Abstract
The actinobacterial cholesterol catabolic gene cluster contains a subset of genes that encode β-oxidation enzymes with a putative role in sterol side chain degradation. We investigated the physiological roles of several genes, i.e., fadD17, fadD19, fadE26, fadE27, and ro04690DSM43269, by gene inactivation studies in mutant strain RG32 of Rhodococcus rhodochrous DSM43269. Mutant strain RG32 is devoid of 3-ketosteroid 9α-hydroxylase (KSH) activity and was constructed following the identification, cloning, and sequential inactivation of five kshA gene homologs in strain DSM43269. We show that mutant strain RG32 is fully blocked in steroid ring degradation but capable of selective sterol side chain degradation. Except for RG32ΔfadD19, none of the mutants constructed in RG32 revealed an aberrant phenotype on sterol side chain degradation compared to parent strain RG32. Deletion of fadD19 in strain RG32 completely blocked side chain degradation of C-24 branched sterols but interestingly not that of cholesterol. The additional inactivation of fadD17 in mutant RG32ΔfadD19 also did not affect cholesterol side chain degradation. Heterologously expressed FadD19DSM43269 nevertheless was active toward steroid-C26-oic acid substrates. Our data identified FadD19 as a steroid-coenzyme A (CoA) ligase with an essential in vivo role in the degradation of the side chains of C-24 branched-chain sterols. This paper reports the identification and characterization of a CoA ligase with an in vivo role in sterol side chain degradation. The high similarity (67%) between the FadD19DSM43269 and FadD19H37Rv enzymes further suggests that FadD19H37Rv has an in vivo role in sterol metabolism of Mycobacterium tuberculosis H37Rv.
INTRODUCTION
Phytosterols are among the most abundant sterols in nature and are mineralized from decaying plant material by soil bacteria, e.g., Rhodococcus strains belonging to the class Actinobacteria (37). Microbial degradation of sterol molecules involves elimination of the alkyl side chain and opening of the polycyclic steroid nucleus (Fig. 1) (1, 13, 26–28). The order of these two processes varies between bacterial genera and even between members of the same genus (23).
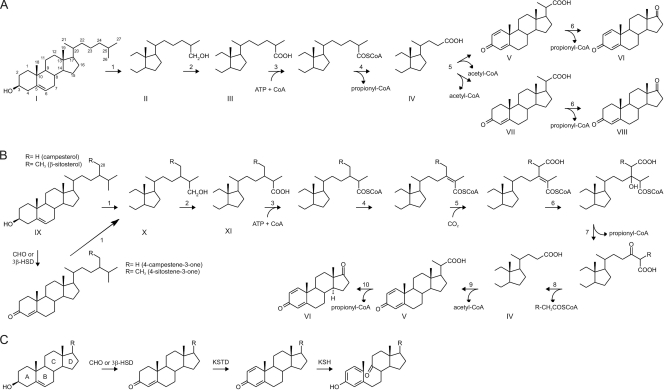
Fig. 1.
Schematic overview of the side chain degradation pathways of cholesterol (A), β-sitosterol and campesterol (B), and steroid ring opening (C) in actinobacteria (6, 10, 36). The arrow numbering indicates reaction steps which are explained in the text. Abbreviations: CoA, coenzyme A; CHO, cholesterol oxidase; 3β-HSD, 3β-hydroxysteroid dehydrogenase; KSTD, 3-ketosteroid Δ1-dehydrogenase; KSH, 3-ketosteroid 9α-hydroxylase.
Prior to their degradation, sterols are actively transported into the actinobacterial cells by the Mce4 steroid transporter (17, 20, 36). Following uptake by Rhodococcus jostii RHA1 cells, the cytochrome P450 monooxygenase CYP125 initiates degradation of the sterol side chain by catalyzing hydroxylation of the C-26 or C-27 carbon (Fig. 1A and B, reaction 1) (5, 15, 19, 23). After complete oxidation of the hydroxylated terminal carbon atom to its carboxylic acid intermediate (Fig. 1, compounds III and XI), the sterol side chain is shortened by a process similar to β-oxidation of fatty acids. This process is initiated by an ATP-dependent sterol/steroid-coenzyme A (CoA) ligase (Fig. 1A and B, reaction 3) (6, 27, 28) catalyzing the CoA activation of the C26 carboxylic acid intermediates (Fig. 1, compounds III and XI). An ATP-dependent steroid-CoA ligase (65 kDa) from Mycobacterium sp. strain NRRL B3805 was purified to near homogeneity and shown to be highly specific toward C26 carboxylic sterols (6). The gene encoding this activity remained unknown, however. Thiolytic cleavage results in shortening of the cholesterol side chain in several rounds of β-oxidation via C24 (Fig. 1A, compound IV) and C22 (Fig. 1A, compounds V and VII) intermediates and concomitant release of propionyl-CoA and acetyl-CoA, respectively. This process has been elucidated at the biochemical level in strains of Mycobacterium and Nocardia and proceeds differently for β-sitosterol than for cholesterol (Fig. 1) (9, 10, 13, 14, 27–29).
Knowledge of genes involved in microbial degradation of the alkyl sterol side chain is extremely limited. Recently, a wealth of information on sterol catabolism was provided by the identification of a cholesterol catabolic gene cluster in R. jostii RHA1, encoding several enzymes with predicted functions in β-oxidation (36). A subset of these genes (i.e., ro04683 to ro04694) is located proximal to cyp125, encoding a steroid 26-hydroxylase involved in sterol side chain degradation (5, 15, 19, 23, 36). The aim of our work is to decipher the roles of these genes in sterol metabolism, particularly those genes encoding steps of the β-oxidation cycle of sterol side chain degradation. The strain RHA1 cholesterol transcriptome was used to identify two candidate genes, i.e., fadD17 (ro04691) and fadD19 (ro04689), encoding putative acyl-CoA ligases, which were highly upregulated during growth on cholesterol (Fig. 2A) (36). Previous studies on FadD17 and FadD19 homologs from Mycobacterium tuberculosis H37Rv showed that in vitro they act as CoA ligases capable of activation of long-chain fatty acids (31). Their in vivo physiological roles remained unknown. Only a few examples of sterol side chain degradation genes have been reported, including baiB of Eubacterium sp. strain VPI 12708 and caiA of Pseudomonas sp. strain Chol1, encoding a CoA ligase and an acyl-CoA dehydrogenase, respectively, involved in cholic acid degradation (4, 12). Also, fadA5 of M. tuberculosis H3Rv was shown to encode a thiolase with a role in cholesterol side chain degradation (18).
Fig. 2.
(A) β-Oxidation gene cluster comprised of ro04677 to ro04695 within the cholesterol catabolic gene cluster of R. jostii RHA1 (16, 36). The relative fold change of expression previously observed during growth on cholesterol compared to pyruvate is also shown (36). (B and C) Genetic organization of homologs of ro04688 to ro04690 and ro04691 to ro04695 of R. rhodochrous DSM43269, respectively.
