Abstract
This study identified 431 psychrophilic or psychrotrophic isolates from commercial Irish beef abattoir environments and “blown packs” of vacuum-packed beef, using PCR and 16S rRNA sequencing, and estimated their intraspecies genetic diversity using restriction fragment length polymorphism (RFLP) analysis and spacer region PCR (SR-PCR). Twenty-five species were identified in the 431 isolates, with the most frequently recovered species being Clostridium gasigenes (n = 315), Clostridium estertheticum (n = 17), and a potentially novel species designated strain TC1 (n = 52). These species were previously found to be associated with a particular type of spoilage known as blown-pack spoilage (BPS), which occurs in chilled-stored (i.e., −1.5°C to 4°C) vacuum-packaged meat within 2 to 4 weeks and involves the production of large volumes of gas. Overall, the study demonstrates the considerable and not previously reported diversity of the anaerobic microflora in abattoirs and the presence of a wide range of organisms capable of causing BPS at chilled temperatures.
INTRODUCTION
Psychrophilic and psychrotrophic anaerobic clostridia have been associated with food poisoning (17) and the spoilage of vacuum-packaged chilled meats (7). Such bacteria have been detected in beef abattoirs and their environments, including animal feces, hides, gastrointestinal tracts of animals, and soil, as well as sewage and water (1, 5, 7, 25). During common abattoir operations, i.e., slaughter and dressing processes, there is a potential for these organisms to contaminate carcasses and derived meat products.
Preliminary studies on the prevalence of psychrophilic and psychrotrophic bacteria in beef abattoirs focused on the PCR detection of the 16S rRNA gene of specific spoilage organisms, e.g., Clostridium gasigenes and Clostridium estertheticum (7, 25). These species have been associated with “blown-pack” spoilage (BPS), which occurs in chilled (i.e., −1.5°C to 4°C) batches of vacuum-packaged meat within 2 to 4 weeks of storage and involves the production of large volumes of gas sufficient to produce severe pack distension, a putrid smell, and a metallic sheen on the affected meat (4, 15, 21, 25, 37). However, the prevalence of other pathogenic/spoilage psychrophilic or psychrotrophic anaerobic bacteria in beef abattoirs remains unknown.
Analysis of the 16S rRNA gene was also previously applied in the investigation of the interrelationships among psychrophilic spoilage clostridia and their relative significances in meat spoilage (18, 33, 35). Other molecular tools have also been widely used for differentiation within other bacterial groups (22, 32), to determine genetic diversity (16, 30), and to investigate bacterial epidemiology (26, 31). Some studies have used restriction fragment length polymorphism (RFLP) analysis and spacer region PCR (SR-PCR) to discriminate between clostridia (5, 8, 9). However, those studies used a relatively limited number of isolates (n = 20 to 22). A larger study employing a range of advanced analytical methods is necessary to gain a more adequate understanding of the genetic diversity of the cold-tolerant bacteria in beef abattoir environments.
The aim of this study was to identify 431 previously recovered (25) cold-tolerant anaerobic bacteria from commercial beef abattoirs and their environments using species-specific PCR and 16S rRNA sequencing and to investigate their intraspecies diversity using RFLP analysis and SR-PCR (27, 32) to gain a more accurate view of the nature and diversity of such organisms in these environments.
MATERIALS AND METHODS
Bacteria and sources.
The isolates (n = 431) were strictly anaerobic psychrophilic or psychrotolerant bacteria recovered from various sites in four Irish beef abattoirs and their environments and from “blown packs” of beef, using cold (i.e., 4°C), long anaerobic enrichments (approximately 3 weeks) in nonspecific media, i.e., prereduced peptone yeast extract glucose starch (PYGS) medium (23) and Columbia blood agar (CBA; Oxoid Ltd., Basingstoke, United Kingdom) supplemented with 5% defibrinated horse blood, as previously described (25). Strain TC1, a local (Teagasc) isolate recovered from a cattle hide in an Irish beef abattoir, was used for a phylogenetic comparison with its phylogenetically closest related species. Reference strains Clostridium bifermentans DSMZ 14991T, C. estertheticum subsp. estertheticum DSMZ 8809T, C. estertheticum subsp. laramiense DSMZ 14864T, C. gasigenes DSMZ 12272T, C. glycolicum DSMZ 1288T, and C. lituseburense DSMZ 797T were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany).
