Abstract
Identification of specific genes and enzymes involved in conversion of lignocellulosics from an expanding number of potential feedstocks is of growing interest to bioenergy process development. The basidiomycetous wood decay fungi Phanerochaete chrysosporium and Postia placenta are promising in this regard because they are able to utilize a wide range of simple and complex carbon compounds. However, systematic comparative studies with different woody substrates have not been reported. To address this issue, we examined gene expression of these fungi colonizing aspen (Populus grandidentata) and pine (Pinus strobus). Transcript levels of genes encoding extracellular glycoside hydrolases, thought to be important for hydrolytic cleavage of hemicelluloses and cellulose, showed little difference for P. placenta colonizing pine versus aspen as the sole carbon source. However, 164 genes exhibited significant differences in transcript accumulation for these substrates. Among these, 15 cytochrome P450s were upregulated in pine relative to aspen. Of 72 P. placenta extracellular proteins identified unambiguously by mass spectrometry, 52 were detected while colonizing both substrates and 10 were identified in pine but not aspen cultures. Most of the 178 P. chrysosporium glycoside hydrolase genes showed similar transcript levels on both substrates, but 13 accumulated >2-fold higher levels on aspen than on pine. Of 118 confidently identified proteins, 31 were identified in both substrates and 57 were identified in pine but not aspen cultures. Thus, P. placenta and P. chrysosporium gene expression patterns are influenced substantially by wood species. Such adaptations to the carbon source may also reflect fundamental differences in the mechanisms by which these fungi attack plant cell walls.
INTRODUCTION
Efficient and complete degradation of woody plant cell walls is generally ascribed to certain basidiomycetes collectively referred to as white rot fungi (14). Commonly associated with woody debris and forest litter, these fungi can depolymerize, degrade, and fully mineralize all cell wall polymers, including cellulose, hemicelluloses, and the normally rather recalcitrant polymer lignin. Such plant cell wall deconstruction requires complex extracellular oxidative and hydrolytic systems, and these have been studied extensively in the model white rot fungus Phanerochaete chrysosporium (34). Mechanistic aspects of the degradative processes remain uncertain, but the field has attracted interest because woody feedstocks are increasingly viewed as potential sources for high-value low-molecular-weight products (3).
Brown rot wood decay fungi, exemplified by Postia placenta, exhibit patterns of cell wall degradation distinct from those of white rot. In particular, these fungi rapidly depolymerize cellulose early in the decay process, but even after extensive decay, the lignin remains in situ as a modified polymeric residue (45, 65). The involvement of low-molecular-weight, diffusible oxidants, especially hydroxyl radicals, has long been suspected (10, 11). The generation of hydroxyl radicals by a nonenzymatic Fenton reaction (H2O2 + Fe2+ + H+ → H2O + Fe3+ + ·OH) has been implicated repeatedly for brown rot (recent studies include references 9, 40, and 62) and, to a lesser extent, white rot (2).
Irrespective of strikingly different decay patterns, P. placenta and P. chrysosporium are phylogenetically related Polyporales fungi, both lying within the “Phlebia” clade (6, 20, 21). Recent comparative genome analyses (40, 57) are consistent with oxidative depolymerization of cellulose by P. placenta, including a substantial contraction in the number of genes potentially involved in hydrolytic attack on crystalline cellulose. For example, genes encoding extracellular exocellobiohydrolases are absent. In contrast, the P. chrysosporium genome reveals numerous genes encoding cellulases and an array of high-oxidation-potential lignin peroxidases. Viewed together with biochemical analyses, the preponderance of literature strongly supports a conventional hydrolytic mechanism of cellulose degradation by P. chrysosporium. Nevertheless, considerable uncertainty persists, especially with respect to genetic multiplicity, a prominent feature of the P. chrysosporium genome (41). For example, at least 6 sequences are predicted to encode cellobiohydrolase I (CBH), all of which are members of glycoside hydrolase (GH) family 7 (19; http://www.cazy.org/). The role(s) of these genes is poorly understood, but structural diversity within such families may reflect subtle functional differences (for an example, see reference 43) that permit adaptation to a changing substrate composition and/or other environmental conditions.
Substrate preferences among certain wood decay fungi are well known, with brown rot species often associated with gymnosperms and white rot fungi more typically isolated from angiosperms (14, 61). First reported by Holzbaur and Tien for lignin peroxidase (25), numerous studies involving defined media have demonstrated the substantial influence of substrate on gene expression by P. chrysosporium (reviewed in references 15 and 31) and P. placenta (40). Although differential transcriptional regulation and shifting secretome patterns have also been observed in comparing defined media to cultures containing a woody substrate (46, 47, 57), side-by-side quantitative comparisons are lacking. To address this issue, we show here that differences in the lignocellulosic substrate dramatically alter the transcript and secretome profiles of both P. chrysosporium and P. placenta.
