Abstract
The ancestral role of the Hox gene family is specifying morphogenetic differences along the main body axis. In vertebrates, HoxD genes were also co-opted along with the emergence of novel structures such as limbs and genitalia. We propose that these functional recruitments relied on the appearance, or implementation, of regulatory sequences outside of the complex. Whereas transgenic human and murine HOXD clusters could function during axial patterning, in mice they were not expressed outside the trunk. Accordingly, deletion of the entire cluster abolished axial expression, whereas recently acquired regulatory controls were preserved.
Keywords: Hox cluster, colinearity, BAC, remote enhancers
During vertebrate development, Hox genes are activated in a spatio-temporal sequence that leads to partially overlapping transcript domains along the trunk axis. These expression domains generate various combinations of HOX proteins at different anterior–posterior positions, instructing cohorts of cells about their fate (e.g. Krumlauf 1994). In most cases, there exists a correspondence between the order of the genes in the genome and their domains of expression, a phenomenon referred to as colinearity. This feature is very ancient, as it seems to operate in all animals with a bilateral body plan; hence, it is likely that colinearity relies on a conserved mechanism.
In addition to this function, Hox genes were recruited during evolution to carry out a number of other tasks. For example, a given subset of vertebrate Hox genes is required for limb development (Davis and Capecchi 1996; Rijli and Chambon 1997; Zakany et al. 1997), gut morphogenesis (Zakany and Duboule 1999), or hair follicle development (Godwin et al. 1998). In some cases, the colinear process was also recruited, whereas elsewhere groups of genes acquired shared enhancer sequences acting independently of colinearity. For instance, the early phase of Hoxd gene expression in limb buds is regulated in a colinear fashion, whereas expression of the same genes in digits is concurrent, rather than colinear (Nelson et al. 1996).
In the HoxD complex, gene recruitment involved in many instances the design of potent enhancer sequences, which regulate several genes at once. We proposed earlier that expression of four genes in developing digits was controlled by a unique enhancer that displays poor promoter specificity as it influenced foreign promoters when targeted to the locus (van der Hoeven et al. 1996; Hérault et al. 1999). Targeted deletions in the posterior HoxD complex placed this enhancer somewhere upstream of Evx2, outside the cluster (Kondo and Duboule 1999). Likewise, several genes respond to a gut enhancer sequence that is required to form the ileo-coecal sphincter (Zakany and Duboule 1999) and is localized either in the first 30 kb of the complex (around Hoxd1) or outside of the complex (Kmita et al. 2000b).
Regulation inside Hox clusters is complex because of a high density of genes with embedded and shared regulatory elements, making it difficult to assign a control sequence to one individual gene, rather than to a series of genes (Gérard et al. 1996; Hérault et al. 1998; Sharpe et al. 1998). To assess which features of Hoxd gene regulation are intrinsic to the complex and which are located at a distance (i.e., act in a global scale over the locus) we produced transgenic mice carrying additional HoxD loci. We used a human PAC extending from HOXD3 to 30 kb upstream EVX2 and a mouse BAC containing a tagged HoxD. We compared the regulatory potentials of these transgenic clusters with that of targeted deletions of the mouse HoxD complex, including a knock-in replacement of the cluster. We show that both approaches mapped regulatory elements responsible for colinear expression within the cluster or close to it, whereas non-colinear expression in appendicular structures is dictated by regulatory elements located at remote positions. We also show that the presence of the complex is required for the maintenance of Hox gene expression profiles throughout development.
Results
Rescue of Hoxd mutations by a human HOXD complex
To study the regulatory potential of a Hox complex, we produced transgenic mice carrying a 120 kb large human PAC (Fig. 1A) extending from the HOXD3–HOXD1 region to ∼30 kb 5′ upstream of EVX2. Five founder animals were recovered. One carried a partial PAC and thus was not studied further. Among the others, three lines with either one or two copies of the transgene were established (TgN[HOXD]1–3). One high-copy number animal was recovered but the line could not be established due to perinatal lethality.
