Abstract
The symbiotic association between aphids (Homoptera) and Buchnera aphidicola (Gammaproteobacteria) started about 100 to 200 million years ago. As a consequence of this relationship, the bacterial genome has undergone a prominent size reduction. The downsize genome process starts when the bacterium enters the host and will probably end with its extinction and replacement by another healthier bacterium or with the establishment of metabolic complementation between two or more bacteria. Nowadays, several complete genomes of Buchnera aphidicola from four different aphid species (Acyrthosiphon pisum, Schizaphis graminum, Baizongia pistacea, and Cinara cedri) have been fully sequenced. C. cedri belongs to the subfamily Lachninae and harbors two coprimary bacteria that fulfill the metabolic needs of the whole consortium: B. aphidicola with the smallest genome reported so far and “Candidatus Serratia symbiotica.” In addition, Cinara tujafilina, another member of the subfamily Lachninae, closely related to C. cedri, also harbors “Ca. Serratia symbiotica” but with a different phylogenetic status than the one from C. cedri. In this study, we present the complete genome sequence of B. aphidicola from C. tujafilina and the phylogenetic analysis and comparative genomics with the other Buchnera genomes. Furthermore, the gene repertoire of the last common ancestor has been inferred, and the evolutionary history of the metabolic losses that occurred in the different lineages has been analyzed. Although stochastic gene loss plays a role in the genome reduction process, it is also clear that metabolism, as a functional constraint, is also a powerful evolutionary force in insect endosymbionts.
INTRODUCTION
Aphids (Homoptera, Aphididae) feed on phloem sap, which has an unbalanced nitrogen/carbon content and is deficient in a number of nutrients, mainly amino acids, which insects, like other animals, cannot synthesize and are provided by their primary endosymbiont Buchnera aphidicola (5, 9). After their association, which took place about 100 to 200 million years ago according to the fossil record, host and symbiont lineages have evolved strictly in parallel (29). The relationship is mutualistic, since aphids need B. aphidicola for normal growth and reproduction, whereas the bacterium cannot live outside the host. Occasionally, the host tolerates secondary symbionts, defined as facultative bacterial endosymbionts that coexist with Buchnera. As they are facultative, they are considered nonessential to the host, although positive effects have been shown in some cases, such as rescuing the host from heat damage, providing defense against natural enemies, and participating in host specialization (27).
At present, the B. aphidicola genome has been sequenced from four aphid species, two from bacteria harbored by aphids belonging to the subfamily Aphidinae, B. aphidicola BAp (44) and B. aphidicola BSg (47), the primary endosymbionts of Acyrthosiphon pisum and Schizaphis graminum, respectively. The other two belong to two different aphid lineages, B. aphidicola BBp from Baizongia pistaciae (52) and B. aphidicola BCc from Cinara cedri (37), members of the subfamily Eriosomatinae and Lachninae, respectively. In addition, the genomes of seven strains of B. aphidicola from A. pisum have been sequenced (28). The comparative analysis of the B. aphidicola genomes revealed an extreme case of evolutionary stasis with nearly perfect gene order conservation. Thus, the gene order in extant Buchnera can be considered a gene order fossil that has practically been preserved since the last common symbiotic ancestor (LCSA) of all present B. aphidicola lineages (37, 47, 52). However, all these bacteria possess different genome sizes, with the B. aphidicola genome from C. cedri (with 425 kb) the smallest B. aphidicola genome known, up to 200 kb smaller than the others. In this genome, gene losses were especially dramatic in biosynthesis of nucleotides, metabolism of cofactors and vitamins, and cell envelope and transport (37). However, it has conserved a simplified metabolism that uses glucose as an energy source through substrate-level phosphorylation. Regarding the main role of B. aphidicola in aphid symbiosis as a provider of amino acids, B. aphidicola BCc is unable to fulfill this role, as it has lost the ability to synthesize tryptophan.
A particular feature of C. cedri is the massive presence of the secondary endosymbiont “Candidatus Serratia symbiotica,” which has been reported to have become an obligate symbiont in this aphid (17, 22), whereas it is a facultative symbiont in other aphid species (30, 34). Indeed, there are recent reports of a close endosymbiotic consortium that involves B. aphidicola and “Ca. Serratia symbiotica” in C. cedri (18). Thus, both bacteria are involved in the synthesis of tryptophan. Therefore, the obligate biochemical interdependence between the two endosymbionts can represent an evolutionary seal of bacterial metabolic complementation and establishment of a stable consortium (31). In the other B. aphidicola strains analyzed thus far, the first two genes of the tryptophan pathway (trpEG) are either in a plasmid or in the main chromosome, but they are always separate from the rest of the genes in this pathway (trpDCBA), which remain in the chromosome. In B. aphidicola BCc, there is a plasmid containing trpEG, but the rest of the genes for the tryptophan biosynthesis pathway (trpDCBA) are located on the chromosome of “Ca. Serratia symbiotica” (18). The case of B. aphidicola BCt, primary endosymbiont of the aphid Cinara tujafilina, is very striking. In this strain, there is a chimeric pLeu/Trp plasmid that contains the first two genes of the tryptophan pathway (trpEG) and the structural genes for leucine synthesis (14, 23).
The B. aphidicola BCt genome has been estimated to be about 25 kb bigger than the B. aphidicola BCc genome by pulsed-field gel electrophoresis (13). On the other hand, C. cedri and C. tujafilina are closely related both phylogenetically and entomologically (40); they also live on closely related plant hosts (cedar and thuja trees, respectively), and both harbor “Ca. Serratia symbiotica” as a second endosymbiont. However, the bacterial phylogenetic analysis carried out with members of the subfamily Lachninae showed two different and very divergent “Ca. Serratia symbiotica” lineages. One lineage encompasses “Ca. Serratia symbiotica” from aphids belonging to different subfamilies of the family Aphididae, including C. tujafilina, while the other lineage only comprises species from the subfamily Lachninae, including C. cedri (6, 22).
In this work, we report the complete genome sequencing of B. aphidicola BCt and the results of a phylogenetic and comparative genomic analysis with the other sequenced B. aphidicola genomes, and more specifically with the closely related B. aphidicola BCc. This has enabled us to reconstruct the gene composition of the LCSA, as well as the history of gene losses, but also and more importantly, the role played by metabolism as a functional constraint in the evolution of genome reduction of bacterial endosymbionts in insects.