Molecular characterization studies on sterol side chain degradation are generally hampered by the lack of a stable, genetically accessible strain with inactivated steroid ring degradation but capable of selective sterol side chain degra- dation. Interestingly, Rhodococcus rhodochrous IFO3338 (=DSM43269) is amenable to genetic manipulation and able to selectively degrade the sterol side chain in the presence of iron chelators, which inhibit 3-ketosteroid 9α-hydroxylase (KSH) activity (2, 23). KSH is a two-component enzyme system, consisting of a terminal oxygenase, KshA, and a ferredoxin-reductase, KshB, and a key enzymatic step in steroid ring opening during microbial steroid catabolism (22, 33, 35). We created a genetically stable, multiple kshA gene deletion null mutant strain of R. rhodochrous DSM43269 (strain RG32) lacking KSH activity. Analogously to the chemical inhibition of strain DSM43269 by iron chelators, mutant strain RG32 was completely blocked in steroid ring degradation and capable of selective sterol side chain degradation, thereby accumulating 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (1,4-BNC) and 1,4-androstadiene-3,17-dione (ADD) (Fig. 1, compounds V and VI) from sterols. The strain RG32 was used as a tool to assess the role of various genes in sterol side chain degradation. We identified fadD19 as an essential gene for the degradation of the C-24 branched-chain sterols campesterol and β-sitosterol but not cholesterol. Direct enzyme activity measurement of heterologously produced FadD19 of DSM43269 conclusively identified its function as a steroid-CoA ligase in view of its ability to activate steroid carboxylic acid side chains.
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and chemicals.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains and R. rhodochrous DSM43269 were grown in Luria-Bertani (Sigma, Zwijndrecht, Netherlands) medium at 37°C and 30°C, respectively. Liquid cultures were grown with shaking (220 rpm). Ampicillin (100 μg ml−1), kanamycin (25 μg ml−1), or apramycin (50 μg ml−1) was added to the medium if appropriate after autoclaving. For growth on solid medium, Difco agar (Becton, Le Pont de Claix, France) was added at a final concentration of 1.5% (wt/vol).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | Host for expression of T7-based plasmids | Novagen |
| DH5α | General host for cloning purposes | Bethesda Research Laboratories |
| S17-1 | Host strain for conjugal mobilization of pK18mobsacB-derived mutagenic plasmids to Rhodococcus strains | DSMZ culture collection |
| Rhodococcus rhodochrous | ||
| DSM43269 | Wild-type strain; potent sterol degrader; identical to strain IFO3338 | DSMZ culture collection |
| RG32 | Mutant strain of DSM43269 capable of selective sterol side chain degradation; carries 5-fold unmarked kshA gene deletion | This study |
| RG32Δro04690DSM43269 | Gene deletion mutant of ro04690DSM43269 in R. rhodochrous strain RG32 | This study |
| RG32Δro04690DSM43269 ΔfadE26 | Double deletion mutant of ro04690DSM43269 and fadE26 in R. rhodochrous strain RG32 | This study |
| RG32Δro04690DSM432690 ΔfadE26 ΩfadE27 | Gene inactivation mutant of fadE27 in RG32Δro04690DSM43269 ΔfadE26 | This study |
| RG32ΔfadD19 | Gene deletion mutant of fadD19 in R. rhodochrous strain RG32 | This study |
| RG32 ΩfadD17 | Gene inactivation mutant of fadD17 in R. rhodochrous strain RG32 | This study |
| RG32ΔfadD19 ΩfadD17 | Gene inactivation mutant of fadD17 in RG32ΔfadD19 | This study |
| Plasmids | ||
| pK18mobsacB | Conjugative plasmid for gene mutagenesis in Rhodococcus; aphII sacB oriT (RP4) lacZ | 25, 34 |
| pBluescript(II)KS | General E. coli cloning vector; bla lacZ | Stratagene |
| pRESQ | E. coli-Rhodococcus shuttle vector; aphII lacZ-ccdB rep (pMVS301) | 35 |
| pZeRO2.1 | General cloning vector | Invitrogen |
| pBs-Pkan | pBluescript(II)KS containing aphII promoter region | 32 |
| pET15b | T7 promoter-based expression plasmid; bla lacI | Novagen |
| pET15bfadD19DSM43269 | pET15b containing fadD19DSM43269 | This study |
| pDELBox | pK18mobsacB-derived mutagenic plasmid for deletion of ro04683 to ro04694 in RHA1 | This study |
| pDELfadD19DSM43269 | pK18mobsacB-derived mutagenic plasmid for deletion of fadD19DSM43269 in RG32 | This study |
| pDELfadE26DSM43269 | pK18mobsacB-derived mutagenic plasmid for deletion of fadE26DSM43269 in RG32 | This study |
| pDELro04690DSM43269 | pK18mobsacB-derived mutagenic plasmid for deletion of ro04690DSM43269 in RG32 | This study |
| pΩfadE27DSM43269 | pK18mobsacB-derived mutagenic plasmid for disruption of fadE27DSM43269 in RG32 | This study |
| pΩfadD17DSM43269 | pK18mobsacB-derived mutagenic plasmid for disruption of fadD17DSM43269 in RG32 | This study |
| pRESQ4690 | Genomic library clone DSM43269 carrying ro04690DSM43269 | This study |
| pRESQ4693 | Genomic library clone DSM43269 carrying fadE26DSM43269 | This study |
| pCOMPfadD19DSM43269 | pRESQ-derived plasmid carrying fadD19DSM43269 behind the aphII promoter for functional complementation of RG32ΔfadD19 | This study |
Cholesterol, campesterol, and coenzyme A trilithium salt were obtained from Sigma-Aldrich. β-Sitosterol was obtained from Acros Organics. 5-Cholestene-26-oic acid-3β-ol was synthesized at Schering-Plough, Oss, Netherlands (23). 4-Androstene-3,17-dione (AD), 1,4-androstadiene-3,17-dione (ADD), 23,24-bisnorchol-5-en-22-oic acid-3β-ol, 3-oxo-23,24-bisnorchol-4-en-22-oic acid (4-BNC), 4-cholestene-3-one, 1,4-cholestadiene-3-one, 5-cholenic acid-3β-ol, and 3-oxo-4-cholestene-26-oic acid were obtained from Steraloids. ATP was purchased at Duchefa, Haarlem, Netherlands.
General molecular techniques.
Recombinant DNA techniques were performed according to standard protocols (24). DNA-modifying enzymes (restriction enzyme ligases and DNA polymerases) were purchased from Roche (Mannheim, Germany), New England BioLabs (Beverly, MA), or Fermentas (St. Leon-Rot, Germany) and were used according to the manufacturer's protocols. All PCR products were obtained using Pwo polymerase (Roche) using standard conditions: 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. DNA fragments were purified from an agarose gel using a Sigma Genelute gel extraction kit.
Gene cloning of four kshA homologs of R. rhodochrous strain DSM43269 and construction of kshA null mutant R. rhodochrous strain RG32.