DNA extraction.
All isolates and reference strains were revived anaerobically in prereduced PYGS medium and subcultured on CBA supplemented with 5% defibrinated horse blood. Colonies were picked off with a sterile loop, mixed with 180 μl of a cell lysis buffer (20 mM Tris-Cl [pH 8.0], 2 mM sodium EDTA, 1.2% Triton X-100, 20 mg ml−1 lysozyme), and incubated at 37°C for 2 h. Genomic DNA was extracted by using a DNeasy blood and tissue kit (Qiagen Ltd., Crawley, United Kingdom) according to the manufacturer's recommended protocol for DNA extraction from Gram-positive bacteria.
Identification of isolates.
All isolates were screened for the presence of specific fragments of the 16S-23S rRNA internal transcribed spacer and the 16S rRNA gene of the “blown-pack” spoilage bacteria C. estertheticum and C. gasigenes, respectively. PCR was conducted by using primers EISRF and EISRR for the detection of 16S-23S rRNA internal transcribed spacer fragments of C. estertheticum subsp. estertheticum and C. estertheticum subsp. laramiense. Primers 16SDBF and 16SDBR were used to detect 16S rRNA gene fragments of C. gasigenes according to previously reported protocols (6). Primers were purchased from MWG Biotech, Martinsried, Germany. All isolates that did not give a positive reaction using species-specific PCR amplification (including two isolates identified as C. estertheticum and two isolates identified as C. gasigenes) were subjected to PCR amplification using universal (eu)bacterial primers 8FPL and 806R (29), annealing to positions 8 to 27 and 806 to 787 of the Escherichia coli numbering of the 16S rRNA gene, respectively. Successful PCR from these primers gave an 800-bp product. Prior to the addition of DNA templates, any residual DNA in the Taq DNA polymerase of the PCR mix was inactivated by the addition of the restriction enzyme AluI (Roche Diagnostics, Basel, Switzerland) and incubation at 37°C for 20 min, followed by heating at 80°C for 2 min and cooling to 4°C. Products were separated on a 1.5% (wt/vol) agarose gel containing 0.5 mg ml−1 ethidium bromide at 90 V for 1 h, visualized on a UV transilluminator, and excised by using a scalpel. The QIAquick gel extraction kit (Qiagen Ltd.) was used to purify the PCR products according to the manufacturer's protocol. Purified PCR products were dissolved in 5 mM Tris-HCl (pH 8.0). The nucleic acid concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and adjusted to a final concentration of 5 to 30 ng μl−1. Sequencing reactions were performed by using the BigDye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA), and products were purified by using the BigDye XTerminatorT purification kit (Applied Biosystems). Sequencing was performed by using an ABI Prism 3130XL genetic analyzer (Applied Biosystems). Primers for the sequencing of both strands of DNA were 8FPL-SHORT (5′-GAGTTTGATCCTGGCTCAG-3′) and 806R-SHORT (5′-GCGGCCGCGGACTACCAG-3′). Forward and reverse sequences from each microorganism were aligned by using an online BLAST alignment tool (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi), and mismatches were corrected manually following a visual inspection of the electropherograms. Sequences were compared with the NCBI sequence database by using the BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and results with the highest similarity values were assigned to species.
Restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA genes of the isolates.
PCR amplification of the isolates was performed by using universal primers 63F and 1387R (27), corresponding to nucleotides 63 to 1387 of the 16S rRNA gene, according to previously reported protocols (27). Following PCR, 5-μl samples containing the amplified 16S rRNA gene were digested with the restriction enzyme CfoI (Roche Diagnostics) according to the manufacturer's recommendations. Digestion products and the molecular weight marker (DNA ladder, 100 bp; Promega, Southampton, United Kingdom) were run on a 1.5% (wt/vol) agarose gel containing 0.5 mg ml−1 ethidium bromide at 90 V for 1 h. Fingerprints were saved as tagged image file format (TIFF) files and imported into BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). Band positions on each gel were normalized against the molecular weight marker.
SR-PCR of the isolates.