MATERIALS AND METHODS
Culture conditions and characterization.
RNA and protein were obtained from P. chrysosporium strain RP78 and P. placenta strain MAD-698-R (Forest Mycology Center, Forest Products Lab) grown in Highley's basal salt medium (23) containing 0.5% (wt/vol) ball-milled bigtooth aspen (Populus grandidentata) (BMA), ball-milled white pine (Pinus strobus) (BMP), or glucose as the sole carbon source. Each 2-liter Erlenmeyer flask contained 250 ml medium and was inoculated with approximately 107 P. chrysosporium spores or with P. placenta mycelia scraped from the surface of potato dextrose agar. P. chrysosporium and P. placenta cultures were incubated for 5 days on a rotary shaker (150 rpm) at 37°C and room temperature, respectively.
Consistent with previous analysis of P. chrysosporium grown for 5 days in medium containing ball-milled aspen, standard assays for lignin peroxidase (52), manganese peroxidase (60), glyoxal oxidase (32, 33), and cellobiose dehydrogenase (4) showed no activity. None of these activities were detected in any P. placenta cultures, a predictable result given the absence of those genes. Carbohydrate compositions of inoculated and uninoculated media are listed in Table S3 in the supplemental material.
For RNA analysis, mycelia from triplicate cultures were collected by filtration through Miracloth (Calbiochem, EMD Biosciences, Gibbstown, NJ), squeeze dried, and snap-frozen in liquid nitrogen. Pellets were stored at −80°C until use. For mass spectroscopic analysis, culture filtrates were processed after 5 and 14 days of incubation.
Expression microarrays.
P. chrysosporium and P. placenta Roche NimbleGen array designs are available under platforms GPL8022 and GPL13673, respectively, within the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/index).
Total RNA was purified from frozen mycelial pellets, converted to Cy3-labeled cDNA, hybridized to microarrays, and scanned as described previously (57). The 12 arrays used in these experiments were scanned on an Axon 4000B scanner (Molecular Dynamics), and data were extracted using NimbleScan v2.4. Quantile normalization and robust multiarray averaging (RMA) (26) were applied to the raw data by using DNAStar ArrayStar v4 (Madison, WI). Expression levels were based on log2 signals, and significant differences in expression were determined using the moderated t test (48), with the false discovery rate (FDR) (5) threshold set at P values of <0.05. The MIAME-compliant (8) microarray expression data were deposited in NCBI's Gene Expression Omnibus. The newly acquired data can be viewed/downloaded together with previously deposited data, which include results from glucose-grown, microcrystalline cellulose-grown, and BMA-grown arrays (40, 56, 57).
Competitive reverse transcription-PCR (RT-PCR) was used to quantify transcripts of P. placenta genes encoding an aryl alcohol dehydrogenase and three putative P450s. Gene-specific primers and amplicon information for these genes are listed in Table S1 in the supplemental material. The quantitative RT-PCR methodology was used previously for confirmation of P. chrysosporium (56) and P. placenta (57) microarrays.
Mass spectrometry.
Soluble extracellular protein was precipitated from culture filtrates by direct addition of solid trichloroacetic acid (TCA) to 10% (wt/vol), and trypsin-generated peptides were analyzed by nano-liquid chromatography–tandem mass spectrometry (nano-LC-MS/MS), using an Agilent 1100 nanoflow system (Agilent, Palo Alto, CA) connected to a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific, San Jose, CA) equipped with a nanoelectrospray ion source as described previously (57). Using protein databases for P. placenta (http://jgi.doe.gov/Postia; nonredundant haploid set) and P. chrysosporium (http://jgi.doe.gov/whiterot; BestModels_2.1), the MS/MS spectra were analyzed using an in-house Mascot search engine (version 2.2.07; Matrix Science, London, United Kingdom). Mascot searches were done with a fragment ion mass tolerance of 0.6 Da, a parent ion tolerance of 15 ppm, and methionine oxidation as a variable modification. Scaffold (version Scaffold_3_00_6; Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Protein identifications were accepted if they contained at least 2 uniquely identified peptides and if protein probabilities exceeded 95.0%, as determined by the Protein Prophet algorithm (44).