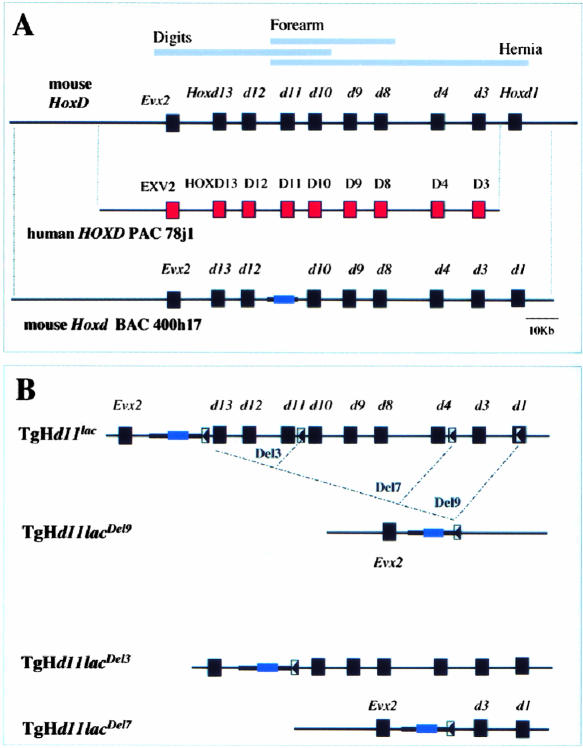
Figure 1.
The mouse HoxD cluster, the human PAC, and nested deletions. (A) The HoxD complex (top) with genes as black boxes (red for human). Below is the human PAC 78j1, with the human HOXD3 to EVX2 genes and 30 kb of DNA upstream, and the mouse BAC 400h17, with the murine cluster and 100 kb of DNA upstream. (B) Scheme of nested deletions. (Top) The HoxD complex with a Hoxd11/lacZ reporter transgene inserted upstream of Hoxd13. After recombination of a loxP site within Hoxd1 (arrowhead), Cre-mediated deletion removed the cluster leaving the reporter gene construct TgHd11/lacDel9 (below). The bottom lines are intermediate configurations obtained by using other loxP sites within the cluster. After recombination, partial deletions of either three (Del3) or seven (Del7) genes are obtained, with the reporter transgene at the same position.
Mice with the human PAC showed abnormal vertebral formulae with a high penetrance of five lumbar vertebrae (L5) instead of the normal L6 (Fig. 2, top). Besides this anteriorization, no alteration was observed, suggesting that the human genes were expressed during mouse trunk development with the appropriate specificity. This was verified further when the transgene was combined with a triple inactivation in cis of Hoxd13, Hoxd12, and Hoxd11 (Zakany and Duboule 1996). Mice (16 out of 18) lacking these functions (HoxDDel3/Del3) displayed an L7 formula (a supernumerary lumbar vertebra; Fig. 2, top). The introduction of the human PAC into this background reinstated the predominant L6 formula (Fig. 2, top) with a substantial incidence of L5 animals. This showed a robust rescue of the mouse defect by the human proteins.
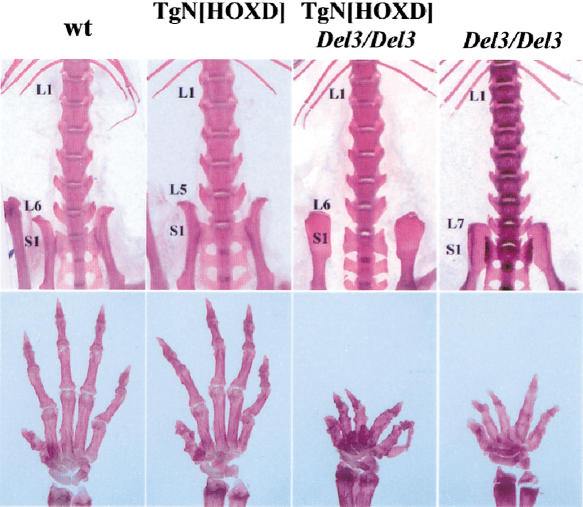
Figure 2.
Vertebral and limb phenotypes. (Top) Skeletal preparations of lumbo–sacral transitions. Wild-type mice have six lumbar vertebrae (L6). Mice transgenic for human HOXD PAC (TgN[HOXD]) had an anteriorized lumbo–sacral transition with L5. In contrast, mice carrying a deletion of Hoxd13 to Hoxd11 showed seven lumbar vertebra (L7). When the human HOXD transgene was added to this latter configuration (TgN[HOXD]; Del3/Del3), the L7 phenotype was rescued to L6.
Nevertheless, rescue was not detected in the limb skeleton and no difference was scored between wild-type and transgenic hand skeletons (Fig. 2, bottom). Because human HOXD proteins could functionally rescue vertebral alterations, lack of rescue in limbs was likely due to abnormal expression of the transgenes in these structures. Thus, we analyzed the expression of human HOXD genes during mouse development.