MATERIALS AND METHODS
Aphid material and DNA isolation.
Cinara tujafilina aphids were collected from thuja trees from a natural population in Valencia, Spain, during May and June 2009.
The bacterial endosymbionts were extracted from aphids as described previously (15). An enriched fraction of bacteriocytes was then obtained and used to extract total DNA following treatment with CTAB (cetyltrimethylammonium bromide) (3).
Genome sequencing and gene annotation.
The complete genome sequence of Buchnera aphidicola was obtained performing half a run of shotgun sequencing using GS FLX titanium single protocol (GATC Biotech). To close the chromosome, two PCRs were performed with two pairs of specific primers (primers BCtu_200 [5′-CCCTCCCTTAGAGATGCGTA] and BCtu_202 [5′-AGTTCGTTCTAGTGTTGTTAAAGATTC] and primers Bctu_170 [5′-TTAGCCTTGAAGAATTTTCTGTTTT] and BCtu171 [5′-TTTAGCGCTTTGTTTAAATTACCA]). Gap4.8b1 from Staden Package (46) was used for total assembly. The putative coding regions were identified with the GLIMMER v3.02 software (8). ARTEMIS was used to verify the determined start and stop codons (42). Final annotation was performed using BLASTP (E value to 10−5) comparison (2) with the four previously sequenced B. aphidicola genomes. Noncoding RNAs were identified by different approaches. tRNAs as well as other small RNAs (transfer-messenger RNA [tmRNA] or the RNA component of the RNase P) were predicted by tRNAscan (25). Signal recognition particle (SRP) RNA was identified with SRPscan, as well as running the Rfam database search (12, 39). To locate pseudogenes that were not found with GLIMMER, intergenic regions were manually analyzed by BLASTX and BLASTN (best hit and E value to 10−5) (2). The G+C content was calculated online with GeeCee (http://inn-temp.weizmann.ac.il/cgi-bin/emboss/geecee).
The putative protein-coding genes were compared to orthologous groups in the COG (Clusters of Orthologous Groups) database (48).
Phylogenetic analyses and reconstruction of the last common symbiotic ancestor (LCSA).
The alignment of the 310 orthologous genes of the five B. aphidicola genomes used in this study and the corresponding genes from the Escherichia coli K-12 (RefSeq accession no. NC_000913) genome, used as the outgroup, was performed with Clustal X (49). Phylogenetic reconstruction was carried out by maximum likelihood (ML) using PHYML v2.4.4 with the evolutionary model GTR and gamma correction for heterogeneity of substitution rates (19). Other useful parameters considered in the present study were the number of invariant sites derived by MODELTEST 3.7 (38) and the Akaike information criterion (1). To test the reliability of the clades, bootstrapping was carried out with 1,000 pseudoreplicates. A Bayesian phylogenetic inference analysis was also performed with the same evolutionary model using BEAST software (10). Phylogenetic trees were generated from two runs of 1,000,000 generations, sampling every 100 generations and discarding the first 20,000 generations as “burn in.”
We obtained the gene composition of the LCSA as the addition of all the genes present in B. aphidicola strains BAp, BSg, BBp, BCc, and BCt. Then, the tree obtained was used to reconstruct the different gene loss events. According to reference 16, a gene loss in current B. aphidicola genomes was defined either as the absence of a gene present in the ancestral LCSA or as any mutational event disrupting gene functionality and producing a pseudogene.
Metabolic inference.
A classification based on the nonredundant functional categories used for the Aquifex aeolicus genome (7) with modifications (15) was obtained. This classification was used to derive the metabolic pathway reconstruction using the EcoCyc database (21).
Nucleotide sequence accession number.
The genome sequence was deposited in the GenBank database under accession number CP001817.
RESULTS
Genome features of Buchnera aphidicola BCt.
The 397,353 pyrosequencing reads were assembled into 103,968 contigs. Then, the sequences were compared to the B. aphidicola genome sequences from the databases (B. aphidicola BAp [NC_011833], B. aphidicola BSg [NC_004061], B. aphidicola BBp [NC_004545], and B. aphidicola BCc [NC_008513] [RefSeq accession numbers shown in brackets]) using BLASTN (E value to 10−5). All sequences with positive matches were selected, obtaining a total of 29,808 contigs. Then, by manual assembly taking into account the genomic synteny among the B. aphidicola chromosomes (47, 52), the number of contigs was finally reduced to two of 400,475 and 44,451 bp. The genome was finally closed by PCR using the two sets of specific primers shown in Materials and Methods. The chimeric pLeu/Trp plasmid previously characterized in this strain (14) was also obtained as a single contig.
The general features of the B. aphidicola BCt genome and comparison with those of the other sequenced genomes of B. aphidicola are shown in Table 1 (see Fig. S1 in the supplemental material). The genome is composed of a circular chromosome of 444,930 bp, slightly smaller than the value previously determined by pulsed-field gel electrophoresis, and a plasmid of 8,069 bp, similar to that previously characterized (13, 14). Thus, this is the second smallest B. aphidicola genome studied so far. A total of 404 putative genes have been assigned, of which 367 are protein-coding genes (361 in the main chromosome and six in the chimeric plasmid) and 37 RNA-specifying genes (31 tRNAs, three rRNAs, and three small RNAs). Nine pseudogenes have been found, six more than in B. aphidicola BCc. The G+C content is 25%, higher than the 20.20% for B. aphidicola BCc, but similar to the content for the other three B. aphidicola strains. It is worth mentioning that although the genome size of B. aphidicola BCt was 27,770 bp bigger than that of B. aphidicola BCc, it contains only two more genes (404 versus 402). However, different genes have been retained in the two strains, as there are 26 and 28 different gene losses in the lineages of BCt and BCc, respectively (Fig. 1). Finally, as the total length of the coding sequences (CDS) is slightly smaller in B. aphidicola BCt than in B. aphidicola BCc (11,916 bp), the extra base pairs of the B. aphidicola BCt chromosome compared to that of B. aphidicola BCc are located in the intergenic regions and in the pseudogenes.
Table 1.