To access detailed information, P. chrysosporium and P. placenta protein model identification numbers are preceded by “Pchr” and “Ppl,” respectively, and the corresponding data can be accessed directly via their respective JGI genome portals. The protein pages include information from the Gene Ontology (GO) database for each InterPro domain. Function or “putative” function was assigned when it was supported by direct experimental evidence or when comparisons to known proteins revealed conserved catalytic features and/or significant alignment scores (bit scores of >150) to known proteins within the SwissProt database. All other proteins were designated “hypothetical.”
Microarray data accession numbers.
The MIAME-compliant (8) microarray expression data were deposited in NCBI's Gene Expression Omnibus and are accessible through GEO accession number GSE29659.
RESULTS
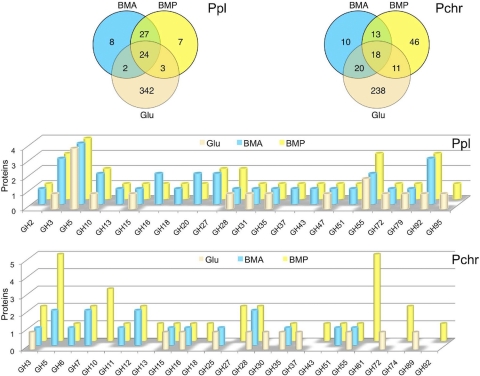
On media containing glucose, BMA, or BMP, we identified peptides corresponding to 356 P. chrysosporium genes (Fig. 1, upper right panel; see Table S2 in the supplemental material). Focusing our analyses on the woody substrates, 31 proteins were identified in both media, and all were previously detected in BMA (57). These included well-characterized glycoside hydrolases, such as a family 6 (GH6) cellobiohydrolase II (51), two family 7 CBH1s (43, 54, 55), two family 12 and one family 5 endo-1,4-β-glucanase (18, 53), a family 28 polygalacturonase and a rhamnogalacturonase, a family 55 1,3-β-glucosidase (27), a family 16 likely laminarinase (30), a family 51 putative arabinofuranosidase, and a family 35 putative β-galactosidase. A total of 30 proteins were identified in BMA but not in BMP (Fig. 1, upper right panel). With two exceptions, all had been reported earlier (57). The two exceptions were a putative mitochondrial ATP carrier protein (Pchr134565) and a serine protease (Pchr3855). The former was also present in glucose medium and was likely the result of hyphal autolysis, but the latter was detected exclusively in BMA and featured a clear secretion signal. The proportion of proteins of likely intracellular origin was especially high in glucose medium and after 14 days in wood cultures, when starvation-induced lysis is expected (see Table S2).
Fig. 1.
Distribution of P. placenta (Ppl) and P. chrysosporium (Pchr) proteins identified by LC-MS/MS in BMA and BMP culture filtrates after 5 days of growth. Upper Venn diagrams show the partitioning of glucose-, BMA-, and BMP-derived proteins. In the lower panels, the numbers of proteins identified within glycoside hydrolase families are illustrated.
Fifty-seven P. chrysosporium proteins were confidently detected in BMP but not in BMA. Among these, only two proteins, both of unknown function (Pchr8221 and Pchr134789), had been reported in an earlier BMA analysis (57). We could not confidently assign these to BMA medium here because only a single peptide was detected for each.
Carbohydrate-active enzyme (CAZy)-encoding genes represented 33% (20 of 61 genes) and 45% (40 of 88 genes) of the total P. chrysosporium protein genes identified in BMA and BMP, respectively, but the number and distribution of medium-specific proteins better illustrate the influence of substrate (Fig. 1, lower bar graph). Of 30 P. chrysosporium genes confidently identified in BMA but not in BMP, only three were predicted to encode CAZys. Possibly reflecting the relative abundance of O-acetylgalactoglucomannans in softwoods, P. chrysosporium proteins found exclusively in BMP included two family GH74 proteins (28), a GH92 1,2-α-mannosidase, two GH27 α-galactosidases (17), and three GH10 endo-1-4-β-xylanases (13). Five GH61 proteins, a grouping of considerable recent interest (16), were also observed only in BMP. Twenty-nine of the glycoside hydrolase proteins identified after 5 days of growth were also observed after 14 days (see Table S2 in the supplemental material). Nine new GHs were identified in these older cultures, all in BMP (GH27 protein Pchr4422, GH28 protein Pchr4449, GH31 protein 125462, GH43 proteins Pchr333 and Pchr133070, GH7 protein Pchr137216, GH78 protein Pchr122292, GH92 protein Pchr3431, and the possible GH74 protein Pchr28013).
