Abstract
The retinoblastoma protein-interacting zinc finger gene RIZ (PRDM2) is a member, by sequence homology, of a nuclear protein–methyltransferase (MTase) superfamily involved in chromatin-mediated gene expression. The gene produces two protein products, RIZ1 that contains a conserved MTase domain and RIZ2 that lacks the domain. RIZ1 gene expression is frequently silenced in human cancers, and the gene is also a common target of frameshift mutation in microsatellite-unstable cancers. We now report studies of mice with a targeted mutation in the RIZ1 locus. The mutation inactivates RIZ1 but not RIZ2. These RIZ1 mutant mice were viable and fertile but showed a high incidence of diffuse large B-cell lymphomas (DLBL) and a broad spectrum of unusual tumors. RIZ1 deficiency also accelerated tumorigenesis in p53 heterozygous mutant mice. Finally, several missense mutations of RIZ1 were found in human tumor tissues and cell lines; one of these was particularly common in human DLBL tumors. These missense mutations, as well as the previously described frameshift mutation, all mapped to the MTase functional domains. All abolished the capacity of RIZ1 to enhance estrogen receptor activation of transcription. These data suggest a direct link between tumor formation and the MTase domain of RIZ1 and describe for the first time a tumor susceptibility gene among methyltransferases.
Keywords: RIZ1, MTase, mutation, tumor formation, human cancer, DLBL
Inactivation of key cellular genes involved in cell proliferation, differentiation, survival, and genome integrity plays an important role in cancer formation and progression. These genes are broadly defined here as tumor suppressor genes (TSGs). Loss of function of TSGs may result through either genetic or epigenetic mechanisms. Although the importance of genetic mutations in cancer has long been recognized, the appreciation of epigenetic inactivation is more recent (Jones and Laird 1999; Baylin and Herman 2000; Eng et al. 2000). DNA methylation of promoters of TSGs commonly occurs in human tumors. In recent studies, methylation was analyzed directly and firmly established as one of the modes for TSG inactivation in tumors (Jones and Laird 1999). The frequency with which promoter methylation contributes to TSG inactivation ranges from ∼9% of Rb1 in retinoblastoma to 33% of VHL in Von Hippel Lindau disease to 84% of MLH1 in microsatellite-unstable colorectal tumors. Therefore, epigenetic silencing represents a significant contributor to human carcinogenesis.
The retinoblastoma protein-interacting zinc finger gene RIZ (PRDM2) was isolated by us in a functional screening for Rb-binding proteins (Buyse et al. 1995). It has also been isolated independently as a DNA-binding protein MTB-Zf (Muraosa et al. 1996), a GATA3 transcription factor binding protein G3B (Shapiro et al. 1995), and a coactivator of estrogen receptor (ER) (Abbondanza et al. 2000). RIZ contains two important peptide motifs, the canonical Rb-binding motif LXCXE and the nuclear hormone receptor-binding motif LXXLL. Furthermore, RIZ is a member of a gene family that shares a ∼130 amino acid sequence called the PR (PRDI-BF1 and RIZ) domain (also termed SET for Suvar3–9, Enhancer-of-zeste, Trithorax); this family is known to play an important role in chromatin-mediated regulation of gene expression, in development, and in cancer (Huang et al. 1998b). The detection of the PR/SET domain in plant protein lysine methyltransferases (MTases) suggested a MTase function for the domain (Rea et al. 2000). The human genome is estimated to contain ∼50 PR/SET containing genes that can be divided into the PR subfamily (20–25 members) and the SET subfamily (20–25 members) on the basis of sequence identity (Huang et al. 1998b; S. Huang, unpubl.). The PR subfamily has the gene symbol PRDMs (e.g., RIZ is PRDM2) and most members have a characteristic C2-H2 class of DNA-binding zinc finger motifs. The SET subfamily has yet to be given an official gene symbol and most members do not contain obvious DNA-binding motifs. The human genome lacks apparent orthologs of any of the plant lysine MTases and the plant genes cannot be divided into subfamilies and are equally related to human PR and SET subfamilies (S. Huang, unpubl.).
Recent data suggest that nuclear protein MTases play a role in cell cycle control and serve as transcriptional cofactors. The human SET-containing gene SUV39H1 functions as a histone H3 MTase and is important in chromatin condensation during mitosis (Rea et al. 2000). A role in transcription has been shown for the H3 MTase CARM1 that functions as a coactivator of nuclear hormone receptors (Chen et al. 1999). Two members of the PR/SET MTase family, NSD1 and RIZ, can also function as coactivators or corepressors of nuclear hormone receptors (Huang et al. 1998a; Abbondanza et al. 2000). These studies have begun to reveal a novel class of transcriptional coregulators that function as protein MTases.
A large number of observations suggest a role for RIZ in human cancers. The RIZ gene maps to the distal short arm of chromosome 1 or 1p36 that is frequently deleted in many types of human cancers, including lymphomas/leukemias and solid tumors (Weith et al. 1996). Common deletion of the gene has been shown to occur in breast, liver, and familial and sporadic colon cancers (Chadwick et al. 2000; Fang et al. 2000, 2001). The RIZ gene produces two mRNA and protein products through alternative promoters, RIZ1 that contains the PR domain, and RIZ2 that lacks this domain (Liu et al. 1997). Except for the PR domain and its neighboring regions, RIZ1 and RIZ2 are identical. Decreased or lost expression of RIZ1 mRNA, but not of RIZ2, is found in all types of human cancers examined—including those of breast, liver, bone, skin (melanoma), lung, colon, and neuroendocrine tissues—suggesting a selective epigenetic silencing of RIZ1 (He et al. 1998; Jiang et al. 1999; Chadwick et al. 2000). The expression of two RIZ proteins and the selective inactivation of the PR+ product in tumors are remarkably similar to features of another member of the family, the MDS1-EVI1 gene (Fears et al. 1996). The PR+ product MDS1-EVI1 is disrupted by chromosomal translocations and the PR− product EVI1 is overexpressed in myeloid leukemia. Together, these observations are consistent with an antioncogenic role of the PR+ product and an oncogenic role of the PR− product (Jiang and Huang 2000).
In addition to epigenetic silencing, genetic frameshift mutations of RIZ are common in microsatellite-unstable cancers of the colon, stomach, endometrium, and pancreas (Chadwick et al. 2000; Piao et al. 2000; Sakurada et al. 2001). The mutation is a 1- or 2-bp deletion in the (A)9 or (A)8 tract of the RIZ coding region resulting in frameshift and production of C terminus-truncated RIZ1 and RIZ2 proteins. Although the frameshift mutation affects both RIZ1 and RIZ2 proteins, the truncation of the C terminus is likely to affect RIZ1 more seriously, because the C terminus can bind to the PR-domain (Huang et al. 1998b).
Consistent with inactivation of RIZ1 in a broad spectrum of human cancers, recombinant adenovirus-mediated RIZ1 expression can induce G2/M cell cycle arrest, apoptosis, or both in several tumor cell lines (He et al. 1998; Jiang et al. 1999; Chadwick et al. 2000). Moreover, preclinical animal studies showed that RIZ1 could suppress the growth of xenograft colorectal cancers (Jiang and Huang 2001).
Although RIZ1 is unusual in that it has many different qualities related to human cancer, a causal relationship between RIZ1 and carcinogenesis has not been established. We addressed this issue by using mouse models in which RIZ1 but not RIZ2 is inactivated. We found that RIZ1-deficient mice develop an unusual spectrum of tumors. Also, in a genetic background compromised for p53, they suffer accelerated tumor formation and mortality. Missense mutations of RIZ1 were also found in human tumor tissues and cell lines. These mutations, as well as the frameshift mutation, abolished the capacity of RIZ1 to act as a coactivator of the estrogen receptor. These data provide evidence for a direct link between RIZ1 inactivation and tumor formation in mammals.
