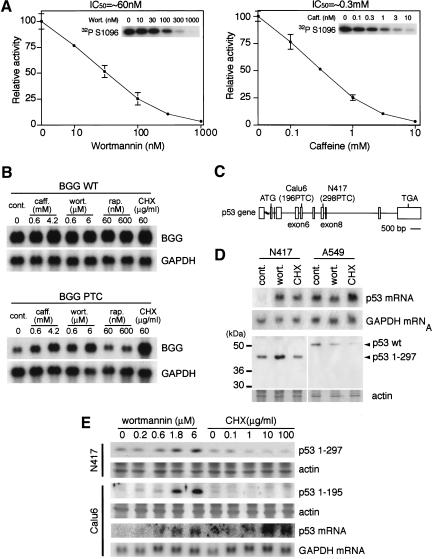
Previous studies on the PIKK family have identified inhibitors acting on this family of kinases. These include wortmannin and caffeine. To evaluate the role of hSMG-1 in mammalian NMD and to determine a pharmacological strategy for studying the specific inhibition of NMD, we next examined the effects of these inhibitors on the kinase activity of hSMG-1. As shown in Figure 7A, both wortmannin and caffeine inhibited the kinase activity of hSMG-1 with IC50 values of ∼60 nM and 0.3 mM, respectively. In addition, treatment of HeLa or MEF Tet-Off cells with wortmannin inhibited the kinase activity of endogenous hSMG-1 with an IC50 value of ∼1 μM (data not shown). On the other hand, staurosporine, and rapamycin in the presence of purified recombinant FKBP12, did not inhibit the kinase activity of hSMG-1 (data not shown). The effects of the two hSMG-1 inhibitors on the phosphorylation of endogenous hUPF1/SMG-2 in HeLa cells were also examined. Both wortmannin and caffeine but not rapamycin inhibited the upward shift of hUPF1/SMG-2 in HeLa cells (data not shown), consistent with the results obtained using the purified system. If the protein kinase activity of hSMG-1 is essential for mammalian NMD, these inhibitors of hSMG-1 should suppress NMD. To test this, we first employed the reporter BGG system used in experiments shown in Figure 3. As shown in Figure 7B, the protein synthesis inhibitor, cycloheximide (CHX), inhibits NMD and induces the accumulation of BGG-39PTC mRNA but not BGG-WT, consistent with the previous observation (Carter et al. 1995). In particular, the inhibitors of hSMG-1, caffeine and wortmannin, induced the accumulation of BGG-39PTC mRNA. This provides pharmacological evidence that hSMG-1 is involved in mammalian NMD.
Figure 7.
Inhibitors of hSMG-1 suppress NMD and cause accumulation of truncated p53 proteins in human cancer cell lines with PTC mutations. (A) Wortmannin or caffeine inhibits protein kinase activity of hSMG-1 in vitro. In vitro kinase assays were performed using immunoprecipitated His-hSMG-1 and a GST- hUPF1/SMG-2 fusion protein, hUPF1/SMG-2 S1096, as a substrate in the presence of an indicated concentration of wortmannin or caffeine. Autoradiograms of hUPF1/SMG-2 S1096 phosphorylation are indicated in the corresponding insets. (B) The BGG reporter mRNA with a PTC is specifically accumulated by inhibitors of hSMG-1. The MEF Tet-Off cells were transfected with a wild-type (upper) or mutant (lower) BGG reporter gene (Fig. 4C) and reseeded. Cells were treated for 2 h with caffeine (caff.), wortmannin (wort.), rapamycin (rap.), or cycloheximide (CHX) in the absence of doxycycline. Total RNAs were analyzed by Northern blotting using a BGG probe (top) or a GAPDH probe (bottom). (C) Schematic representation of p53 gene structure with PTC mutations in human lung cancer cell lines, Calu6 (196PTC) and N417 (298PTC). Boxes show exons. (D) Wortmannin induces the accumulation of transcripts and their corresponding proteins from the p53 gene with PTC. N417 and A549 cells were treated with or without (cont.) 10 μM wortmannin (wort.) or 100 μg/mL cycloheximide (CHX) for 4 h. Total RNA and cell lysates were analyzed by Northern blotting using a p53 probe and by Western blotting using an anti-p53 antibody, DO-1, respectively. The CBB image showing actin staining is also shown (bottom). (E) Wortmannin induces expression of p53 transcripts and p53 proteins of lower molecular weights in a dose-dependent manner. Calu6 or N417 cells were treated with an indicated concentration of wortmannin or CHX for 2 h. The total RNA and cell lysate were prepared and analyzed, as in E, by Northern blotting using a p53 probe and by Western blotting using DO-1, respectively.