When a mutant strain of P. chrysosporium was cultured on the softwood species Pinus nigra (black pine), eight CAZys were detected (46), five of which were also found in our BMP cultures (GH3 protein Pchr129849, GH5 protein Pchr5115, GH74 protein Pchr134556, and GH7 proteins Pchr137372 and Pchr127029). In contrast to the results with black pine, we did not detect peptides matching GH88 protein Pchr840, GH37 protein Pchr140267, or GH92 protein Pchr3431. (The latter protein was detected in 14-day BMP medium.) Recent LC-MS/MS and transcriptome analyses of Phanerochaete carnosa-colonized Picea glauca (white spruce) support the importance of common extracellular CAZys (37, 38), but it is unclear whether these comparisons are meaningful given the substantial differences in our experimental design.
P. chrysosporium oxidoreductases detected in BMP included the copper radical oxidase CRO2 (59), cellobiose dehydrogenase (36), and an FAD-dependent oxidoreductase of unknown function, whereas BMA cultures contained a glucose oxidase-like GMC oxidoreductase, a catalase, and an alcohol oxidase (see Table S2 in the supplemental material). The latter protein (Pchr126879) is 89% identical to the methanol oxidase of the brown rot fungus Gloeophyllum trabeum (12). In black pine, oxidoreductase identification was limited to a single CRO2 peptide (46). Based on peptide matches to the P. chrysosporium database, five putative oxidoreductases were detected in P. carnosa-colonized spruce, but none corresponded to those observed here in BMP (38).
A total of 413 P. placenta proteins were identified, of which a total of 71 were identified in either BMA or BMP, and 51 of these were present in both BMA and BMP (Fig. 1; see Table S2 in the supplemental material). With one exception, a putative phosphodiesterase (Ppl127047), all proteins detected in BMA were observed previously (57). Manual inspection suggests that the gene model corresponding to Ppl127047 is inaccurate, possibly containing 2 or more genes. Twenty-eight of the proteins common to the two media corresponded to glycoside hydrolase-encoding genes, and all had homologous sequences in P. chrysosporium (see Table S2). Among these common enzymes was a putative GH5 endo-1,4-β-glucanase (Ppl115648). The Ppl115648 protein lacks a cellulose binding module, but the catalytic domain is similar to those of known endoglucanases. In addition to BMP and BMA, the protein has been detected in medium containing microcrystalline cellulose (40). No peptides corresponding to Postia's two GH61 genes were detected.
Compared to P. chrysosporium, relatively few P. placenta proteins were found exclusively in BMP (see Table S2 in the supplemental material). Among the glycoside hydrolases, no clear trends were obvious. Certain members of the GH95, GH92, GH55, and GH28 families were identified only in BMP, but other members of these families, potentially having similar functions, were also observed in BMA (see Table S2). Two proteins of unknown function (hypothetical) were confined to BMP cultures. Three had no P. chrysosporium homolog, but Ppl114327 was 73% identical to P. chrysosporium Pchr139777. The protein contains a highly conserved domain of unknown function (DUF1237) and was detected only in BMP for both fungal species.
Twenty-nine of the P. placenta glycoside hydrolase proteins identified after 5 days of growth were also observed after 14 days (see Table S2 in the supplemental material). Eight new GHs were identified in these older cultures. Among these, a GH5 endo-1,4-β-glucanase (Ppl117690), a GH51 α-arabinofuranosidase (Ppl94557), and a GH16 enzyme (Ppl115248) were present in BMA and in BMP. Peptides corresponding to GH18 proteins (Ppl107968 and Ppl119925) and a GH28 protein (Ppl105117) were detected in BMA but not in BMP after 14 days, whereas GH2 (Ppl114395) and GH5 (Ppl121713) enzymes were observed only in BMP.
Microarray analyses revealed large numbers of differentially regulated genes in glucose- versus BMA- or BMP-grown cultures. Transcript levels in BMA cultures were largely consistent with those in previous reports (57). Two hundred thirty-six P. placenta genes accumulated >2-fold (P < 0.05) higher levels in BMP than in glucose, while transcripts of 136 genes were more abundant in glucose than in BMP. Fewer P. chrysosporium genes were upregulated in BMP than in glucose (27), but 174 were upregulated in glucose relative to BMP.
