Abstract
The eastern Mediterranean Sea represents an ultraoligotrophic environment where soluble phosphate limits the growth of bacterioplankton. Correspondingly, genes coding for high-affinity phosphate uptake systems and for organophosphonate utilization are highly prevalent in the plankton metagenome. Chemotaxis toward inorganic phosphate constitutes an alternative strategy to cope with phosphate limitation, but so far has only been demonstrated for two bacterial pathogens and an archaeon, and not in any free-living planktonic bacterium. In the present study, bacteria affiliated with the genus Thalassospira were found to constitute a regular, low-abundance member of the bacterioplankton that can be detected throughout the water column of the eastern Mediterranean Sea. A representative (strain EM) was isolated in pure culture and exhibited a strong positive chemotaxis toward inorganic phosphate that was induced exclusively in phosphate-starved cultures. Phosphate-depleted cells were 2-fold larger than in exponentially growing cultures, and 43% of the cells retained their motility even during prolonged starvation over 10 days. In addition, Thalassospira sp. strain EM was chemotactically attracted by complex substrates (yeast extract and peptone), amino acids, and 2-aminoethylphosphonate but not by sugar monomers. Similarly to the isolate from the eastern Mediterranean, chemotaxis toward phosphate was observed in starved cultures of the other two available isolates of the genus, T. lucentensis DSM 14000T and T. profundimaris WP0211T. Although Thalassospira sp. represents only up to 1.2% of the total bacterioplankton community in the water column of the eastern Mediterranean Sea, its chemotactic behavior potentially leads to an acceleration of nutrient cycling and may also explain the persistence of marine copiotrophs in this extremely nutrient-limited environment.
INTRODUCTION
Previous studies of oligotrophic open ocean environments have indicated that the growth of bacterioplankton is often limited by the availability of inorganic phosphate (8, 15, 23, 38). So far, bacteria have been shown to cope with low ambient phosphate concentrations by the expression of high-affinity uptake systems for phosphate (e.g., the membrane-bound ABC transporter PstCAB) (16); by mobilizing phosphate bound in organophosphate esters or organophosphonates using alkaline phosphatase (in particular PhoA or PhoX) (47) or specific transporters and C-P lyases (18), respectively; or by replacing phospholipids with membrane-forming lipids devoid of phosphorus, such as sulfolipids (63).
Even the seemingly homogenous oceanic open waters are known to harbor a multitude of nutrient hot spots (2). As an alternative adaptive strategy toward nutrient limitation, diverse marine bacterial isolates have acquired the capability to chemotactically respond toward such gradients of growth-limiting substrates and thus are capable of exploiting spatial and temporal sources of higher nutrient concentrations more efficiently. Typical chemoattractants that have been identified for marine bacteria so far comprise amino acids (Pseudoalteromonas haloplanktis, Silicibacter, and Vibrio) (3, 36, 39), glucose (Vibrio) (36), different nitrogenous compounds (Synechococcus) (60), dimethylsulfoniopropionate metabolites (Silicibacter) (39), and algal exsudates (P. haloplanktis and Silicibacter) (39, 53).
Phosphate esters constitute the dominant fraction (on average 64%) of total dissolved phosphorus in the marine environment (62). Marine aggregates display intense activities in particular for phosphatase (49) and thus represent important point sources for inorganic phosphate. Though absent in Escherichia coli (31), chemotaxis toward phosphate has been shown to occur in the two bacterial pathogens Pseudomonas aeruginosa (24, 43) and Enterobacter cloacae (31) and, more recently, in the archaeon Halobacterium salinarum strain R1 (59), where it was induced during phosphate starvation. However, it is unknown whether any bacterioplankton members of oligotrophic marine waters are capable of chemotaxis toward inorganic phosphate.
The eastern Mediterranean Sea is characterized by very low ambient nutrient concentrations and an unusual high Redfield ratio due to the depletion in inorganic phosphate to concentrations below 50 nM (10, 64). Correspondingly, microbial growth in the Mediterranean Sea most likely is limited by the availability of soluble phosphate (54, 57, 58, 64, 65). Metagenomic analyses support this conclusion and indicate that the bacterioplankton of the eastern Mediterranean Sea has adapted to the nutrient limitation by employing high-affinity phosphate uptake systems and by utilizing organophosphonates (12). Recent culture-independent studies of the eastern Mediterranean Sea (A. Hütz, K. Schubert, M. Mayer, and J. Overmann, unpublished data) have revealed that bacteria affiliated with the genus Thalassospira exhibit a specific chemotaxis toward inorganic phosphate. The aim of the following study was to further elucidate the chemotaxis toward phosphate of this previously unknown component of oligotrophic bacterioplankton.
MATERIALS AND METHODS
Sampling locations.
Water samples were obtained during cruise M71, leg 3, of the R/V Meteor from Heraklion, Greece, to Istanbul, Turkey, between 17 January and 4 February 2007. The five sampling stations H02 (21°0.046′E, 35°45.014′N), H07 (17°44.969′E, 39°0.994′N), H10 (19°0.063′E, 30°55.142′N), Rho02 (27°42.026′E, 35°37.008′N), and Ier01 (26°12.103′E, 34°26.476′N) were located in the oligotrophic eastern Mediterranean Sea (Fig. 1A). Conductivity, temperature, molecular oxygen concentrations, and chlorophyll a (Chl a) fluorescence were monitored in 1-m intervals (46) employing an SBE911 plus CTD system (Sea-Bird Electronics, Inc., Bellevue, WA). Water samples were recovered from 5 m below the sea surface, from the Chl a maximum and the salinity maximum, and from a water depth of 1,000 m, using an SBE32 rosette equipped with 24 10-liter FreeFlow bottles (Hydrobios).
Fig. 1.
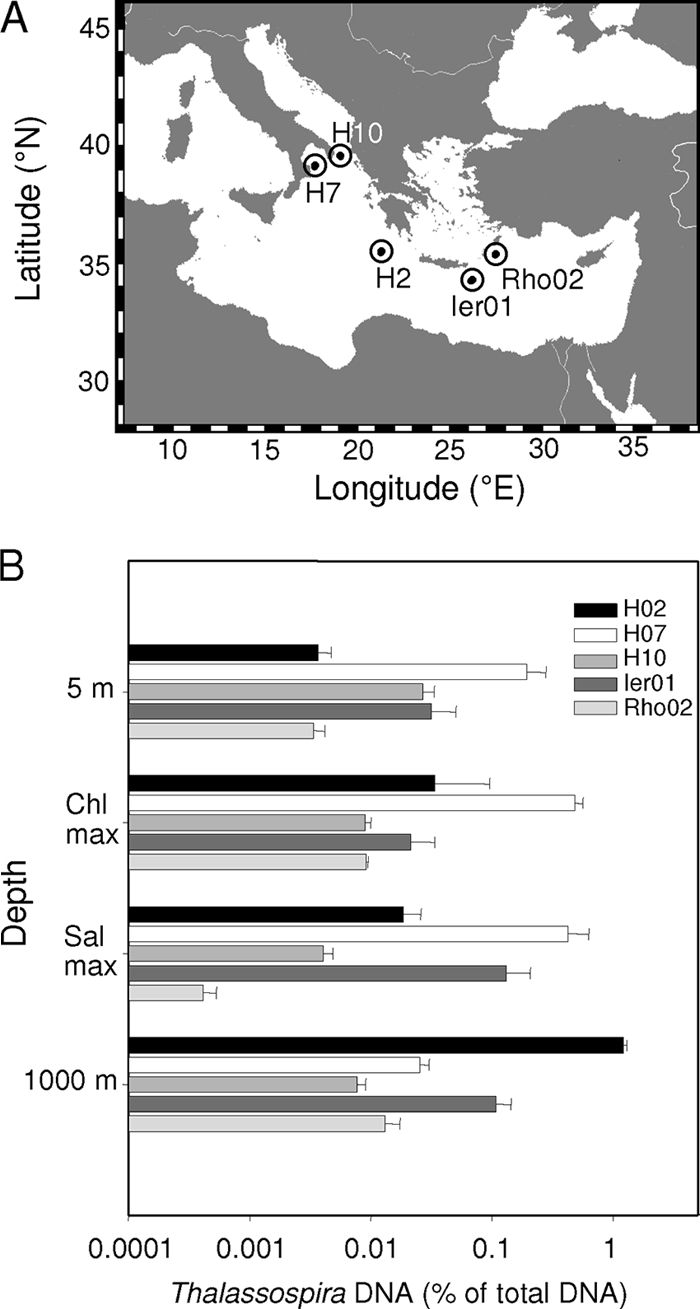
(A) Sampling locations in the eastern Mediterranean Sea. (B) Relative abundance of Thalassospira sp. in bacterioplankton at the five stations as determined by genus-specific qPCR. Error bars represent 1 standard deviation. The detection limit was 0.0001%. Water samples were collected near the surface (5-m depth), at the chlorophyll maxima (positioned at 50-, 40-, 10-, 40-, and 45-m depths at stations H07, H10, H02, Ier01, and Rho02, respectively), at the salinity maximum (positioned at 300-, 300-, 350-, 100-, and 180-m depths at stations H07, H10, H02, Ier01, and Rho02, respectively), and at a depth of 1,000 m.