The method used for spacer region PCR (SR-PCR) was described previously by Song et al. (32), with digital analysis of the fingerprints. PCR products were separated on a 6% bisacrylamide (29:1) gel with 1× Tris-borate-EDTA running buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA [pH 8.3]) for 90 min at a field strength of 12.5 V cm−1. Following electrophoresis, gels were stained for 20 min in a 1.0-μg ml−1 ethidium bromide solution, and the patterns of the amplified products were visualized under UV light. Banding patterns were imported into BioNumerics software as described above.
Intraspecies differentiation of the isolates.
All typeable RFLP and SR-PCR isolates (i.e., having bands in gels) were inserted into BioNumerics software. Fingerprints from strains representing each species type were imported as a group. The similarity matrix for each group was calculated by using Dice's coefficient, and dendrograms were created by using the unweighted-pair group method with arithmetic means (UPGMA) for (i) RFLP on its own, (ii) SR-PCR on its own, and (iii) a combination of both experiments. Cluster cutoff values were calculated as described in the BioNumerics software.
Phylogenetic comparison of TC1 with its most closely related species.
A phylogenetic comparison of isolate TC1 and its closest related species, i.e., C. lituseburense DSMZ 797T, C. bifermentans DSMZ 14991T, and C. glycolicum DSMZ 1288T, was carried out by using 16S rRNA gene sequencing and SDS-PAGE. 16S rRNA gene sequence analysis using universal (eu)bacterial primers 8FPL and 806R was performed as described above. A similarity matrix was created by using BioNumerics software, after discarding unknown bases, with similarity calculation with a gap penalty of 0%, based on pairwise alignment using an open-gap penalty of 100% and a unit gap penalty of 0%. For SDS-PAGE, the preparation of the protein extracts and protein gel electrophoresis were performed according to methods described previously by Pot et al. (28). Normalized and digitized protein patterns were numerically analyzed and compared by using BioNumerics software. A molecular weight marker (DNA ladder, 100 bp; Promega, Southampton, United Kingdom) was included for the normalization of the fingerprints.
RESULTS
Identification of the isolates.
In total, 25 discrete species of strictly anaerobic psychrophilic or psychrotolerant bacteria were identified in the 431 abattoir isolates (Table 1). Thirteen of the 25 species (i.e., species 2, 5, 7, 8, 14, 15, 18, 19, 20, 22, 23, 24, and 25) (Table 1) were assigned to uncultured or not previously described species (as the BLAST database indicated), with similarity values of ≥98%. The majority of the isolates (n = 315 of 431; 73.1%) were C. gasigenes. Fifty-two isolates (12%) had 16S rRNA sequences identical to the sequence of TC1. Seventeen strains (3.9%) were C. estertheticum. Four isolates identified with species-specific PCR (2 of the 17 C. estertheticum and two of the 315 C. gasigenes isolates) were confirmed with 16S rRNA sequencing analysis. Nonclostridial isolates included Bacteroides propionicifaciens (n = 2) and Vagococcus sp. strain YS32 (n = 1). Vagococcus sp. strain YS32 was included in the study because it was not able to grow aerobically within 21 days at 4°C. The sequencing method used failed to produce usable data for eight isolates, which were therefore assigned as “unidentified” species.
Table 1.