Although NMD protects cells against the accumulation of potentially toxic peptides that originate from PTC mRNA, NMD sometimes eliminates mRNAs encoding truncated peptides with residual activity that may partially rescue the disordered phenotype if expressed. Thus, at least in some cases of PTC mutations, the specific inhibition of NMD may provide a novel therapeutic strategy for rescuing the genetic disorder. The specific inhibitor of SMG-1 will serve as a novel tool for such a strategy. As a first step to evaluate this, we examined the ability of the inhibitors of hSMG-1 to specifically rescue the synthesis of the truncated proteins derived from the PTC allele. We selected the p53 gene as a model system to evaluate such a possibility because of the availability of cell lines with characterized mutations. We selected two human cancer cell lines with PTCs. Calu6 contains a PTC at codon 196 and N417 contains a PTC at codon 298 (Fig. 7C). Treatment of N417 cells with wortmannin increases the amounts of p53 298 PTC mRNA and the truncated p53 protein; whereas in the control A549 cells with a wild-type p53 allele, there are no increases in the amounts of the mRNA and the protein (Fig. 7D). The increase in the amount of the truncated p53 was also observed when Calu6 cells were treated with an increasing concentration of wortmannin (Fig. 7E). On the other hand, CHX did not increase the amount of the truncated p53 protein, although it enhanced the accumulation of the corresponding mRNA. These results clearly show that hSMG-1 inhibitors induce p53 PTC mRNA accumulation via a mechanism independent of the inhibition of translation.
Discussion
hSMG-1 is a critical component of mammalian NMD
Here, we have identified the fifth member of the human PIK-related protein kinase family and termed it hSMG-1 based on its overall sequence similarity to CeSMG-1. hSMG-1 is a very large protein kinase with a unique structure and the ability to phosphorylate specific serine residues in the C-terminal SQ motifs of hUPF1/SMG-2, another component of the mammalian NMD. The use of a phospho-specific antibody revealed that this phosphorylation occurs in vivo in an hSMG-1-kinase activity-dependent manner, establishing that hUPF1/SMG-2 is one of the physiological substrates of hSMG-1. The physical interaction of hSMG-1 with hUPF1/SMG-2 in cells further supports this notion. Most importantly, we showed that the overexpression of SMG-1-DA, a point mutant of hSMG-1 lacking kinase activity, markedly suppresses the phosphorylation of hUPF1/SMG-2 in vivo and the PTC-dependent β-globin mRNA degradation; whereas that of wild-type SMG-1 enhances both. In addition, inhibitors of hSMG-1 suppress the phosphorylation of hUPF1/SMG-2 in vivo and the PTC-dependent β-globin mRNA degradation. Furthermore, inhibitors of hSMG-1 cause the accumulation of an endogenous p53 mRNA with PTC and the truncated protein product. These observations provide strong evidence that hSMG-1 is a critical component of mammalian NMD. This also reveals a novel aspect of mammalian NMD in which protein phosphorylation plays a critical role. In C. elegans, SMG-2 is a phosphoprotein whose phosphorylation state depends on the other six smg genes including smg-1. In smg-1, smg-3, and smg-4 mutants, phosphorylation of SMG-2 does not occur; whereas in smg-5, smg-6, and smg-7 mutants, the phosphorylated form of SMG-2 is accumulated (Page et al. 1999). Furthermore, smg-5 has been recently shown to encode a targeting subunit of protein phosphatase 2A (PP2A), and the product interacts with SMG-7, SMG-2, PR65A, the scaffolding subunits of PP2A, and PP2Ac, the catalytic subunit of PP2A (P. Anderson, pers. comm.). In addition, we have shown that the phosphorylation of hUPF1/SMG-2 is induced by treatment of cells with okadaic acid, an inhibitor of PP2A. Taken together with the results of studies on C. elegans smg genes, the results of our present study reveal the general importance of protein phosphorylation in NMD in higher eukaryotes.