Results
RIZ1 gene targeting
We constructed a targeting vector with a neomycin-resistance (neor) gene expression cassette inserted into exon 5 of RIZ1. The insertion maps to the center of the PR domain. A Herpes Simplex Virus thymidine kinase (TK) gene was included for negative selection of nonhomologous recombinants (Fig. 1A). Because the RIZ2 promoter is located at exon 6 (Liu et al. 1997), which is ∼8 kb from where the neor cassette was inserted, this targeting strategy was expected to affect RIZ1 but not RIZ2 mRNA splicing or RIZ1/2 transcription. After transfecting the targeting vector into mouse embryonic stem (ES) cells, nine of twelve G418- and ganciclovir-insensitive colonies analyzed were heterozygous for the mutation at the RIZ locus. We used five heterozygous mutant D3 ES cells (Gossler et al. 1986) to generate chimeric mice and backcrossed chimeras to C57BL/6 mice. Animals bearing the targeted RIZ1 gene were identified by Southern blot (Fig. 1B) or PCR analysis (data not shown). Intercross of the RIZ1+/− mice produced offspring at close to predicted Mendelian ratios. Mating between RIZ1−/− mice also produced apparently normal offspring. No obvious gross or microscopic abnormalities were noted in any of the major organs at 1 to 2 mo of age. Northern blot analysis showed nondetectable RIZ1, but, consistent with the targeting strategy, the RIZ1−/− animals had normal levels of RIZ2 mRNA (Fig. 1C). In situ hybridization analysis further confirmed selective ablation of RIZ1 expression in RIZ1−/− animals (J. Yu and S. Huang, unpubl.). Finally, immunoprecipitation and immunoblot analysis showed that RIZ1 protein was absent but RIZ2 protein expression was normal in homozygous animals (Fig. 1D). These results showed that the targeted RIZ1 allele is an RIZ1-specific null allele.
Figure 1.
RIZ gene targeting. (A) Schematics of murine RIZ gene (top), targeting vector (middle), and targeted allele (bottom) are shown with horizontal bars representing genomic DNA and horizontal lines representing plasmid DNA. The positive and negative selection cassettes are boxes labeled Neo and TK, respectively. Restriction sites listed include: EcoRI (E), Xba I (X), BglII (B), HindIII (H), NsiI (N), KpnI (K), Kpn2I (K2), SacII (S), NotI (No), and XhoI (Xh). (B) Southern blot analysis of genomic DNA of RIZ-targeted mice. Genomic DNA was digested with XbaI and was hybridized with the 5′ probe. (C) Northern blot analysis. Total RNA isolated from adult mouse brain was analyzed by Northern blotting with the RIZ1-specific probe or the common (exon 8) probe, as indicated. A similar amount of loaded RNA was shown by ethidium bromide staining of rRNA. (D) Immunoblot analysis. Protein extracts were prepared from the testis of adult animals and immunoprecipitated with anti-KG7.1S serum against the N terminus of RIZ2. The immunoprecipitated products were resolved on 5% SDS gel followed by immunoblot by use of 1715 serum against the C terminus of RIZ1/2 proteins. (E) RT–PCR analysis. Size in base pairs (bp) of molecular weight standards in lane M are indicated.
Since the insertion mutation alters the structure of exon 5 (153 bp) and increases its size by 1.5 kb, we determined whether the targeted allele generates rare novel messages. RT–PCR of total RNA from wild-type samples yielded the expected 617-bp major product (Fig. 1E), cloning and sequencing confirmed that it represents messages including exons 3 through the 5′ end of exon 8 (data not shown). The one other product from wild-type samples is ∼510 bp in size and is much less abundant. This ∼510-bp product probably represents messages either not including exon 4 (104 bp) and/or exon7 (111 bp) as judged by size (note, exon 6 is 127 bp and is likely to be present in the ∼510-bp product based on size). In contrast, the 617-bp and ∼510-bp products were not generated from RIZ1 homozygous mutant samples, thereby confirming targeted ablation of wild-type RIZ1.
On the other hand, two RT–PCR products unique to the mutated RIZ1 allele were generated. Both a 466-bp and ∼360-bp product were generated from both RIZ1+/− and RIZ1−/− samples, but not from RIZ1+/+ samples. Both products were at very low levels, which was similar to that of the minor 510-bp product seen in wild-type animals. The 466-bp product was cloned and sequenced. It represents a message lacking exon 5 (153 bp) but containing exons 4 and 6 spliced together without alteration of the translation reading frame (data not shown). Based on length, the ∼360 bp product(s) probably represents the same messages as the minor wt allele-unique product, except that it contains exon 5.
The possibility that the RT–PCR products, unique to tissues harboring the mutant RIZ1 allele, represent mature, functional messages is rendered unlikely by the Northern and Western blot data. The RIZ1-specific probe used for Northern blots maps 5′ to exon 5 and therefore should detect a RIZ1 exon 5-deletion mutant message. Similarly, the antibodies employed in Western blots are able to recognize a RIZ1 exon 5-deletion mutant protein (see Fig. 7, below).
Figure 7.
Effects of RIZ gene products on transactivation activities of estrogen receptor. (A) Immunoblot analysis of RIZ proteins in transfected CV1-cells. Nuclear extracts from CV1 cells transfected with the indicated plasmid were analyzed by immunoblot analysis using RIZ serum anti-KG7.1S. (B) Expression vectors of RIZ1 proteins and ER were cotransfected into CV1 cells with the reporter construct ERE-tk-CAT containing a synthetic ER response element (Lee et al. 1995). Cells were treated with or without 17-β-estrodiol or E2 (10−7 M) for 24 h and assayed for CAT activity as described previously (Lee et al. 1995). Expression of β-gal was used to control transfection efficiency. Data shown represent the means (±S.D.) of three independent experiments.
Tumor formation in RIZ1 mutant mice
To determine whether ablation of RIZ1 affects the adult life of mice, a small cohort of 14 RIZ1−/− animals was initially kept for long-term observation. By the age of 20 mo, eight had developed tumors, whereas two died for unknown reasons. The tumors included stomach squamous cell carcinoma, stomach polyp, fibrosarcoma, lung adenocarcinoma, pituitary tumor, lipoma, and hemangiosarcoma. These results encouraged us to maintain a larger cohort of animals for long-term observation. The genetic background of the second cohort was on average 87.5% C57BL/6 and 12.5% 129Sv (the original chimeric mice were backcrossed twice to C57BL/6). The first cohort was on average 50% C57BL/6 and 50% 129Sv. By 21 mo of age, 28% of the RIZ1−/− animals were dead or moribund compared with 20% of RIZ1+/− and 8% of RIZ1+/+ littermates (Fig. 2). Most of the dead animals had tumors, suggesting an increased mortality in RIZ1−/− and RIZ1+/− animals possibly due to increased tumor burden. To determine whether some of the apparently healthy animals actually had tumors, we sacrificed and autopsied 35 RIZ1−/−, 47 RIZ1+/−, and 30 RIZ1+/+ animals at 18 to 22 mo of age. Tumors were found in 80% (29/35) of RIZ1−/− animals relative to 49% (23/47) of RIZ1+/−; animals (P < 0.001 by χ2 test) and 30% (9/30; P < 0.001) of RIZ1+/+ animals (Table 1). The difference in tumor incidence between RIZ1+/− and RIZ1+/+ animals was also significant (P < 0.01). Tumors in RIZ1−/−; and RIZ1+/− animals appeared more aggressive and often occurred in multiple organs. Multiple-organ tumors represented 37% (13/35) of RIZ1−/− animals relative to 19% (9/47; P < 0.05) of RIZ1+/− and 3% (1/30; P < 0.001) of RIZ1+/+ animals (Table 1). No difference in tumor incidence was noted between males and females.
Figure 2.