Human HOXD genes in the mouse
We used RNA probes specific for human HOXD13, HOXD11, and HOXD4 transcripts, in parallel with the mouse counterparts. Analysis of human transgene expression revealed that the HOXD genes were regulated correctly during trunk development, as expression boundaries were similar to those of the corresponding resident mouse genes (Fig. 3A, top, arrows). Human HOXD13 was weakly expressed posteriorly, starting caudal to the hindlimb bud, whereas expression of HOXD11 was at the level of pv25, around the lumbo–sacral transition. As expected, HOXD4 was expressed up to the hindbrain and in somites, similar to the mouse gene. These results indicated that the rescue observed in the vertebrae derived from a faithful expression of the transgenes.
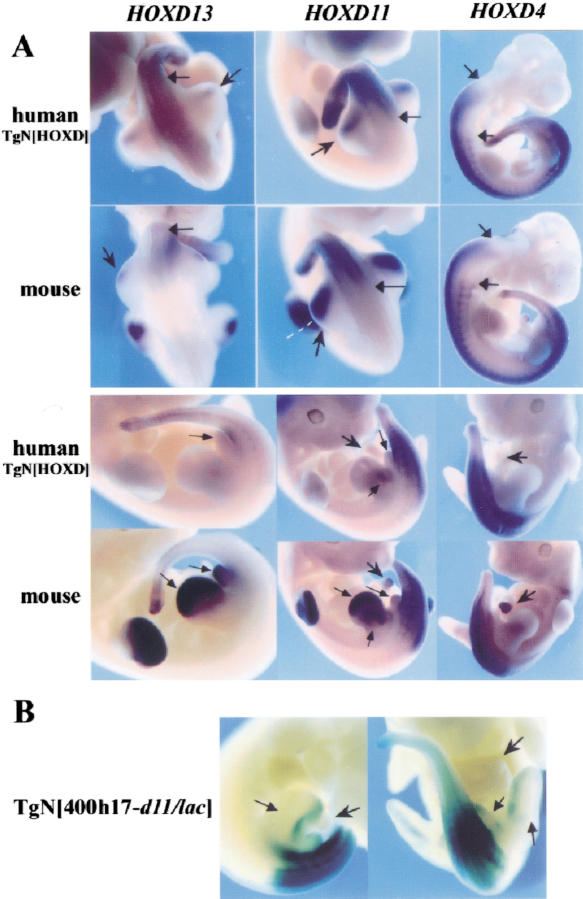
Figure 3.
(A) Human HOXD genes in mice. In E10.5 embryos (top), the human HOXD13, HOXD11, and HOXD4 genes were expressed in the trunk, up to anterior levels similar to those of the corresponding murine genes (black arrows). Human HOXD13 and HOXD11 were also expressed weakly in the developing posterior hindlimbs (arrowheads). In contrast, mouse Hoxd13 and Hoxd11 were expressed in both hindlimb and forelimb buds, with a more anterior extension (black arrowhead). In E11.5 embryos (bottom), the human genes were expressed like their murine counterparts in the trunk. However, neither HOXD13 nor HOXD11 were expressed in the limbs or genital bud (black arrows), whereas the murine genes were strongly active there. Arrowheads indicate the intestinal hernia, which stained with both Hoxd4 and Hoxd11 probes, while the human genes were silent. (B) Hoxd11/lacZ expression in transgenic embryo with a 215 kb mouse BAC with the entire HoxD cluster and lacZ sequences within Hoxd11. The faint expression in posterior hindlimb did not correspond to the signal given by the endogenous gene (cf. with Hoxd11 in A).
In contrast, expression of human genes in developing limbs was deficient. Early on, weak expression of both HOXD13 and HOXD11 was detected in the posterior limb bud (Fig. 3A, top, arrows). At this stage, transcript distribution differed from that of the corresponding mouse genes, which gave stronger signals over a wider domain including distal parts, in addition to the posterior half (Fig. 3A, top). Subsequently (Fig. 3A, bottom), the difference accentuated with an almost complete disappearance of all transcripts from the limbs for both HOXD13 and HOXD11, whereas endogenous genes developed their robust expression patterns in both presumptive digits (Hoxd13 and Hoxd11) and forearms (Hoxd11). Expression of human posterior genes was also absent from the developing genitalia, whereas a strong signal was detected for mouse Hoxd10, Hoxd11, Hoxd12, and Hoxd13. Likewise, expression of the mouse Hoxd4 and Hoxd11 genes in part of the intestinal hernia (Fig. 3A, bottom, arrows) was not recapitulated by the PAC transgene.