General genomic properties of five Buchnera aphidicola strains
| Genomic propertya | Value for property for B. aphidicola strainb: |
||||
|---|---|---|---|---|---|
| BAp | BSg | BBp | BCc | BCt | |
| Genome size (bp) | 652,115 | 653,001 | 618,379 | 425,229 | 452,999 |
| Chromosome size (bp) | 640,681 | 642,454 | 615,980 | 416,380 | 444,930 |
| Plasmid size (bp) | 11,434 | 11,547 | 2,399 | 8,849 | 8,069 |
| Total no. of genesc | 609 | 597 | 546 | 402 | 404 |
| No. of CDS (chromosome + plasmids) | 562 + 9 | 550 + 9 | 505 + 3 | 358 + 7 | 361 + 6 |
| No. of rRNA genes | 3 | 3 | 3 | 3 | 3 |
| No. of tRNA genes | 32 | 32 | 32 | 31 | 31 |
| No. of other RNA genes | 3 | 3 | 3 | 3 | 3 |
| No. of pseudogenes | 12 | 33 | 9 | 3 | 9 |
| Avg length of CDS (bp) | 984 | 979 | 996 | 994 | 966 |
| Avg length of IGR (bp) | 126.90 | 113.30 | 200.50 | 135.80 | 200.15 |
| G+C content (%) | 26.24 | 26.30 | 25.30 | 20.20 | 25.00 |
CDS, coding sequences; IGR, intergenic region.
B. aphidicola strains (RefSeq accession numbers are shown in parentheses) are abbreviated as follows: BAp, B. aphidicola from Acyrthosiphon pisum (NC_011833); BSg, B. aphidicola from Schizaphis graminum (NC_004061); BBp, B. aphidicola from Baizongia pistaciae (NC_004545); BCc, B. aphidicola from Cinara cedri (NC_008513); BCt, B. aphidicola from Cinara tujafilina (CP001817).
In the final number of genes, we have considered only one ribD gene for all genomes. The fliO and fliP genes and the fliM and fliN genes have been considered independent genes even though they are fused in some genomes.
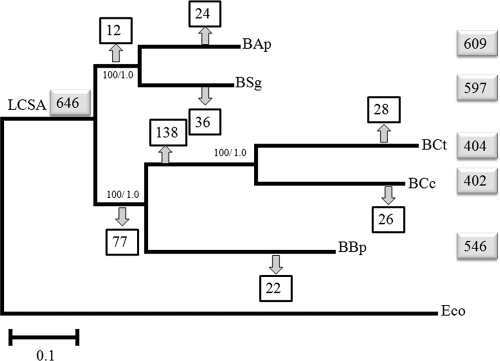
Fig. 1.
Maximum likelihood (ML) phylogeny inferred from the 310 concatenated gene sequences shared by Buchnera strains and E. coli (Eco). The support values for the corresponding inner branch are 100/1.0 in the form of the proportion of bootstrap pseudoreplicates/Bayesian a posteriori probabilities, respectively. The total numbers of genes of each strain, as well as in the last common symbiotic ancestor (LCSA), are indicated in gray boxes. Numbers within squares at the nodes indicate the total number of gene losses. B. aphidicola strain abbreviations are as explained in Table 1, footnote b.
Functional analysis of the predicted protein-coding genes.
The protein-coding genes of B. aphidicola BCt were classified according to COG categories (see Table S1 in the supplemental material) (see Fig. S2 in the supplemental material for a comparative analysis). Genes involved in translation and ribosomal structure and biogenesis (category J) are the most represented (30.57%), followed by genes devoted to transport and amino acid metabolism (category E; 12.29%) and energy production and conservation (category C; 8.29%). The gene losses in B. aphidicola BCt have mainly affected categories F (transport and nucleotide metabolism), H (coenzyme metabolism), and M (biogenesis of cell wall and external membrane). Overall, the analyses indicate that the gene reduction is biased toward the preservation of gene functions involved in the central maintenance of the bacteria and their symbiotic role to the detriment of other categories such as synthesis and transport of cofactors, cell envelope biogenesis, and synthesis of nucleotides, as previously stated (31). In accordance with its role in symbiosis, B. aphidicola BCt strain, like other B. aphidicola strains, devoted a great effort to genes involved in amino acid metabolism (4), and unlike B. aphidicola BCc, this strain has retained the complete set of genes in the tryptophan pathway.
To assess whether gene reduction is associated with a similar loss of genes in all the COG categories or a differential loss in some of the categories, we took the distribution of COG categories in B. aphidicola BCc, the most reduced genome, as the expected distribution for the rest of the genomes. The expected values and the corresponding values by the χ2 test are shown in Table S2 in the supplemental material. As can be observed, B. aphidicola strains BAp, BSg, and BBp do not follow the pattern shown by B. aphidicola BCc, whereas the COG distribution pattern shown by B. aphidicola BCt was statistically similar to the B. aphidicola BCc pattern.
Metabolic analysis.
According to the gene repertoire and the metabolic network inferred (see Materials and Methods), the metabolism of B. aphidicola BCt was dramatically simplified and very similar to the metabolism of B. aphidicola BCc (37). This is due to the massive gene loss that predates the split of the lineages Cinara (Cinara) and Cinara (Cupresobium) to which B. aphidicola BCc and B. aphidicola BCt belong, respectively (see below). The pathways most severely affected by the reduction process are those related to nucleotide biosynthesis, metabolism of cofactors and vitamins, cell envelope synthesis and transport systems. However, there are some differences between these two B. aphidicola strains in glycolysis, the pentose phosphate pathway, and amino acid biosynthesis. B. aphidicola BCt has lost the pgi gene coding for glucose-6-phosphate isomerase. Thus, instead of using glucose-6-phosphate like other B. aphidicola strains, including B. aphidicola BCc, B. aphidicola BCt strain can use only fructose-6-phosphate as a substrate in glycolysis. Associated with this observation, the pentose phosphate pathway preserved in all other B. aphidicola strains is also different in B. aphidicola BCt. This strain has retained the genes to obtain ribose-5-phosphate from fructose-6-phosphate, but it has lost the pgl and zwf genes of the oxidative pentose pathway. This constitutes a typical example of a domino effect in the reductive process in bacterial endosymbionts where once a gene of a given metabolic pathway is lost by chance, the remaining ones are rendered unnecessary (45).
Regarding biosynthesis of essential amino acids, B. aphidicola BCt is the only Buchnera strain to have lost the aroE gene coding for sikimate dehydrogenase, a necessary step in chorismate synthesis, the starting point of synthesis of aromatic amino acids. Finally, B. aphidicola BCt, like B. aphidicola BAp, BSg, and BBp strains, has retained the genes for tryptophan synthesis. The trpEG genes are located in the pLeu/Trp chimeric plasmid (14), and unlike B. aphidicola BCc, B. aphidicola BCt also contains the trpDCBA genes located in the main chromosome.