Significance of phosphorylation of hUPF1/SMG-2 in higher eukaryotes
In yeast, a translation termination event triggers the assembly of an mRNA surveillance complex that searches the 3′ end of the termination codon to detect specific signals indicative of premature termination events (Czaplinski et al. 1998; Gonzalez et al. 2000). The mRNA surveillance complex contains at least Upf1p, Upf2p, Upf3p, and translation release factors Sup35p and Sup45p in which Upf1p plays an essential role. In mammals, hUPF1/SMG-2, hUPF2, and hUPF3 also form a complex (Lykke-Anderson et al. 2000; Serin et al. 2001). Taken together, the finding that hSMG-1 can interact with the three components of the surveillance complex, hUPF1/SMG-2, hUPF2, and hUPF3, suggests that hSMG-1 is a member of the surveillance complex including those three molecules. The presence of hUPF1/SMG-2 in immunoprecipitates of either hUPF2 or hUPF3 supports this notion. The increase in the concentration of the hyperphosphorylated form of hUPF1/SMG-2 in hUPF3 immunoprecipitates may suggest the importance of hUPF3 in the phosphorylation of hUPF1/SMG-2. Collectively, our results strongly support the idea that hSMG-1 is a regulatory component of the mammalian surveillance complex and the phosphorylation occurs in the surveillance complex.
We have provided evidence showing that hSMG-1 directly phosphorylates hUPF1/SMG-2 at at least two serine residues, S1078 and S1096, located at the C terminus in vivo and in vitro. This is consistent with the recent finding that hUPF1/SMG-2 is a phosphoprotein and that the phosphorylation is inhibited by wortmannin (Pal et al. 2001). Notably, the amino acid sequence motif surrounding S1078 and S1096 are conserved at the C terminus of C. elegans SMG-2, supporting the importance of phosphorylation at these sites. Upf1/SMG-2 proteins from yeast, C. elegans, and mammals share a homologous sequence spanning 800 amino acids and containing a zinc finger and an RNA helicase domain. On the other hand, the N-terminal and C-terminal flanking sequences are not conserved between yeast and higher eukaryotes; whereas SMG-2 and hUPF1/SMG-2 share a homologous C-terminal region. Moreover, hSMG-1 and perhaps SMG-1 phosphorylate serine residues in the SQ motifs in this region. These may reflect the basic difference between yeast and higher eukaryotes in terms of the mechanism for regulating the function of Upf1/SMG-2.
A novel member of the PIKK family
Four members of the PIK-related protein kinase family have been identified in human. We identified hSMG-1 as a novel member of this family. The proposed role of members of the PIK-related kinases from yeast to mammals leads us to speculate that this family of kinases is involved in the surveillance of the fundamental steps in gene expression. ATR, ATM, and DNA-PK are involved in the surveillance of the structural integrity of the template DNA itself, FRAP is involved in the regulation of the translation initiation, and SMG-1 is involved in the integrity of mRNA.
Specific inhibition of hSMG-1 as a potential therapeutic strategy
One-fourth of all mutations in human genetic diseases and cancers are of the type that can target the corresponding mRNA for NMD. Although NMD protects cells against deleterious gain-of-function mutations caused by the dominant negative effects of aberrant truncated proteins, there are some cases in which the truncated protein does not show such an effect, rather, it retains residual activity and can compensate for the normal gene function. Thus, the specific inhibition of NMD may provide a novel therapeutic strategy based on the type of mutation rather than on the gene in which the mutation resides. We have shown for the first time that inhibitors of hSMG-1 cause the accumulation of truncated p53 proteins from a PTC allele, as well as the increase in the level of mRNA with PTC, opening the possibility of the above strategy by specifically suppressing NMD through the inhibition of hSMG-1.