From each station and sampling depth, bacterial cells were collected by filtering 500 to 2000 ml of water on sterile 0.1-μm-pore-size polycarbonate filters (type VCTP; Millipore, Eschborn, Germany). Filters were stored at −20°C until analyses. To generate a sufficient inoculum required for gradient agar tubes and for the determination of enzyme activities, bacterial cells in 100 liters of water obtained from the Chl a maxima were concentrated using a tangential flow filtration system (Pellicon 2; 0.1-μm-pore-size filtration cassette; Millipore).
DNA extraction.
Genomic DNA was extracted from membrane filters by the method of Fuhrman et al. (14), using a modified protocol (37). For further purification, DNA was precipitated by adding 0.1 volume of 3 M sodium acetate and 2.5 volumes of 100% ethanol followed by incubation at −20°C overnight. The DNA was sedimented by centrifugation at 18,000 × g for 30 min at 4°C (centrifuge 5417R; Eppendorf, Hamburg, Germany), washed with ice-cold 70% (vol/vol) ethanol, dried, and resuspended in 10 μl of sterile Tris buffer (2 mM Tris-HCl [pH 8.0]). DNA concentrations were determined by fluorescent dye binding with PicoGreen (Invitrogen, Karlsruhe, Germany).
Fingerprinting, sequencing, and phylogenetic analysis of 16S rRNA genes.
Bacterial enrichments were screened by amplification of 16S rRNA gene fragments employing primers for Bacteria, separation of the fragments by denaturing gradient gel electrophoresis (DGGE), and sequencing of the excised bands (17).
For phylogenetic analyses of the isolated bacterial strain, the almost full-length 16S rRNA gene was amplified by colony PCR with universal bacterial primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) (32). After purification (NucleoSpin extract II kit; Machery-Nagel, Düren, Germany), PCR products were sequenced by the dideoxynucleotide method on an ABI Prism 3730 genetic analyzer (Applied Biosystems), employing primers 8f and 1492r and the AmpliTaq FS Big Dye terminator cycle sequencing kit. Sequences were edited and assembled with the Vector NTI computer package (Invitrogen). Additional 16S rRNA gene sequences of Thalassospira spp. were retrieved from the GenBank database (1) and imported into the ARB program package (35). After alignment, phylogenetic trees were constructed with the FastdnaML maximum likelihood algorithm as implemented in the ARB software package.
qPCR.
The relative abundance of bacteria of the genus Thalassospira in the water samples was determined by quantitative PCR (qPCR) targeting its specific 16S rRNA gene sequence. The genus-specific PCR primers Tha585f (5′-CGGTCTTGCCAGTCAGGG-3′) and Tha655r (5′-CACCACCCTCTCCTAGTC-3′) were designed with the ARB software package (35). Primer specificity was checked with the probe match tool of the RDP database (6) and by qPCR trials with genomic DNA of Thalassospira sp. strain EM as a positive control and 5 ng of genomic DNA from the six other Alphaproteobacteria Rhodospirillum rubrum DSM107, Rhizobium radiobacter DSM30147T, Sphingomonas kaistensis DSM16846T, Hyphomonas jannaschiana DSM5153T, Ruegeria pomeroyi DSM15171T, and Erythrobacter citreus DSM14432T as negative controls. Genomic DNA of Thalassospira sp. EM served as a standard for calibration and was employed as a 10-fold dilution series ranging from 5 fg to 500 pg. All reactions were run at least in triplicates using an iCycler iQ multicolor real-time detection system (Bio-Rad, München, Germany). Each 25-μl reaction mixture consisted of 8.5 μl PCR-grade water, 20 μg bovine serum albumin, 12.5 μl IQ SYBR green supermix (Bio-Rad), and 2 μmol each of primers Tha585f and Tha655r and the respective DNA templates. The cycling conditions included an initial denaturation at 94°C for 2 min, followed by 40 cycles of denaturation at 94°C for 15 s, annealing at 65°C for 20 s, and elongation at 72°C for 20 s. After completion, the melting behavior of products was analyzed to assess their specificity.
Cultivation and isolation of Thalassospira sp. EM from the eastern Mediterranean Sea.
Cultivation trials were conducted in substrate gradient agar tubes. The substrates consisted of either an equal mixture of 43 different carbon substrates (including acetate, arabinose, benzoate, butanol, buyrate, citrate, ethanol, formate, glucose, lactate, malate, mannitol, methanol, N-acetylglucosamine, 2-oxoglutarate, propanol, propionate, pyruvate, salicylate, succinate, and valerate; a combination of the 20 canonical amino acids, at 20 μM each; and yeast extract and Tween 80 at 0.1% and 0.01% [wt/vol], respectively). One-milliliter bottom agar plugs contained artificial seawater (ASW) medium, substrates, and 1% (wt/vol) washed agar (Oxoid, Cambridge, United Kingdom) and were overlaid with 4 ml of 1% washed agar without substrates. One liter of ASW basic medium contained 24.4 g NaCl, 10 g MgCl2·6H2O, 1.5 g CaCl2·2H2O, 0.66 g KCl, 4 g Na2SO4 and 2.38 g HEPES (7); trace elements and vitamins were added according to reference 37, while KH2PO4 (P source) and NH4Cl (N source) were omitted in basic medium. Gradients were produced by preincubation over 5 days. Subsequently, the bacterial cell suspension generated by tangential flow filtration was used to inoculate the gradient agar tubes. The gradients were inoculated over their entire depth using a sterile Pasteur pipette and incubated at 15°C in the dark for up to 5 weeks.
After growth became visible, subsamples were streaked onto agar plates composed of ASW amended with carbon substrates (see above), 1 μM KH2PO4, 20 μM KNO3, and 20 μM NH4Cl and solidified with 1.5% (wt/vol) purified agar. Agar plates were incubated at 15°C in the dark until growth became visible. Colonies of different morphologies were then transferred to liquid artificial seawater medium of the same composition. An isolate that showed motility was purified by repeated streaking onto agar plates composed of marine broth (Difco, Becton Dickinson, Heidelberg, Germany) solidified with 1.5% agar. Routine cultivation of this strain was done in marine broth. For chemotaxis experiments, the isolate was cultured in artificial seawater medium containing 5 mM KH2PO4, 15 mM NH4Cl, and 10 mM glucose and 0.005% (wt/vol) yeast extract (ASWGluNPYE). In order to obtain phosphate-depleted bacterial cells, KH2PO4 was omitted (ASWGluNYE).
Chemotaxis assays.
Thalassospira sp. EM isolated from the eastern Mediterranean Sea and Thalassospira lucentensis DSM 14000T and Thalassospira profundimaris WP0211T (= DSM 17430T) were maintained in the artificial seawater medium supplemented with phosphate, ammonia, glucose, and trace amounts of yeast extract as described above. An initial series of cultivation experiments revealed that none of the Thalassospira strains was able to grow in defined mineral medium in the absence of trace amounts of yeast extract (see the Results section). The concentration of yeast extract was minimized to 0.005% (wt/vol) in order to enable growth of the bacteria but at the same time to produce phosphate-starved cultures. Based on a phosphorus content of yeast extract of 1.34% by weight (27), this amount corresponds to starting concentrations of 21 μM phosphate in the cultures. For the chemotaxis experiments, phosphate-depleted cultures were generated by two consecutive passages in ASW lacking free phosphate. In the first passage, cells were starved in phosphate-free ASWGluN to deplete intracellularly stored P. To generate sufficient cell densities for the chemotaxis experiments, the prestarved cells were then inoculated into ASW devoid of phosphate (ASWGluNYE) for phosphate starvation and in parallel into artificial seawater medium containing 5 mM phosphate (ASWGluNPYE) for phosphate-replete cultures.