Phylogenetic reconstruction of B. aphidicola lineages.
In order to derive the evolutionary relationships among the five B. aphidicola strains, phylogenetic reconstructions based on maximum likelihood (ML) and Bayesian phylogenetic inference analyses were carried out with the 310 concatenated orthologous genes, including E. coli as the outgroup. Both approaches yielded similar and well-supported topologies, with two separate clusters, one formed by B. aphidicola from the two members of the subfamily Aphidinae (Acyrthosiphon pisum and Schizaphis graminum) and the other formed by B. aphidicola from the two members of the subfamily Lachninae (Cinara cedri and Cinara tujafilina), which in turn clustered with B. aphidicola from the member of the subfamily Eriosomatinae (Baizongia pistacea) (Fig. 1).
Identification of pseudogenes and gene loss events.
In a previous study, Gómez-Valero and coworkers (16) carried out the reconstruction of the last common symbiotic ancestor (LCSA) by comparison of B. aphidicola BAp, BSg, and BBp genomes. To do so, gene content was determined following the criterion that all genes found in some of the extant strains were originally in the ancestral genome (52). This was based on high genome stability and almost total absence of horizontal gene transfer events (47, 51). We have added five genes to the LCSA previously reconstructed (16), the genes specific to B. aphidicola BCt and BCc (sspA, yebC, zapA, and dusA), and one more due to considering the fliO and fliP genes as independent genes. Thus, the ancestral B. aphidicola genome would contain at least 645 genes, 634 of these genes located in the main chromosome and 11 in the pLeu or pTrp plasmid. Of the 634 genes, 596 would be protein-coding genes and 38 would be genes coding for RNA (see Table S3 in the supplemental material for a list of genes). It is worth mentioning that this is the minimum gene content, as we cannot incorporate the genes that have been lost simultaneously in the five strains. With this knowledge of the LCSA gene composition, we can evaluate the status of each ancestral gene for each B. aphidicola genome, i.e., active or absent genes as well as pseudogenes. The results are reported in Table 2. The logic behind evaluating the status of each gene loss is based on the phylogenetic reconstruction (Fig. 1) and on the following three assumptions. (i) If a functional gene is present in one strain but is a pseudogene or is absent in the other(s), then gene loss took place after divergence in the later lineage(s). (ii) If the genes are absent in some related strains, we consider that gene loss has taken place prior to divergence of the corresponding lineages. (iii) If some unrelated strains present pseudogenes or the genes are absent, this is interpreted as a convergent loss. The results reported in Table 2 enabled us to locate the number of losses on the phylogenetic tree throughout the evolutionary history of the five B. aphidicola lineages (Fig. 1). The substantial number of losses experienced by two inner branches is remarkable, the ancestral one leading to B. aphidicola BBp, BCc, and BCt, and the other one leading to the two Cinara species, 77 and 138, respectively.
Table 2.
Gene repertoire of the LCSA and gene status in the five Buchnera strainsa
| Type of lossb | No. of genes | Gene(s) in the LCSA | Status of gene in Buchnera strainc: |
||||
|---|---|---|---|---|---|---|---|
| BAp | BSg | BBp | BCc | BCt | |||
| U | 3 | yhhF, ung, miaA | + | ψ | + | + | + |
| U | 21 | fliJ, fliM, pmbA, nth, yabI, apaH, dksA, gloB, ychE, trpA, trpB, trpC, trpD, mviN, flgB, flgC, flgG, ycfF, pepA, yhgN, yggH | + | + | + | − | + |
| U | 15 | rnpB, grpE, pyrH, pgl (ybhE), zwf, yoaE, cyoD, mdlA, mdlB, yqgF, yggW, yggX, pgi, cyaY, repA1 | + | + | + | + | − |
| U | 6 | yraL, yfcN, recB, recD, aroE, sbcB | + | + | + | + | ψ |
| U | 2 | tldD, yccK | + | + | + | ψ | + |
| U | 1 | fabD | ψ | + | + | + | + |
| U | 1 | fabZ | − | + | + | + | + |
| U | 4 | himA (ihfA), yaeT, himD (ihfB), ompA | + | + | − | + | + |
| S5 (BAp-BSg)-(BBp-BCc-BCt) | 1 | hemD | ψ | ψ | − | − | − |
| 1 | ansA | − | ψ | − | − | − | |
| S4 (BSg)-(BBp-BCc-BCt) | 1 | ycfM | + | ψ | ψ | − | − |
| 8 | nlpD, cysN, cysD, cysG, cysH, cysI, cysQ, hemC | + | ψ | − | − | − | |
| 2 | hns, cspC | + | − | − | − | − | |
| S4 (BAp-BSg)-(BCc-BCt) | 3 | cvpA, apbE, cmk | ψ | ψ | + | − | − |
| 7 | mutH, norM, bioC, bioF, pal, yqgE, uspA | − | − | + | − | − | |
| S4 (BAp-BSg)-(BBp)-(BCt) | 1 | pcnB | − | − | − | + | ψ |
| S4 (BAp)-(BBp-BCc-BCt) | 7 | bcp,folE, yqcD, znuA, ygcF, ygcM (ptpS), ybaX | − | + | − | − | − |
| 2 | metR, lolE | ψ | + | − | − | − | |
| S3 (BSg)-(BCc-BCt) | 14 | rnfC or rsxC, yfaE, mrcB, murC, mraY, murF, murE, mltE, bioD, phrB, mrsA (glmM), lgt, pyrE, yjeA | + | ψ | + | − | − |
| 6 | ispE, dxr, ispG, ispF, ispD, dxs | + | + | ψ | − | − | |
| 1 | ycfC | + | − | + | − | − | |
| S3 (BBp-BCc-BCt) | 56 | dnaT, argE, argC, secB, rfaE, cysK, priA, queA, tgt, surA, ribF, nadE, smpA, hpt, panC, panB, mutT, coaE, speD, speE, lpcA, topA, lolC, lolD, glpF, pyrC, flgN, flgA, flgD, flgE, flgK, ompF, pncB, pyrD, ycbY, pyrB, pyrI, yfiO, metK, ygbQ, cysC, cysJ, nadD, argA, mltA, thiL, ispA, tig, smg, argD, yeeX, rnr, ibpA, coaD, yba-4, yhiQ | + | + | − | − | − |
| S3 (BAp-BSg)-(BBp) | 4 | sppA, yebC, zapA, dusA | − | − | − | + | + |
| S3 (BAp)-(BCc-BCt) | 1 | ddlB | ψ | + | + | − | − |
| 5 | yidD, yjeK, ygjT, yebA, ygfA | − | + | + | − | − | |
| S2 (BSg)-(BCt) | 1 | yba3 | + | ψ | + | + | ψ |