We previously reported the cloning, heterologous expression, and characterization of a kshA homolog of R. rhodochrous DSM43269 (22), that was renamed kshA4 (see Results). Four additional kshA homologs of R. rhodochrous DSM43269 were cloned, sequenced, and deleted in DSM43269 as follows. A genomic library of R. rhodochrous DSM43269 (22) was introduced by electroporation into ΔkshA mutant Rhodococcus erythropolis RG2 (35) for functional complementation of growth on AD, resulting in a clone carrying 5.4 kb of insert DNA harboring the kshA2 homolog of strain DSM43269. A mutagenic plasmid for unmarked kshA2 gene deletion in strain DSM43269 was constructed as follows. A 2.3-kb BglII DNA fragment containing kshA2 and its upstream region was ligated into pZeRO (pKSH503). A 1.6-kb BglII/HindIII DNA fragment containing kshA2 and its downstream region was ligated into pZeRO (pKSH516). A 1.2-kb BamHI/Asp718I fragment of pKSH516 was ligated into BamHI/Asp718I-digested pKSH503, and a 2.5-kb HindIII/SpeI DNA fragment of the resulting construct carrying the kshA2 deletion was ligated into HindIII/XbaI-digested pK18mobsacB (25), producing pKSH518 used to construct the ΔkshA2 mutant of R. rhodochrous. Next, genomic DNA was isolated from the ΔkshA2 mutant and used as template for PCR with kshA degenerate primers KshAType-F and KshAType-R (33). The obtained 0.3-kb amplicon was cloned and sequenced, identifying the kshA1 homolog from strain DSM43269. Gene-specific primers for kshA1 (KshA1Rho-F and -R, Table 2) were developed and used to isolate a clone from the genomic library of R. rhodochrous DSM43269 carrying kshA1. A mutagenic plasmid for unmarked kshA1 gene deletion was constructed as follows. A 1.6-kb EcoRI (blunt-ended by Klenow)/Asp718I DNA fragment containing the 5′ part of kshA1 and its upstream region and a 2.3-kb BglII (blunt-ended by Klenow)/BamHI DNA fragment containing the 3′ part of kshA1 and its downstream region were ligated stepwise into pBluescript(II)KS. A 3.4-kb XbaI/EcoRI DNA fragment of the resulting construct carrying the kshA1 deletion was ligated into XbaI/EcoRI-digested pK18mobsacB and used to construct a ΔkshA1 ΔkshA2 double gene deletion mutant of the R. rhodochrous ΔkshA2 mutant. Genomic DNA was again isolated from the ΔkshA1 ΔkshA2 mutant and used as template for PCR with kshA degenerate primers. PCR product was obtained both with primer pair KshAType-F and KshAType-R and with primer pair KshA2-F and KshA2-R (33), the latter pair resulting in a 0.6-kb amplicon containing the kshA3 homolog of strain DSM43269. Specific kshA3 primers (KshA3Rho-F and-R, Table 2) were developed and used to isolate a clone (pKSH800) from the genomic library carrying kshA3. A mutagenic plasmid for unmarked kshA3 gene deletion was constructed as follows. A 3.5-kb BamHI fragment of pKSH800 was ligated into BamHI-digested pBluescript(II)KS, which was subsequently digested with BglII and MluI, blunt ended by Klenow treatment, and self ligated. Next, a 1.7-kb BamHI fragment was ligated into BamHI digested pK18mobsacB, rendering the plasmid for kshA3 gene deletion of the R. rhodochrous ΔkshA1 ΔkshA2 mutant strain.
Table 2.
List of primers used in this studya
| No. | PCR amplicon | Size (bp) | Oligonucleotide sequence |
|---|---|---|---|
| 1 | Genomic library clone carrying kshA1 | 227 | KshA1Rho-F, GCCGCTGCAAGAACATCCCGTAC |
| KshA1Rho-R, GATGATCTCGCGGCAGTTGGTGTC | |||
| 2 | Genomic library clone carrying kshA3 | 547 | KshA3Rho-F, CGACCTGTCACAGGGCGAGAT |
| KshA3Rho-R, TACGGATTCGGTCTGGTACC | |||
| 3 | Genomic library clone carrying kshA3 | 161 | KshA5Rho-F, CGCCTGGACGACCCTCGAACGCA |
| KshA5Rho-R, AGTGGGAGCCCTTGATGCGCAG | |||
| 4 | Internal fragment ro04690 homolog (screening genomic library) | 361 | ro04690DEG-F, CACACCGG(C/T)GAG(A/G)T(C/G)GCG(A/T)C(C/G)ATG |
| ro04690DEG-R, TCTGCAGCGGCAT(C/G)(C/G)(C/G)(A/C)AGCGG | |||
| 5 | Internal fragment fadE26 homolog (screening genomic library) | 670 | fadE26DEG-F, (C/G)(A/G)TCAACGG(A/C)CAGAAGATGTGG |
| fadE26DEG-R, A(C/T)(C/T)TCGTTGGT(G/T)CCGCCGCCGAA | |||
| 6 | fadD19DSM43269 gene (heterologous expression in pET15b) | 1,679 | fadD19exp-F, TCATATGGCCCTAAACATCGCAGACC |
| fadD19exp-R, TAGATCTCTATCCCGTCGCCGCGGCGCCG | |||
| 7 | fadD19DSM43269 gene (confirmation ΔfadD19 mutant) | 527 (wt, 1,278) | fadD19Contr-F, GTGAAGACCGTCGTGGTGGTAG |
| fadD19Contr-R, GTTGCCGTCGCCGTTGTTGTAG | |||
| 8 | fadD1943269 gene (complementation ΔfadD19 mutant) | 1,673 | COMPfadD19-F, GGCCCTAAACATCGCAGACCTC |
| COMPfadD19-R, CGGTACCCTATCCCGTCGCCGCGGCGCCG | |||
| 9 | Internal fragment fadD17DSM3269 (disruption construct) | 826 | fadD17-F, GACATCGCGCTCGCCGATTGCC |
| fadD17-R, TGTAGTAGCCGTCGAACAGACC | |||
| 10 | fadD17DSM43269 (confirmation fadD17 disruption) | 1,027 | pK18Contr-F, AATGCAGCTGGCACGACAGGTT |
| fadD17Contr-R, CGAGCATCCTGCGCGAACTCGG | |||
| 11 | Internal fragment fadE27DSM3269 (disruption construct) | 753 | fadE27-F, GCAGCATCGTCGCGGACATC |
| fadE27-R, GGAGCCGAGCAGGAACTCGT | |||
| 12 | fadE27DSM43269 (confirmation fadE27 disruption) | 1,510 | fadE27Contr-F, GCAGCATCGTCGCGGACATC |
| pK18Contr-R, CTGCGTGCAATCCATCTTGTTC | |||
| 13 | Upstream region ro04690DSM43269 (deletion construct) | 1,247 | ro04690UP-F, AGCGCCGACGACATCTACATCC |
| ro04690UP-R, TCATATGCGTGAATCCGAAGATCGGATAC | |||
| 14 | Downstream region ro04690DSM43269 (deletion construct) | 1,236 | ro04690DOWN-F, TCATATGGAGATCATGGCCGAACTCGTC |
| ro04690DOWN-R, CTGCAGGATCACGGCAACGAC | |||
| 15 | ro04690DSM43269 gene (confirmation Δro04690 mutant) | 129 (wt, 1,113) | ro04690Contr-F, ATGCAGACCGGCCTCAGCAAGA |
| ro04690Contr-R, CTAGCGGGCCTCGTTCAGCCGTTC | |||
| 16 | Upstream region fadE26DSM43269 (deletion construct) | 1,234 | fadE26UP-F, TCGAGCAGTTCGGTAACGGTGAG |
| fadE26UP-R, AGATCTCGAGGTCCAGCGCGACATCATC | |||
| 17 | Downstream region fadE26DSM43269 (deletion construct) | 1,230 | fadE26DOWN-F, TTAGATCTTTCCTCGCGGAGGGCTTGTTGC |
| fadE26DOWN-R, GGCGAGATAGGCGACCAGATTC | |||
| 18 | fadE26DSM43269 gene (confirmation ΔfadE26 mutant) | 520 (wt, 1,593) | fadE26Contr-F, GCTTCACGCACGGCGTCCTCAC |
| fadE26Contr-R, GCGGTCCTCGGAGGCATCGAGT |
Introduced restriction sites are underlined. wt, wild type.