Chemotactic responses of the bacterioplankton were analyzed by capillary assays (13, 44). After reaching stationary growth phase, 15-ml aliquots of the Thalassospira cultures were transferred into sterile Meplat bottles that had 12 bore holes drilled through one side wall. Flat rectangular glass capillaries (length, 50 mm; inside diameter, 0.1 by 2.0 mm) (VitroCom, Mountain Lakes, NJ) were filled by capillary action with the stock solutions, sealed at one end with plasticine (Münchner Künstler Plastilin, Munich, Germany), inserted into the holes of the Meplat bottles such that the open ends of the capillaries were immersed in the seawater sample inside, and fixed in the holes with plasticine. Chemotaxis assays lasted for 1.5 h. Different concentrations of KH2PO4, 2-aminoethylphosphonate (2 mM), a mixture of amino acids (l-threonine, l-arginine, l-histidine, l-methionine, l-proline, glycine, l-lysine, l-valine, l-serine, l-alanine, l-glutamic acid, l-leucine, l-cysteine, l-asparagine, l-tryptophan, l-aspartic acid, and l-glutamine at 2 mM each), a mixture of sugars [α-d(+)-glucose, α-mannitol, l(+)-arabinose, d(+)-trehalose, and (+)-xylose at 2 mM each], and yeast extract (0.1% [vol/vol]) or peptone (0.1% vol/vol) were assessed as chemoattractants and for this purpose were dissolved in culture supernatant. Capillaries filled with culture supernatant alone served as a negative control. Experiments were routinely carried out in two separate Meplat bottles as parallels. The entire contents of three capillaries per Meplat bottle were recovered, and their contents were combined and fixed with glutardialdehyde (2% vol/vol) for cell counting.
Determination of swimming speed and cell size.
The mean swimming speed of Thalassospira sp. EM under various growth conditions was determined as described by Eschemann et al. (11). Bacterial cultures were diluted as required, and a drop of the cell suspension was spotted onto a glass slide. The movement of cells was immediately traced under an inverted microscope (Axiovert; Zeiss) employing a dark field at 400-fold magnification. Tracks of swimming cells were measured over an exposure time of 500 ms with a digital camera (Hamamatsu Orca R2, C10500; Olympus, Munich, Germany) and the software package cellF^ (Olympus, Munich, Germany). Depending on the variability, the mean swimming speed for each growth condition was determined based on measurements of 16 to 109 individual cells. The fraction of motile cells was determined based on the number of trajectories of motile cells and numbers of point-shaped traces of nonmotile cells.
Bacterial cell sizes in phosphate-starved and phosphate-replete Thalassospira sp. cultures were determined by phase-contrast microscopy using fixed cells and 1,250-fold magnification. Images of were recorded digitally (Hamamatsu Orca R2, C10500; Olympus) and analyzed with the software package cellF^.
Analytical procedures.
Growth was monitored by observing the optical density of subsamples at 580 nm (OD580) (Genesys 20; Thermo Scientific, Dreieich, Germany) and cell counting in a Neubauer counting chamber (Marienfeld, Lauda-Königsdorf, Germany). For cell counting, 500-μl aliquots of the cultures were fixed with glutardialdehyde (final concentration, 2% [vol/vol]). Numbers of CFU were determined by plating on marine broth agar plates.
Total protein content was determined in triplicate with 2-ml culture aliquots. Cells were harvested by centrifugation at 20,800 × g for 15 min. The pellets were washed once with artificial seawater medium devoid of glucose, resuspended in 250 μl of 0.1 N NaOH, and lysed by boiling for 2 min. Subsequently, 1 to 2 μl of the solution was used for measurements of protein concentrations with a bicinchoninic acid assay (50).
The presence of 19 different exoenzymes was assessed with API ZYM test strips (API Systems, bioMérieux, Nürtingen, Germany). Bacterial cells from 1 ml of cultures were collected by centrifugation at 20,800 × g for 15 min, and the cells were resuspended in 1 ml of 1 M NaCl and transferred into the API ZYM chambers. Test strips were incubated for 4 h at room temperature. The activity of the different exoenzymes in seawater samples from the eastern Mediterranean Sea was assessed by using the bacterial cell concentrates obtained by tangential flow filtration (see above). In this case, the incubation time was extended to 16 h, and the incubation temperature was adjusted to the in situ value of 15°C.
Nucleotide sequence accession number.
The 16S rRNA gene sequence obtained in this study has been deposited in the GenBank database under accession no. JF292454.
RESULTS
Isolation of Thalassospira sp. EM from eastern Mediterranean seawater.
Recent investigations had revealed that bacteria affiliated with the genus Thalassospira are constituents of the oligotrophic eastern Mediterranean bacterioplankton that exhibit a selective chemotactic response toward phosphate (A. Hütz, K. Schubert, M. Mayer, and J. Overmann, unpublished data). In order to isolate a representative strain for detailed investigations of the chemotactic behavior, gradients of multiple organic carbon substrates were inoculated with concentrated bacterioplankton samples. The enrichments were screened by DGGE fingerprinting and sequencing of the separated 16S rRNA gene fragments. Enrichments of motile bacteria that were affiliated with the genus Thalassospira were chosen for subsequent isolation. Strain EM could be isolated from station Ier01 after several rounds of purification on agar-solidified artificial seawater medium supplemented with a combination of 43 organic carbon substrates at low concentrations. Cells of this strain were short, highly motile spirilla (see Fig. S2 in the supplemental material).
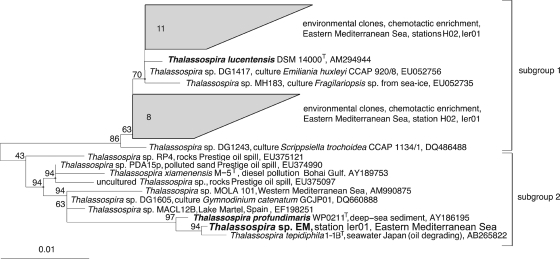
Phylogenetic analyses of the 1,349 bp-long 16S rRNA gene sequence placed strain EM within the radiation of the genus Thalassospira (Fig. 2). At a sequence similarity of >99.5%, the closest relatives were Thalassospira tepidiphila 1-1BT, isolated from petroleum-contaminated seawater (29), and Thalassospira profundimaris WP0211T (33), isolated from deep-sea sediment of the west Pacific Ocean. The 16S rRNA gene sequence of Thalassospira lucentensis DSM 14000T clusters separately (subgroup 1 in Fig. 2) together with environmental clones from the eastern Mediterranean bacterioplankton.
Fig. 2.
Maximum likelihood phylogenetic tree of 16S rRNA gene sequences of cultured and not-yet-cultured bacteria affiliated with the genus Thalassospira. Clusters of sequences obtained in a recent investigation of bacterioplankton chemotaxis in the eastern Mediterranean are included for comparison. Strains included in the present study are marked in boldface. The size bar depicts 0.01 fixed point mutation per nucleotide position. Bootstrap values derived from 100 resamplings are indicated at the nodes.
Abundance of Thalassospira sp. in situ.
So far, Thalassospira sp. has never been reported to occur in marine bacterioplankton assemblages but has only been recovered from other environments, in particular the marine littoral, algal culture, or hydrocarbon-affected environments (Fig. 2). Recent investigations of chemotactically active bacterioplankton yielded strong and specific enrichments of Thalassospira sp. in capillaries containing phosphate (A. Hütz, K. Schubert, M. Mayer, and J. Overmann, unpublished data). Therefore, the in situ abundance of bacteria belonging to the genus Thalassospira in the water column of the eastern Mediterranean Sea was determined by a quantitative PCR assay employing newly developed genus-specific primers. Five different sampling locations were studied (Fig. 1A). Measurements were performed for the same water masses at all locations for better comparability. Accordingly, water samples were recovered from 5 m below the sea surface within the modified Atlantic surface water, and from the chlorophyll (Chl) maximum positioned at depths of 50, 40, 10, 40, and 45 m at stations H7, H10, H02, Ier01, and Rho02, respectively. Deeper layers represented the salinity maximum of the Levantine intermediate water that forms during wintertime and were sampled at depths of 300, 300, 350, 100, and 180 m at stations H7, H10, H02, Ier01, Rho02, respectively. The deepest samples originated from a water depth of 1,000 m within the homogenous eastern Mediterranean deep water that forms in the Adriatic Sea (46).