Isolates from beef abattoirs and their environmentsb
| Species | Microorganism | No. of isolates from source |
Total no. of isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | T | L | BA | HP | ET | F | H | HV | C | BH | P | |||
| 1 | C. gasigenes | 5 | 3 | 6 | 3 | 5 | 3 | 72 | 208 | 2 | 4 | 2 | 2 | 315 |
| 2 | TC1 | 1 | 1 | 11 | 36 | 1 | 1 | 1 | 52 | |||||
| 3 | C. estertheticum | 1 | 1 | 1 | 1 | 1 | 4 | 6 | 2 | 17 | ||||
| 4 | C. algidixylanolyticum | 1 | 3 | 3 | 2 | 2 | 2 | 13 | ||||||
| 5 | Clostridium sp. CYP7 | 2 | 1 | 3 | ||||||||||
| 6 | B. propionicifaciens | 1 | 1 | 2 | ||||||||||
| 7 | Uncultured bacterium clone HTE5 | 1 | 1 | 2 | ||||||||||
| 8 | Uncultured bacterium clone HDBW-WB50 | 1 | 1 | 2 | ||||||||||
| 9 | C. propionicum | 1 | 1 | |||||||||||
| 10 | C. bowmanii | 1 | 1 | |||||||||||
| 11 | C. beijerinckii | 1 | 1 | |||||||||||
| 12 | C. botulinum type E | 1 | 1 | |||||||||||
| 13 | C. glycolicum | 1 | 1 | |||||||||||
| 14 | Clostridium sp. strain CYP11 | 1 | 1 | |||||||||||
| 15 | Clostridium sp. strain V13 | 1 | 1 | |||||||||||
| 16 | C. thiosulfatireducens | 1 | 1 | |||||||||||
| 17 | C. xylanolyticum | 1 | 1 | |||||||||||
| 18 | Swine effluent bacterium CHNDP10 | 1 | 1 | |||||||||||
| 19 | Swine effluent bacterium CHNDP4 | 1 | 1 | |||||||||||
| 20 | Uncultured bacterium clone AKIW891 | 1 | 1 | |||||||||||
| 21 | C. botulinum type F | 1 | 1 | |||||||||||
| 22 | Uncultured bacterium clone Hg16 | 1 | 1 | |||||||||||
| 23 | Uncultured bacterium clone ORSPEP_b09 | 1 | 1 | |||||||||||
| 24 | Uncultured rumen bacterium clone BS40 | 1 | 1 | |||||||||||
| 25 | Vagococcus sp. YS32a | 1 | 1 | |||||||||||
| Unidentified | 2 | 6 | 8 | |||||||||||
| Total | 8 | 6 | 10 | 9 | 10 | 4 | 101 | 266 | 7 | 4 | 3 | 3 | 431 | |
Facultative anaerobic isolate.
BLAST analysis using the NCBI nucleotide sequence database identified each isolate with similarity values of ≥98%. S, soil; T, transport; L, lairage; BA, bleeding area; HP, hide puller; ET, evisceration table; F, feces; H, hides; HV, hooves; C, carcasses; BH, boning hall; P, purge from blown packs.
Intraspecies differentiation using RFLP and SR-PCR.
RFLP analysis identified up to five bands in the examined isolates, although most isolates had between two and four bands. SR-PCR banding patterns were more complex than RFLP patterns. Fragments of the 16S-23S rRNA spacer region yielded between 5 and 15 bands, ranging in size from 45 to 1,000 bp. One hundred twenty-four isolates were not typeable by RFLP, and 15 were not typeable by SR-PCR analysis. Separately and in combination, RFLP and SR-PCR analyses revealed intraspecies differentiation. For each species type, RFLP analysis gave more clusters than did SR-PCR. The similarity values as well as the number of clusters obtained for each of the most frequently isolated species (C. gasigenes, TC1, C. estertheticum, and C. algidixylanolyticum) are shown in Table 2. These data show that the greater the number of isolates within each species type, the more clusters were obtained. For example, C. gasigenes isolates were the most diverse in terms of the number of clusters obtained (seven with RFLP, five with SR-PCR, and seven with a combination of both methods). There was no correlation between the memberships of RFLP and SR-PCR clusters (i.e., isolates “clustered” by RFLP were not clustered by SR-PCR).
Table 2.
Intraspecies differentiation of C. gasigenes, TC1, C. estertheticum, and C. algidixylanolyticum using RFLP, SR-PCR, and a combination of the two methodsa
| Microorganism | RFLP |
SR-PCR |
RFLP and SR-PCR |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of typeable isolates | SV (%) | C (%) (no. of clusters) | No. of typeable isolates | SV (%) | C (%) (no. of clusters) | No. of typeable isolates | SV (%) | C (%) (no. of clusters) | |
| C. gasigenes | 208 | 53.3 | 75 (7) | 300 | 54.6 | 80 (5) | 208 | 54.2 | 76.5 (7) |
| TC1 | 52 | 50.9 | 82.9 (5) | 51 | 61.3 | 92.1 (3) | 51 | 63.1 | 85.9 (2) |
| C. estertheticum | 18 | 60.8 | 80 (3) | 19 | 75.7 | 86.3 (2) | 18 | 72.3 | 80.7 (2) |
| C. algidixylanolyticum | 2 | 33.3 | 33.3 (1) | 13 | 72.3 | 89.9 (2) | 2 | 71.4 | 71.4 (1) |
SV, similarity value (percent); C, percent cluster cutoff.