A mutagenic plasmid for kshA4 gene deletion was constructed as follows. A 4.2-kb XbaI/HindIII DNA fragment harboring kshA4 was subcloned into XbaI/HindIII-digested pZeRO. The resulting plasmid was digested by XhoI, treated with Klenow fragment, and digested with XbaI to give a 4.5-kb fragment harboring pZeRO and the downstream flanking region of kshA4. The upstream region of kshA4 was subsequently ligated as a 1.9-kb fragment liberated following digestion by XhoI, treatment with Klenow fragment, and digestion by XbaI. Next, a 2.8-kb PstI/BamHI fragment was cloned into PstI/BamHI-digested pK18mobsacB, resulting in pKSH631 used for kshA4 gene deletion to construct ΔkshA1 ΔkshA2 ΔkshA3 ΔkshA4 mutant strain RG31. Genomic DNA was isolated from mutant strain RG31 and used as template for PCR with kshA degenerate primers KshAType-F and KshAType-R (33), resulting in the kshA5 homolog of DSM43269. Specific kshA5 primers (KshA5Rho-F and -R, Table 2) were developed to isolate a clone from the genomic library carrying kshA5. A 1.5-kb Eco47III fragment carrying the downstream region of kshA5 was cloned into EcoRV-digested pBluescript(II)KS and subsequently ligated as an EcoRI/Asp718I fragment into EcoRI/Asp718I-digested pBluescript(II)KS carrying a 1.6-kb Asp718I (Klenow-treated) fragment harboring kshA5 and its upstream region. A 1.05-kb BglII/XhoI fragment of the resulting construct was ligated into BamHI/SalI-digested pK18mobsacB, resulting in pKSH912 used for kshA5 gene deletion to construct ΔkshA1 ΔkshA2 ΔkshA3 ΔkshA4 ΔkshA5 mutant strain R. rhodochrous RG32. Genomic DNA was isolated from mutant strain RG32 and used as template for PCR with kshA degenerate primers KshAType-F and KshAType-R (33), which did not reveal the presence of additional kshA genes.
Screening of a R. rhodochrous DSM43269 genomic library for ro04690 and fadE26 homologs.
Degenerate ro04690DEG primers (Table 2) were based on conserved amino acid sequences HTGE(I/V)A(S/T)M (amino acids [aa] 193 to 200) and PLPMPLQ (aa 306 to 312) in Ro04690. Degenerate fadE26DEG primers (Table 2) were based on conserved amino acid sequences V(I/V)NGQKMW (aa 153 to 160) and FGGGTNE(V/I) (aa 369 to 376) in FadE26RHA1. A genomic library of R. rhodochrous DSM43269 (22) was screened by PCR using the ro04690DEG and fadE26DEG primers for clones containing the full-length ro04690 and fadE26 genes, respectively.
Targeted disruption and unmarked gene deletions in Rhodococcus strains.
Disruption and unmarked gene deletion mutants of R. rhodochrous DSM43269 were constructed using the sacB counterselection system (34).
Mutagenic plasmids for targeted gene deletion were constructed by PCR amplification of up- and downstream regions of the target genes using the primers listed in Table 2. The obtained amplicons were separately subcloned into EcoRV-digested pBluescript(II)KS. The amplicons were then ligated together using a native restriction site from pBluescript(II)KS (EcoRI or XbaI) and the restriction site that was introduced by PCR, being either BglII or NdeI. The genomic fragment was then isolated by digestion with either EcoRI or XbaI and HindIII and cloned into pK18mobsacB.
A construct for deletion of fadD19DSM43269 in strain RG32 was made as follows: pRESQ-4690 was digested with EcoO119I and PstI, giving a 3.0-kb fragment, whose 5′ overhang was filled in with Klenow fragment and 3′ overhang was removed by T4 DNA polymerase. Plasmid pK18mobsacB was digested with BamHI and made blunt ended by Klenow fill-in and used for ligation with the blunt-ended 3.0-kb fragment, giving pK18fadD19. A 751-bp deletion was introduced by digesting pK18fadD19 with NspV and BamHI, followed by 5′ overhang fill-in using Klenow fragment and self-ligation, yielding mutagenic plasmid pDELfadD19DSM43269.
Mutagenic plasmids for targeted gene disruptions were obtained by PCR amplification of internal fragments of the target genes, using the primers listed in Table 2. The amplicons were subsequently ligated into SmaI-digested pK18mobsacB.
Mutagenic plasmids were transferred to Escherichia coli S17-1 by transformation and subsequently mobilized to R. rhodochrous DSM43269 by conjugational transfer as described previously (34). All mutants were verified by PCR using specific primers (Table 2) to confirm deletion or disruption of the target gene(s).
Functional complementation of RG32ΔfadD19.
The fadD19DSM43269 gene was amplified by PCR using the PCR primers compfadD19-F and compfadD19-R (Table 2). The obtained PCR product of 1,673 bp was digested with Acc65I and ligated into EcoRV/Acc65I-digested pBs-Pkan, yielding plasmid pBs-Pkan-fadD19DSM43269. The pBs-Pkan-fadD19DSM43269 plasmid was digested with SpeI/Acc65I to obtain the 2.1-kb Pkan-fadD19DSM43269 cassette that was subse- quently ligated into SpeI/Acc65I-digested pRESQ, resulting in plasmid pCOMPfadD19DSM43269 that was used to transform electrocompetent cells of RG32ΔfadD19 as described previously (8).
Whole-cell biotransformation of R. rhodochrous RG32-derived mutants and steroid analyses.
Cell cultures of R. rhodochrous RG32 and derived mutant strains were grown overnight in liquid LB medium at 30°C with shaking (220 rpm) until mid-exponential phase. Stock solutions of sterols were prepared by dissolution in acetone at a concentration of 25 mM and added to cultures at a final concentration of 0.5 mM. After further incubation for 3 days under the same conditions, samples were taken for high-pressure liquid chromatography (HPLC) analysis.
To analyze sterol conversions, 1-ml culture samples were taken and centrifuged (1 min at 16,000 × g) and 0.5 ml of supernatant was mixed with 2 ml methanol and filtered prior to analysis by HPLC. HPLC was performed on an Alltima C18 column (250 by 4.6 mm; 35°C; Alltech, Deerfield, IL) using a mobile phase consisting of methanol-water-formic acid (80:19:1) at a flow rate of 1 ml min−1 with UV detection at 254 nm. Authentic 1,4-androstadiene-3,17-dione and 3-oxo-23,24-chola-1,4-dien-22-oic acid, obtained by incubating authentic 3-oxo-23,24-bisnorchol-4-en-22-oic acid with purified 3-ketosteroid dehydrogenase enzyme (Δ1-KSTD1) (11), were used as references to enable quantification of the conversion rates. Steroids were extracted from culture broth using 2 volumes of ethyl acetate. Samples were directly applied onto the same HPLC and detection system as described above, except that the mobile phase consisted of acetonitrile-tetrahydrofuran (75:25) and the flow rate was set at 2 ml min−1. Authentic β-sitosterol (a mixture of β-sitosterol and campesterol) was incubated with cholesterol oxidase (Sigma) to obtain 4-sitostene-3-one and 4-campestene-3-one and used as reference sample.
Production of CoA ligase FadD19DSM43269.