These observations implied that enhancers regulating Hoxd genes in limbs, genitalia, and intestinal hernia were absent from the 120-kb human PAC. This was further investigated with transgenic mice carrying a 215-kb large BAC with the murine HoxD complex, starting 100 kb 5′ of Hoxd13 and extending up to 10 kb downstream of Hoxd1. Thus, it contained an additional 95 kb of flanking DNA when compared to the human PAC. By using recE/T dependent recombination (Zhang et al. 1998; Muyrers et al. 1999), lacZ reporter sequences were introduced in the Hoxd11 gene to follow its expression in mice transgenic for the BAC. Three founders were recovered, stained for β-gal activity and compared with the conventional Hoxd11/lacZ transgene patterns (Gérard et al. 1993; Fig. 3B). In all cases, the BAC Hoxd11 was expressed with the expected anterior boundary in the trunk. However, as for the human PAC, Hoxd11/lacZ failed to be expressed in distal limbs, genitalia, and intestinal hernia. The transgenic pattern resembled that described for the short conventional transgene (Gérard et al. 1993), suggesting that no major Hoxd11 regulatory elements were localized in the 215-kb piece of DNA, other than those already present at the Hoxd11 locus.
Deletion of the mouse HoxD complex
To confirm the remote locations of these enhancers, we engineered a full deletion of HoxD, from Hoxd1 to upstream Hoxd13 (Del9). This deletion was made by placing a Hoxd1/lacZ fusion along with a loxP site into a chromosome carrying a Hoxd11/lacZ reporter transgene (TgH[d11/lac]Ge) between Hoxd13 and Evx2 (Fig. 1B). As this latter transgene also contains a loxP site, treatment of positive embryonic stem (ES) cells with Cre recombinase led to the replacement of the entire complex by a Hoxd11/lac reporter transgene. Once the cluster had been removed, β-gal detection was used as an indicator of the remaining regulatory influences. This expression (Fig. 4; Del9) was compared with that of the same transgene, either when recombined upstream of Hoxd13 (Fig. 4; d11/lac), or after partial deletions (Fig. 4; Del3 and Del7, respectively). In this way, the transcription of the same reporter gene was monitored at four different positions in the cluster, along with a progressive reduction of the gene complex.
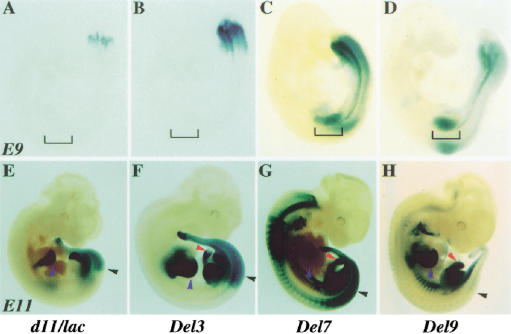
Figure 4.
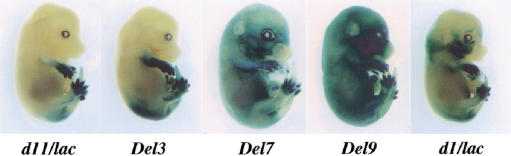
Hoxd11/lacZ gene expression in HoxD deletions. (Left to right) TgH[d11/lac]Ge, Del3, Del7, and Del9 embryos. (A–D) In E9 embryos, the TgH[d11/lac]Ge locus was expressed around the proctodeum (A), like the neighboring Hoxd13 gene. The Del3 allele showed a more anterior domain, mimicking Hoxd11 (B). The brackets emphasize the absence of staining in limb buds. In the right two panels (C,D), the Del7 and Del9 alleles showed expression profiles expanded anteriorly, involving the forelimb buds. (E–H) In E11 embryos, the TgH[d11/lac]Ge locus (E) showed the most posterior expression. In the trunk, the anterior limit was located at pre-vertebra 27, the future lumbo–sacral transition (black arrowhead in E–H). Blue arrowheads indicate expression in digit primordia. In Del3 embryos, the limit of expression was at pre-vertebra 25 (F). Expression in hernial gut (F, red arrowhead) was maintained in the other configurations to the right (G–H). In Del7 embryos, the anterior limit of expression in the central nervous system was reminiscent of Hoxd3 (G). In Del9 embryos, few cells only showed expression in either paraxial mesoderm or spinal cord (H). The strong staining in cervical pre-vertebrae detected from the Del7 locus was lost. In ventral regions such as the hyoid, rib primordia, and ventral tail mesoderm, expression was seen as anterior as the first branchial arch. Expression in both the digit primordia and hernial gut were preserved in the absence of the HoxD cluster (H).