Phylogenetic comparison of TC1 with its most closely related species.
The phylogenetic analysis of TC1, based on its 16S rRNA sequence, separated this isolate from its closest relatives, i.e., C. lituseburense (97.3% similarity), C. bifermentans (95.8%), C. bartlettii (95.8%), and C. glycolicum (95.7%). SDS-PAGE profiles (Fig. 1) clearly differentiated TC1 from its closest neighbors within the genus Clostridium (C. lituseburense, C. bifermentans, and C. glycolicum). Overall, comparisons of the phylogenetic analysis (16S rRNA sequence) and SDS-PAGE profile similarity values for TC1 and related species suggest that TC1 may represent a new and separate species of the genus Clostridium.
Fig. 1.
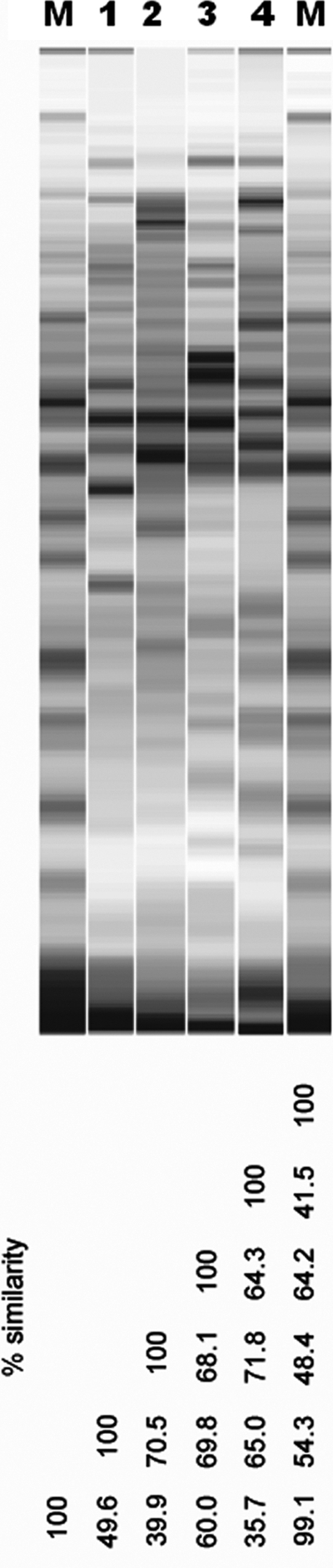
SDS-PAGE profiles of strains and similarity values (percent). Lanes: M, molecular weight marker; 1, TC1; 2, Clostridium lituseburense (DSMZ 797); 3, C. bifermentans (DSMZ 14991); 4, C. glycolicum (DSMZ 1288).
DISCUSSION
Clostridium was the most frequently isolated genus within the anaerobic psychrophilic and/or psychrotrophic bacterial populations recovered from beef abattoir environments and/or samples. These organisms were recovered from a wide range of pre- and postslaughter locations and/or samples, with beef hides and feces being identified as the most highly contaminated sites. Contamination of meat with such microorganisms may occur as a result of contact either with clostridial spores surviving passage through the digestive system of slaughter animals or with spores of environmental origin, e.g., carried on the coats of animals entering the abattoir. Alternatively, such contamination may be indirect, via meat plant ventilation or drainage systems.
The most frequently isolated species were C. gasigenes, TC1, C. estertheticum, and C. algidixylanolyticum, all of which were previously detected in various locations in beef abattoirs in New Zealand (1, 5) and, more recently, in Ireland (25). The current study is, however, the first to show that these species make up the majority of the psychrophilic and/or psychrotolerant anaerobic bacterial population in beef abattoirs and their environments. These species have been frequently associated with the spoilage of vacuum-packed chilled lamb and beef (3, 4, 13, 24), and their relatively high incidence presents a considerable commercial threat to the meat industry. It also highlights the need for effective measures to reduce their incidence, persistence, and dissemination within abattoir locations and their transfer to, and persistence in, meat products likely to be stored under conditions which favor their growth.