| S2 (BCc-BCt) | 100 | atpB, atpE, atpF, atpH, atpA, atpG, atpD, atpC, glmS, glmU, yigL, purH, hupA, murB, yibN, cysE, ribB, bacA, crr, ptsI, ptsH, tRNAAla gene, fliE, fliK, ytfN, dcd, ribE, rnfA or rsxA, rnfB, rnfD or rsxD, rnfG or rsxG, rnfE or rsxE, ydiK, yajC, folA, lytB (ispH), lspA, folC, nrdB, nrdA, yba-2, ppnK, sodA, guaC, pfs, murG, ftsW, murD, ftsI (pbpB), ftsL, htrA, uppS, gpt, yfjF, purB, pyrF, ribA, cls, yciB, sohB, bioB, bioA, znuB, znuC, flgJ, rne, tmk, ptsG, murA, gshA, endA, pyrG, mutS, dsbA, gmk, thyA, ybeX, ribH, ribD, yajR, yrdC, fkpA, yhfC, deoD, deoB, prfC, gshB, yggS, murI, purA, hflC, hflK, mutL, mtlD, mtlA, amiB, pitA, ynfM, yqhA, repA2 | + | + | + | − | − |
| 1 | ppiD | + | + | + | ψ | ψ | |
| S2 (BBp)-(BCt) | 1 | secG | + | + | ψ | + | − |
| 2 | argB, fis | + | + | − | + | − | |
| S2 (BBp)-(BCc) | 1 | yedA | + | + | ψ | − | + |
| 2 | uup, yggJ | + | + | − | − | + | |
| S2 (BAp-BSg) | 1 | bioH | − | ψ | + | + | + |
| S2 (BAp)-(BCc) | 1 | rnhA | ψ | + | + | − | + |
B. aphidicola strains are abbreviated as explained in Table 1, footnote b.
U, unique loss; S2, S3, S4, and S5, losses in two, three, four, and five strains, respectively. The strains with the gene losses are indicated in parentheses. The number of parentheses indicates the number of probable convergent losses, according to the phylogeny shown in Fig. 1.
The status of the gene(s) is indicated as follows: +, gene present; −, gene absent, ψ, pseudogene.
Finally, as already mentioned, during the evolution of B. aphidicola BCt and BCc lineages, 26 and 28 independent gene losses occurred, respectively, affecting different pathways (see Table S4 in the supplemental material). As can be seen, B. aphidicola BCt and BCc strains have losses affecting similar functional categories, though different genes have been lost: information storage and processing (8 and 6, respectively), protein processing folding and secretion (3 and 2, respectively), cellular processes (2 and 3, respectively) and metabolism (7 and 7, respectively). In addition, the BCt and BCc strains show losses affecting poorly characterized genes (6 and 4, respectively), and in B. aphidicola BCc, six genes in the cell envelope category have been lost. These genes are involved in the flagellar apparatus and are responsible for the extremely simplified flagellum found in B. aphidicola BCc, as previously reported (31, 50).
Metabolic gene losses.
We will now examine how the loss of a particular gene involved in a given metabolic pathway at a particular moment of the aphid phylogeny can affect the history of the metabolic pathway (Fig. 2). Metabolic genes have been considered based on the nonredundant classification by Deckert and coworkers (7) (see Table S5 in the supplemental material).
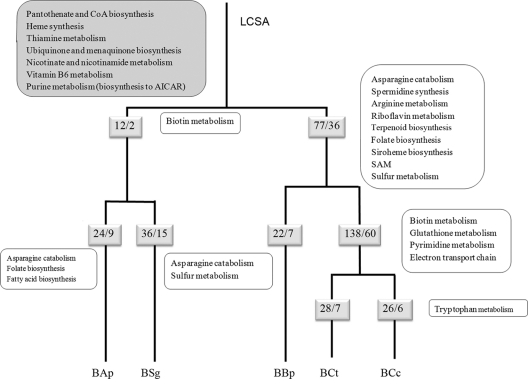
Fig. 2.
Evolutionary diagram of metabolic gene losses occurring in the five strains of B. aphidicola since the LCSA. The inferred pathways that had already been lost in the LCSA are indicated in the large gray rounded box in the top left corner of the figure. The small shadowed boxes show the number of total gene losses/number of gene losses involved in metabolism in each lineage. The adjacent boxes to the right of the lineages indicated only the new metabolic pathways affected by these gene losses. B. aphidicola strains are abbreviated as explained in Table 1, footnote b. CoA, coenzyme A; AICAR, amino-imidazole carboxamide riboside 5′-phosphate; SAM, S-adenosylmethionine.
By assuming that the genes of the metabolic pathways absent in the five strains were lost in the common ancestor, the LCSA had already lost the ability to synthesize cofactors and vitamins (heme group, thiamine, ubiquinone, menaquinone, nicotine, nicotinamide, panthothenate, coenzyme A [CoA], and vitamin B6). In addition, the purine synthesis pathway underwent considerable shrinkage, and therefore, purines must be synthesized from amino-imidazole carboxamide riboside 5′-phosphate (AICAR), coming from histidine biosynthesis.
In the common ancestor of the two members of the subfamily Aphidinae, A. pisum and S. graminum, two metabolic losses involved in the biotin biosynthesis pathway would have occurred. During the evolution of B. aphidicola BAp and BSg lineages, nine and 15 losses are involved in metabolism, respectively, some involved in the already inactive biotin pathway and some involved in new metabolic pathways (Fig. 2). All the pathways affected are also affected in other lineages, with the exception of fatty acid biosynthesis. Thus, B. aphidicola BAp is the only Buchnera strain unable to synthesize fatty acids, even with the provision of malonyl-CoA from the host. With respect to the inner branch of the clade formed by Eriosomatinae (BBp) and Lachninae (BCc and BCt) lineages, we have detected 36 metabolic losses affecting many more new pathways than in the Aphidinae clade (Fig. 2). However, there is a great contrast with the situation in the Erisosomatinae and Lachninae lineages. Whereas the highest number of metabolic losses (60 out of 138) is detected in the Lachninae ancestor, before the divergence of B. aphidicola BCc and BCt, only seven losses (none of them affecting a new pathway) are detected in the outer branch leading to B. aphicola BBp. Concerning the two Cinara species, seven and six gene losses involved in metabolism have occurred in C. cedri and C. tujafilina, respectively, although the only new pathway affected is the one involved in the loss of tryptophan biosynthesis in B. aphidicola from C. cedri.