Survival of RIZ1 mutant animals in p53 wild-type or mutant background. (A) The graph summarizes the viability of 110 RIZ1−/− (filled circles) animals relative to 138 RIZ1+/− (open circles) and 68 RIZ1+/+ (filled triangles) animals. Error bars, S.E.M. (B) Graph summarizing the viability of 24 RIZ1−/−p53−/− (filled circles) animals relative to 32 RIZ1+/−p53−/− (open circles) and 21 RIZ1+/+p53−/− (closed triangles) animals. (C) Graph summarizing the viability of 34 RIZ1−/−p53+/− (open circles) animals relative to 32 RIZ1+/−p53+/−, 40 RIZ1+/+p53+/−, and 26 RIZ1−/−p53+/+ (open diamond) animals. The graph was drawn with the Prism statistics software program (GraphPad Software) on the basis of the Kaplan-Meyer theory.
Table 1.
Tumor incidence in animals autopsied at ∼20 months
|
|
RIZ1−/−
|
RIZ1+/−
|
RIZ1+/+
|
|---|---|---|---|
| Mean age of mice | 20.3 ± 1.0 | 20.8 ± 1.4 | 20.0 ± 1.5 |
| Mice with tumors | 28 (80%) | 23 (49%) | 9 (30%) |
| Mice with tumors in multiorgans | 13 (37%) | 9 (19%) | 1 (4%) |
| DLBL | 13 (37%) | 9 (19%) | 0 |
| Lung adenocarcinoma | 13 (37%) | 10 (21%) | 5 (17%) |
| FCC lymphoma | 5 (12%) | 6 (13%) | 5 (17%) |
| Hemangiosarcoma | 2 (5%) | 1 (2%) | 0 |
| Pituitary adenoma | 1 (3%) | 1 (2%) | 0 |
| Fibrosing large cell lymphoma | 1 (3%) | 1 (2%) | 0 |
| Hepatoma | 1 (3%) | 0 | 1 (3%) |
35 RIZ1−/−, 47 RIZ1+/−, and 30 RIZ1+/+ animals at 18–22 months of age were randomly sacrificed for autopsy analysis. Numbers in parentheses indicate the percentage of 35 homozygote, 47 heterozygote, or 30 wild-type animals that developed tumors or a particular type of tumor. (DLBL) Diffuse large B-cell lymphoma; (FCC) follicular center cells.
To investigate tumor formation in a tumor-prone genetic background and possible cooperative effects on tumor formation of mutations in the RIZ1 and p53 genes, we mated animals homozygous for the RIZ1 null mutation with animals heterozygous for a p53 null mutation (Jacks et al. 1994). Once animals heterozygous for both mutations were generated, they were intercrossed to yield animals of all possible genotypes. The RIZ1 mutation did not alter the survival and tumor spectrum of p53−/− animals (Fig. 2B). However, a cooperative effect of RIZ1 and p53 mutations was observed in RIZ1−/−p53+/− animals. The mean age of survival with this genotype was 415 d (Fig. 2C). In contrast, animals heterozygous for the p53 mutation alone or homozygous for the RIZ1 mutation alone survived on average >550 d. Most of the moribund or dead RIZ1−/−p53+/− animals had tumors, many of which were of the same type as the tumors of the animals homozygous for the RIZ1 mutation alone (Table 2). The mean age of survival of doubly heterozygous animals was also slightly less than p53 singly heterozygous mutant animals (Fig. 2C). The tumor types of the doubly heterozygous animals were primarily sarcomas, similar to p53 singly mutant animals (Table 2).
Table 2.
Tumor spectrum of RIZ1 and p53 doubly mutant mice
| Tumors
|
RIZ1−/−p53+/−
|
RIZ1+/−p53+/−
|
RIZ1+/+p53+/−
|
|---|---|---|---|
| DLBL | 9 (69%) | 1 (12.5%) | 0 |
| sarcoma | 1 (8%) | 5 (62.5%) | 2 (50%) |
| thymus lymphoma | 2 (16%) | 1 (12.5%) | 1 (25%) |
| squamous cell carcinoma | 0 | 1 (12.5%) | 0 |
| lung adenocarcinoma | 1 (8%) | 0 | 0 |
| testis tumor | 0 | 0 | 1 (25%) |
| Total | 13 | 8 | 4 |
Tumors were found in moribund or dead animals in a cohort of 34 RIZ1−/−p53+/−, 32 RIZ1+/−p53+/−, and 40 RIZ1+/+p53+/− animals. Numbers in parentheses indicate the percentage of the total number of tumor incidences.
The predominant tumor in RIZ1−/−, RIZ1+/−, and RIZ1−/−p53+/− animals was malignant B-cell lymphoma in the Peyer's patch, mesenteric lymph nodes, and other lymphoid tissues. This lymphoma was commonly found in RIZ1−/− (37%) and RIZ1 −/− (19%) animals but was absent in the RIZ1+/+ littermates (Tables 1 and 2). The gross appearance of the Peyer's patch lymphoma was a round glistening mass that bulged from the gut and measured 0.5–3.0 cm in greatest dimension (Fig. 3A,D). The affected lymphoid tissue showed complete loss of architecture and diffuse replacement by malignant lymphoma cells, which were irregularly shaped and showed great variation in size, elongated, bizarre-shaped nuclei and prominent nucleoli (Fig. 3B,E). This histological appearance is similar to that of the centroblastic diffuse large B-cell lymphoma (DLBL), which is the most common type of B-cell non-Hodgkin's lymphoma (B-NHL) in humans (Gaidano and Dalla-Favera 1993). The B-cell origin of the lymphomas was confirmed by the uniform reaction of the neoplastic cells with an anti-B-cell antibody B220 (Fig. 3C,F).
Figure 3.
Diffuse large B cell lymphomas (DLBL) in RIZ1 mutant animals. (A) Gross appearance of a lymphoma of the Payer's patch of RIZ1−/− animals. (B) Hematoxylin and eosin stained sections (40 × 10) of a lymphoma in RIZ1−/− animals. Note that normal lymph node architecture is completely effaced and diffusely replaced by lymphoma cells. Neoplastic large cells are irregularly shaped; some are angulated, with elongated, bizarre-shaped nuclei, some of which have prominent nucleoli. (C) B220 antibody staining of the tumor section as shown in B (20 × 10). Note that neoplastic cells uniformaly interact with B220 antibody (counter stained with hematoxylin). (D) Gross appearance of lymphoma involving two enlarged lymphoid tissues in RIZ1−/− animals, including the Peyer's patch (arrow) and the mesenteric lymph node (arrowhead). (E) Hematoxylin and eosin-stained section (40 × 10) of the enlarged mesenteric lymph node as shown in C. Note that the large neoplastic cells are mixed with reactive small lymphocytes. (F) B220 antibody staining of the tumor section as shown in E (counter-stained with hematoxylin, 20 × 10). (G) Presence of clonal tumor cells in the enlarged lymphoid tissues of RIZ mutant animals. Genomic DNAs isolated from the enlarged lymphoid tissues were digested with BamHI or EcoRI, followed by Southern blot analysis by use of immunoglobulin heavy-chain joining segments J3 and J4 as probe. The germ-line band (G) is present in normal tissue (1255N) as well as in tumor samples (T). The presence of the germ-line band is consistent with the admixture of nonneoplastic lymphocytes in tumors.
To confirm the cancer diagnosis on the basis of histopathology, and to rule out any possibility of reactive, nontumor lymphoid tissue, the presence of clonal tumor cells was examined by Southern blot analysis by use of the immunoglobulin heavy chain J region as probe. Genomic DNAs isolated from the enlarged lymphoid tissues were restricted by BamHI or EcoRI. As shown in Figure 3G, 10 of 12 sample DNAs examined showed distinct bands representing rearranged immunoglobulin genes when digested by EcoRI. Six of these ten DNAs also showed rearranged bands when digested by BamHI. The germ-line band detected in tumor DNAs appeared to originate from non-neoplastic lymphocytes because its intensity relative to that of the rearranged bands roughly reflected the amount of neoplastic tissues in each tumor. It is clear from Figure 3G that the proportion of neoplastic cells varied and was smaller than that of non-neoplastic cells in most of these tumors, which was consistent with the results of histopathology analysis. The apparent absence of rearranged bands in two cases (1117T and 1296T) was likely due to a low number of neoplastic cells; the presence of tumor cells in these samples was concluded from histopathology analysis.