The relative abundance of Thalassospira sp. ranged from very low values of 0.0004% ± 0.0001% of total genomic DNA at station Rho02 (salinity maximum at 180-m water depth) to 1.2% ± 0.08% at station H02 (deep water at 1,000-m depth). In the upper part of the water column, the relative abundance of Thalassospira was highest at station H07, but no apparent pattern with respect to distance from the coast or depth was detected. Based on these results, members of the genus Thalassospira sp. are present throughout the eastern Mediterranean Sea and in some layers constitute a well-discernible component of the bacterioplankton.
Response of Thalassospira strain EM to phosphate starvation.
Bacterioplankton growth in the eastern Mediterranean is assumed to be limited by the availability of inorganic phosphate. Since cells of Thalassospira sp. EM were observed to rapidly lose their motility during centrifugation, phosphate-starved cultures could not be obtained by simply concentrating, washing, and starving a dense suspension of routinely grown laboratory cultures. Instead, phosphate-depleted and phosphate-replete laboratory cultures were generated in parallel, and their chemotactic responses were analyzed directly in order to elucidate the effect of phosphate starvation on chemotaxis. While the eastern Mediterranean isolate Thalassospira sp. EM grew rapidly in marine broth, it was not capable of growing in defined minimal artificial seawater medium ASWGluNP containing glucose as the sole organic carbon source. However, the strain resumed growth upon transfer in defined mineral medium supplemented with small amounts of yeast extract (ASWGluNPYE; see Fig. S1 in the supplemental material), indicating a requirement of growth factors in addition to the 10 vitamins and 12 trace elements that are present in ASW. Contrary to previous reports (29, 33, 34), this obligate dependence on unidentified growth factors was also observed for Thalassospira lucentensis DSM 14000T and Thalassospira profundimaris WP0211T. Consequently, all cultures for subsequent chemotaxis experiments had to be routinely grown in the presence of 0.005% (vol/vol) yeast extract.
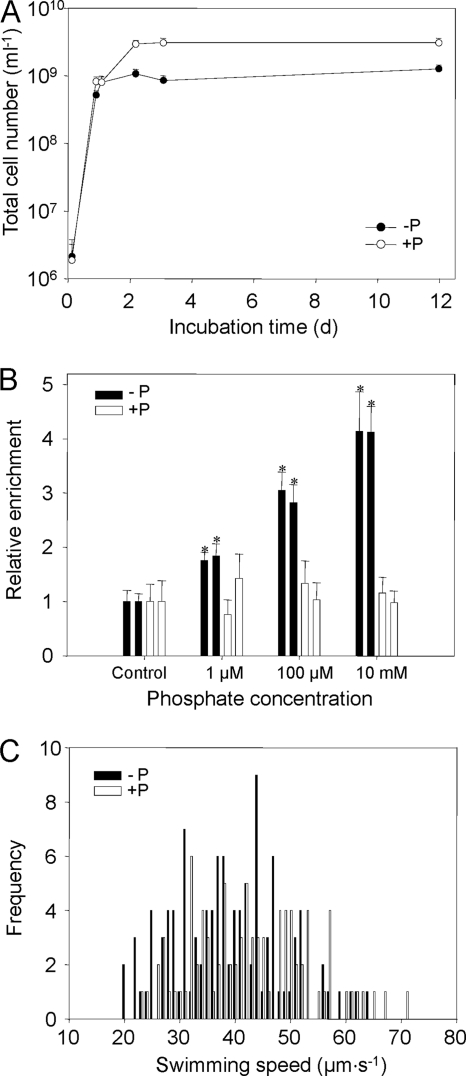
Phosphate-depleted (ASWGluNYE) and phosphate-replete (ASWGluNPYE) laboratory cultures of Thalassospira sp. EM reached doubling times of 6.2 h during the exponential growth phase that lasted for 22 h under the cultivation conditions chosen (Fig. 3A). In phosphate-replete media, total cell numbers reached in the stationary phase significantly surpassed those in phosphate-depleted cultures by a factor of almost 3, and the protein content was 1.7-fold higher than that of phosphate-depleted cultures (compare the last three time points in Fig. 3A and Table 1). However, the OD580 values in the stationary phase remained similar in the two types of cultures (Table 1). Taken together with the initially similar growth rates observed in both types of cultures, our results thus indicate that P-depleted cultures reached the P limitation of growth within 1 day of incubation. Individual cells in phosphate-depleted cultures were significantly elongated compared to cells in phosphate-replete media (see Fig. S2 in the supplemental material), whereas the cell diameter did not change (0.86 ± 0.08 μm in P-replete cells and 0.82 ± 0.15 μm in P-depleted cells, respectively). As a result, mean cell volumes reached 2.8 ± 0.8 μm3 for phosphate-starved cells and hence were 2-fold larger than the volumes of cells grown under phosphate-replete conditions, which attained a volume of only 1.4 ± 0.2 μm3. Correspondingly, the cellular protein content of phosphate-starved cells was 187 ± 76 fg·cell−1, compared to 94 ± 59 fg·cell−1 for phosphate-replete cells. This cellular protein content of Thalassospira falls into the range determined for various bacterial cultures (60 to 330 fg·cell−1), but surpasses that of bacterioplankton (average, 24 fg·cell−1) (66). The numbers of CFU were about 1 order of magnitude lower than the total cell numbers under all growth conditions, but this fraction of culturable cells did not change with the amount of available phosphate (Table 1).
Fig. 3.
(A) Growth curves of Thalassospira sp. EM grown in phosphate-depleted artificial seawater medium ASWGluNP (−P; solid circles) and phosphate-replete medium ASWGluNPYE (+P; open circles). The time course of total cell numbers is shown. Cultures were inoculated with prestarved starter cultures (see Materials and Methods). Vertical bars represent 1 standard deviation. (B) Accumulation of phosphate-starved (black columns) and phosphate-replete (white columns) cells of Thalassospira sp. EM in chemotaxis capillaries loaded with artificial seawater containing different concentrations of KH2PO4. Incubation lasted for 1.5 h. Control capillaries were devoid of phosphate. Error bars represent 1 standard deviation. Asterisks indicate values that differ significantly (P < 0.001) from negative controls based on a standard t test. (C) Frequency distribution of swimming speeds determined for phosphate-starved (black columns) and phosphate-replete (white columns) cells of Thalassospira sp. EM determined after 52.5 h of incubation. Frequencies per class of 1 μm·s−1 are given. The resulting mean swimming speeds are listed in Table 1.
Table 1.
Characteristics of Thalassospira sp. strain EM cultivated in phosphate-depleted ASWGluNYE medium and phosphate-replete ASWGluNPYE mediuma
| Time (h) | OD580 |
Total cell count/ml (108) |
CFU/ml (107) |
Mean swimming speed (μm·s−1) |
Protein content (μg·ml−1) |
Cellular protein content (fg·cell−1) |
Motile cells (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +P | −P | +P | −P | +P | −P | +P | −P | +P | −P | +P | −P | +P | −P | |
| 0 | 0.002 | 0.001 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 3.0 | 0.004 | 0.004 | 0.018 (0.013) | 0.021 (0.017) | 0.014 (0.003) | 0.015 (0.0012) | 35.1 (11.2) | 41.1 (12.8) | ND | ND | ND | ND | 87.2 (0.4) | 89.0 (2.5) |
| 21.8 | 0.307 | 0.187 | 8.2 (1.4) | 5.2 (0.83) | 7 (0.63) | 3.6 (0.25) | 45.1 (14.9) | 39.4 (14.2) | 58.7 (11.7) | 22.1 (10.0) | 71.6 (18.8) | 42.5 (20.4) | 82.9 (1.7) | 85.1 (0.3) |
| 26.0 | 0.460 | 0.253 | 8.0 (1.6) | 7.9 (1.9) | ND | 3.1 (0.99) | 51.4 (15.9) | 41.4 (14.1) | 78.9 (11.8) | 71.5 (23.6) | 98.6 (24.6) | 90.5 (37.0) | 83.4 (4.5) | 84.2 (0.8) |
| 52.5 | 2.48 | 0.785 | 29 (4.4) | 11 (1.9) | 28 (17) | 11 (4.8) | 42.4 (11.0) | 38.3 (9.6) | 228 (66.2) | 109 (47.9) | 78.6 (25.8) | 99.1 (46.8) | 50.7 (4.1) | 77.7 (1.8) |
| 74.0 | 2.30 | 1.27 | 30 (4.8) | 8.5 (1.5) | 16 (6.6) | 2.2 (0.62) | 62.6 (19.4) | 62.2 (14.3) | 294 (209) | 172 (56.9) | 98.0 (71.4) | 202.4 (75.9) | 37.3 (11.3) | 46.5 (2.5) |
| 170.8 | 2.08 | 1.86 | ND | ND | ND | ND | ND | ND | 361 (25.2) | 190 (56.9) | ND | ND | ND | ND |
| 287.5 | 2.02 | 2.12 | 31 (5.3) | 12 (1.8) | 26 (4.2) | 19 (0.19) | 60.1 (14.4) | 53.8 (12.9) | 281 (135) | 205 (85.9) | 90.6 (46.2) | 170.8 (76.0) | 27.3 (10.3) | 42.5 (1.8) |
Standard deviations are given in parentheses. OD580, optical density at 580 nm; +P, phosphate-replete medium; −P, phosphate-depleted medium; ND, not determined.