Evolution of the repair systems.
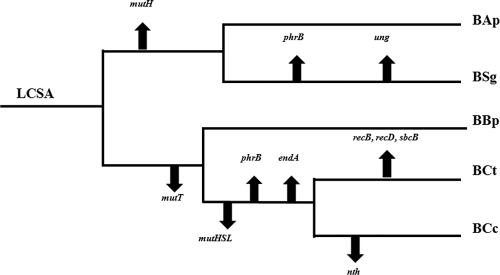
A very striking result is the loss of genes involved in the repair machinery in B. aphidicola BCt. We have evaluated the degree of gene loss in the repair apparatus of the different B. aphidicola lineages, according to the phylogenetic reconstruction obtained (Fig. 3; see Table S6 in the supplemental material). Although these bacteria lack recA, involved in the repair system by recombination, they could maintain adequate replication and repair processes due to alternative strategies involving the combined action of RecBCD and SbcB exonucleases (43). On the other hand, it has been reported that Ruthia magnifica and Vesicomysocius okutanii, endosymbionts of deep-sea clams, lack RecBCD but retain homologues of SbcB, RecG, and RecJ recombinases (43). Surprisingly, B. aphidicola BCt genome analysis revealed that it has lost the sbcB gene, while the genes coding for RecB and RecD subunits are pseudogenes. In addition, like B. aphidicola BCc, it has lost the complete MutSLH system and endA and phrB genes. However, it has retained the base excision repair system (nfo, nth, ung, and polA), which could partially counteract a variety of lesions, as occurs in some phytoplasma that lack recombination functions (43) and the mutY gene involved in G and A mismatch repair. Moreover, B. aphidicola BCt possesses a completely active DNA polymerase I that could fill in the gapped duplex. It is worth mentioning that despite B. aphidicola BCt having the most reduced repair system, the G+C content of B. aphidicola BCt is higher than in B. aphidicola BCc and is similar to those of other B. aphidicola strains. This would seem to indicate that the A+T bias of the bacterial endosymbionts is due to other factors and not simply the loss of repair genes.
Fig. 3.
Phylogenetic representation of gene loss events in each Buchnera strain for repair machinery. B. aphidicola strains are abbreviated as explained in Table 1, footnote b.
DISCUSSION
Genome reduction in endosymbiotic bacteria is a continuous process derived from their adaptation to intracellular life. The first step toward the establishment of an obligate endosymbiosis takes place when a free-living bacterium infects the host. After the first step, both organisms coevolve to adapt to the new situation. The bacterium suffers an evolutionary genome reductive process, thus saving energy through the removal of unnecessary redundant genes. The limit to gene loss is not well understood, although the endosymbiotic genome sequences obtained in recent years provide new clues to understanding the genome reduction process. The system has been particularly well studied in insects, where with the sole exception of Carsonella ruddi, the primary endosymbiont of psylids (33), all endosymbionts with the smallest bacterial genomes have been found to coexist with other symbionts, thus establishing a metabolic complementation that keeps the system alive (6, 34).
Specific losses in the two members of the subfamily Lachninae.
The comparative genomic analysis of B. aphidicola BCt with other previously sequenced B. aphidicola strains (37, 44, 47, 52) has shown that the genome features of the two members of the subfamily Lachninae are the most similar, although the B. aphidicola BCt genome is 28 kb bigger than that of B. aphidicola BCc. Thus, B. aphidicola BCt has only two more coding genes (a total of 404) than B. aphidicola BCc. This size difference is due to the larger size of the intergenic regions and to a higher number of pseudogenes (9 versus 3). In terms of the reduction process, these data could be interpreted to mean that this genome is in a previous step to that of B. aphidicola BCc. Thus, with time, both the pseudogenes and the intergenic part that is still undergoing nucleotide erosion will disappear (16, 28).
At a functional level, B. aphidicola BCt shows nonsignificant differences with B. aphidicola BCc regarding the number of genes belonging to different COG functional categories (see the observed values for both strains in Table S2 in the supplemental material). These results can be explained by the large number of shared gene losses (138) that occurred in the ancestors of both Lachninae. However, the functions are not carried out by the same genes because independent gene inactivation (26 and 28 in B. aphidicola BCt and BCc, respectively) has occurred throughout the diversification process of both lineages, revealing a predominant role played by chance in the genome reduction process. In fact, the loss of functional genes is also occurring on a recent evolutionary time scale, as found by comparing the genomes of seven B. aphidicola strains from A. pisum strains (28). Interestingly, the 16 inactivated genes found in these strains were also inactivated in some of the other lineages studied.
Metabolic particularities of B. aphidicola BCt genome.
B. aphidicola BCt, as expected due to the low number of retained genes, was found to have a simplified metabolism, very similar to that of B. aphidicola BCc. However, there are two main differences with the other B. aphidicola strains in carbohydrate metabolism and chorismate synthesis. B. aphidicola BCt is the only B. aphidicola strain to use fructose instead of glucose in glycolysis. A similar situation has been observed in Blattabacterium, the endosymbionts of cockroaches (24), and “Candidatus Sulcia muelleri,” the cosymbiont of the sharpshooters (26), again indicating a convergent adaptation to the loss of essential (or at least beneficial) genes in the reduced genomes. In relation to the synthesis of essential amino acids, it is tempting to speculate that the loss of the essential aroE gene may have been possible if other dehydrogenases [FAD/NAD(P)-binding Rossmann fold superfamily] could carry out the same metabolic reaction, provided there is a loss of substrate specificity. Putative enzymes that could replace the shikimate dehydrogenase are encoded by the genes gapA, gnd, thrA, asd, and folD.
Phylogenetic reconstruction and gene losses.