The fadD19DSM43269 gene was amplified by PCR from genomic DNA of R. rhodochrous DSM43269 using primer pair fadD19exp-F and fadD19Exp-R (Table 2). After subcloning of the amplicon in pBluescript(II)KS, the gene was cloned into pET15b using NdeI and BamHI, yielding pET15bfadD19DSM43269, and introduced into competent E. coli BL21(DE3) cells for expression. Overexpression of FadD19DSM43269 was obtained by growing 50-ml cultures in LB medium, supplemented with 0.5 M sorbitol, 100 mM ampicillin, and 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 24 h at 25°C with shaking (220 rpm). Cells were harvested by centrifugation (4,600 × g, 15 min, 4°C). Cell pellets were resuspended in 50 mM Tris buffer, pH 8.0, and cells were disrupted by sonication (10 times for 30 s each at 10 μm with 30-s intervals) on ice. To obtain cell extracts, cell debris was removed by centrifugation (40,000 × g, 20 min, 4°C). Proteins were analyzed by SDS-PAGE analysis as described previously (24) and quantified using the Bradford method, using the Bio-Rad reagent and bovine serum albumin (BSA) as a standard.
Enzymatic activity of FadD19DSM43269.
The activity of FadD19DSM43269 on various carboxylic acid sterols was analyzed by thin-layer chromatography (TLC). Reaction mixtures consisted of 20 mM Tris, pH 8.0, 5 mM MgCl2, 0.5 mM substrate, and 2 mM ATP and/or 1 mM CoA, where applicable, in a final volume of 10 μl. Reactions were initiated by the addition of 1 μl of cell extract (CFE) containing FadD19DSM43269 (∼4 μg of total protein). Care was taken to use freshly produced enzyme from the same production batch in all experiments. CFE of E. coli BL21(DE3) carrying empty pET15b was used as a negative control. After 4 h of incubation at 30°C, total reaction volumes were directly applied to F254 silica gel TLC plates (Merck, Darmstadt, Germany) and separated using a mobile phase consisting of 1-butanol–acetic acid–water (80:25:40) for 30 min. Spots were visualized using Cer reagent consisting of 1% (wt/vol) Ce(SO4)2, 2.5% (wt/vol) H3[P(Mo3O10)4]·H2O, and 8% (vol/vol) concentrated H2SO4 and developed by heating with a hot air gun.
Phylogenetic tree construction.
Protein sequences of CoA ligases from R. jostii RHA1 were aligned using ClustalW; MEGA4.1 was used for phylogenetic tree construction (30). Amino acid sequences, annotated as acyl-CoA ligases or acyl-CoA synthetases, were obtained from the RHA1 genome website (www.rhodococcus.ca) and used for phylogenetic tree construction. Furthermore, BaiB of Eubacterium sp. strain VPI 12708 (accession number P19409) was included in the phylogenetic tree.
Nucleotide sequence accession numbers.
DNA nucleotide sequencing was performed by Agowa (Berlin, Germany). The R. rhodochrous DSM43269 sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers HM588719 (pRESQ4690), HM588720 (pRESQ4693), HQ425873 (kshA1), HQ425874 (kshA2), HQ425875 (kshA3), HQ425876 (kshA4), and HQ425877 (kshA5).
RESULTS
Cloning and molecular characterization of a total of five kshA homologs from Rhodococcus rhodochrous DSM43269.
We previously reported the cloning, heterologous expression, and characterization of the kshA gene of R. rhodochrous DSM43269 (22). In the current work, four additional kshA gene homologs from DSM43269 were cloned and sequenced as described in detail in Materials and Methods. Thus, a total of five kshA genes were identified in R. rhodochrous DSM43269. The gene homologs were designated kshA1 through kshA5 based on the similarities of the encoded KshA protein sequences to those of the three known kshA genes (kshA1 to kshA3) of R. erythropolis SQ1 (33, 35). The previously reported KshA from DSM43269 (22) and the fifth KshA were less similar in amino acid sequence to KshA1, KshA2, or KshA3 of strain SQ1 than were the other newly identified KshA homologs of strain DSM43269 and were therefore renamed KshA4 and KshA5, respectively.
Bioinformatic analysis revealed that the typical Rieske Fe2S2 binding domain (C-X-H-X16,17-C-X2-H) and the nonheme Fe2+ motif (D-X3-D-X2-H-X4-H) (35) were present in all five KshA homologs.
Construction and characterization of kshA null mutant strain R. rhodochrous RG32 performing selective sterol side chain degradation.
The cloned DNA fragments of wild-type R. rhodochrous DSM43269 were used to construct a 5-fold kshA null mutant, designated strain RG32 (see Materials and Methods). Whole-cell biotransformations of 4-androstene-3,17-dione (AD, Fig. 1, compound VIII) by strain RG32 resulted in 69% molar conversion into 1,4-androstadiene-3,17-dione (ADD, Fig. 1, compound VI) (Table 3 and Fig. 1C, KSTD activity), which was not accumulated in detectable amounts by wild-type strain DSM43269 (data not shown), confirming impaired KSH activity (35). Next, mutant strain RG32 was tested in whole-cell biotransformations for its ability to perform selective side chain degradation on a range of sterols/steroids. All compounds tested were converted into ADD (Fig. 1, compound VI) and 3-oxo-23,24-bisnorchola-1,4-dien-22-oic acid (1,4-BNC; Fig. 1, compound V) in molar ratios of 1 to 7% and 50 to 80%, respectively, depending on the substrate used (Table 3). Thus, mutant strain RG32 is completely blocked in steroid ring degradation and capable of selective sterol side chain degradation, enabling analysis of putative sterol side chain degradation genes.
Table 3.
Whole-cell bioconversion of sterols/steroids into ADD and 1,4-BNC by R. rhodochrous mutant strain RG32 after 72 h of incubationa
| Steroid substrate | ADD (molar %) | 1,4-BNC (%) |
|---|---|---|
| 4-Androstene-3,17-dione | 69 (±9) | NA |
| 23,24-Bisnorchol-5-en-22-oic acid-3β-ol | − | 60 (±21) |
| Campesterol | 1 (±0) | 49 (±9) |
| 5-Cholenic acid-3β-ol | 1 (±0) | 77 (±4) |
| 4-Cholestene-3-one | 4 (±1) | 79 (±2) |
| 1,4-Cholestadiene-3-one | 3 (±1) | 71 (±4) |
| Cholesterol | 3 (±1) | 73 (±12) |
| β-Sitosterol | 7 (±2) | 67 (±7) |
NA, not applicable; −, not detected. The data represent the molar % conversion as averages of triplicate experiments; standard deviations are shown in parentheses.
Cloning homologous cholesterol catabolic genes from R. rhodochrous DSM43269.