Cells in stationary cultures of phosphate-starved and phosphate-replete Thalassospira sp. EM exhibited a similar spectrum of exoenzyme activities that included alkaline phosphatase, acid phosphatase, C4 esterase, and leucine arylamidase (Table 2). In addition, the phosphate-starved cells showed C8 esterase lipase activity, whereas weak activities of valine arylamidase and naphthol-AS BI-phosphohydrolase were observed for phosphate-replete cultures. In comparison, concentrated bacterioplankton samples from the eastern Mediterranean Sea also exhibited alkaline phosphatase as the dominant exoenzyme activity and in addition clear naphthol-AS BI-phosphohydrolase activity (Table 2). Only weak activities were determined for C8 esterase lipase, valine arylamidase, and acid phosphatase at all stations, whereby C8 esterase lipase activities were higher at the easternmost stations, Ier01 and Rho02.
Table 2.
Exoenzyme activities in stationary-phase cultures of Thalassospira EM and in bacterioplankton concentrated from water samples from four sampling stations in the eastern Mediterranean as determined by the API ZYM test systema
| Enzyme | Activity for: |
|||||
|---|---|---|---|---|---|---|
| EM +P | EM −P | H10 | H02 | IER01 | Rho02 | |
| Control | − | − | − | − | − | − |
| Alkaline phosphatase | ++ | +++ | ++ | ++ | ++ | ++ |
| Esterase (C4) | (+) | + | − | − | − | − |
| Esterase lipase (C8) | − | + | (+) | (+) | + | + |
| Lipase (C14) | − | − | − | − | − | − |
| Leucine arylamidase | ++ | + | − | − | − | − |
| Valine arylamidase | (+) | − | (+) | (+) | (+) | + |
| Cystine arylamidase | − | − | − | − | − | − |
| Trypsin | − | − | − | − | − | − |
| α-Chymotrypsin | − | − | − | − | − | − |
| Acid phosphatase | +++ | ++ | (+) | (+) | (+) | (+) |
| Naphthol-AS-BI-phosphohydrolase | (+) | − | + | + | + | + |
| α-Galactosidase | − | − | − | − | − | − |
| β-Galactosidase | − | − | − | − | − | − |
| β-Glucuronidase | − | − | − | − | − | − |
| α-Glucosidase | − | − | − | − | − | − |
| β-Glucosidase | − | − | − | − | − | − |
| N-Acetyl-β-glucosaminidase | − | − | − | − | − | − |
| α-Mannosidase | − | − | − | − | − | − |
| α-Fucosidase | − | − | − | − | − | − |
+P, cells grown in ASWGluNPYE; −P, cells grown in ASWGluNYE; −, no activity; (+), weak activity; +, activity; ++, intense activity; +++, very intense activity.
Differential chemotactic response of Thalassospira spp. toward inorganic phosphate.
Stationary-phase cells of Thalassospira sp. EM cultivated in ASWGluNP and ASWGluNPYE were assessed for their chemotactic behavior toward different concentrations of KH2PO4 (Fig. 3B). In these assays, phosphate-replete Thalassospira cells never accumulated in capillaries containing inorganic phosphate irrespective of the concentrations offered. Cells of Thalassospira sp. EM exhibited a broad range of swimming speeds (Fig. 3C). Maximum values of 63 μm·s−1 were reached in the stationary phase (Table 1). Mean swimming speeds did not vary significantly between phosphate-starved and phosphate-saturated cells. On the contrary, a significant accumulation relative to control capillaries containing artificial seawater medium was only observed for the phosphate-starved cells. The minimum phosphate concentration evoking a positive response was determined to be as low as 1 μM. The accumulation of Thalassospira cells observed in the capillaries has to be attributed to chemotaxis but not growth because of the following reasons. First, the Thalassospira strains employed in the present study are not capable of growing without yeast extract, which was omitted in the chemotaxis assays. Second, cell numbers increased in the capillaries by >4-fold within 1.5 h, whereas the doubling time in this medium (if supplemented with yeast extract) was 6.2 h. Third, phosphate-replete Thalassospira cells should have entered the capillaries by chance and then grown, but no increase in cell numbers was observed in chemotaxis assays with these cells.
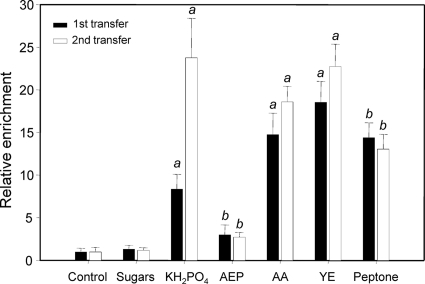
In a subsequent series of experiments, the chemotactic responses of Thalassospira sp. EM toward phosphate, organic phosphate, and carbon substrates were compared. Cells from two consecutive passages in ASWGluN were tested for their chemotactic response toward KH2PO4 (2 mM), 2-aminoethyl phosphonate (AEP; 2 mM), a mixture of amino acids (l-threonine, l-arginine, l-histidine, l-methionine, l-proline, glycine, l-lysine, l-valine, l-serine, l-alanine, l-glutamic acid, l-leucine, l-cysteine, l-asparagine, l-tryptophan, l-aspartic acid, l-glutamine at 2 mM each), a mixture of sugars [α-d(+)-glucose, α-mannitol, l(+)-arabinose, d(+)-trehalose and (+)-xylose at 2 mM each], and yeast extract (0.1% [vol/vol]) or peptone (0.1% vol/vol) (Fig. 4). Thalassospira sp. EM showed no response toward sugars and only weakly responded toward the organic phosphate compound AEP. In contrast, strong responses toward inorganic phosphate, amino acids, yeast extract, and peptone could be observed. Whereas the chemotactic responses toward inorganic phosphate were significantly stronger after the second transfer in ASWGluN and increased slightly but significantly for the amino acid mixture and yeast extract, an increase in enrichment factors was not observed for any of the other three chemoattractants. These results suggest that chemotaxis particularly toward inorganic phosphate is selectively induced under conditions of phosphate starvation.
Fig. 4.
Chemotaxis of Thalassospira sp. EM toward different phosphorus and organic carbon compounds. AEP, 2-aminoethyl phosphonate; AA, mixture of amino acids; YE, yeast extract. The compositions of the sugar and amino acid mixtures are given in the text. The relative chemotactic enrichment of bacterial cells from the first (black columns) and the second (white columns) transfer compared to that of controls without substrates is given. Error bars represent 1 standard deviation. a, enrichment factors significantly different between the first and second transfers and from the control; b, enrichment factors similar between the first and second transfers but significantly different from the control (P < 0.001, t test).