The first phylogenetic reconstruction based on both morphological and molecular characteristics considered the Lachninae subfamily a sister group of Aphidinae and, thus, very divergent from the Eriosomatinae (20, 53). However, the position of this subfamily remains controversial. Recent molecular phylogenetic analyses of nuclear and mitochondrial aphid genes as well as B. aphidicola genes do not support the relatedness of Lachninae and Aphidinae and place the subfamily Lachninae in a basal position (35, 36). Our analysis with the concatenated orthologous genes supports the notion that both lineages are highly divergent. However, the two members of Lachninae are not basal but form a monophyletic group with the member of the Eriosomatinae subfamily. In the present study, the analysis of the number of losses clearly indicates that the tree obtained is the most parsimonious regarding gene loss events. In Fig. 1, the topology shows that 77 gene losses occurred in the ancestor of the Lachninae and Eriosomatinae lineages, whereas other topologies would need to consider these 77 losses as convergent ones in the ancestor of the two different lineages (Lachninae and Eriosomatinae).
Metabolic reconstruction: from the LCSA to the extant Buchnera.
The metabolic reconstruction of the LCSA from the gene content of the extant Buchnera shows us how the transition from a free-living environment has led to the inactivation of many genes involved in metabolic pathways, mainly those involved in cofactor and vitamin synthesis. This indicates that in the early stages of adaptation to intracellular life, the necessary metabolites that were lost must be obtained either from the plant sap or from the aphid. These first losses in the ancestors, leading to the inactivation of certain genes corresponding to a particular metabolic pathway, can explain the high number of convergent losses in the same metabolic pathways in different lineages, in turn inactivating other genes of that pathway. For example, the 12 gene loss events identified in the ancestor of the two members of Aphidinae are all convergent losses with at least some of the other genomes analyzed.
The action of the reductive evolution has been more dramatic in the ancestor of the subfamily Lachninae and Eriosomatinae, with 78 gene losses, of which 36 are metabolic. Again, a study of the convergent gene losses indicates that 21 out of the 78 gene losses have also been lost in B. aphidicola BAp and/or B. aphidicola BSg lineages. The number of newly inactivated metabolic pathways that took place preceding the divergence of the Lachninae and Eriosomatinae lineages is remarkable. At this point, the losses affect amino acid metabolism and, again, cofactor and vitamin synthesis. It is the case of riboflavin, described as essential for aphid survival (32), terpenoids, folate, arginine, and inactivation of sulfur metabolism, affecting cysteine synthesis and S-adenosylmethionine production, needed for protein methylation. There are two possible explanations why these losses do not affect the survival of the host or the bacteria itself; the aphid can afford them, or there is a second bacterium doing the job.
In the Lachninae, the massive gene loss was probably due to gene redundancy because of the association with secondary endosymbionts (6, 22). Thus, a total of 138 genes involved in the inactivation of biotin synthesis, glutathione metabolism, pyrimidine synthesis, and reduction of the electron chain transport and the ATP synthase complex were lost. Finally, during the evolution of the B. aphidicola BCt linage, 28 more gene losses occurred, seven of which were metabolic but none affecting new metabolic pathways. We detected 26 losses in B. aphidicola BCc, with the loss of genes involved in tryptophan biosynthesis being the most striking.
The history of gene losses in B. aphidicola reveals the interplay with other actors.
A careful analysis of the history of gene losses in Buchnera provides some important clues about the role played by other actors in the symbiotic consortia (mainly different aphid hosts and a plethora of different secondary endosymbionts, most, but not all of which are considered facultative [34, 41]). The presence of secondary or facultative symbionts in members of the subfamily Aphidinae is well-known (34), and thus, we postulate that in the common ancestor of the members of this subfamily, Buchnera alone played a symbiotic role. Similarly, secondary endosymbionts are not found in members of the Eriosomatinae (41). Thus, although the ancestor of the Eriosomatinae and Lachninae lineages underwent more losses than the ancestor of the Aphidinae, which involved the inactivation of many new metabolic pathways, we postulate that B. aphidicola was the only endosymbiont, indicating that this ancestor could survive without synthesizing the missing metabolites. In the Eriosomatinae lineage, the number of gene losses is low, and more interestingly, no new pathways are affected. This situation clearly contrasts with the massive gene losses that occurred in the other derived clade, the ancestor of Lachninae. A possible explanation may again be associated with the lack of secondary endosymbionts in this lineage and the need to keep essential metabolic symbiotic functions in B. aphicola BBp, which contrasts with the presence of secondary endosymbionts, such as “Ca. Serratia symbiotica” in the lachnid host (6, 22). Previous studies into the presence of facultative symbionts from the aphid subfamily Lachninae covering the Cinarini and Lachnini tribes showed that almost all aphids studied were infected with a bacterial symbiont in addition to Buchnera. Many of these endosymbionts were “Ca. Serratia symbiotica,” although bacteria belonging to other lineages were present in some aphids. Thus, it was proposed that facultative symbionts such as “Ca. Serratia symbiotica” may be beneficial in Lachninae by compensating for inadequate nutrient provisioning by Buchnera (6). We postulate that this association with facultative symbionts occurred in the common ancestor of the Lachninae lineage, thus allowing the massive gene loss that took place at that point in its evolution. Subsequently, new bacterial infections could have occurred, some of which replaced the ancestral one.
It has previously been shown that “Ca. Serratia symbiotica” is also present in C. tujafilina but it belongs to a different clade than that found in C. cedri (6, 22). According to the phylogenetic reconstructions with 16S rRNA genes, “Ca. Serratia symbiotica” from C. tujafilina is similar to the one found in A. pisum, where it is a facultative endosymbiont (30). Moreover, both “Ca. Serratia symbiotica” isolates showed similar bacillary morphology, which differed greatly from the round shape of “Ca. Serratia symbiotica” from C. cedri (11, 17, 22). These facts could be related to the differences found in tryptophan provision in B. aphidicola BCt and B. aphidicola BCc. B. aphidicola BCt does not need metabolic complementation with “Ca. Serratia symbiotica” to make this essential amino acid, as it occurs in B. aphidicola BCc, which is a very important difference regarding its symbiotic role.
In summary, in spite of the stochastic factors that may be responsible for particular gene losses in different B. aphidicola lineages, it seems that the evolving metabolic network formed by the different actors also plays an important selective role in keeping it as a whole.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grant FU2009-12895-CO2-01 from the Ministerio de Ciencia e Innovación (Spain) and Prometeo 92/2009 from Generalitat Valenciana (Spain).