The clostridia identified in beef abattoirs included C. botulinum types E and F, isolated from cattle hooves and feces, respectively, and C. glycolicum, isolated from cattle feces. Clostridium botulinum is a very significant human pathogen, producing neurotoxin types E and F, the agents of botulism (17), while C. glycolicum is a potential human pathogen associated with wound infections (20). Although nonproteolytic C. botulinum types E and F have been detected (by PCR) in cattle feces (14), the detection of C. glycolicum from beef abattoir environments has not been previously reported. These pathogens are capable of germination, growth, and/or toxin production under the low-temperature, anaerobic conditions applied in commercial meat storage. Their presence, as demonstrated in this study, highlights the importance of the development and implementation of proper control practices to prevent their access to, and persistence in, derived meat products.
A comparison of the results obtained in this study with data in the BLAST database indicated that a significant number of the isolates could not be easily allocated to known species, suggesting continuing uncertainty as to the specific nature of some elements of the psychrophilic and psychrotrophic microflora in the abattoir environments.
The diversity of RFLP and SR-PCR data obtained in this study reconfirms previous reports on the scale of diversity within the genus Clostridium and reinforces the need for an improved understanding of the relationships within this genus (19, 34). This study has shown that even though molecular fingerprinting techniques and/or analyses of spacer region sequences between the 16S and the 23S rRNA genes can demonstrate a greater range of diversity than phenotypic methods (2, 10), these molecular methods are not sufficiently discriminatory to resolve bacterial microdiversity (11, 36) within such a complex genus. That being said, the species and/or subspecies relevance of the observed diversity remains unclear.
The phylogenetic study of TC1, based on 16S rRNA gene sequencing and SDS-PAGE, clearly differentiated this organism from its most closely related neighbors, suggesting that it may be a novel species within the genus Clostridium. Further studies are necessary to fully characterize this organism, which is of clear commercial significance, as it has been demonstrated to cause BPS (24). The recognition of TC1 as a potentially important causative agent of BPS in vacuum-packed chilled meats highlights the need for more information on its incidence in abattoir environments and for an urgent review of its relative significance within the range of the causative agents of BPS in vacuum-packed meats.
In conclusion, this study has established the considerable diversity of the anaerobic microflora present in, and recoverable from, beef abattoirs and their environments. The psychrophilic and psychrotrophic clostridial isolates did not form a homologous cluster by RFLP and SR-PCR analyses and exhibited levels of diversity similar to those reported previously for the clostridia in clusters I and XIVa (12). The study identified considerable overall intrastrain diversity and a number of isolates which may represent new species. Further investigations using a polyphasic approach (combination of phenotypic and genotypic characterizations) may be worthwhile and necessary to help characterize such new species and their interrelationships. The results of such studies would be of particular interest and value to the meat industry in its efforts to better understand and control current and emerging agents of psychrophilic and psychrotrophic spoilage of meat.
ACKNOWLEDGMENTS
We acknowledge the Food Institutional Research Measure (FIRM), administered by the Department of Agriculture, Fisheries and Food (DAFF), Ireland, for funding this research.
Footnotes
Published ahead of print on 15 April 2011.