The hallmark of a two-hit hypothesis for TSGs is the loss of the wild-type allele during tumor formation (Knudson 1971). To examine the status of RIZ1 wild-type allele in tumors of RIZ1+/− animals, we studied solid tumors developed in RIZ1 and p53 doubly heterozygous mutant mice. These animals developed mostly sarcomas, which were relatively free of normal tissues and suitable for Southern blot analysis. A loss or decreased level of the RIZ1 wild-type allele was found in three of five tumors examined (Fig. 4).
Figure 4.
Status of wild-type allele in tumors from RIZ1+/− animals. Genomic DNAs were digested with XbaI and analyzed by Southern blot analysis using the 5′ probe of RIZ1 as described in Fig. 1. DNAs from normal livers of wild-type (+/+) and homozygous mutant (−/−) animals were loaded in lanes 1 and 2. DNAs from normal liver (N1) and matched osteosarcoma (T1) of a RIZ1+/−p53+/− animal were loaded in lanes 3 and 4. DNAs from osteosarcomas of four other RIZ1+/−p53+/− animals (T2–T5) were loaded in lanes 5–8.
RIZ1 mutation in human tumors
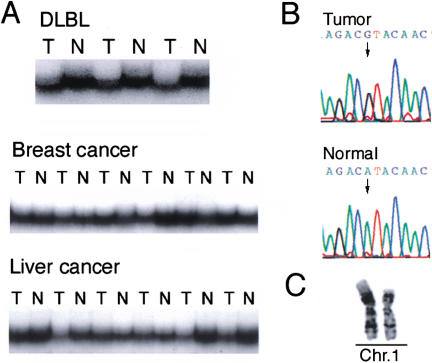
The above studies suggest that RIZ1 can be considered a tumor susceptibility gene in mice. To show the relevance of this finding to humans, we next examined whether human tumors, particularly DLBLs, might carry RIZ1 mutations. We focused our mutational analysis on the RIZ1-specific PR domain region because the remaining RIZ1 region overlaps with RIZ2. Single strand conformation polymorphism (SSCP) analysis was performed on 55 cases of B-cell lymphoma tumor DNAs. When analyzed by use of the PCR primer pair RP263 and RP264, an abnormal SSCP band was detected in 10 of 55 B-lymphoma cases. Remarkably, all of these 10 cases turned out to be DLBLs, even though only 35 of the 55 cases examined were DLBLs. Thus, this abnormal band was significantly associated with DLBLs (10/35) versus non-DLBL B-cell lymphomas (0/20, P < 0.05). The abnormal band was absent in matched normal tissue DNAs of three of the DLBL cases (the rest of the cases had no matched normal tissues available for study), suggesting a somatic mutation (Fig. 5A). We also analyzed tumor and normal tissue DNAs of 412 solid tumor cases but did not detect any abnormalities in the RP263/264 band (Fig. 5A; Table 3).
Figure 5.
Mutational analysis of human DLBL tumors. (A) SSCP analysis. Genomic DNAs from tumor (T) and matched normal tissue DNA (N) were analyzed by SSCP with primers RP263 (5′-AACACATGAGGGAATGAATGAATG-3′) and RP264 (5′-ACACCCAATTCAGACCAAGCC-3′) as described by Fang et al. (2000). (B) Sequencing analysis. Genomic DNAs from tumor and matched normal tissues were amplified by PCR with primers RP263 and RP264. The PCR products were subcloned and sequenced. (C) Karyotyping analysis of DLBL tumors. A representative chromosome 1 karyotype of a tumor with RIZ mutation is shown. The picture shows deletion of the region from 1p32 to 1pter.
Table 3.
Analysis of the A563G missense mutation in human cancers
| Tumors
|
A563G mutation
|
|---|---|
| DLBL | 10/35 (28.6%) |
| Other B-lymphomas | 0/20 (0%) |
| Burkitt's | 0/4 |
| Immunoblastic large cell | 0/8 |
| Myeloma | 0/2 |
| Small cleaved cell | 0/1 |
| Plasmacytoid | 0/3 |
| Other non-Hodgkin's | 0/2 |
| Breast cancers | 0/61 (0%) |
| Colon cancers | 0/90 (0%) |
| Gastric carcinomas | 0/66 (0%) |
| Hepatocellular carcinomas | 0/143 (0%) |
| Normal individuals | 0/52 (0%) |
| Total non-DLBL cases | 0/432 (0%) |
Sequence analysis was performed on the 10 cases of DLBLs that showed the abnormal SSCP band. In each case, a single base substitution was found that changed A563 in the coding sequence to a G that corresponds to amino acid Ile188 (CAT) changing to Val (CGT) (Fig. 5B). This mutation is located in a region that is immediately C-terminal to the PR domain and is not part of the conserved core sequence of the PR/SET MTase (Fig. 6C). Deletion in chromosome band 1p36 was found by karyotyping in three of eight DLBL cases that carried the mutation (Fig. 5C). Molecular genetic analysis, such as loss of heterozygosity (LOH), will be required to identify whether the remaining cases also lost the wild-type allele of RIZ1. However, LOH analysis was not possible because most of the samples available to use lacked matched normal controls, and their tumor cell population was not sufficiently homogeneous.
Figure 6.
Missense mutation in human cancer cell lines. (A) Genomic DNA from various cancer cell lines were PCR amplified with the primers RP97 and RP99. PCR product (50 ng) was dotted onto a nitrocellulose filter, which was than hybridized with either the wild-type oligo or mutant oligo. The sole spot that was negative for wild-type oligo but positive for mutant oligo was dotted with Saos2 cell DNA. The other spots were DNAs from 10 hepatoma, 10 breast cancer, 5 lung cancer, 1 osteosarcoma, and 5 melanoma cell lines. (B) PR/SET domain alignment. Part of the RIZ1 PR/SET domain is aligned with those of HRX/MLL, SET1, SUV39H1, and ASH1. The C106 residue of RIZ1 is highlighted in bold. (C) Schematic diagram of the RIZ1 mutations found in human cancers. Full-length RIZ1 peptide (from residue 1 to 1719) and RIZ2 peptide (from residue 201 of RIZ1 to 1719) are shown as horizontal bars. The motifs shown (not to scale) include the PR domain (residue 35–150), Rb-binding motif LXCXE (residue 311–315), zinc-finger motif ZNF (residue 362–510; residue 1130–1480), ER-binding motif LXXLL (residue 962–966), and PR-binding motif (residue 1514–1680). The arrows indicate the three missense mutations described in this study and the frameshift mutation.
In addition to the mutations in DLBLs, missense mutations of RIZ1 were discovered in human cancer cell lines. While performing cDNA cloning and sequencing of RIZ1, we discovered a homozygous/hemizygous missense mutation changing nucleotide G317 to A, which corresponds to a change of amino acid Cys106 (TGC) to Tyr (TAC) in Saos2 human osteosarcoma cells (Fig. 6A). This mutation was not found in 200 samples of normal tissue by oligo hybridization assay, suggesting that it is not likely a polymorphism. The mutation altered one of the most conserved residues of the core sequence of the PR/SET MTase (Fig. 6B). Also, Saos2 cells express RIZ1 transcript at normal levels (He et al. 1998).
The 1p36 chromosomal region is best known for harboring a neuroblastoma (NB) suppressor gene(s). To investigate a possible role of RIZ1 in NB, a total of 32 NB cell lines were examined for mutations in the PR domain by direct sequencing of RT–PCR products. A single homozygous/hemizygous base-pair substitution was found in the SMS–KCNR cell line. This substitution changes coding nucleotide C476 to a T, resulting in an alanine to valine substitution at codon 159 (data not shown). This mutation is located immediately C-terminal of the PR domain, outside the conserved core sequences (Fig. 6C). To confirm that this base-pair change was a bona fide mutation and not a polymorphism, DNA samples obtained from 100 normal individuals were screened for this particular base-pair change by restriction enzyme digestion. All 100 individuals showed the same restriction pattern, which was unlike SMS–KCNR (data not shown). In addition, real time quantitative RT–PCR analysis revealed normal levels of RIZ1 transcript in SMS–KCNR cells (M. Van Gele and F. Speleman, unpubl.).