Sequence data on the cholesterol catabolic gene cluster of R. rhodochrous DSM43269, necessary to perform mutational analysis of genes involved in sterol side chain degradation in strain RG32, were not available yet. Therefore, homologs of R. jostii RHA1 genes predicted to be involved in sterol side chain degradation were cloned from strain DSM43269. First, a genomic library of strain DSM43269 (22) was screened using degenerate PCR primers based on amino acid sequences that were highly conserved among actinobacterial homologs of strain RHA1 Ro04690 and FadE26 (Ro04693) (Fig. 2A). Library screening by PCR resulted in the isolation of two separate clones that were sequenced and analyzed (Fig. 2B and C). A 5.4-kb genomic fragment (Fig. 2B) carried ro04690DSM43269, encoding a protein displaying 85% amino acid sequence identity with Ro04690RHA1. Two genes located immediately upstream of ro04690DSM43269 encode proteins sharing 74% and 76% amino acid sequence identity with the deduced amino acid sequences of echA19 (partial sequence) and fadD19 in strain RHA1, respectively. Three genes located downstream of ro04690DSM43269 encode proteins showing the highest similarities to Ro03510 (23%), Ro04422 (25%), and Ro01580 (56%) of strain RHA1, respectively. Bioinformatic analysis of the sequence of a 5.5-kb insert (Fig. 2C) confirmed the presence of fadE26DSM43269, whose gene product displayed 86% amino acid sequence identity to its counterpart in strain RHA1. Moreover, the fragment contained genes that encode proteins with 46 to 70% amino acid sequence identities to fadD17 (partial sequence), fadE27, fdxD, and hsd4A of strain RHA1, exhibiting identical genetic organizations (Fig. 2). The two cloned genomic fragments of strain DSM43269 did not show sequence overlap.
FadD19 is essential for C-24 branched-chain sterol side chain degradation.
To substantiate a role of fadD17DSM43269, fadD19DSM43269, fadE26DSM43269, fadE27DSM43269, and ro04690DSM43269 in sterol side chain degradation, gene inactivation mutants of strain RG32 were constructed. Biotransformations of cholesterol with whole-cell cultures of these mutants revealed that all mutants displayed parental phenotypes, with similar amounts of ADD and 1,4-BNC accumulating from cholesterol as observed with parent strain RG32 (Table 4). Although the putative CoA ligases encoded by fadD17 and fadD19 show only a relatively low sequence identity at the amino acid level (21 and 23% for strain RHA1 and strain DSM43269, respectively), the possibility that they are functional homologs able to cross-complement each other during cholesterol catabolism could not be ruled out. Therefore, a ΩfadD17 inactivation mutant was constructed in strain RG32ΔfadD19 and its ability to degrade cholesterol was tested by whole-cell biotransformation. Cholesterol degradation also was unaffected in RG32ΔfadD19 ΩfadD17, displaying conversion rates similar to that of strain RG32 (Table 4).
Table 4.
Whole-cell bioconversion of sterols by R. rhodochrous RG32 and mutants thereof into ADD and 1,4-BNC, measured after 72 h of incubationa
| Strain | % conversion of sterol: |
|||||
|---|---|---|---|---|---|---|
| Cholesterol |
β-Sitosterol |
Campesterol |
||||
| ADD | 1,4-BNC | ADD | 1,4-BNC | ADD | 1,4-BNC | |
| RG32 | 3 (±1) | 73 (±12) | 7 (±2) | 67 (±7) | 1 (±0) | 49 (±9) |
| RG32Δro04690DSM43269 | 4 (±1) | 74 (±8) | 7 (±4) | 54 (±9) | ND | ND |
| RG32Δro04690DSM43269 ΔfadE26 | 3 (±1) | 72 (±4) | 6 (4) | 61 (±9) | ND | ND |
| RG32Δro04690DSM43269 ΔfadE26 ΩfadE27 | 3 (±2) | 70 (±6) | 7 (±2) | 65 (±6) | ND | ND |
| RG32ΩfadD17 | 3 (±2) | 74 (±9) | 8 (±2) | 64 (±5) | ND | ND |
| RG32ΔfadD19 | 6 (±2) | 64 (±4) | − | − | − | − |
| RG32ΔfadD19+pCOMPfadD19DSM43269 | ND | ND | 6 (±1) | 60 (±4) | ND | ND |
| RG32ΔfadD19 ΩfadD17 | 6 (±2) | 69 (±1) | − | − | ND | ND |
ND, not determined; −, not detected. The data represent averages of duplicate experiments; standard deviations are shown in parentheses.
FadE26, FadE27, and Ro04690 all belong to the superfamily of acyl-CoA dehydrogenases (7). We therefore also constructed a double gene inactivated mutant strain, RG32Δro04690DSM43269 ΔfadE26, and a triple mutant, RG32Δro04690DSM43269 ΔfadE26 ΩfadE27, to prevent possible cross-complementation. However, all these mutants were unaffected in cholesterol side chain degradation and displayed parent strain RG32 phenotypes (Table 4). Although clearly upregulated in strain RHA1 cells grown on cholesterol (Fig. 2A), either these R. rhodochrous DSM42369 genes are not involved in cholesterol side chain degradation or the reaction can be performed by additional isoenzymes encoded by strain DSM42369.
Next, all mutants were tested in whole-cell biotransformations with β-sitosterol, a C-24–ethyl branched phytosterol (Fig. 1B). Interestingly, strain RG32ΔfadD19 was blocked in its ability to convert the side chain of β-sitosterol (Table 4). All other mutants displayed parental strain phenotypes, accumulating similar amounts of ADD and 1,4-BNC from β-sitosterol as did strain RG32 (Table 4). Further analysis of the RG32ΔfadD19 mutant revealed that also the degradation of campesterol, a C-24–methyl branched sterol, was blocked (Table 4).
When incubated with β-sitosterol, strain RG32ΔfadD19 accumulated metabolites that were not detected in strain RG32 (Fig. 3). These metabolites corresponded to ring-oxidized derivatives of the β-sitosterol mixture, i.e., 4-sitostene-3-one and 4-campestene-3-one, as was confirmed by reference samples. Mutant strain RG32ΔfadD19 regained parental phenotype when complemented with fadD19DSM43269 under the control of the aphII promoter, with β-sitosterol as the substrate (Table 4). This excluded the possibility that β-sitosterol side chain degradation in RG32ΔfadD19 was blocked by polar effects rather than by inactivation of fadD19 directly.
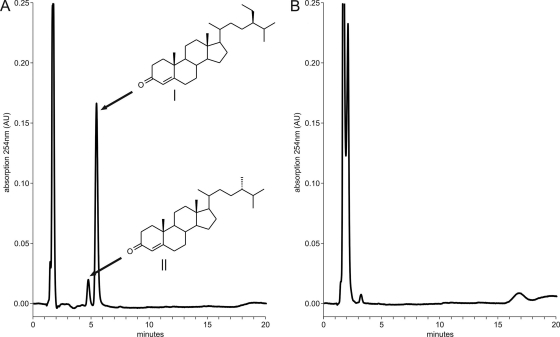
Fig. 3.
HPLC graphs of steroid extracts from β-sitosterol bioconversions after 3 days of incubation. (A) Profile of mutant strain RG32ΔfadD19 showing the accumulation of metabolites (degradation pathway intermediates) that, based on identical HPLC retention times, were identified as 4-sitostene-3-one (I) and 4-campestene-3-one (II). (B) Profile of parent strain RG32.
FadD19DSM43269 displays sterol-CoA ligase activity toward C-26-oic acid steroids.