REFERENCES
- 1. Boerema J. A., Broda D. M., Bell R. G. 2003. Abattoir sources of psychrophilic clostridia causing blown pack spoilage of vacuum-packed chilled meats determined by culture-based and molecular detection procedures. Lett. Appl. Microbiol. 36:406–411 [DOI] [PubMed] [Google Scholar]
- 2. Brambilla E., Hippe H., Hagelstein A., Tindall B. J., Stackebrandt E. 2001. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23–33 [DOI] [PubMed] [Google Scholar]
- 3. Broda D., Saul D., Bell R., Musgrave D. 2000. Clostridium algidixylanolyticum sp. nov., a psychrotolerant, xylan-degrading, spore-forming bacterium. Int. J. Syst. Evol. Microbiol. 50:623–631 [DOI] [PubMed] [Google Scholar]
- 4. Broda D., Saul D., Lawson P., Bell R., Musgrave D. 2000. Clostridium gasigenes sp. nov., a psychrophile causing spoilage of vacuum-packed meat. Int. J. Syst. Evol. Microbiol. 50:107–118 [DOI] [PubMed] [Google Scholar]
- 5. Broda D. M., Bell R. G., Boerema J. A., Musgrave D. R. 2002. The abattoir source of culturable psychrophilic Clostridium spp. causing ‘blown pack’ spoilage of vacuum-packed chilled venison. J. Appl. Microbiol. 93:817–824 [DOI] [PubMed] [Google Scholar]
- 6. Broda D. M., Boerema J. A., Bell R. G. 2003. PCR detection of psychrophilic Clostridium spp. causing ‘blown pack’ spoilage of vacuum-packed chilled meats. J. Appl. Microbiol. 94:515–522 [DOI] [PubMed] [Google Scholar]
- 7. Broda D. M., Boerema J. A., Brightwell G. 2009. Sources of psychrophilic and psychrotolerant clostridia causing spoilage of vacuum-packed chilled meats, as determined by PCR amplification procedure. J. Appl. Microbiol. 107:178–186 [DOI] [PubMed] [Google Scholar]
- 8. Broda D. M., Musgrave D. R., Bell R. G. 2003. Molecular differentiation of clostridia associated with ‘blown pack’ spoilage of vacuum-packed meats using internal transcribed spacer polymorphism analysis. Int. J. Food Microbiol. 84:71–77 [DOI] [PubMed] [Google Scholar]
- 9. Broda D. M., Musgrave D. R., Bell R. G. 2000. Use of restriction fragment length polymorphism analysis to differentiate strains of psychrophilic and psychrotrophic clostridia associated with ‘blown pack’ spoilage of vacuum-packed meats. J. Appl. Microbiol. 88:107–116 [DOI] [PubMed] [Google Scholar]
- 10. Clarridge J. E., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clementino M. M., et al. 2001. PCR analyses of tRNA intergenic spacer, 16S-23S internal transcribed spacer, and randomly amplified polymorphic DNA reveal inter- and intraspecific relationships of Enterobacter cloacae strains. J. Clin. Microbiol. 39:3865–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins M. D., et al. 1994. The phylogeny of the genus Clostridium—proposal of five new genera and eleven new species combinations. Int J. Syst. Bacteriol. 44:812–826 [DOI] [PubMed] [Google Scholar]
- 13. Collins M. D., Rodrigues U. M., Dainty R. H., Edwards R. A., Roberts T. A. 1992. Taxonomic studies on a psychrophilic Clostridium from vacuum-packed beef: description of Clostridium estertheticum sp. nov. FEMS Microbiol. Lett. 75:235–240 [DOI] [PubMed] [Google Scholar]
- 14. Dahlenborg M., Borch E., Rådström P. 2003. Prevalence of Clostridium botulinum types B, E and F in faecal samples from Swedish cattle. Int. J. Food Microbiol. 82:105–110 [DOI] [PubMed] [Google Scholar]
- 15. Dainty R. H., Edwards R. A., Hibbard C. M. 1989. Spoilage of vacuum-packed beef by a Clostridium sp. J. Sci. Food Agric. 49:473–486 [Google Scholar]
- 16. Garcia-Martinez J., Acinas S. G., Anton A. I., Rodriguez-Valera F. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55–64 [DOI] [PubMed] [Google Scholar]
- 17. Gupta A., Sumner C. J., Castor M., Maslanka S., Sobel J. 2005. Adult botulism type F in the United States, 1981-2002. Neurology 65:1694–1700 [DOI] [PubMed] [Google Scholar]
- 18. Helps C. R., Harbour D. A., Corry J. E. L. 1999. PCR-based 16S ribosomal DNA detection technique for Clostridium estertheticum causing spoilage in vacuum-packed chill-stored beef. Int. J. Food Microbiol. 52:57–65 [DOI] [PubMed] [Google Scholar]