RIZ1 has been shown to stimulate ER function in activating target gene promoters (Medici et al. 1999; Abbondanza et al. 2000). We used it as a functional assay on the RIZ1 mutants. We also tested the function of RIZ2 protein, which lacks the PR/SET MTase domain. RIZ2 lacked any activity in stimulating ER transactivation function (Fig. 7), suggesting an important role of the PR domain in the coactivator function of RIZ1. Consistently, all of the mutations found in human cancers appear to affect the PR/SET domain and all were unable to act as coactivators of ER (Fig. 7). Also as expected, an in-frame deletion mutant lacking exon 5 (del-Ex5) did not activate ER transcription function (Fig. 7). Furthermore, this mutant, as well as RIZ2 and other naturally occurring mutants, inhibited RIZ1 function on ER (Fig. 7; T. Carling and S. Huang, unpubl.).The results suggest that the three missense mutations, as well as the previously described frameshift mutation in microsatellite-unstable tumors, are functional mutations.
Discussion
RIZ1 appears to function as a transcriptional coregulator. RIZ1 gene silencing is common in human cancers. Here, we show that genetic mutation of RIZ1 is linked with tumorigenesis in both mice and humans. Mice deficient in RIZ1 were prone to develop tumors, and this was the case on two different genetic backgrounds (wild-type and mutant p53). The results suggest that RIZ1 inactivation (whereas RIZ2 is unaffected) can cause tumor formation in mice, and that RIZ1 gene silencing in human cancers is linked causally with carcinogenesis. Furthermore, we found several missense mutations of RIZ1 in human cancers, providing additional evidence of a genetic link between RIZ1 and human tumorigenesis first suggested by the discovery of frameshift mutations in microsatellite-unstable colorectal cancers.
The RIZ1−/− animals generated here are deficient in wild-type RIZ1 but normal for RIZ2. The phenotypes of these animals are therefore primarily caused by the deficiency in RIZ1. However, due to the inherent limitations of the gene-targeting technology, we cannot exclude the possibilities of chromosomal positional effects by the neor cassette insertion, or of some undetectable mutant products caused by the insertion. But these possibilities are not likely to be real. Although our RT–PCR analysis did detect exon 5 skipping at some level, it does not necessarily reveal expression of full-length mRNAs of an exon 5-deletion mutant. Indeed, such a mutant could not be detected by Northern blot, immunoprecipitation, or immunoblot analysis. Importantly, deletion of exon 5 appeared to abolish RIZ1-specific function, at least in activating ER (Fig. 7). Thus, even if the exon 5-deletion mutant is expressed at some low levels, the conclusion still stands that the knock-out animals generated in this study are deficient in RIZ1 functions. An exon 5-deletion mutant, as expected, appears to have similar activities as RIZ2, such as inhibiting RIZ1 function on the ER (Fig. 7). However, in view of the normal presence of RIZ2 in the knock-out animals, the potential contribution of the exon 5-deletion mutant to the cellular pool of RIZ2 is unlikely to be significant given the low levels of the mutant. Overall, regardless of its functions, the low or undetectable level of expression makes it highly unlikely that this exon 5-deletion mutant plays a role in vivo in the RIZ1−/− animals.
The tumor spectrum of RIZ1 mutant mice included tumors, such as DLBL and squamous carcinoma of the stomach, which were not seen in our control mice and which have been rarely seen in wild-type mice of any genetic background (Bronson and Lipman 1991; Jacks et al. 1994). The unusual tumor spectrum suggests that the tumor-prone phenotype of the RIZ1 mutant mice is robust despite the relatively long latency of tumor development in these mice. One might expect a germ-line mutation in a true TSG to be associated with an early onset tumor phenotype. However, a review of the literature suggests that this is the case primarily for the types of tumors that wild-type animals are inherently prone to develop at old age. For example, testis tumors are common in the 129Sv mouse strain, and the latency of this tumor is significantly shortened by p53 deficiency (Muller et al. 2000). In contrast, testis tumors are rare in C57BL/6 mice, and a p53 mutation in these mice does not affect the frequency of testis tumors. Also, carcinoma formation is inherently rare in mice, and is not accelerated by a p53 deficiency. Hence, the development of unusual tumors in an engineered mutant mouse could be regarded as a robust tumor phenotype: That a particular tumor occurs in a mutant mouse but not in a wild-type mouse clearly represents a significant enhancement in tumor development or an acceleration in latency for that particular tumor. Finally, it is remarkable that RIZ1 heterozygous mutant animals show a tumor phenotype, because heterozygous mutations in several important familial TSGs are not able to enhance tumor frequency in mice at any age (e.g., WT1, VHL, BRCA1, and BRCA2). Carcinogenesis is now well established to be a multistep process; a need to accumulate changes in other genes is likely to explain the late development of tumors in RIZ1 mutant mice in particular, and in genetically modified animals in general.
Tumor formation in the RIZ1 mutant mice appears to be cell autonomous rather than a general systemic defect. We found loss of the wild-type allele in tumors that developed in RIZ1 heterozygous mutant mice. Furthermore, the RIZ1 mutation did not increase the incidence of certain common mouse tumors such as T-cell lymphoma, which would be unexpected if the animals would be inherently tumor prone due to a systemic defect.
The accelerated DLBL tumor formation and decreased viability of RIZ1−/−p53+/− mice relative to RIZ1−/− or p53+/− animals further showed the tumor-prone phenotype of RIZ1-deficient animals. We also showed that RIZ1 and p53 deficiencies could cooperate in tumor formation. It remains to be determined by future investigations whether this finding indicates a specific functional collaboration or merely reflects a generic effect that one is expected to observe when crossing any TSG mutation onto a tumor-prone background, be it p53+/− or any other tumor-prone mouse strain. Mice carrying heterozygous p53 mutations are known to be tumor prone, and this has been linked to both haploinsufficiency and LOH (Venkatachalam et al. 1998). Therefore, we expect that both haploinsufficiency and LOH may underlie the accelerated tumor formation associated with RIZ1 on the p53+/− background. It is a technically challenging task to examine the status of p53 in DLBL tumors because these tumors are composed of heterogeneous cell populations. Regardless of the molecular mechanism, however, our studies here suggest that p53 and RIZ1 cooperation does occur in animals and may be expected to also occur in human cancers. Many sporadic human cancers carry both a p53 mutation and a silenced RIZ1 gene (He et al. 1998). The present study shows a further example of this, that is, RIZ1 is mutated in the Saos2 human osteosarcoma cell line in which p53 is also known to be mutated.
Many previous studies suggest a role for RIZ1 as a human TSG. This is clearly supported by the present finding of RIZ1 as a tumor susceptibility gene in mice. In addition, the present study provides further genetic evidence of a TSG role of RIZ1 in human cancers. We showed that RIZ1 mutations occurred in human cancers. The mutation of RIZ1 in the Saos2 cell line, together with the previous finding of RIZ1 silencing in the U2OS cell line (He et al. 1998), suggests a role for RIZ1 in human osteosarcoma. The fact that only 1 of 32 human NB cell lines carried a mutation suggests that mutations in RIZ1 may be uncommon in NB. Given a previous finding that RIZ1 expression is frequently reduced or lost in NBs (He et al. 1998), it is likely that epigenetic silencing may be the more common way of inactivating RIZ1 in this tumor. TSGs do tend to be preferentially inactivated by a preferred mechanism, for example, p53 is mostly by point mutations, p16INK4A is largely by homozygous deletion, and MLH1 is by silencing. We also note that in both of the cell lines that carry RIZ1 missense mutations, RIZ1 transcript is expressed at normal levels. This result is consistent with the expectation that tumors carrying RIZ1 mutations should not also carry a silenced RIZ1 gene, because there would be no selection for mutations to occur in silenced genes and vice versa. One would further expect that mutations would be rare in tumor types in which RIZ1 silencing is common and vice versa.