Transcript levels of 47 P. chrysosporium genes were >2-fold (P < 0.5) higher in BMA cultures than in BMP cultures (see Table S2 in the supplemental material). Proteins corresponding to 17 of these regulated genes were identified in BMA and/or BMP (Table 1), and among these, 6 had no clear homolog in P. placenta. These P. chrysosporium genes included genes for an aldose epimerase (Pchr138470) and a hypothetical protein containing a cellulose binding domain (Pchr131440). Ten of the 17 genes were assigned to CAZy domains.
Table 1.
P. chrysosporium genes encoding transcripts with >2-fold accumulation and detectable proteins in BMA and/or BMP culturesa
| ID | Description |
P. chrysosporium gene |
P. placenta homolog |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal (log2) |
A/P | P value | No. of peptides |
ID | Allele | Signal (log2) |
A/P | Prob | No. of peptides |
||||||||||
| G | A | P | G | A | P | G | A | P | A | P | G | ||||||||
| 121077 | Oxalate decarboxylase | 12.82 | 14.99 | 11.83 | 8.91 | 0.04 | 14 | 15 | 8 | 46778 | 61132 | 11.87 | 12.33 | 11.90 | 1.35 | 0.10 | |||
| 140501 | GH5 (man5D) | 9.41 | 13.07 | 9.96 | 8.64 | 0.02 | 0 | 3 | 4 | 121831 | 134772 | 13.50 | 14.61 | 14.72 | 0.93 | 0.08 | 2 | 2 | 2 |
| 131440 | Hypothetical protein with CBM1 | 10.04 | 13.56 | 10.65 | 7.53 | 0.02 | 0 | 1 | 2 | ||||||||||
| 131217 | Oxalate decarboxylase | 14.75 | 13.49 | 10.93 | 5.87 | 0.02 | 9 | 9 | 0 | 43635 | 127692 | 9.19 | 9.18 | 9.13 | 1.04 | 0.22 | |||
| 6458 | GH5, endo-1,4-β-glucanase | 11.94 | 14.91 | 12.37 | 5.81 | 0.02 | 0 | 0 | 5 | 117690 | 103675 | 13.97 | 13.85 | 13.31 | 1.45 | 0.01 | |||
| 4361 | GH5, endo-1,4-β-glucanaseb | 10.47 | 14.20 | 11.67 | 5.76 | 0.03 | 0 | 2 | 4 | ||||||||||
| 6482 | CE15, glucuronyl esterase, GE2 | 10.69 | 13.71 | 11.39 | 5.00 | 0.04 | 0 | 2 | 4 | 92582 | 101999 | 9.60 | 10.09 | 9.80 | 1.22 | 0.01 | |||
| 138479 | Aldose 1-epimerase (ale1) | 11.48 | 14.54 | 12.23 | 4.96 | 0.01 | 0 | 0 | 5 | ||||||||||
| 134001 | GH27 α-galactosidase | 11.19 | 13.49 | 11.20 | 4.89 | 0.01 | 0 | 0 | 2 | 128150 | 98662 | 9.63 | 9.69 | 9.81 | 0.92 | 0.18 | 2 | 3 | 1 |
| 41650 | GH61b | 9.71 | 12.35 | 10.28 | 4.19 | 0.02 | 0 | 0 | 2 | ||||||||||
| 8466 | GH12, endo-1,4-β-glucanase | 11.32 | 14.04 | 12.18 | 3.63 | 0.02 | 0 | 2 | 2 | 121191 | 112685 | 14.48 | 13.73 | 14.13 | 0.76 | 0.01 | |||
| 140079 | Glutaminase (gta1) | 10.85 | 12.83 | 10.99 | 3.57 | 0.01 | 1 | 0 | 14 | 47839 | None | 11.38 | 13.04 | 13.28 | 0.85 | 0.03 | 2 | 2 | 0 |
| 138266 | GH74 (gly74A) | 10.47 | 12.91 | 11.11 | 3.48 | 0.01 | 0 | 0 | 17 | ||||||||||
| 10607 | GDSL lipolytic enzyme | 9.23 | 10.65 | 9.25 | 2.64 | 0.02 | 0 | 0 | 5 | ||||||||||
| 133585 | GH92 α-1,2-mannosidase | 11.99 | 13.68 | 12.49 | 2.28 | 0.02 | 0 | 0 | 7 | 62385 | 48716 | 11.34 | 10.82 | 10.80 | 1.01 | 0.93 | [4] | [2] | [0] |
| 126191 | Lipase | 12.25 | 13.66 | 12.51 | 2.22 | 0.01 | 0 | 0 | 5 | 50078 | 20448 | 10.39 | 11.77 | 11.73 | 1.02 | 0.83 | [2] | [3] | [0] |
| 3328 | Hypothetical protein | 11.54 | 11.89 | 10.86 | 2.04 | 0.02 | 0 | 0 | 2 | 117005 | None | 11.01 | 11.02 | 11.30 | 0.83 | 0.03 | |||
Normalized microarray data are presented as average log2 signal strengths for three fully replicated experiments. Significant accumulation (A/P ratio) of transcripts in ball-milled aspen (A) relative to ball-milled pine (P) cultures was determined using the moderated t test (48) and associated FDR values (5) (P values). Data are ranked according to A/P ratios. The number of unique peptides was detected by LC-MS/MS after 5 days of growth on glucose (G), BMA (A), or BMP (P) medium. Numbers in brackets indicate the number of peptides assigned to an allelic variant.