In yeast, three upf genes have been shown to be involved in efficient translation termination. Mutations in either of these genes result in elevated rates of read-through of translation stop codons producing a full-length protein, a phenomenon called nonsense suppression (Wang et al. 2001). Moreover, several observations suggest that in C. elegans, the rate of nonsense suppression increases in the smg (−) background (Page et al. 1999). These suggest that smg genes are responsible for ensuring the fidelity of translation termination. In mammals, it is likely that SMG/UPF proteins, including hSMG-1, have the same function. Nonsense suppression has also been proposed to provide a possible therapeutic approach for diseases associated with PTC mutations, and several aminoglycoside antibiotics have been shown to cause nonsense suppression. Although whether these inhibitors affect NMD remains to be clarified, it is plausible to speculate that the inhibitors of SMG-1 can cause nonsense suppression. If this is the case, the inhibitors of SMG-1 can rescue the synthesis of mature proteins through two independent mechanisms (i.e., the inhibition of NMD to increase the mRNA level and the suppression of translational termination that leads to the synthesis of a read-through mature protein product). In this sense, the specific inhibitors of hSMG-1 will be of potential therapeutic importance for all the genetic diseases associated with PTC mutations.
Materials and methods
Molecular cloning and DNA manipulation
During the identification of the reported protein, LIP, an atypical PKCλ-interacting protein (Diaz-Meco et al. 1996), we failed to identify the mRNA and the protein product but found that the overlapping cDNA clone, KIAA0421, encodes a very large protein, hSMG-1. The nature of the reported ORF of LIP remains to be established. KIAA0421 covers the 3′ portion of hSMG-1. Successive back-screenings of human cDNA libraries and RT–PCR from HeLa cell total RNA (see Fig. 1B) were performed. The ORF was open to 5′ even when all the clones were assembled. Thus, we searched and identified the human EST clone AI005513 (Research Genetics). This EST clone has an in-frame ATG codon. To construct an hSMG-1 expression vector, SR6H hSMG-1, an hSMG-1 cDNA fragment encoding amino acids (aa) 107–3567, was inserted into SR6H, a modified SRD vector with a sequence encoding the 6 × His tag upstream of its MCS. A kinase-deficient mutant of hSMG-1 (SR6H-hSMG-1-DA) in which an aspartic acid residue at position 2331 was substituted with alanine was generated following the standard procedures.
The KIAA0221 cDNA fragment encoding aa 6–1118 was inserted into SRHA, a modified SRD with the HA tag sequence upstream of its MCS, generating SRHA-hUPF1/SMG-2. A phospho-deficient mutant of hUPF1/SMG-2 (SRHA-hUPF1/SMG-2-SSAA) in which serine residues at positions 1078 and 1096 were substituted with A was constructed following the standard procedure. We identified human cDNA clones, KIAA1408 and AA356414 (ATCC), similar to Saccharomyces cerevisiae Upf2p. These two clones span the full-length ORF of the human homolog of S. cerevisiae Upf2p. For SRmyc-hUPF2, KIAA1408, and AA356414 chimeric cDNA fragment encoding aa 1–1272 was inserted into SRmyc, a modified SRD vector, that has the myc tag sequence upstream of its MCS. We identified human EST clones, AA211131, AW197446, AA034360, and AA865368 (Research Genetics), similar to S. cerevisiae Upf3p. The 5′ region of the cDNA was amplified and cloned by RT–PCR. Those clones span the full-length ORF of the human homolog of UPF3Al/As. For SRSV-hUPF3As, a hUPF3As cDNA fragment encoding aa 1–443 was inserted into SRSV, a modified SRD vector with a sequence encoding the HSV tag upstream of its MCS.
hSMG-1 mRNA was probed with the radiolabeled cDNA (nucleotides 6255–7048). FISH mapping was carried out as previously described (Izumi et al. 1998) using the full-length cDNA encoded by KIAA0421 as a probe.