We acknowledge J. J. Abellan for helpful assistance with the statistical analysis. We thank Silvia Ramos for technical help.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Akaike H. 1974. New look at statistical-model identification. Trans. Automat. Control 19:716–723 [Google Scholar]
- 2. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–33402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ausubel F. M. 1999. Short protocols in molecular biology: a compendium of methods from Current protocols in molecular biology, 4th ed. Wiley, New York, NY [Google Scholar]
- 4. Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 59:155–189 [DOI] [PubMed] [Google Scholar]
- 5. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY [Google Scholar]
- 6. Burke G. R., Normark B. B., Favret C., Moran N. A. 2009. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl. Environ. Microbiol. 75:5328–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deckert G., et al. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353–358 [DOI] [PubMed] [Google Scholar]
- 8. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Douglas A. E., Wilkinson T. L. 1998. Host cell allometry and regulation of the symbiosis between pea aphids, Acyrthosiphon pisum, and bacteria, Buchnera. J. Insect Physiol. 44:629–635 [DOI] [PubMed] [Google Scholar]
- 10. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukatsu T., Nikoh N., Kawai R., Koga R. 2000. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 66:2748–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gardner P. P., et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gil R., Sabater-Munoz B., Latorre A., Silva F. J., Moya A. 2002. Extreme genome reduction in Buchnera spp.: toward the minimal genome needed for symbiotic life. Proc. Natl. Acad. Sci. U. S. A. 99:4454–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gil R., Sabater-Munoz B., Perez-Brocal V., Silva F. J., Latorre A. 2006. Plasmids in the aphid endosymbiont Buchnera aphidicola with the smallest genomes. A puzzling evolutionary story. Gene 370:17–25 [DOI] [PubMed] [Google Scholar]
- 15. Gil R., et al. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. U. S. A. 100:9388–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gomez-Valero L., Latorre A., Silva F. J. 2004. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 21:2172–2181 [DOI] [PubMed] [Google Scholar]
- 17. Gomez-Valero L., et al. 2004. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J. Bacteriol. 186:6626–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gosalbes M. J., Lamelas A., Moya A., Latorre A. 2008. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J. Bacteriol. 190:6026–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 20. Heie O. E. 1987. Paleontology and phylogeny, p. 367–391 In Minks A. K., Harrewijn P. (ed.), Aphids: their biology, natural enemies and control, vol. 2A World crop pests series Elsevier, Amsterdam, The Netherlands [Google Scholar]
- 21. Keseler I. M., et al. 2009. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 37:D464–F470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamelas A., et al. 2008. Evolution of the secondary symbiont “Candidatus Serratia symbiotica” in aphid species of the subfamily Lachninae. Appl. Environ. Microbiol. 74:4236–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Latorre A., Gil R., Silva F. J., Moya A. 2005. Chromosomal stasis versus plasmid plasticity in aphid endosymbiont Buchnera aphidicola. Heredity 95:339–347 [DOI] [PubMed] [Google Scholar]
- 24. Lopez-Sanchez M. J., et al. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 5:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCutcheon J. P., Moran N. A. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392–19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran N. A., McCutcheon J. P., Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 28. Moran N. A., McLaughlin H. J., Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382 [DOI] [PubMed] [Google Scholar]
- 29. Moran N. A., Munson M. A., Baumann P., Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. Roy. Soc. Lond. 253:167–171 [Google Scholar]
- 30. Moran N. A., Russell J. A., Koga R., Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moya A., Pereto J., Gil R., Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218–229 [DOI] [PubMed] [Google Scholar]
- 32. Nakabachi A., Ishikawa H. 1999. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J. Insect Physiol. 45:1–6 [DOI] [PubMed] [Google Scholar]
- 33. Nakabachi A., et al. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. [DOI] [PubMed] [Google Scholar]
- 34. Oliver K. M., Degnan P. H., Burke G. R., Moran N. A. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55:247–266 [DOI] [PubMed] [Google Scholar]
- 35. Ortiz-Rivas B., Martinez-Torres D. 2010. Combination of molecular data support the existence of three main lineages in the phylogeny of aphids (Hemiptera: Aphididae) and the basal position of the subfamily Lachninae. Mol. Phylogenet. Evol. 55:305–317 [DOI] [PubMed] [Google Scholar]
- 36. Ortiz-Rivas B., Moya A., Martinez-Torres D. 2004. Molecular systematics of aphids (Homoptera: Aphididae): new insights from the long-wavelength opsin gene. Mol. Phylogenet. Evol. 30:24–37 [DOI] [PubMed] [Google Scholar]
- 37. Perez-Brocal V., et al. 2006. A small microbial genome: the end of a long symbiotic relationship? Science 314:312–313 [DOI] [PubMed] [Google Scholar]
- 38. Posada D., Crandall K. A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 39. Regalia M., Rosenblad M. A., Samuelsson T. 2002. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 30:3368–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Remaudière G., Remaudière M. 1997. Catalogue des Ahididae du monde. Institut National de la Recherche Agronomique, Paris, France [Google Scholar]
- 41. Russell J. A., Latorre A., Sabater-Munoz B., Moya A., Moran N. A. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12:1061–1075 [DOI] [PubMed] [Google Scholar]
- 42. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 43. Sharples G. J. 2009. For absent friends: life without recombination in mutualistic gamma-proteobacteria. Trends Microbiol. 17:233–242 [DOI] [PubMed] [Google Scholar]
- 44. Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 45. Silva F. J., Latorre A., Moya A. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615–618 [DOI] [PubMed] [Google Scholar]
- 46. Staden R., Beal K. F., Bonfield J. K. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115–130 [DOI] [PubMed] [Google Scholar]
- 47. Tamas I., et al. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379 [DOI] [PubMed] [Google Scholar]
- 48. Tatusov R. L., et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toft C., Fares M. A. 2008. The evolution of the flagellar assembly pathway in endosymbiotic bacterial genomes. Mol. Biol. Evol. 25:2069–2076 [DOI] [PubMed] [Google Scholar]
- 51. Van Ham R. C., et al. 2000. Postsymbiotic plasmid acquisition and evolution of the repA1-replicon in Buchnera aphidicola. Proc. Natl. Acad. Sci. U. S. A. 97:10855–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. van Ham R. C., et al. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. U. S. A. 100:581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von Dohlen C. D., Moran N. A. 2000. Molecular data support a rapid radiation of aphids (Aphididae) in the Cretaceous and multiple origins of host alternation. Biol. J. Linn. Soc. 71:689–717 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.