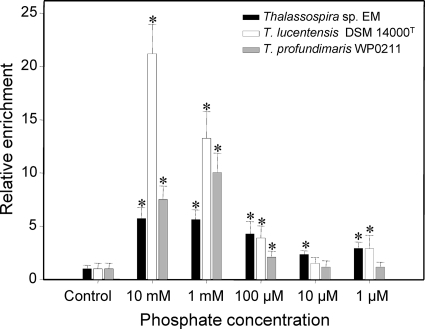
In a final set of chemotaxis experiments, the chemotactic responses of the three available representatives of the genus Thalassospira toward inorganic phosphate were compared. Thalassospira lucentensis (34), Thalassospira profundimaris (33), and the newly obtained isolate Thalassospira sp. EM were starved for phosphate in ASWGlucN for 1 week. Cells of all three strains accumulated in capillaries containing 10 mM, 1 mM, or 100 μM KH2PO4 (Fig. 5). In addition, Thalassospira sp. EM showed a significant positive response toward 10 μM as well as 1 μM KH2PO4, and Thalassospira lucentensis showed a positive response toward 1 μM KH2PO4.
Fig. 5.
Chemotactic accumulation of three different Thalassospira strains in capillaries loaded with ASW containing different concentrations of phosphate. Prior to chemotaxis assays, all cultures were starved for phosphate. Error bars represent 1 standard deviation. Asterisks indicate significant differences from negative controls without phosphate (P < 0.001, t test).
DISCUSSION
Response of Thalassospira to phosphate starvation.
The phosphate-replete growth medium ASWGluNPYE contained C, N, and P in a molar ratio of 62:16:5 (as calculated from the concentrations and elemental composition of the ingredients) when accounting also for the amounts of nutrients added with the small amount of yeast extract. Assuming that half of the organic carbon substrate is respired, the C/N/P ratio of nutrients available for biomass formation in this medium is 31:16:5. Based on the average composition of the bacterial cells of C43.5N11.4P1.0 (4), the ASWGluNPYE medium thus provides limiting amounts of organic carbon for bacterial growth and produces cells starved for organic carbon.
The amount of yeast extract added had to be minimized in a manner to obtain phosphate-starved cells but at the same time to generate cell numbers that were sufficient for the chemotaxis experiments. The protein content of phosphate-depleted cultures in the stationary phase amounted to 189 μg·ml−1 (Table 1; mean of last three measurements). Based on the typical protein/dry weight ratio of 0.55 (42) and a phosphorus content of 15 mg P·(g dry weight)−1 of Escherichia coli cells (9), the demand of phosphorus to synthesize the biomass observed for Thalassospira sp. EM is expected to be 161 μM. However, the concentration of phosphate available in the culture medium in the form of yeast extract amounted to only 21 μM (see Materials and Methods). The decreased biomass determined in ASWGluNYE thus is commensurate with the conclusion that growth of Thalassospira in this medium was limited by phosphate and resulted in the formation of phosphate-starved cells. Compared to E. coli, the cellular phosphorus demand of Thalassospira sp. EM is lower, which suggests that the novel isolate is capable of decreasing its intracellular phosphorus concentrations and/or capable of substituting for part of the phosphorus-containing molecules during its adaptation to phosphate starvation.
The pronounced elongation of the cells and the 2-fold increase in cell volume in the Thalassospira sp. EM cultures exposed to phosphate limitation are in line with morphological changes reported for other copiotrophic marine bacteria under these conditions. Starvation of Vibrio angustum strain S14 for phosphorus also leads to markedly enlarged cells (28), whereas starvation of bacteria for carbon substrates typically induces a reductive division and dwarfing due to degradation of endogenous cell material, eventually resulting in the formation of small coccoid cells (28, 41). Similarly, phosphate starvation of Thiobacillus ferrooxidans induces a filamentation of the cells that has been attributed to inhibition of DNA synthesis and, as a consequence, of cell division (48). It remains to be determined whether cell elongation in Thalassospira sp. EM represents a developmental program, as in Caulobacter crescentus, where stalk elongation under phosphate limitation is under the control of the transcriptional activator PhoB (19). By comparison, obligately or facultatively oligotrophic bacteria undergo no or only a limited reduction of their cell volume during starvation (21).
It has previously been observed that low nutrient conditions lead to high swimming velocities in natural assemblages of marine bacteria (40). In the cyanobacterium Synechococcus, phosphate stress induces genes encoding cell-surface proteins required for swimming motility (56). During prolonged starvation of Thalassospira sp. EM, mean swimming speeds increased by about 20 μm·s−1 compared to the speed at the onset of starvation. However, mean swimming speeds of Thalassospira sp. EM did not differ between cultures starved for organic carbon or inorganic phosphate and thus do not seem to be differentially regulated by the limiting nutrient. In cultures of Vibrio angustum S14, motility is almost completely lost over the first 24 h of starvation for organic carbon (36), and starvation also strongly decreases the fraction of motile cells in cultures of Silicibacter sp. strain TM1040 (39). The loss of motility in Thalassospira sp. EM cultures was much less pronounced. Even after 10 days of starvation, the fraction of motile cells still amounted to 43% in phosphate-depleted cultures. Maintaining motility ensures that the cells remain capable of tracing point sources of phosphorus even after prolonged periods of phosphate shortage.
Natural bacterial communities throughout the eastern basin of the Mediterranean Sea exhibited high alkaline phosphatase activities. The activity of this enzyme has been shown to be inversely correlated with inorganic phosphate concentrations in situ (58). Induction of alkaline phosphatase (in particular PhoA or PhoX) is a typical response of planktonic microorganisms to phosphate limitation (52) and has also been observed in other marine Alphaproteobacteria of the Roseobacter clade (47). The induction of extracellular alkaline phosphatase allows the cells to scavenge phosphate from organic sources, is repressed by inorganic phosphate, and hence represents a strategy to cope with low-availability inorganic phosphate. In a similar fashion, acid phosphatase in Thiobacillus ferrooxidans is induced by phosphate limitation (48). It was therefore unexpected that starvation of Thalassospira cells for organic carbon produced high levels of alkaline and acid phosphatase activity similar to those of phosphate-starved cells. Under both conditions, alkaline and acid phosphatase exhibited the highest activity among all 19 exoenzymes tested and therefore may be expressed constitutively in Thalassospira sp. EM. Similar to Thalassospira sp. EM, extracellular phosphatase activity is not regulated by inorganic phosphate in other bacteria (e.g., Mycobacterium bovis) (5).
Taken together, the results of the present study suggest that the specific adaptative response of Thalassospira sp. EM to phosphate-limiting conditions involves other mechanisms than increased motility or increased hydrolysis of organophosphate esters.
Chemotaxis toward phosphate as an adaptive strategy.
Phosphate concentrations in the eastern Mediterranean Sea are in the nanomolar range (10, 64). Recent metagenomic analyses indicate that certain members of the bacterioplankton community of the eastern Mediterranean Sea have adapted to this limitation by employing high-affinity phosphate uptake systems as well as by utilizing organophosphonates (12). The majority of the corresponding functional genes were affiliated with the Alphaproteobacteria, including members of the SAR11 group. The latter represent oligotrophic bacteria that are known to harbor a disproportionately large number of substrate binding proteins for phosphate and phosphonate (51), do not grow at high nutrient concentrations, and typically are nonmotile like, e.g., Pelagibacter ubique, which also has long generation times of 29 h (45). In comparison, the marine copiotrophs Alteromonas, Pseudoalteromonas, and Vibrio typically occur at low abundance, yet are widespread and continuously present in ultraoligotrophic oceans, where they have also been demonstrated to occur in an active metabolic state (55). Obviously, even marine copiotrophic bacteria find appropriate ecological niches within such oligotrophic marine environments.
All Thalassospira strains isolated so far are adapted to higher nutrient concentrations and grow well in complex media (29, 33, 34). Based on its consistent presence throughout the eastern Mediterranean, the comparatively short doubling times of 6.2 h, and its pronounced changes in cell morphology during starvation determined in the present study, Thalassospira represents a previously unrecognized but widely distributed genus of marine copiotrophs. Our results indicate that chemotaxis represents a mechanism that permits members of this genus to populate the eastern Mediterranean despite the pronounced phosphate limitation in this environment.