Antibodies
Anti-hSMG-1 antisera designated as P1, C3, L2, and N1 were raised in rabbits. Antigen for P1 is a peptide (CDNLAQLYEGWTAWV) conjugated to KLH (keyhole limpet hemocyanin). cDNA fragments corresponding to hSMG-1 aa 3076–3542, 864–1054, and 1–106 were inserted into pGEX6P (Amersham Pharmacia Biotech) for C3, L2, and N1, respectively. GST-fusion proteins produced in Escherichia coli were used as antigens. For the antibody against hUPF1/SMG-2, a peptide (EEDEEDTYYTKDLPIHAC) conjugated to KLH was used as the antigen. Affinity purification of the antibody was performed following standard procedures. For the phosphospecific antibody against hUPF1/SMG-2 S1078, a peptide (LSQPELS[-P]QDSYLG) conjugated to KLH was used as an antigen. Affinity purification of the phospho-specific antibody was performed following standard procedures. Shortly, the antiserum was negatively selected against unphosphorylated peptide, followed by affinity purification using the phosphopeptide.
Anti-His (His-tag) and anti-HSV antibodies were purchased from Novagen. Anti-HA (12CA5) and anti-myc (9E10) antibodies were from Boehringer Mannheim and Calbiochem, respectively.
NMD analysis
To construct the reporter gene plasmid, pTRE BGG WT, from a human genomic library (Clontech), a human β-globin gene fragment was amplified by PCR and inserted into the pTRE vector (Clontech). A nonsense mutation of the human β-globin gene at codon 39 was introduced following standard procedures, generating pTRE BGG PTC. Half-life assays for BGG mRNA species were performed as follows: HeLa Tet-Off cells (Clontech) were transiently transfected by polyfectin (QIAGEN) with either of the reporter constructs (1.5 μg) in combination with SR6H-hSMG-1-WT or DA (3 μg). After 24 h, cells were reseeded in six 10-cm dishes and further cultured in the absence of doxycycline for 24 h. At an indicated time point (0, 30, 60, 90, 120, or 180 min) after addition of 50 ng/mL doxycycline to suppress the transcription from the reporter, cells were harvested and total RNA was isolated. The abundance of BGG mRNA was evaluated by Northern blot analysis using a BGG probe. The relative steady-state level of the mRNA was normalized to that of GAPDH.
For the BGG mRNA accumulation assay, HeLa Tet-Off or MEF Tet-Off (Clontech) cells were transiently transfected with either of the BGG reporters (1.5 μg) in combination with a CAT construct (0.5 μg) as a transfection marker. After 2 d of culture without doxycycline, cells were harvested and total RNA was isolated. Northern blotting was performed using a BGG or a CAT probe.
To evaluate the effect of hSMG-1 kinase inhibitors on p53 expression, three cell lines were used. Calu6, a lung adenocarcinoma cell line, and N417, a small cell lung carcinoma cell line, have a nonsense mutation within the p53 gene at codons 196 and 298, respectively. A549, a lung adenocarcinoma cell line, expresses wild-type p53 (Lehman et al. 1991; Bodner et al. 1992). After the cells were cultured with the indicated concentration of wortmannin or cycloheximide for 4 h, total RNA was isolated and analyzed by Northern blotting using p53 cDNA as a probe. At the same time, the cell lysate was prepared and analyzed by Western blotting using an anti-p53 antibody (DO-1, Calbiochem).
Substrates for in vitro kinase assay
Recombinant proteins of hUPF1/SMG-2 were prepared as follows and used as exogenous substrates for hSMG-1. cDNA fragments corresponding to hUPF1/SMG-2 aa 1–462, 463–800, and 801–1118 were inserted into the pMal-c2 vector (New England Biolabs) for the production of MBP (maltose-binding protein)-hUPF1/SMG-2 N, M, and C, respectively. These MBP-fusion proteins were purified from inclusion bodies as previously described (Hirai et al. 1994) with minor modifications.