The most interesting findings with regard to RIZ1 mutation in human cancers were observed with DLBL tumors. Not only was the gene a DLBL susceptibility gene in mice, RIZ1 mutation appeared frequent in human DLBLs. Although a rare tumor in mice, DLBL is common in humans, accounting for ∼40% of B-NHL, which is the fifth most common cancer in the USA (Landis et al. 1999). The pathogenesis of DLBL is poorly understood. Several translocation breakpoint genes are associated with DLBL, including Bcl2, Bcl6, and Myc; rearrangements of these genes occur in 30%, 20%, and 10% of cases, respectively (Ladanyi et al. 1991; Lo Coco et al. 1994). A role for p53 mutation and p16INK4A deletion in DLBL histologic transformation has also been suggested (Lo Coco et al. 1993; Dreyling et al. 1998). Although cytogenetic deletions in DLBL suggest the presence of several novel TSGs including one on chromosome 1p36 (Cigudosa et al. 1999), none has been identified so far. Our results here suggest that RIZ1 is a strong candidate for the DLBL suppressor on chromosome 1p36. We found a somatic missense mutation (A563G or I188V) of the gene in human DLBL tumors. The fact that the mutation clustered on a single site suggests a mutational hot spot. Whereas the I188V mutation is common in DLBL tumors (10/35), it is rare in other tumors or normal controls (0/432; P < 10−8). The mutation was associated with chromosome 1p36 deletions in several human DLBL cases examined. The I188V mutation abolished the capacity of RIZ1 as a transcriptional coactivator of ER. It will be interesting to determine whether RIZ1 gene silencing also occurred in human DLBLs, but experiments addressing RNA and protein expression status are not possible at the present time because of technical limitations. Nevertheless, the results of this study suggest that RIZ1 may play an important role in the pathogenesis of human DLBL.
This study provides evidence for a member of the MTase family as a TSG, and suggests a direct link between the PR/SET domain and tumor suppression. The coactivator function of RIZ1 on ER, as well as its potential activity as a MTase, suggests that transcriptional coregulation may be the main mode of action of RIZ1. Given that RIZ1 has been shown to bind to several transcription factors (e.g., Rb and GATA3), ER may represent only one of the transcription factors for which RIZ1 can serve as a cofactor. Although a direct protein MTase activity remains to be established for RIZ1, it seems likely that a MTase-based transcription cofactor activity of RIZ1 is important for tumor suppression. Consistent with this notion, the RIZ2 protein that lacks the PR MTase domain is uniformly present in human tumors and we found that RIZ2 lacked any ER coactivator activity. The RIZ1 deficiency in our knockout mouse is essentially a deficiency in PR/SET or MTase function, as RIZ2 is expressed normally in these animals. Furthermore, the mutations found so far in human cancers, including missense and frameshift mutations, appear to affect the PR domain and the ER coactivator function. The I188V and A159V mutations occur in the vicinity of PR; the regions adjacent to the PR/SET domain have been shown to be required for the MTase activity (Rea et al. 2000). The C106Y mutation changes the highly conserved Cys residue within the PR domain. As mutating the corresponding Cys residue in the SUV39H1 protein to Ala abolishes the MTase activity of SUV39H1 (Rea et al. 2000), the RIZ1 mutation may have the same effect. Finally, the frameshift mutation found in microsatellite-unstable tumors truncated the PR-binding motif. These combined observations suggest a novel link between carcinogenesis and MTase/transcription cofactor activities of the PR/SET family.
A role for protein methylation in carcinogenesis has not been considered seriously in the past, leaving it the least understood posttranslational modification in tumor biology. However, a link between protein MTases and tumor suppression is consistent with several previous observations. For example, an inhibitor of MTases, methylthioadenosine (MTA), commonly accumulates in cancer cells as a result of homozygous deletion of MTA phosphorylase (MTAP) (Nobori et al. 1996). MTA is a natural substrate of MTAP and inhibits the aminopropyltransferases that synthesize polyamines from putrescine and decarboxylated S-adenosylmethionine, and also impairs S-adenosylmethionine-dependent transmethylation reactions (Williams-Ashman et al. 1982). Although not a strong inhibitor of MTases in vitro, MTA can cause accumulation of a more potent MTase inhibitor, S-adenosylhomocysteine (SAH), by inhibiting the SAH hydrolase. MTA has been recently shown to inhibit the protein MTase PRMT1 in vitro and in vivo (Mowen et al. 2001). MTAP is abundant in normal tissues and prevents inhibition by cleaving MTA to adenine and 5′-methylthioribose L-phosphate, which are recycled to adenine nucleotides and methionine, respectively (Backlund and Smith 1981). MTAP and p16INK4A (CDKN2) are ∼100 kb apart on chromosome 9p21 and the two genes are usually deleted together (Nobori et al. 1996). Of particular interest to the present study, codeletion of MTAP and p16INK4A has been specifically correlated with progression of human DLBLs (Dreyling et al. 1998). Moreover, deletions that eliminate MTAP, but not p16INK4A, are more frequent than p16INK4A deletion alone (Schmid et al. 1998). These findings, taken together with the fact that deletion, rather than mutation, is the primary mechanism of inactivating the p16INK4A locus, support the suggestion that there might be a selective advantage for tumors to lose MTAP in addition to p16INK4A (Nobori et al. 1996). Although Nobori and colleagues speculated on MTA inhibition of DNA MTases, it is not clear how decreased DNA methylation could facilitate carcinogenesis. Our findings suggest that MTAP deletion could facilitate tumor formation and/or progression by causing accumulation of the MTase inhibitor, MTA, which in turn could inactivate RIZ1. We are presently examining the effects of MTA and MTAP on the tumor-suppressive functions of RIZ1.
We have suggested previously that the PR/SET superfamily may share a conserved function in tumor suppression (Jiang and Huang 2000). Whereas most members of the superfamily have yet to be studied in the context of cancer, several members were originally discovered as human cancer genes, for example, MDS1–EVI1 and ALL1/MLL1. The way in which these genes are altered in cancer is consistent with loss of their PR/SET domain or MTase functions. Given that a TSG function has now been established for the first time for a member of the family, we expect more members to be accorded TSG status in the future. Although protein methylation is yet to reach the prominent role of other posttranslational modifications such as phosphorylation and ubiquitination, it appears poised to become an important new frontier of cancer research.
In summary, RIZ1 is a tumor-susceptibility gene in mice, which in turn suggests that RIZ1 gene silencing in human cancers may in fact be causally linked with carcinogenesis. Although epigenetic silencing may be the common way of inactivating RIZ1 in human cancers, genetic missense mutations also occur and may be common in certain human tumors.
Materials and methods
RIZ1 gene targeting and Southern blot analysis
A PR-domain-encoding cDNA fragment of rat RIZ1 cDNA was used to screen a mouse 129Sv genomic library (Stratagene) to obtain mouse genomic DNA. The targeting construct was designed to insert the PGK–neor–BpA positive selection gene expression cassette (Soriano et al. 1991) into exon 5 at the Kpn2I site and to place the Herpes Simplex Virus thymidine kinase gene expression cassette from PMC1–TK (Mansour et al. 1988) external and adjacent to the region of homology. The targeting plasmid was linearized by XhoI digestion and electroporated into D3 ES cells (Gossler et al. 1986). The ES clones were selected in medium containing G418 (150 μg/mL) and ganciclovir (2.0 μM). Homologous recombination events were screened by Southern blot analysis with XbaI digestion and hybridization to the 0.5-kb XbaI–BglII 5′ flanking probe or with NotI plus EcoRI digestion and hybridization to the 0.5-kb SacII–NotI 3′ flanking probe. Five RIZ1+/− ES cell-lines were microinjected into C57BL/6 blastocysts that were implanted subsequently into the uteri of Balb/c foster mothers. Male chimeric mice generated from two RIZ1+/− ES clones transmitted the RIZ mutation into germ line. The presence of the RIZ1 mutant allele in the F1 animals was confirmed by Southern blotting and PCR analysis of genomic DNA from the tail of animals. RIZ1+/− F1 animals were bred together to generate RIZ1−/− mice.