In embryonic day (E)9.5 embryos, the TgH[d11/lac]Ge transgene was severely suppressed, giving a delayed signal restricted to the posterior part of the embryo (Fig. 4A), thus resembling that of the neighboring gene Hoxd13 (van der Hoeven et al. 1996). When the transgene was placed near Hoxd10, along with a deletion of Hoxd13 and Hoxd12, expression extended anteriorly, to recapitulate the Hoxd11 pattern (Del3; Fig. 4B). After deletion of seven genes (Del7) or of the entire complex (Del9), the early expression profile was not much different from the Del3 pattern. However, lacZ expression was detected at a more anterior body level, including the intermediate plate mesoderm and the emerging forelimb buds (Fig. 4C–D). Subsequently, well-established Hox gene profiles were observed (Fig. 4E–G), with a posterior restriction for the TgH[d11/lac]Ge locus in both spinal cord and somitic mesoderm. A rostral extension of these expression domains was observed in Del3 animals, reaching the lumbar region (Fig. 4F, black arrowhead), whereas expression in Del7 was wider, resembling that of Hoxd3 or Hoxd4. In this latter case, a “pan–Hoxd” pattern was recovered, in which all expression sites for Hoxd genes were stained. In developing limbs, a strong staining was seen in the distal parts, the presumptive digits, with additional staining in the future forearm for both Del3 and Del7 configurations (Fig. 4F–G). At this stage, expression was also observed in the genital bud for all three lines, whereas only the Del3 and Del7 fetuses displayed staining in their intestinal hernia (Fig. 4F–G).
The staining of Del9 embryos revealed two remarkable features. First, staining was absent from most of the developing CNS and somitic mesoderm of E11.5 fetuses (Fig. 4H). Therefore, the deletion of the cluster not only removed elements necessary for colinear expression, but also prevented expression of the Hoxd11/lacZ transgene in the posterior regions, as expected from its behavior at random genomic positions (Gérard et al. 1993). Second, the three major sites of transgene expression in the Del3 and Del7 loci, besides the developing trunk (i.e., the distal limbs, genital eminence, and intestinal hernia), still stained strongly, even though none of these expression specificities was detected on random integration of the same transgene. From these deletions, we confirmed that while the presence of the complex is required for the colinear process in the trunk, regulatory elements responsible for additional expression features were located mostly outside the cluster.
Maintenance of expression
Once Hox genes have been activated, their expression needs to be maintained (Deschamps et al. 1999). We used our set of deletions to assess the importance of the cluster in this mechanism by looking for expression maintenance of the same transgene within a Hox complex progressively reduced in size. At 12.5 days, the patterns were maintained and staining was absent from the heads of all four fetuses (Fig. 4E-H). At 15.5 days, both the TgH[d11/lac]Ge and Del3 embryos showed a tight restriction of the expression patterns to the posterior parts, indicating maintenance of the early pattern (Fig. 5). However, a diffuse staining appeared throughout Del7 embryos, including the head (Fig. 5), suggesting that large deletions in the cluster had an impact on maintenance. Accordingly, Del9 fetuses showed strong reporter activity throughout the embryos, in contrast to the pattern observed three days earlier (Fig. 5). This progressive deregulation of maintenance along with size-reduction of the cluster indicated that qualitative as well as quantitative parameters are involved in this process.
Figure 5.
Maintenance of Hoxd gene expression. (Left to right) TgH[d11/lac]Ge, Del3, Del7, and Del9 E15 fetuses are shown. Both TgH[d11/lac]Ge and Del3 fetuses showed tight maintenance of the Hoxd11/lacZ expression pattern at E11. Cells anterior to the early expression boundaries showed no staining at a later stage. In Del7 late fetuses, however, some diffuse staining appeared in anterior regions, i.e., the early pattern was no longer maintained. In Del9 fetuses, expression spread throughout the entire specimen, indicating a deficient maintenance. The right panel (d1/lacZ) shows that expression at the Hoxd1 locus in a fully preserved complex was maintained as well, indicating that the nonmaintenance of the Del9 configuration was not caused by sequences in 3′ of the Hoxd1 locus, but rather to the deletion the cluster.
At the same stage, expression of a knock-in Hoxd1/lac allele was absent from most structures that showed nonmaintained Hoxd11/lac expression in Del9 (Fig. 5; right), suggesting that the inability of Del9 to maintain expression correctly correlated with the absence of the cluster rather than with the position of the reporter gene.