The P. placenta genome contains distantly related sequences within these families, but their similarity lies below our E value threshold (>10−15).
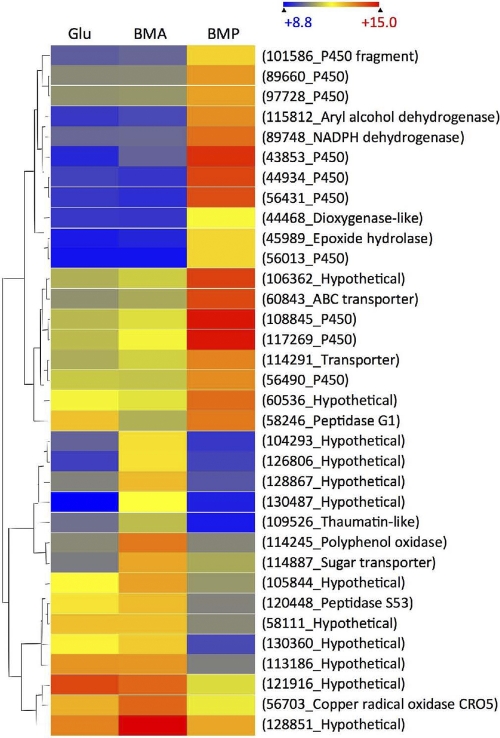
Eighty-five P. placenta genes showed >2-fold accumulation (P < 0.05) in BMP relative to that in BMA, and 79 showed the reverse pattern (see Table S2 in the supplemental material). Nine of these regulated genes were associated with proteins, including 2 GH28 family members (Ppl111730 and Ppl58192), whose transcripts showed significant accumulation in BMP relative to that in BMA (Table 2). Genes of unknown function predominated (63%) among those upregulated in BMA, and 24 of these hypothetical genes had no clear homolog in P. chrysosporium. Oxidoreductase transcripts accumulating in BMA included those encoding polyphenol oxidase (Ppl114245), amine oxidase (Ppl98543), peroxidase (Ppl111839), a laccase (Ppl46931), alcohol dehydrogenase (Ppl55493), two P450s (Ppl97939 and Ppl128850), and a copper radical oxidase, CRO5 (Ppl56702). Peptides corresponding to the P. placenta cro5 gene were identified in BMA and BMP.
Table 2.
P. placenta genes encoding transcripts with >2-fold accumulation and detectable extracellular proteinsa
| ID | Description |
P. placenta |
P. chrysosporium homologs |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal (log2) |
A/P ratio | P value | No. of unique peptidesc |
No. of homologs | ID | Description | Signal (log2) |
A/P ratio | P value | No. of unique peptides |
|||||||||
| G | A | P | A | P | G | A | P | A | P | G | |||||||||
| 128867 | Hypothetical | 127765 | 12.73 | 9.88 | 8.37 | <0.01 | 1 | 4 | 5 | ||||||||||
| 113186b | Hypothetical | 130016 | 13.17 | 10.40 | 6.80 | <0.01 | 10 [14] | 11 [13] | 11 [12] | 0 | |||||||||
| 121916b | Hypothetical | None | 13.77 | 11.51 | 4.79 | <0.01 | 10 | 11 | 11 | 0 | |||||||||
| 56703 | CRO5 | 99632 | 13.79 | 11.70 | 4.25 | <0.01 | 6 | 7 | 0 | 3 | 8882 | cro4 | 12.45 | 11.83 | 1.54 | 0.23 | 0 | 0 | 0 |
| 129055 | Hypothetical | 13.18 | 11.58 | 3.04 | <0.01 | 2 | 3 | 9 | 0 | ||||||||||
| 64088 | Peptidase A | 57301 | 11.54 | 10.10 | 2.71 | <0.01 | [1] | [2] | [1] | 18 | 135608 | asp1 | 10.84 | 10.60 | 1.18 | 0.59 | 9 | 9 | 12 |
| 112047 | GH92 | 116992 | 13.07 | 11.96 | 2.16 | <0.01 | 10 [2] | 10 [2] | 9 [0] | 3 | 3431 | GH92 | 13.18 | 12.87 | 1.24 | 0.43 | 0 | 0 | 0 |
| 111730 | GH28 | 43189 | 12.11 | 13.17 | 0.48 | <0.01 | [2] | [5] | [2] | 1 | 3805 | epg28 | 14.59 | 11.93 | 6.30 | 0.07 | 10 | 6 | 4 |
| 58192 | GH28 | 45730 | 11.57 | 12.64 | 0.48 | <0.01 | [0] | [2] | [0] | 1 | 29397 | rhg28 | 13.43 | 12.27 | 2.23 | 0.25 | 5 | 3 | 0 |