Northern blot, RT–PCR, and immunoblot analysis of RIZ gene expression
Total RNAs were prepared from mouse tissues or fibroblast cultures by use of RNAzol. Northern blot analysis was performed using a PCR fragment of exon 8 from residue 782–1115 to detect RIZ1 plus RIZ2 or a PCR fragment (321 nucleotides in length) from residue 1–76 (exons 2–4) plus 93 nucleotides of the 5′-untranslated region to detect RIZ1 only. The primers used for amplifying such a fragment were RP45 (5′-CGGCTGAGGCTC TGGGCT-3′) and RP157 (5′-CTCCCACATGTACACATTAT TC-3′); the template was the plasmid pBSK5.4. For RT–PCR analysis, first, strand cDNA was prepared using MuLV reverse transcriptase and random oligos as primers. For PCR of RIZ1, the primers are KO5 (5′-AGACTCTGGCTGAGGTACCT-3′) and RP170 (5′-GAAGCCAAAGGCCTCTCATC-3′). This PCR fragment includes exon 3 through exon 8 (617 bp, from amino acid residue 13 to 219). For immunoblot analysis, animal tissues were lysed in RIPA lysis buffer using a homogenizer as described previously (Liu et al. 1997). Tissue extracts were then immunoprecipitated with RIZ serum anti-KG7.1S against the N terminus of RIZ2 or preimmune serum (Liu et al. 1997). The immunoprecipitated products were resolved on 5% SDS gel followed by immunoblot using 1715 rabbit serum against the C terminus of RIZ1/2 proteins.
Clonal analysis by use of immunoglobulin heavy-chain gene rearrangement
Genomic DNAs were isolated from DLBL tumors removed physically from the animals (no microdissection was done on the tumors). DNAs were restricted with BamHI or EcoRI and processed for Southern blot analysis. The probe used for detecting immunoglobulin heavy-chain gene rearrangement was a 0.63-kb PCR product of the mouse germ-line immunoglobulin heavy-chain joining segments J3 and J4. The complete sequence of this probe is available at GenBank, accession no. V00770. The primers used were J3F (5′-GTCACAATGTGCCTGGTTTG-3′) and J4R (5′-AGACGGTGACTGAGGTTCCT-3′).
Mutational and karyotyping analysis of human tumors
Genomic DNA isolated from normal and lymphoma tissues were PCR amplified with primers that amplified each exon in the N-terminal region of RIZ1. Primer sequences and SSCP analysis of the PCR products were as described previously (Fang et al. 2000). PCR products were either sequenced directly or subcloned into plasmids followed by DNA sequencing.
For oligo hybridization to detect the C106Y mutation, PCR products (50 ng) were dotted onto nitrocellulose filters (primers RP97, 5′-ATGGATGTGCATTGATGCCAC-3′ and RP99, 5′-TAGTAAATGGCTCTGTTG-3′). The filters were hybridized with either 32P-labeled wild-type oligo RP104 (5′-TTGGGCTT GCTCAGGAG-3′) or mutant oligo RP103 (5′-TTGGGCTTAC TCAGGAG-3′) overnight at 42°C in 6× SSC, 0.1% SDS, 5× Denhardt's solution, and 0.1 mg/mL salmon sperm DNA. The filters were then washed twice for 10 min at room temperature in 6× SSC and for 15 min at 59°C in 2× SSC.
For analysis of neuroblastoma cell lines, a 5′-M13 tailed forward primer 5′-GTAAAACGACGGCCAGTACTACTGAGCC TGTGGCGGC-3′ and a reverse primer 5′-TCCCTTTCCG GCTCTTGG-3′ were used for RT–PCR amplification of RIZ1's PR domain region (coding nucleotide 15–514). The PCR products were purified with the QIAquick PCR purification kit (QIAGEN). The purified fragments were then sequenced directly with an universal M13 primer and the same reverse primer as described above on an ABI PRISM™ 377 DNA sequencing system by use of the Big Dye Terminator Sequencing Kit (Applied Biosystems). Samples with nucleotide changes were resequenced and confirmed with restriction enzyme digestions.
Cytogenetic methods were as described in Fletcher et al. (1991). Briefly, metaphase cells were obtained from lymph-node biopsy specimens by exposure to Colcemid for 14 h. Karyotypes were made after hypotonic treatment with 0.075 M potassium chloride for 2 min, fixation with 3:1 methanol:acetic acid, and trypsin-Giemsa banding.
Assay of RIZ1 effects on the transactivation function of the estrogen receptor
The human RIZ1 protein expression vector p3RIZ1RH4.1 was constructed by cloning the full-length wild-type human RIZ1 cDNA into pcDNA3 vector (Invitrogen). Missense mutations and frameshift mutations were introduced by Quick-Change mutagenesis kit (Stratagene), and the correct sequence of the mutant RIZ1 expression vector was confirmed by DNA sequencing. To generate the exon 5-deletion mutant, the ExSite mutagenesis kit (Stratagene) was used according to the manufacturer's instructions. Briefly, two oligo-primers flanking exon 5 were used to PCR amplify the plasmid pBSKY7912PS containing human RIZ1 cDNA 5′ region, in order to generate a mutant cDNA plasmid lacking exon 5 (pBSKY7912PSΔ5). The primers were RP153 (5′-CCAATCGCGCCGGGCGA-3′) and RP300 (5′-CTCCCACATGTATACATTATTC-3′). In frame deletion of exon 5 and correct sequence in the mutant cDNA was confirmed by DNA sequencing analysis. Next, the mutant cDNA fragment (1.5 kb) was restricted out from pBSKY7912PSΔ5 by KpnI and BsmBI and was used to replace the corresponding region in the wild-type cDNA expression vector p3RIZRH4.1 to generate the exon 5-deletion mutant expressing plasmid p3RIZ1RHΔ5. Expression vectors of RIZ1 proteins and ER were cotransfected into CV1 cells with the reporter construct ERE-tk-CAT containing a synthetic ER response element (Lee et al. 1995). Cells were grown in phenol-red free DMEM supplemented with 10% charcoal-treated fetal bovine serum. Cells were treated with or without 17-β-estrodiol or E2 (10−7 M) for 24 h and assayed for CAT activity as described previously (Lee et al. 1995). Expression of β-gal was used to control transfection efficiency. At least three independent experiments were performed.
Acknowledgments
We thank Drs. Jack Yu and Guang-Liang Jiang for technical assistance in animal autopsy analysis, Drs. Zhe Piao and Manuel Perucho for nonlymphoma tumor DNA specimens, and Dr. K. Quynh Pham for help with library screening and sequencing. We also thank the Burnham and Scripps ES and transgenic cores for assistance. We thank Dr. Erkki Ruoslahti for critical reading of the manuscript. G.S.-P. was a recipient of a Postdoctoral Fellowship from the University of California Tobacco-Related Disease Research Program (TRDRP). This work was supported by NIH grants CA76146 to S.H. and CA60988 to X.-k.Z., and by grants to S.H. from the TRDRP, the Cancer Research Program of the University of California, and the Pew Scholars Program in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shuang@burnham.org; FAX (858) 646-3192.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.870101.