Chemotaxis toward inorganic phosphate is absent in E. coli and Salmonella enterica serovar Typhimurium and to date has only been demonstrated for the two facultatively pathogenic bacteria Pseudomonas aeruginosa (24) and Enterobacter cloacae (31). Among the archaea, only Halobacterium salinarum has been shown to exhibit phosphate chemotaxis (59). Our finding of phosphate chemotaxis in a marine planktonic alphaproteobacterium suggests that this chemotactic behavior occurs in typical environmental bacteria and may also be present in other bacterial lineages. Besides the type species T. lucentensis, subgroup 1 of the genus Thalassospira mostly comprises isolates from various algal cultures that are likely to be adapted to exploitation of nutrients in the algal phycosphere. In addition, subgroup 1 encompasses novel phylotypes that were recently shown to exhibit phosphate-directed chemotaxis by cultivation-independent methods (A. Hütz, K. Schubert, M. Mayer, and J. Overmann, unpublished data). Most members of Thalassospira subgroup 2 originate from hydrocarbon-affected environments, but this group also encompasses the isolate of the present study. The previous isolation of a close relative of Thalassospira strain EM, T. profundimaris WP0211T, from west Pacific deep-sea sediment suggests a broader distribution also of this subgroup. The present comparative study, together with culture-independent evidence for additional Thalassospira phylotypes, thus demonstrates that chemotaxis toward phosphate occurs across the entire phylogenetic breadth of the genus Thalassospira and may therefore be a characteristic of this genus.
In other marine bacteria, the chemotactic response may either decrease significantly within only a few hours of starvation (36) or, alternatively, may be increased by prior starvation or during the starvation phase (39). Our results show that Thalassospira sp. EM follows the second pattern. In Vibrio angustum S14, the genes that are upregulated during starvation for phosphate differ from those induced by carbon starvation. Correspondingly, the proteins of the phosphate starvation stimulon are distinctly different from that of carbon-starved cells (28). In Enterobacter cloacae, a genetic knockout of phosphate chemotaxis does not affect the chemotaxis toward peptone (31). Phosphate-directed chemotaxis of Pseudomonas aeruginosa PAO1 is induced by phosphate starvation (43) and involves two novel chemoreceptors (61). Our finding that P starvation but not C starvation elicits the chemotactic response of cells toward inorganic phosphate suggests that the chemotaxis genes also in Thalassospira sp. EM are differentially regulated and form part of a specific P starvation response. In Pseudomonas aeruginosa PAO1, both phosphate chemotaxis and alkaline phosphatase expression are controlled by the same negative regulator, phoU (25). In contrast, our results suggest that the regulation of phosphate chemotaxis in Thalassospira sp. EM is independent of the expression of alkaline phosphatase. Detailed genetic analysis of the regulation of chemotaxis genes in Thalassospira sp. has to await the availability of the genome sequence of a representative of this genus.
Besides organic carbon substrates, 2-aminoethylphosphonate elicited a chemotactic response of Thalassospira sp. EM that proved to be independent of the phosphate supply of the cultures. To our knowledge, this is the first report of bacterial chemotactic behavior toward a phosphonate. In the marine environment, phosphonates represent a considerable fraction of 25% of organic phosphorus compounds (30). Besides the hydrolysis of organophosphate esters and the chemotaxis toward phosphate, chemotactic accumulation and subsequent utilization of phosphonates may thus represent an additional adaptation of Thalassospira sp. EM to phosphate-depleted ocean surface waters.
Ecological relevance of phosphate-directed chemotaxis.
Modeling has revealed that chemotaxis permits motile bacteria to take advantage of point sources of organic carbon substrates and also to use plumes of dissolved organic matter formed in the wake of sinking organic particles. Ultimately, chemotaxis toward organic carbon compounds accelerates their turnover in the natural environment since it significantly increases the exposure of bacterial cells to their substrates (53). In a similar fashion, chemotaxis toward point sources of phosphate is likely to constitute an alternative strategy of motile bacteria to enhance their phosphate acquisition in an ultraoligotrophic and inhomogeneous planktonic environment. In fact, our data indicate a selective advantage of Thalassospira sp. EM for rapid accumulation at phosphate sources in a heterogenous environment. First, the threshold concentrations of phosphate-directed chemotaxis in Thalassospira spp. were 10- to 50-fold lower than those of Pseudomonas aeruginosa PAO1 or the eukaryotic alga Chattonella antiqua (20). Second, Thalassospira sp. EM swims faster than other marine copiotrophic bacteria like Pseudoalteromonas, Alteromonas, or Vibrio, which only reach mean velocities of 11 to 38 μm·s−1 (22). Thalassospira sp. thus is expected to accumulate at microscale phosphate sources which would enable the cells to scavenge inorganic phosphate more efficiently under the ultraoligotrophic phosphate-limiting conditions that prevail in the Mediterranean Sea (64). Future research will reveal whether Thalassospira actually colonizes point sources such as phosphate-liberating marine snow or just remains in and exploits the nutrient plumes caused by exoenzymatic activity of other, particle-attached bacteria (26) or by the egesting of food vacuoles by protists (2).
The chemotactic response observed in the present study is likely to accelerate the cycling of phosphorus in the eastern Mediterranean and indirectly suggests the existence of phosphate hot spots analogously to the well-established hot spots of organic carbon (2). Since phosphorus limitation of the bacterioplankton may also occur in other oligotrophic open ocean environments like the Sargasso Sea (8), the Caribbean Sea (38), the subtropical north Pacific (23), or the Red Sea (15), bacterial chemotaxis toward inorganic phosphate may affect nutrient cycling and productivity also in other oceanic provinces.
Supplementary Material
ACKNOWLEDGMENTS
We thank Evelyn Lage-Sonntag for help with the qPCR measurements.
This work was supported by grants OV 20/12-1 and OV 20/16-1 of the Deutsche Forschungsgemeinschaft to J. Overmann.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Azam F., Malfatti F. 2007. Microbial structuring of marine ecosystems. Bacterioplankton growth in the Sargasso Sea. Nat. Rev. Microbiol. 5:782–791 [DOI] [PubMed] [Google Scholar]
- 3. Barbara G. M., Mitchell J. G. 2003. Marine bacterial organisation around point-like sources of amino acids. FEMS Microbiol. Ecol. 43:99–109 [DOI] [PubMed] [Google Scholar]
- 4. Battley E. H. 1995. An apparent anomaly in the calculation of ash-free dry weights for the determination of cellular yields. Appl. Environ. Microbiol. 61:1655–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braibant M., Content J. 2001. The cell surface associated phosphatase activity of Mycobacterium bovis BCG is not regulated by environmental inorganic phosphate. FEMS Microbiol. Lett. 195:121–126 [DOI] [PubMed] [Google Scholar]
- 6. Cole J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coolen M. J. L., Overmann J. 2000. Functional exoenzymes as indicators of metabolically active bacteria in 124,000-year-old sapropel layers of the eastern Mediterranean Sea. Appl. Environ. Microbiol. 66:2589–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotner J. B., Ammerman J. W., Peele E. R., Bentzen E. 1997. Phosphorus-limited bacterioplankton growth in the Sargasso Sea. Aquat. Microb. Ecol. 13:141–149 [Google Scholar]
- 9. Damoglou A. P., Dawes E. A. 1968. Studies on the lipid content and phosphate requirement of glucose- and acetate-grown Escherichia coli. Biochem. J. 110:775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emeis K.-C., et al. 2010. External N inputs and internal N cycling traced by isotope ratios of nitrate, dissolved reduced nitrogen, and particulate nitrogen in the eastern Mediterranean Sea. J. Geophys. Res. 115:G04041 [Google Scholar]
- 11. Eschemann A., Kühl M., Cypionka H. 1999. Aerotaxis in Desulfovibrio. Environ. Microbiol. 1:489–494 [DOI] [PubMed] [Google Scholar]
- 12. Feingersch R., et al. 2010. Microbial community genomics in eastern Mediterranean Sea surface waters. ISME J. 4:78–87 [DOI] [PubMed] [Google Scholar]
- 13. Fröstl J. M., Overmann J. 1998. Physiology and tactic response of the phototrophic consortium “Chlorochromatium aggregatum”. Arch. Microbiol. 169:129–135 [DOI] [PubMed] [Google Scholar]
- 14. Fuhrman J. A., Comeau D. E., Hagström Å., Chan A. M. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuller N. J., et al. 2005. Dynamics of community structure and phosphate status of picocyanobacterial populations in the Gulf of Aqaba, Red Sea. Limnol. Oceanogr. 50:363–375 [Google Scholar]