The gene inactivation studies in strain RG32 showed that fadD19 is essential for side chain degradation of branched-chain sterols. To substantiate that fadD19 encodes a steroid-CoA ligase, FadD19DSM43269 activity was tested. Heterologous expression of the protein was achieved using E. coli strain BL21(DE3) carrying pET15bfadD19DSM43269. Cell extracts (CFE) derived from E. coli cultures expressing FadD19DSM43269 were active toward 5-cholestene-26-oic acid-3β-ol, using ATP and CoA as cosubstrates (Fig. 4A). Since CoA ligases require the presence of a carboxylic acid for their activity, FadD19DSM43269 must have been active toward the side chain of 5-cholestene-26-oic acid-3β-ol. A negative control, consisting of the same components but with CFE obtained from cultures of the same E. coli strain carrying empty pET15b vector, did not show product formation (Fig. 4B). Incubations of CFE containing FadD19DSM43269 also resulted in the formation of reaction products with 3-oxo-4-cholestene-26-oic acid or 5-cholenic acid-3β-ol in a steroid substrate-, ATP-, and CoA-dependent manner (Fig. 4A). Omitting any of these reaction components resulted in loss of enzyme activity, indicating that these are all essential components. Therefore, the products formed are most likely the CoA esters of 5-cholestene-26-oic acid-3β-ol and 3-oxo-4-cholestene-26-oic acid, confirming that FadD19DSM43269 functions as a steroid-CoA ligase. Activity toward C-24 branched-chain steroids with a terminal carboxylic acid could not be tested, since these compounds were not (commercially) available.
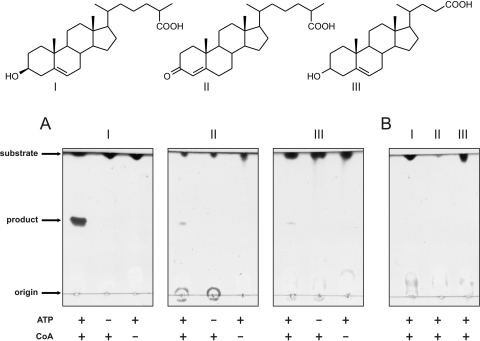
Fig. 4.
(A) TLC analysis of the reactions of cell extracts (CFE) of E. coli BL21(DE3) cells expressing fadD19DSM43269, incubated with 5-cholestene-26-oic acid-3β-ol (I), 3-oxo-4-cholestene-26-oic acid (II), and 5-cholenic acid-3β-ol (III). (B) TLC analysis of reactions of CFE of BL21 cells containing empty plasmid pET15b, using the same sterol substrates (I to III). All incubations were performed for 4 h at 30°C and contained the cofactor Mg2+. ATP and CoA were either included (+) or omitted (−) as a negative control.
Bioinformatic analysis and phylogeny of R. jostii RHA1 CoA ligases.
Database searches served to identify homologs of both FadD19RHA1 and FadD17RHA1 in closely related sterol-degrading actinobacteria, including Nocardia farcinica IFM 10152 (Nfa5290 and Nfa24170; with 75 and 47% identity, respectively), Mycobacterium smegmatis strain MC2 155 (MSMEG_5914 and MSMEG_5908; with 67 and 57% identity, respectively), M. tuberculosis H37Rv (Rv3515c and Rv3506; with 67 and 56% identity, respectively), and Rhodococcus equi 103S and ATCC 33707 (HMPREF0724_4865 and HMPREF0724_4863; with 77 and 72% identity, respectively). A comparison of FadD19RHA1 and FadD17RHA1 with the amino acid sequence of the cholic acid-CoA ligase, encoded by baiB, revealed relatively low similarities (24 and 20% identity, respectively). All sequences in the strain RHA1 genome annotated as CoA ligase or CoA synthetase were compared and depicted in a phylogenetic tree (see Fig. S1 in the supplemental material). This revealed that the strain RHA1 genome contains closely related homologs of FadD17RHA1 and FadD19RHA1, i.e., Ro05822 (44% identity) and Ro04675 (49% identity), respectively. These putative enzymes thus may have a similar steroid-CoA ligase activity, resulting in a metabolic redundancy for activation of the side chain of 5-cholestene-26-oic acid-3β-ol metabolites.
DISCUSSION
This study identified FadD19 of R. rhodochrous DSM43269 as a steroid-CoA ligase, essential for the degradation of C-24 branched-chain sterols in vivo. An unmarked fadD19 deletion mutant in kshA null mutant strain RG32 was blocked in side chain degradation of the C-24 branched sterols β-sitosterol and campesterol but not cholesterol (Table 4). In addition, mutant RG32ΔfadD19 accumulated ring-oxidized derivatives from β-sitosterol (Fig. 3), indicating that in this mutant no sterol side chain degradation occurred, but only steroid ring oxidation. From these results, we conclude that FadD19DSM43269 catalyzes an essential step in the side chain degradation of C-24 branched sterols in R. rhodochrous DSM43269.
In R. jostii RHA1, fadD17 is located proximal to fadD19 within the cholesterol catabolic gene cluster and highly upregulated during growth on cholesterol. Thus, fadD17 is a likely candidate to encode a steroid-CoA ligase involved in cholesterol side chain activation (36). Mutant strains with an inactivated fadD17DSM43269 gene in both RG32 and RG32ΔfadD19, however, were still capable of transformation of the cholesterol side chain. Therefore, it remains elusive whether side chain activation during cholesterol catabolism is performed by multiple redundant CoA ligases or whether a yet-unidentified, highly specific enzyme catalyzes this reaction in vivo.
Direct enzyme activity measurements showed that FadD19DSM43269 was able to activate the side chains of various steroid carboxylic acids (Fig. 4). The intensities of the reaction product spots suggest that FadD19DSM43269 has substrate preference for steroids possessing longer side chains over those with shorter ones. Furthermore, substrates with a 3-hydroxy-5-ene ring structure may be better substrates than their oxidized forms. However, it is difficult to quantitatively compare the reaction products due to the discrepancy in spot intensity between the 3-hydroxy-5-ene and 3-oxo-4-ene substrates at the same substrate concentration. Additional biochemical studies are thus required to assess the substrate specificity of FadD19DSM43269.
Despite the fact that FadD19DSM43269 was able to activate 5-cholestene-26-oic acid-3β-ol and derivatives thereof in vitro, deletion of fadD19DSM43269 in strain RG32 did not affect cholesterol degradation. Therefore, it is most plausible that, besides fadD19 and possibly fadD17, other isoenzymes are encoded by the strain DSM43269 genome that can complement 5-cholestene-26-oic acid-3β-ol activation in this mutant. Orthologs of Ro05822 and Ro04675 (see Fig. S1 in the supplemental material) may be present in strain DSM43269 and could account for the lack of phenotype found with our mutants in whole-cell biotransformations with cholesterol. CoA ligases are known to be promiscuous enzymes, which often have overlapping substrate specificities (3, 21), further supporting the hypothesis that the genomes of strain RHA1 and strain DSM43269 may encode multiple enzymes with the ability to activate the side chain of 5-cholestene-26-oic acid-3β-ol or derivatives thereof (e.g., its 3-oxo-4-ene metabolite).
Similarly, inactivation of multiple genes encoding putative acyl-CoA dehydrogenases (i.e., fadE26DSM43269, fadE27DSM43269, and ro04690DSM43269) in strain RG32 did not hamper cholesterol nor β-sitosterol side chain degradation (Table 4). Rhodococcal genomes are very rich in genes encoding β-oxidation enzymes, making it very likely that enzymatic redundancy for these functions exists. However, the possibility that the fadE26DSM43269, fadE27DSM43269, and ro04690DSM43269 genes in fact do not have a role in sterol side chain degradation cannot be ruled out at the moment.