References
- Abbondanza C, Medici N, Nigro V, Rossi V, Gallo L, Piluso G, Belsito A, Roscigno A, Bontempo P, Puca AA, et al. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc Natl Acad Sci. 2000;97:3130–3135. doi: 10.1073/pnas.050015697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund PS, Jr, Smith RA. Methionine synthesis from 5′-methylthioadenosine in rat liver. J Biol Chem. 1981;244:1533–1535. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Bronson RT, Lipman RD. Reduction in rate of occurrence of age related lesions in dietary restricted laboratory mice. Growth Dev Aging. 1991;55:169–184. [PubMed] [Google Scholar]
- Buyse IM, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick RB, Jiang G-L, Bennington GA, Yuan B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S, de la Chapelle A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci. 2000;97:2662–2667. doi: 10.1073/pnas.040579497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Cigudosa JC, Parsa NZ, Louie DC, Filippa DA, Jhanwar SC, Johansson B, Mitelman F, Chaganti RS. Cytogenetic analysis of 363 consecutively ascertained diffuse large B-cell lymphomas. Genes Chromosomes Cancer. 1999;25:123–133. [PubMed] [Google Scholar]
- Dreyling MH, Roulston D, Bohlander SK, Vardiman J, Olopade OI. Codeletion of CDKN2 and MTAP genes in a subset of non-Hodgkin's lymphoma may be associated with histologic transformation from low-grade to diffuse large-cell lymphoma. Genes Chromosomes Cancer. 1998;22:72–78. [PubMed] [Google Scholar]
- Eng C, Herman JG, Baylin SB. A bird's eye view of global methylation. Nat Genet. 2000;24:101–102. doi: 10.1038/72730. [DOI] [PubMed] [Google Scholar]
- Fang W, Piao Z, Simon D, Sheu J-C, Huang S. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer. 2000;28:269–275. [PubMed] [Google Scholar]
- Fang W, Piao Z, Buyse IM, Simon D, Sheu J-C, Perucho M, Huang S. Preferential loss of a polymorphic RIZ allele in human hepatocelluar carcinoma. Brit J Can. 2001;84:743–747. doi: 10.1054/bjoc.2000.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley JD, Nucifora G. Intergenic splicing of MDS1 and EVII occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc Natl Acad Sci. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JA, Kozakewich HP, Hoffer FA, Lage JM, Weidner N, Tepper R, Pinkus GS, Morton CC, Corson JM. Diagnostic relevance of clonal cytogenetic aberrations in malignant soft-tissue tumors. New Eng J Med. 1991;324:436–442. doi: 10.1056/NEJM199102143240702. [DOI] [PubMed] [Google Scholar]
- Gaidano G, Dalla-Favera R. Biologic and molecular characterization of non-Hodgkin's lymphoma. Curr Opin Oncol. 1993;5:776–784. doi: 10.1097/00001622-199309000-00002. [DOI] [PubMed] [Google Scholar]
- Gossler A, Doetschman T, Korn R, Serfling E, Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Yu JX, Liu L, Buyse IM, Wang M-S, Yang Q-C, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2–M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998a;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998b;273:15933–15940. doi: 10.1074/jbc.273.26.15933. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jiang G-L, Huang S. The yin-yang of PR-domain family genes in tumorigenesis. Histol Histopathol. 2000;15:109–117. doi: 10.14670/HH-15.109. [DOI] [PubMed] [Google Scholar]
- ————— Adenovirus expressing RIZ1 in tumor suppressor gene therapy of microsatellite-unstable colorectal cancers. Cancer Res. 2001;61:1796–1798. [PubMed] [Google Scholar]
- Jiang G-L, Liu L, Buyse IM, Simon D, Huang S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer. 1999;83:541–547. doi: 10.1002/(sici)1097-0215(19991112)83:4<541::aid-ijc17>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Offit K, Jhanwar SC, Filippa DA, Chaganti RS. MYC rearrangement and translocations involving band 8q24 in diffuse large cell lymphomas. Blood. 1991;77:1057–1063. [PubMed] [Google Scholar]
- Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1999. CA Cancer J Clin. 1999;49:8–31. doi: 10.3322/canjclin.49.1.8. [DOI] [PubMed] [Google Scholar]
- Lee MO, Liu Y, Zhang XK. A retinoic acid response element that overlaps an estrogen response element mediates multihormonal sensitivity in transcriptional activation of the lactoferrin gene. Mol Cell Biol. 1995;15:4194–4207. doi: 10.1128/mcb.15.8.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Shao G, Steele-Perkins G, Huang S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain lacking product through an internal promoter. J Biol Chem. 1997;272:2984–2991. doi: 10.1074/jbc.272.5.2984. [DOI] [PubMed] [Google Scholar]
- Lo Coco F, Gaidano G, Louie DC, Offit K, Chaganti RSK, Dalla-Favera R. P53 mutations are associated with histologic transformation of follicular lymphoma. Blood. 1993;82:2289–2295. [PubMed] [Google Scholar]
- Lo Coco F, Ye BH, Lista F, Corradini P, Offit K, Knowles DM, Chaganti RS, Dalla-Favera R. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood. 1994;83:1757–1759. [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Medici N, Abbondanza C, Nigro V, Rossi V, Piluso G, Belsito A, Gallo L, Roscigno A, Bontempo P, Puca AA, et al. Identification of a DNA binding protein cooperating with estrogen receptor as RIZ (retinoblastoma interacting zinc finger protein) Biochem Biophys Res Commun. 1999;264:983–989. doi: 10.1006/bbrc.1999.1604. [DOI] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Muller AJ, Teresky AK, Levine AJ. A male germ cell tumor-susceptibility-determining locus, pgct1, identified on murine chromosome 13. Proc Natl Acad Sci. 2000;97:8421–8426. doi: 10.1073/pnas.140208197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraosa Y, Takahashi K, Yoshizawa M, Shibahara S. cDNA cloning of a novel protein containing two zinc-finger domains that may function as a transcription factor for the human heme-oxygenase-1 gene. Eur J Biochem. 1996;235:471–479. doi: 10.1111/j.1432-1033.1996.00471.x. [DOI] [PubMed] [Google Scholar]
- Nobori T, Takabayashi K, Tran P, Orvis L, Batova A, Yu AL, Carson DA. Genomic cloning of methylthioadenosine phosphorylase: A purine metabolic enzyme deficient in multiple different cancers. Proc Natl Acad Sci. 1996;93:6203–6208. doi: 10.1073/pnas.93.12.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Z, Fang W, Malkhosyan S, Kim H, Horii A, Perucho M, Huang S. Frequent frameshift mutations of RIZ in human gastrointestinal and endometrial carcinomas with microsatellite instability. Cancer Res. 2000;60:4701–4704. [PubMed] [Google Scholar]
- Rea S, Elsenhaber F, O'Carroll D, Strahl B, Zu-Wen S, Manfred S, Opravil S, Mechtler K, Ponting C, Allis C, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sakurada K, Furukawa T, Kato Y, Kayama T, Huang S, Horii A. RIZ, the retinoblastoma protein interacting zinc finger gene, is mutated in genetically unstable cancers of the pancreas, stomach, and colorectum. Genes Chromosomes Cancer. 2001;30:207–211. [PubMed] [Google Scholar]
- Schmid M, Malicki D, Nobori T, Rosenbach MD, Campbell K, Carson DA, Carrera CJ. Homozygous deletions of methylthioadenosine phophorylase (MTAP) are more frequent than p16INK4A (CDKN2) homozygous deletions in primary non-small cell lung cancers (NSCLC) Oncogene. 1998;17:2669–2675. doi: 10.1038/sj.onc.1202205. [DOI] [PubMed] [Google Scholar]
- Shapiro VS, Lee P, Winoto A. Identification and cloning of the G3B cDNA encoding a 3′ segment of a protein binding to GATA-3. Gene. 1995;163:329–330. doi: 10.1016/0378-1119(95)00420-b. [DOI] [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, Donehower LA. Retention of wild-type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weith A, Brodeur GM, Bruns GA, Matise TC, Mischke D, Nizetic D, Seldin MF, van Roy N, Vance J. Report of the second international workshop on human chromosome 1 mapping 1995. Cytogenet Cell Genet. 1996;72:114–144. [PubMed] [Google Scholar]
- Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]