- 16. Gebhard S., Ekanayaka N., Cook G. M. 2009. The low-affinity phosphate transporter PitA is dispensable for in vitro growth of Mycobacterium smegmatis. BMC Microbiol. 9:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gich F., Schubert K., Bruns A., Hoffelner H., Overmann J. 2005. Specific detection, isolation and characterization of selected, previously uncultured members of freshwater bacterioplankton. Appl. Environ. Microbiol. 71:5908–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilbert J. A., et al. 2009. Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ. Microbiol. 11:111–125 [DOI] [PubMed] [Google Scholar]
- 19. Gonin M., Quardokus E. M., O'Donnol D., Maddock J., Brun Y. V. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182:337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikegami S., Imai I., Kato J., Ohtake H. 1995. Chemotaxis toward inorganic phosphate in the red tide alga Chattonella antiqua. J. Plankton Res. 17:1587–1591 [Google Scholar]
- 21. Janssen P. H., Schuhmann A., Mörschel E., Rainey F. A. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johansen J. E., Pinhassi J., Blackburn N., Zweifel U. L., Hagström Å. 2002. Variability in motility characteristics among marine bacteria. Aquat. Microb. Ecol. 28:229–237 [Google Scholar]
- 23. Karl D. M. 2002. Nutrient dynamics in the deep blue sea. Trends Microbiol. 10:410–418 [DOI] [PubMed] [Google Scholar]
- 24. Kato J., Ito A., Nikata T., Ohtake H. 1992. Phosphate taxis in Pseudomonas aeruginosa. J. Bacteriol. 174:5149–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kato J., Sakai Y., Nikata T., Ohtake H. 1994. Cloning and characterization of a Pseudomonas aeruginosa gene involved in the negative regulation of phosphate taxis. J. Bacteriol. 176:5874–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiørboe T., Jackson G. A. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46:1309–1318 [Google Scholar]
- 27. Kirkbright G. F., Marshall M. 1973. Direct determination of phosphorus by atomic absorption flame spectrometry. Anal. Chem. 45:1610–1613 [Google Scholar]
- 28. Kjelleberg S., et al. 1993. How do non-differentiating bacteria adapt to starvation? Antonie Van Leeuwenhoek 63:333–341 [DOI] [PubMed] [Google Scholar]
- 29. Kodama Y., Stiknowati L. I., Ueki A., Ueki K., Watanabe K. 2008. Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 58:711–715 [DOI] [PubMed] [Google Scholar]
- 30. Kolowith L. C., Ingall E. D., Benner R. 2001. Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46:309–320 [Google Scholar]
- 31. Kusaka K., Shibata K., Kuroda A., Kato J., Ohtake H. 1997. Isolation and characterization of Enterobacter cloacae mutants which are defective in chemotaxis toward inorganic phosphate. J. Bacteriol. 179:6192–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley, Chichester, United Kingdom [Google Scholar]
- 33. Liu C., Wu Y., Li L., Ma Y., Shao Z. 2007. Thalassospira xiamenensis sp. nov. and Thalassospira profundimaris sp. nov. Int. J. Syst. Evol. Microbiol. 57:316–320 [DOI] [PubMed] [Google Scholar]
- 34. López-López A., et al. 2002. Thalassospira lucentensis gen. nov., sp. nov., a new marine member of the α-Proteobacteria. Int. J. Syst. Evol. Microbiol. 52:1277–1283 [DOI] [PubMed] [Google Scholar]
- 35. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malmcrona-Friberg K., Goodman A., Kjelleberg S. 1990. Chemotactic responses of marine Vibrio sp. strain S14 CCUG 15956 to low-molecular-weight substances under starvation and recovery conditions. Appl. Environ. Microbiol. 56:3699–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manske A. K., Glaeser J., Kuypers M. M. M., Overmann J. 2005. Physiology and phylogeny of green sulfur bacteria forming a monospecific phototrophic assemblage at a depth of 100 meters in the Black Sea. Appl. Environ. Microbiol. 71:8049–8060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martiny A. C., Huang Y., Li W. 2009. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ. Microbiol. 11:1340–1347 [DOI] [PubMed] [Google Scholar]
- 39. Miller T. R., Hnilicka K., Dziedzic A., Desplats P., Belas R. 2004. Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70:4692–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitchell J. G., et al. 1995. Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl. Environ. Microbiol. 61:877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Navarro Llorens J. M., Tormo A., Martínez-García E. 2010. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 34:476–495 [DOI] [PubMed] [Google Scholar]
- 42. Neidhardt F. C., Umbarger H. E. 1996. Chemical composition of Escherichia coli, p. 13–16 In Neidhardt F. C., et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC [Google Scholar]
- 43. Ohtake H., Kato J., Kuroda A., Wu H., Ikeda T. 1998. Regulation of bacterial phosphate taxis and polyphosphate accumulation in response to phosphate starvation stress. J. Biosci. 23:491–499 [Google Scholar]
- 44. Overmann J. 2005. Chemotaxis and behavioral physiology of not-yet-cultivated microbes. Methods Enzymol. 397:133–147 [DOI] [PubMed] [Google Scholar]
- 45. Rappé M. S., Connon S. A., Vergin K. L., Giovannoni S. J. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 46. Rubino A., Hainbucher D. 2007. A large abrupt change in the abyssal water masses of the eastern Mediterranean. Geophys. Res. Lett. 34:L23607 [Google Scholar]
- 47. Sebastian M., Ammerman J. W. 2009. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 3:563–572 [DOI] [PubMed] [Google Scholar]
- 48. Seeger M., Jerez C. A. 1993. Phosphate-starvation induced changes in Thiobacillus ferrooxidans. FEMS Microbiol. Lett. 108:35–41 [DOI] [PubMed] [Google Scholar]
- 49. Smith D. C., Simon M., Alldredge A. L., Azam F. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139–142 [Google Scholar]
- 50. Smith P. K., et al. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 51. Sowell S. M., et al. 2009. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3:93–105 [DOI] [PubMed] [Google Scholar]
- 52. Stihl A., Sommer U., Post A. F. 2001. Alkaline phosphatase activities among populations of the colony-forming diazotrophic cyanobacterium Trichodesmium spp. (Cyanobacteria) in the Red Sea. J. Phycol. 37:310–317 [Google Scholar]
- 53. Stocker R., Seymour J. R., Samadani A., Hunt D. E., Polz M. F. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. U. S. A. 105:4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tanaka T., et al. 2009. Determining the availability of phosphate and glucose in P-limited mesocosms of NW Mediterranean surface waters. Aquat. Microb. Ecol. 56:81–91 [Google Scholar]
- 55. Taniguchi A., Hamasaki K. 2008. Community structures of actively growing bacteria shift along a north-south transect in the western North Pacific. Environ. Microbiol. 10:1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tetu S. G., et al. 2009. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 3:835–849 [DOI] [PubMed] [Google Scholar]
- 57. Thingstad T. F., et al. 2005. Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science 309:1068–1071 [DOI] [PubMed] [Google Scholar]
- 58. Van Wambeke F., Christaki U., Giannakourou A., Moutin T., Souvemerzoglou K. 2002. Longitudinal and vertical trends of bacterial limitation by phosphorus and carbon in the Mediterranean Sea. Microb. Ecol. 43:119–133 [DOI] [PubMed] [Google Scholar]
- 59. Wende A., Furtwangler K., Oesterhelt D. 2009. Phosphate-dependent behavior of the archaeon Halobacterium salinarum strain R1. J. Bacteriol. 191:3852–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Willey J. M., Waterbury J. B. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl. Environ. Microbiol. 55:1888–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu H., et al. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 182:3400–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Young C. L., Ingall E. D. 2010. Marine dissolved organic phosphorus composition: insights from samples recovered using combined electrodialysis/reverse osmosis. Aquat. Geochem. 16:563–574 [Google Scholar]
- 63. Zavaleta-Pastor M., et al. 2010. Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc. Natl. Acad. Sci. U. S. A. 107:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zohary T., Robarts R. D. 1998. Experimental study of microbial P limitation in the eastern Mediterranean. Limnol. Oceanogr. 43:387–395 [Google Scholar]
- 65. Zohary T., et al. 2005. P-limited bacteria but N and P co-limited phytoplankton in the Eastern Mediterranean—a microcosm experiment. Deep Sea Res. Part II Top. Stud. Oceanogr. Stud. Oceanogr. 52:3011–3023 [Google Scholar]
- 66. Zubkov M. V., Fuchs B. M., Eilers H., Burkill P. H., Amann R. 1999. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl. Environ. Microbiol. 65:3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.