Interestingly, there is apparently no redundancy for activation of C-24 branched sterol side chains, since deletion of fadD19DSM4269 alone in strain RG32 resulted in impaired degradation of C-24 branched-chain sterols. FadD19DSM43269 may be the only enzyme able to activate the side chains of C-24 branched phytosterols, which possess a bulkier side chain than that of cholesterol. However, it cannot be excluded that a homolog (e.g., Ro04675) is encoded by strain DSM43269 that is able to catalyze the same reaction but that it is not expressed in mutant RG32ΔfadD19 under the conditions used.
The results described in our current study are consistent with FadD19 of R. rhodochrous DSM43269 performing a steroid-CoA ligase reaction with an in vivo role in the degradation of C-24 branched sterol side chains. FadD19H37Rv of the human pathogen M. tuberculosis H37Rv, encoded by rv3515c, possesses CoA ligase activity and catalyzes the formation of acyl-CoA from long-chain fatty acids (31). BLAST searches with FadD19DSM43269 and FadD19RHA1 against the H37Rv genome showed that they both have the highest sequence identity with FadD19H37Rv; FadD19RHA1 and FadD19H37Rv also were best reciprocal hits. The high similarity (67%) between these enzymes suggests an in vivo role for FadD19H37Rv in sterol metabolism of M. tuberculosis H37Rv.
The study furthermore described the construction of mutant strain R. rhodochrous RG32, blocked in sterol ring degradation, and demonstrated its successful use as a tool to study the process of microbial sterol side chain degradation specifically, allowing the future characterization of other putative sterol side chain degradation genes.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by grants of the IBOS (M.H.W.) and B-Basic (M.P.) programs of ACTS (Advanced Chemical Technologies for Sustainability), NWO (Netherlands Organization for Scientific Research).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Arima K., Nagasawa M., Bae M., Tamura G. 1969. Microbial transformation of sterols. Part I. Decomposition of cholesterol by microorganisms. Agric. Biol. Chem. 33:1636–1643 [Google Scholar]
- 2. Arima K., Nakamatsu T., Beppu T. 1978. Microbial production of 3-oxobisnorchola-1,4-dien-22-oic acid. Agric. Biol. Chem. 42:411–416 [Google Scholar]
- 3. Arora P., Vats A., Saxena P., Mohanty D., Gokhale R. S. 2005. Promiscuous fatty acyl CoA ligases produce acyl-CoA and acyl-SNAC precursors for polyketide biosynthesis. J. Am. Chem. Soc. 127:9388–9389 [DOI] [PubMed] [Google Scholar]
- 4. Birkenmaier A., et al. 2007. Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 189:7165–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capyk J. K., et al. 2009. Mycobacterial cytochrome P450 125 (CYP125) catalyzes the terminal hydroxylation of C27 steroids. J. Biol. Chem. 284:35534–35542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C. S. 1985. Ph.D. thesis. University of Wisconsin—Madison, Madison, WI. [Google Scholar]
- 7. Daubner S. C., Gadda G., Valley M. P., Fitzpatrick P. F. 2002. Cloning of nitroalkane oxidase from Fusarium oxysporum identifies a new member of the acyl-CoA dehydrogenase superfamily. Proc. Natl. Acad. Sci. U. S. A. 99:2702–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernandes P. J., Powell J. A., Archer J. A. 2001. Construction of Rhodococcus random mutagenesis libraries using Tn5 transposition complexes. Microbiology 147:2529–2536 [DOI] [PubMed] [Google Scholar]
- 9. Fujimoto Y., Chen C. S., Szeleczky Z., Ditullio D., Sih C. J. 1982. Microbial degradation of the phytosterol side chain. I. Enzymic conversion of 3-oxo-24-ethylcholest-4-en-26-oic acid into 3-oxochol-4-en-24-oic acid and androst-4-ene-3,17-dione. J. Am. Chem. Soc. 104:4718–4720 [Google Scholar]
- 10. Fujimoto Y., Chen C. S., Gopalan A. S., Sih C. J. 1982. Microbial degradation of the phytosterol side chain. II. Incorporation of NaH14CO3 onto the C-28 position. J. Am. Chem. Soc. 104:4720–4722 [Google Scholar]
- 11. Knol J., Bodewits K., Hessels G. I., Dijkhuizen L., van der Geize R. R. 2008. 3-Keto-5α-steroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem. J. 410:339–346 [DOI] [PubMed] [Google Scholar]
- 12. Mallonee D. H., Adams J. L., Hylemon P. B. 1992. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J. Bacteriol. 174:2065–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marsheck W. J., Kraychy S., Muir R. D. 1972. Microbial degradation of sterols. Appl. Microbiol. 23:72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin C. K. A., Wagner F. 1976. Microbial transformation of β-sitosterol by Nocardia sp. M29. Eur. J. Appl. Microbiol. 2:243–255 [Google Scholar]
- 15. McLean K. J., et al. 2009. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 284:35524–35533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McLeod M. P., et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohn W. W., et al. 2008. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 283:35368–35374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nesbitt N. M., et al. 2010. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 78:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ouellet H., et al. 2010. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 77:730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pandey A. K., Sassetti C. M. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pei Z., et al. 2003. The acyl-CoA synthetase “bubblegum” (lipidosin): further characterization and role in neuronal fatty acid beta-oxidation. J. Biol. Chem. 278:47070–47078 [DOI] [PubMed] [Google Scholar]
- 22. Petrusma M., Dijkhuizen L., van der Geize R. 2009. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 75:5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosłoniec K. Z., et al. 2009. Cytochrome P450 125 (CYP125) catalyzes C26-hydroxylation to initiate sterol side chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 74:1031–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 26. Sih C. J. 1962. Mechanisms of steroid oxidation by microorganisms. Biochim. Biophys. Acta 62:541–547 [DOI] [PubMed] [Google Scholar]
- 27. Sih C. J., Tai H. H., Tsong Y. Y., Lee S. S., Coombe R. G. 1968. Mechanisms of steroid oxidation by microorganisms. XIV. Pathway of cholesterol side-chain degradation. Biochemistry 7:808–818 [DOI] [PubMed] [Google Scholar]
- 28. Sih C. J., Wang K. C., Tai H. H. 1968. Mechanism of steroid oxidation by microorganisms. XIII. C22 acid intermediates in the degradation of the cholesterol side chain. Biochemistry 7:796–807 [DOI] [PubMed] [Google Scholar]
- 29. Szentirmai A. 1990. Microbial physiology of sidechain degradation of sterols. J. Ind. Microbiol. 6:101–116 [Google Scholar]
- 30. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 31. Trivedi O. A., et al. 2004. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428:441–445 [DOI] [PubMed] [Google Scholar]
- 32. van der Geize R., et al. 2008. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 36:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Geize R., Hessels G. I., Nienhuis-Kuiper M., Dijkhuizen L. 2008. Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9α-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB. Appl. Environ. Microbiol. 74:7197–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Geize R., Hessels G. I., van Gerwen R., van der Meijden P., Dijkhuizen L. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197–202 [DOI] [PubMed] [Google Scholar]
- 35. van der Geize R., Hessels G. I., Van Gerwen R., Van der Meijden P., Dijkhuizen L. 2002. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45:1007–1018 [DOI] [PubMed] [Google Scholar]
- 36. van der Geize R., et al. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 104:1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe K., Aihara H., Tachi N., Nakamura R. 1987. Degradations of 4-cholesten-3-one and 1,4-androstadiene-3,17-dione by cholesterol-degrading bacteria. J. Appl. Bacteriol. 62:151–155 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.