- 19. Hippe H., Andreesen J. R., Gottschalk G. 1992. The genus Clostridium—nonmedical, p. 1800–1866 In Balows A., et al. (ed.), The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. Springer, New York, NY [Google Scholar]
- 20. Jiang W., Abrar S., Romagnoli M., Carroll K. C. 2009. Clostridium glycolicum wound infections: case reports and review of the literature. J. Clin. Microbiol. 47:1599–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalchayanand N., Ray B., Field R. A., Johnson M. C. 1989. Spoilage of vacuum-packaged refrigerated beef by Clostridium. J. Food Prot. 52:424–426 [DOI] [PubMed] [Google Scholar]
- 22. Kawasaki S., Fratamico P. A., Wesley I. V., Kawamotol S. 2008. Species-specific identification of campylobacters by PCR-restriction fragment length polymorphism and PCR targeting of the gyrase B gene. Appl. Environ. Microbiol. 74:2529–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lund B. M., Graham A. F., George S. M., Brown D. 1990. The combined effect of incubation temperature, pH and sorbic acid on the probability of growth of nonproteolytic, type B Clostridium botulinum. J. Appl. Bacteriol. 69:481–492 [DOI] [PubMed] [Google Scholar]
- 24. Moschonas G., Bolton D. J., Sheridan J. J., McDowell D. A. 2009. The effect of storage temperature and inoculum level on the time of onset of ‘blown pack’ spoilage. J. Appl. Microbiol. 108:532–539 [DOI] [PubMed] [Google Scholar]
- 25. Moschonas G., Bolton D. J., Sheridan J. J., McDowell D. A. 2009. Isolation and sources of ‘blown pack’ spoilage clostridia in beef abattoirs. J. Appl. Microbiol. 107:616–624 [DOI] [PubMed] [Google Scholar]
- 26. Mullane N. R., Whyte P., Wall P. G., Quinn T., Fanning S. 2007. Application of pulsed-field gel electrophoresis to characterise and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility. Int. J. Food Microbiol. 116:73–81 [DOI] [PubMed] [Google Scholar]
- 27. Pond M. J., Stone D. M., Alderman D. J. 2006. Comparison of conventional and molecular techniques to investigate the intestinal microflora of rainbow trout (Oncorhynchus mykiss). Aquaculture 261:194–203 [Google Scholar]
- 28. Pot B., Vandamme P., Kersters K. 1994. Analysis of electrophoretic whole organism protein fingerprints, p. 493–521 In Goodfellow M., O'Donnell A. G. (ed.), Chemical methods in prokaryotic systematics, vol. 5 John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 29. Relman D. A. 1993. Universal bacterial 16S rDNA amplification and sequencing, p. 489–496 In Persing D. H., Smith T. F., Tenover F. C., White T. J. (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC [Google Scholar]
- 30. Roth A., et al. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seurinck S., Verstraete W., Siciliano S. D. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song Y. L., et al. 2002. Use of 16S-23S rRNA spacer-region (SR)-PCR for identification of intestinal clostridia. Syst. Appl. Microbiol. 25:528–535 [DOI] [PubMed] [Google Scholar]
- 33. Spring S., et al. 2003. Characterization of novel psychrophilic clostridia from an Antarctic microbial mat: description of Clostridium frigoris sp. nov., Clostridium lacusfryxellense sp. nov., Clostridium bowmanii sp. nov. and Clostridium psychrophilum sp. nov. and reclassification of Clostridium laramiense as Clostridium estertheticum subsp. laramiense subsp. nov. Int. J. Syst. Evol. Microbiol. 53:1019–1029 [DOI] [PubMed] [Google Scholar]
- 34. Stackebrandt E., Hippe H. 2005. Taxonomy and Systematics, p. 19–48 In Bahl P. D. H. (ed.), Clostridia. Biotechnology and medical applications. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 35. Tindall B. J., et al. 2000. Cultivatable microbial biodiversity: gnawing at the Gordian knot. Environ. Microbiol. 2:310–318 [DOI] [PubMed] [Google Scholar]
- 36. Woo P. C. Y., Lau S. K. P., Teng J. L. L., Tse H., Yuen K. Y. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]
- 37. Yang X. Q., Gill C. O., Balamurugan S. 2010. Products of glucose and lactate fermentation, and utilization of amino acids by Clostridium estertheticum subsp. laramiense and estertheticum growing in meat juice medium. J. Food Prot. 73:1348–1352 [DOI] [PubMed] [Google Scholar]