As for GST-hUPF1/SMG-2 14mer fusion proteins, designated as S10, T28, T44, S97, S215, S227, T325 S474, S681, S951, S1078, S1078A, S1096, S1096A, Q1097N, S1116, and GST-p53 S15, the corresponding oligonucleotides were inserted into the pGEX 6P vector. Recombinant proteins were purified following the standard glutathione beads method. For the full-length hUPF1/SMG-2 and hUPF1/SMG-2 SSAA recombinant proteins, 293T cells were transfected with SRHA hUPF1/SMG-2 or hUPF1/SMG-2 SSAA. Two days after transfection, harvested cells were lysed in lysis buffer F (20 mM Tris-HCl at pH 7.5, 0.25 M sucrose, 1.2 mM EGTA, 20 mM β-mercaptoethanol, 1 mM Na orthovanadate, 1 mM Na pyrophosphate, 1 mM NaF, 1% Triton X-100, 0.5% Nonidet P-40, 150 mM NaCl and protease inhibitors). Anti-HA affinity beads (Roche) were added to the precleared lysate. An hour later, the beads were washed three times with lysis buffer F and three times with a washing buffer (20 mM Tris-HCl at pH 7.5, 0.1 M NaCl, 0.1 mM EDTA and 0.05% Tween 20). The recombinant protein was eluted in a washing buffer containing 1 mg/mL HA peptide (YPYDVPDYA) at 37°C and then dialyzed against 1× PBS with 10% glycerol and 1 mM DTT.
In vitro kinase assay
HeLa cell hSMG-1 was purified by immunoprecipitation. Cells were lysed in lysis buffer F. The hSMG1 protein was collected on protein A beads (Amersham Pharmacia Biotech) to which the indicated hSMG-1 antibody was preabsorbed. The immnunocomplex was washed with lysis buffer F containing 0.25 M LiCl and with 1× kinase reaction buffer (10 mM Hepes-KOH at pH 7.5, 50 mM β-glycerophosphate, 50 mM NaCl, 1 mM DTT, 10 mM MnCl2). Reaction was carried out in 50 μL of 1× kinase reaction buffer containing 5 μM ATP and 10 μCi [γ-32P] ATP at 30°C for 15 min with or without the indicated substrate. After development by SDS-PAGE, the phosphoproteins were visualized by autoradiography. The extent of phosphorylation of the proteins was evaluated using an imaging analyzer (BAS2000, Fuji Film).
For recombinant hSMG-1 kinase assay, cDNA-transfected 293T cells were prepared. The assay was performed following essentially the same methods as those for HeLa endogenous hSMG-1, except for the use of an anti-His antibody and protein G beads (Amersham Pharmacia Biotech) for immunoprecipitation.
Analysis of hUPF1/SMG-2 phosphorylation in vivo
To observe hUPF1/SMG-2 phosphorylation in HeLa cells, cells were treated with the indicated concentration of okadaic acid (OA; Calbiochem). Cells harvested were lysed in 1× SDS sample buffer. The upward shift of hUPF1/SMG-2 was detected by Western blotting using the anti-hUPF1/SMG-2 antibody, after 6% SDS-PAGE. To show that the OA-induced upward shift of hUPF1/SMG-2 is caused by phosphorylation, immunopurified hUPF1/SMG-2 was treated with alkaline phosphatase and its mobility in SDS-PAGE was examined as follows. HeLa cells treated with 50 nM OA for 4.5 h were harvested and lysed in lysis buffer F containing 1 μM mycrocystin LR (Calbiochem) and 10 nM OA, followed by immunoprecipitation with the anti-hUPF1/SMG-2 antibody. After washing in lysis buffer F and dephosphorylation buffer (50 mM Tris-HCl at pH 9.0 with 1 mM MgCl2), the immunoprecipitate was suspended in 50 μL of a dephosphorylation buffer. The reaction was primed by adding 60 units of calf intestine alkaline phosphatase (CIAP). After incubation at 37°C for 1 h, the reaction was terminated by adding SDS-sample buffer. The hUPF1/SMG-2 upward shift was detected by Western blotting using the anti-hUPF1/SMG2 antibody. For the analysis of overexpressed hUPF1/SMG-2, 293T cells were transfected with SRHA hUPF1/SMG-2 (0.5 μg) in combination with an SR6H vector (i.e., SR6H SMG-1-WT or DA [10 μg]). Cells were cultured with or without 50 nM OA for 4 h. The upward shift of hUPF1/SMG-2 was detected by Western blotting using with anti-HA antibody.