Calculations are as described in the footnotes to Table 1. To identify P. chrysosporium homologs, the 12,438 P. placenta protein models represented on microarrays were aligned with all 10,048 v2 P. chrysosporium protein models by using Timelogic (Active Motif, Carlsbad, CA) hardware accelerated double-affinity Smith-Waterman alignments as previously described (57).
Gene models lie adjacent on scaffold 67 and are predicted to encode identical proteins. Transcripts differed only slightly, suggesting a possible assembly error.
Complete MS/MS results are listed in Table S2 in the supplemental material. Only proteins with 2 or more peptides in BMP and/or BMA are listed.
P. placenta genes upregulated >2-fold in BMP relative to BMA were particularly rich in genes encoding oxidoreductases, including 15 cytochrome P450s, and transcript levels of 10 of these P450s were upregulated >4-fold (Fig. 2). All but 2 of the 85 BMP-upregulated genes, hypothetical proteins Ppl121538 and Ppl106710, were matched to a P. chrysosporium homolog (see Table S2 in the supplemental material).
Fig. 2.
Heat map showing hierarchical clustering of P. placenta genes with >4-fold (P < 0.05) transcript accumulation in BMP relative to BMA (first 19 genes) and in BMA relative to BMP (last 15 genes). The scale above the map shows log2-based signals.
DISCUSSION
Our results show that gene expression profiles of P. chrysosporium and P. placenta are significantly influenced by the wood substrate. Analyses of transcript levels in all three pairwise comparisons (BMA-glucose, BMP-glucose, and BMP-BMA) showed combined totals of 378 and 513 regulated genes in P. chrysosporium and P. placenta, respectively. More specifically, 240, 201, and 47 P. chrysosporium genes showed differential accumulation in BMA-glucose, BMP-glucose, and BMA-BMP comparisons, respectively. For P. placenta, the same comparisons showed 250, 372, and 162 regulated genes, respectively. In this connection, it should be noted that unless otherwise specified (e.g., Fig. 2), a threshold of >2-fold transcript accumulation (P < 0.05) was imposed.
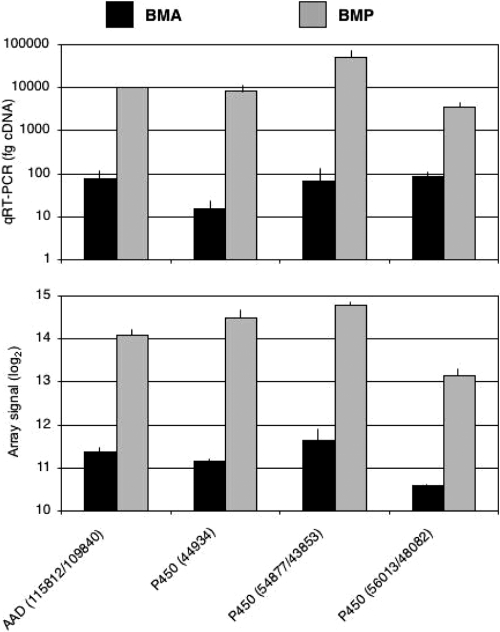
Biological and technical variation was low under the conditions employed (GEO accession number GSE29659), and as in earlier studies (56–58), qRT-PCR results affirmed the relative transcript abundances (Fig. 3). However, it should be emphasized that we were examining steady-state transcript levels, with no information regarding stability/turnover. Furthermore, responses may be indirect, and given the current limitations in automated gene annotation, we have surely underestimated the number of regulated genes. Beyond this, time course experiments, which are prohibitively expensive at current microarray costs, would likely reveal additional genes involved in early colonization and during advanced decay.