Discussion
Human HOXD genes in transgenic mice
Mice with a human HOXD complex expressed these genes during trunk development with the expected specificity, showing that control mechanisms responsible for determining both the tissue type and positioning of the expression boundaries are identical in mouse and human (Tuggle et al. 1990). Expression domains of human genes, such as HOXD11, were established with appropriate anterior boundaries, even when corresponding mouse endogenous functions were abrogated, and human products could restore a normal vertebral column, indicating that auto- and cross-regulatory interactions (Maconochie et al. 1997) between HOXD proteins were not likely behind this observation. However, the exact role of paralogous genes from other Hox clusters in this process will have to be determined.
Rescue was not unexpected as protein sequences are essentially identical between mouse and human. However, mice rescued by the human complex often showed five lumbar vertebrae instead of six, even though expression of the human genes was like that of the mouse counterparts. A given Hox expression boundary is often defined as the body level wherein transcription becomes robust. Nonetheless, low levels of transcripts are routinely found immediately anterior to such defined boundaries. Therefore, overexpression of a gene in its normal expression domain may increase the level of transcript to reach a functional threshold in an anterior adjacent metamere. In this view, a quantitative difference would be translated into an anterior shift of the functional domain (i.e., a mere increased quantity of normally expressed transcripts may lead to an apparent homeotic transformation; Charité et al. 1995).
In contrast to the trunk, the human HOXD complex was unable to rescue the limb phenotype induced by the deletion of Hoxd11, Hoxd12, and Hoxd13, due to the absence of human transcripts in the developing presumptive digit area. Previous work suggested that an enhancer sequence responsible for expression of mouse Hoxd genes in digits was located outside the cluster, upstream of Hoxd13. Our human PAC contained 40 kb of DNA upstream of HOXD13, yet posterior genes were silent in digits, indicating that this enhancer was located further upstream from the cluster. The minimal distance was extended to 100 kb after the mouse BAC transgene experiment. Interestingly, human posterior genes were expressed transiently in an early phase of limb bud development, with a posterior restriction. This particular phase was observed during the budding of zebrafish pectoral fins and was hypothesized to be an ancient feature of Hox gene expression in appendices, perhaps related to their expression in the trunk (Sordino et al. 1995). The fact that an isolated human HOXD complex could reproduce this trait further suggests that the early and transient posterior expression in appendages is linked to the cluster itself, rather than to sequences that developed subsequently during tetrapod evolution.
Deletion of the HoxD complex
A targeted HoxD deletion and its replacement with a Hoxd11/lacZ reporter gene confirmed and extended the results obtained through the transgenic approach. Mice carrying this deficiency showed phenotypic alterations resulting mostly from the combination of single loss-of-function alleles; therefore, the phenotype was somewhat related to the Del7 mutant condition (Zakany and Duboule 1999). Upon deletion of the cluster, modifications in the expression pattern of the reporter construct were seen. However, expression in limbs was maintained. Prior observations on random integration of transgenes away from the HoxD locus, as well as the inability of the PAC and BAC transgenes to implement this regulation, position these regulations outside the cluster.
The expression of the Hoxd11/lacZ reporter transgene, after deletion of the cluster, was complementary to that observed with the human PAC transgene, such that summation of both profiles would produce a pan-Hox pattern, similar to the Del7 configuration (Zakany and Duboule 1999). Consequently, Hox expression domains can be separated between those controlled by sequences located within the complex itself and those depending on enhancer sequences located distal to the cluster. In the first case, these controls likely illustrate the ancient function of this gene family, elaborated through the amplification of a few original genes, which were then used to specify anterior to posterior information in a colinear fashion. This ancestral regulatory module was maintained in most metazoans displaying a rostral–caudal axis and bilateral symmetry. Subsequently, subsets of these genes were recruited for novel functions, through changes either in cis-DNA or in those factors that could recognize a preexisting cis-acting DNA sequence turning it into an active control element. The fact that Hox clusters are tightly organized, with a gene about every 10 kb and embedded regulatory elements, suggests that the emergence of these enhancers from inside the cluster might have disturbed the ongoing ancient gene regulatory circuitry with pleiotropic consequences.
Maintenance of expression
In Drosophila, expression of Hox genes follows two phases. After their activation, transcription is maintained via the action of activating and repressing factors. The Polycomb group of genes (Pc-G) maintains genes silenced wherever they have not been activated, therefore, Pc-G loss-of-function mutations derepress Hox gene transcription. Pc-G-mediated repression is achieved through the interaction between a protein complex and Polycomb response elements (Pirrotta 1999). Inactivation of murine Pc-like genes suggested that vertebrate Hox gene expression may also rely on such a system (e.g., Gould 1997).