Coimmunoprecipitation assay using 293T cells
cDNA-transfected 293T cells were suspended and incubated for 30 min at 4°C in a lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.4% Nonidet-P40, 200 μg/mL RNase A, and protease inhibitors. The precleared lysates were incubated with antibodies preabsorbed on Protein G-Sepharose or 1 h at 4°C. The immunocomplexes were washed with a washing buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.05% Nonidet-P40, 0.1% Triton X-100 and protease inhibitors and then analyzed by Western blotting using the indicated antibodies.
Acknowledgments
We thank Drs. P. Anderson for providing unpublished information on C. elegans smg genes, P. Chambon for providing the HeLa cDNA library, T. Tanaka for cell lines, and T. Nagase and N. Nomura for providing KIAA clones. This work was supported in part by grants from the Japan Society for the Promotion of Science and from the Ministry of Education, Culture, Sports, Science and Technology of Japan. A.Y. is a Research Fellow of the Japan Society for the Promotion of Science. T.O. is a Research Fellow of the Kihara Memorial Yokohama Foundation for the Advancement of Life Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ohnos@med.yokohama-cu.ac.jp; FAX 81-45-785-4140.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.913001.
References
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodner SM, Minna JD, Jensen SM, D'Amico D, Carbone D, Mitsudomi T, Fedorko J, Buchhagen DL, Nau MM, Gazdar AF, et al. Expression of mutant p53 proteins in lung cancer correlates with the class of p53 gene mutation. Oncogene. 1992;7:743–749. [PubMed] [Google Scholar]
- Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- Cali BM, Anderson P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol Gen Genet. 1998;260:176–184. doi: 10.1007/s004380050883. [DOI] [PubMed] [Google Scholar]
- Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Carter MS, Li S, Wilkinson MF. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes & Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Sanchez P, Lozano J, Moscat J. Lambda-interacting protein, a novel protein that specifically interacts with the zinc finger domain of the atypical protein kinase C isotype λ/ι and stimulates its kinase activity in vitro and in vivo. Mol Cell Biol. 1996;16:105–114. doi: 10.1128/mcb.16.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the β-globin gene are not associated with a reduction in β-mRNA accumulation: A mechanism for the phenotype of dominant β-thalassemia. Blood. 1994;83:2031–2037. [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Hilleren P, Parker R. Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet. 1999;33:229–260. doi: 10.1146/annurev.genet.33.1.229. [DOI] [PubMed] [Google Scholar]
- Hirai S, Izumi Y, Higa K, Kaibuchi K, Mizuno K, Osada S, Suzuki K, Ohno S. Ras-dependent signal transduction is indispensable but not sufficient for the activation of AP1/Jun by PKCd. EMBO J. 1994;13:2331–2340. doi: 10.1002/j.1460-2075.1994.tb06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra MF. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Lehman TA, Bennett WP, Metcalf RA, Welsh JA, Ecker J, Modali RV, Ullrich S, Romano JW, Appella E, Testa JR, et al. p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- Loflin PT, Chen CY, Xu N, Shyu AB. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods. 1999;17:11–20. doi: 10.1006/meth.1998.0702. [DOI] [PubMed] [Google Scholar]
- Lykke-Anderson J, Shu M-D, Steiz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upf1 protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes & Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae upf2 protein and upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and ATR: Networking cellular responses to DNA damage. 2001. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- Sun X, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL, Hesketh C, Taylor P, Temperley IJ, Hutchinson RM, Old JM, Wood WG, Clegg JB, Weatherall DJ. Molecular basis for dominantly inherited inclusion body β-thalassemia. Proc Natl Acad Sci. 1990;87:3924–3928. doi: 10.1073/pnas.87.10.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Hall MN. TOR signaling and control of cell growth. Curr OpinCell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteinsin modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: A possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998a;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of b-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998b;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]