Here, we show that expression of a Hox reporter transgene is maintained differentially depending on the presence or absence of the surrounding complex. Tight maintenance was observed when most of the cluster was present, whereas maintenance was abolished when the cluster was reduced in size. Interestingly, deletion of two-thirds of the cluster gave an intermediate picture with a somewhat reduced maintenance ability. This progressive loss of maintenance may indicate an additive effect of multiple sites in operating the silencing process.
Materials and methods
Transgenic mice, ES cells, and targeted deletions
The human PAC78J1 was obtained using a PAC library (Genome Systems; catalog no. FPAC-3387) and a human HOXD9 probe. Restriction analysis and hybridization showed that this PAC contained almost the entire HOXD complex, from 30 kb upstream of EVX2 to 10 kb downstream of HOXD3. The PAC was linearized with Sgf1 and injected in C57BL/6 × DBA F1 fertilized mouse eggs as in Schedl et al. (1993). G0 mice were screened by Southern blot with human probes specific for HOXD13 and the HOXD8 to HOXD4 intergenic region. The integrity of the PAC was verified using several probes and by PCR. Copy number was estimated by comparison with a mouse HoxD probe.
ES cell culture, electroporation, chimera production, skeletal preparations and X-gal staining were as described previously (van der Hoeven et al. 1996). ES cells carrying the TgH[d11/lac]Ge allele (van der Hoeven et al. 1996) were targeted in further experiments to introduce a second loxP site (Del3, Zakany and Duboule 1996; Del7, Zakany and Duboule 1999; Del9, this study). To produce this latter chromosome, a loxP site was introduced into a HindIII site in the second exon of Hoxd1 (Frohman and Martin 1992) with the same polarity as the loxP site in the TgH[d11/lac]Ge ES cell. Clones from secondary targeting event were treated with Cre recombinase to induce deletions, which were passed through the germ line of mice.
BAC recombination and transgenesis
A mouse BAC (RPCI23–400h17) was identified in the GenBank database and obtained from Roswell Park Cancer Institute (Buffalo, NY). To introduce LacZ reporter sequences into the Hoxd11 gene, we used ET recombination (Zhang et al. 1998; Muyrers et al. 1999; Nefedov et al. 2000). The kanamycin resistance gene from pUC4Km (gift of S. Lin, Medical College of Georgia, Altanta) was inserted downstream of the polyadenation sequence of the LacZ reporter gene, in an Afl2–Avr2 subclone of the pGemE/ElacZpA construct (Gérard et al. 1993). The resulting plasmid was digested by NcoI and PpuMI and the 5.8 kb Hoxd11/lacZpAKanR fragment purified by gel electrophoresis. This targeting fragment was electroporated in DH10B cells containing both the target BAC and the pGETrec plasmid, after 40 min induction with 0.2% (w/v) L-arabinose. Eleven recombinant colonies that grew on 12.5 μg/mL chloramphenicol and 20 μg/mL kanamycin were analyzed. All had the correct integration of lacZ in Hoxd11. The integrity of the BACs was verified by restriction enzyme fingerprinting with EcoRI, HincII, HindIII, and XhoI. No change between the original 400h17 BAC and the 400h17–d11/lac BAC was detected, except those caused by the lacZ insertion. The BAC was linearized by PI–SceI and injected as for the PAC.
Whole-mount in situ hybridization
WISH was performed using human probes derived from the 3′ UTRs of HOXD13, HOXD11, and HOXD9. The HOXD13 antisense probe was obtained from the EST nj13h05 and the HOXD11 probe from EST nh27c09. Both EST clones were obtained from the IMAGE consortium. The 3′ UTR of HOXD4 was amplified by PCR using the PAC 78j1 as template. The murine probes were described previously (Hoxd13, Dollé et al. 1993; Hoxd11, Gérard et al. 1996; Hoxd4, Featherstone et al. 1988).
Acknowledgments
We thank members of the Duboule laboratory for sharing reagents and comments, M. Friedli for technical help, F. Stewart and Y. Zhang for help with ET recombination, R. Skoda for the human PAC, and S. Lin and P. Ioannou for plasmids. F.S. was supported by EMBO and HFSPO fellowships. This work was supported by the Swiss National Research Fund and the Claraz fund (D.D.) and by an ACS grant (T.F.V.).
The publication costs of this article were defrayed in part by payment of page charges.This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Denis.Duboule@zoo.unige.ch; FAX 41-22-702-6795.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.205701.
References
- Charité J, de Graaff W, Deschamps J. Specification of multiple vertebral identities by ectopically expressed Hoxb-8. Dev Dyn. 1995;204:13–21. doi: 10.1002/aja.1002040103. [DOI] [PubMed] [Google Scholar]
- Davis AP, Capecchi MR. A mutational analysis of the 5′ HoxD genes: Dissection of genetic interactions during limb development in the mouse. Development. 1996;122:1175–1185. doi: 10.1242/dev.122.4.1175. [DOI] [PubMed] [Google Scholar]
- Deschamps J, van den Akker E, Forlani S, De Graaff W, Oosterveen T, Roelen B, Roelfsema J. Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int J Dev Biol. 1999;43:635–650. [PubMed] [Google Scholar]
- Featherstone MS, Baron A, Gaunt SJ, Mattei GM, Duboule D. Hox-5.1 defines a homeobox-containing gene locus on mouse chromosome 2. Proc Natl Acad Sci. 1988;85:4760–4764. doi: 10.1073/pnas.85.13.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Martin GR. Isolation and analysis of embryonic expression of Hox-4.9, a member of the murine labial-like gene family. Mech Dev. 1992;38:55–67. doi: 10.1016/0925-4773(92)90038-l. [DOI] [PubMed] [Google Scholar]
- Gérard M, Chen JY, Gronemeyer H, Chambon P, Duboule D, Zakany J. In vivo targeted mutagenesis of a regulatory element required for positioning the Hoxd-11 and Hoxd-10 expression boundaries. Genes & Dev. 1996;10:2326–2334. doi: 10.1101/gad.10.18.2326. [DOI] [PubMed] [Google Scholar]
- Gérard M, Duboule D, Zakany J. Structure and activity of regulatory elements involved in the activation of the Hoxd-11 gene during late gastrulation. EMBO J. 1993;12:3539–3550. doi: 10.1002/j.1460-2075.1993.tb06028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes & Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- Gu H, Zou Y R, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre–loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Hérault Y, Beckers J, Kondo T, Fraudeau N, Duboule D. Genetic analysis of a Hoxd-12 regulatory element reveals global versus local modes of controls in the HoxD complex. Development. 1998;125:1669–1677. doi: 10.1242/dev.125.9.1669. [DOI] [PubMed] [Google Scholar]
- Hérault Y, Beckers J, Gérard M, Duboule D. Hox gene expression in limbs: Colinearity by opposite regulatory controls. Dev Biol. 1999;208:157–165. doi: 10.1006/dbio.1998.9179. [DOI] [PubMed] [Google Scholar]
- Kmita M, Kondo T, Duboule D. Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat Genet. 2000a;26:451–454. doi: 10.1038/82593. [DOI] [PubMed] [Google Scholar]
- Kmita M, van der Hoeven F, Zakany J, Krumlauf R, Duboule D. Mechanisms of Hox gene colinearity: Transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes & Dev. 2000b;14:198–211. [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell. 1999;97:407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Maconochie MK, Nonchev S, Studer M, Chan SK, Popperl H, Sham MH, Mann R, Krumlauf R. Cross-regulation in the mouse HoxB complex: The expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes & Dev. 1997;11:1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedov M, Williamson R, Ioannou PA. Insertion of disease-causing mutations in BACs by homologous recombination in Escherichia coli. Nucleic Acids Res. 2000;28:E79. doi: 10.1093/nar/28.17.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycomb silencing and the maintenance of stable chromatin states. Results Probl Cell Differ. 1999;25:205–228. doi: 10.1007/978-3-540-69111-2_10. [DOI] [PubMed] [Google Scholar]
- Rijli FM, Chambon P. Genetic interactions of Hox genes in limb development: Learning from compound mutants. Curr Opin Genet Dev. 1997;7:481–487. doi: 10.1016/s0959-437x(97)80074-3. [DOI] [PubMed] [Google Scholar]
- Schedl A, Larin Z, Montoliu L, Thies E, Kelsey G, Lehrach H, Schutz G. A method for the generation of YAC transgenic mice by pronuclear microinjection. Nucleic Acids Res. 1993;21:4783–4787. doi: 10.1093/nar/21.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Tuggle CK, Zakany J, Cianetti L, Peschle C, Nguyen-Huu MC. Region-specific enhancers near two mammalian homeo box genes define adjacent rostrocaudal domains in the central nervous system. Genes & Dev. 1990;4:180–189. doi: 10.1101/gad.4.2.180. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven F, Zakany J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- ————— Hox genes and the making of sphincters. Nature. 1999;401:761–762. doi: 10.1038/44511. [DOI] [PubMed] [Google Scholar]
- Zakany J, Fromental-Ramain C, Warot X, Duboule D. Regulation of number and size of digits by posterior Hox genes: A dose-dependent mechanism with potential evolutionary implications. Proc Natl Acad Sci. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]