Fig. 3.
Transcript levels of P. placenta genes encoding cytochrome P450s and aryl alcohol dehydrogenase. Protein model numbers are shown in parentheses, with slashes separating allelic variants. The upper panel indicates amounts of cDNA determined by competitive RT-PCR, whereas the lower panel shows log2 microarray signals. Black and gray bars represent results for triplicate BMA and BMP cultures, respectively. PCR primers are listed in Table S1 in the supplemental material.
Protein identifications supported an important role for many genes, particularly those associated with high transcript levels and/or differential regulation. However, the nonquantitative shotgun approach reported here favors identification of soluble and stable proteins. Important proteins with high turnover rates or those bound to the substrate or of low molecular weight may be overlooked. For example, peptides corresponding to P. placenta glp1 were not detected, even though very high transcript signals were observed in both BMA (log2 = 14.78) and BMP (log2 = 14.90). The predicted mature protein of 21.5 kDa (Ppl128976) features a secretion signal, and previous studies of P. chrysosporium (50) support a role for this protein in extracellular Fenton chemistry via iron reduction. In short, the absence of detectable extracellular peptides should be interpreted cautiously.
On the other hand, unambiguous protein identifications, especially those associated with high transcript levels, whether differentially regulated or not, surely support an important role of proteins in lignocellulose degradation. Affirming previous investigations of the P. chrysosporium cellulolytic system, elevated transcript levels and proteins were observed for genes encoding CBH1s (CEL7C [Pchr127029] and CEL7D [Pchr137372]) (see Table S2 in the supplemental material), CBH2 (CEL6 [Pchr133052]) (Table 3), and endoglucanases (CEL5A [Pchr6458] and CEL12A [Pchr8466]) (Table 1). Possibly enhancing cellulolytic activity by nonhydrolytic mechanisms (16), significant (P < 0.01) transcript accumulation (>4-fold) and peptides were detected for GH61 proteins Pchr41650 (Table 2) and Pchr121193 (Table 3). Peptides corresponding to GH61 proteins Pchr41123, Pchr4691, and Pchr122129 were also detected, but their transcript accumulation relative to that in glucose medium was modest, at 1.05-fold, 1.81-fold, and 1.27-fold, respectively.
Table 3.
Highly expressed P. placenta and P. chrysosporium genes encoding proteins detected in culture filtrates by LC-MS/MSa
| Description |
P. placenta |
P. chrysosporium |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Allele | Signal (log2) |
No. of unique peptides |
ID | Gene | Signal (log2) |
No. of unique peptides |
|||||||
| A | P | A | P | G | A | P | A | P | G | |||||
| Highly expressed P. placenta proteins and P. chrysosporium homologs | ||||||||||||||
| GH5 endo-1,4-β-glucanase | 115648 | 108962 | 15.09 | 15.05 | 4 | 4 | 3 | 6458* | cel5A, EG38 | 14.91 | 12.37 | 0 | 5 | 0 |
| GH16 endo-1,3(4-β-glucanase | 112941 | 61809 | 14.64 | 14.10 | 3 [2] | 3 [2] | 0 [0] | 10833* | 13.69 | 11.82 | ||||
| GH5 endo-β-1,4-mannosidase | 121831 | 134772 | 14.61 | 14.72 | 2 | 2 | 2 | 140501* | man5D | 13.07 | 9.96 | 3 | 4 | 0 |
| GH72 1,3-β-glucanosyltransferase | 117860 | 118950 | 14.58 | 14.43 | 3 | 5 | 2 | 6433 | 13.46 | 13.00 | 0 | 0 | 2 | |
| CE10 carboxylesterase | 43588 | 14.32 | 14.81 | 3 | 11 | 0 | 38233 | 9.82 | 9.85 | |||||
| GH55 glucan 1,3-β-glucosidase | 116267 | 108648 | 14.28 | 14.24 | [6] | [5] | [6] | 8072 | exg55A | 14.21 | 12.02 | 12 | 8 | 9 |
| GH55 glucan 1,3-β-glucosidase | 105490 | 119394 | 14.24 | 13.43 | [9] | [9] | [3] | 132568 | 11.19 | 11.39 | ||||
| Phosphodiesterase | 127047 | 128697 | 14.21 | 14.56 | 2 | 2 | 0 | 3652 | 12.63 | 12.72 | ||||
| GH35 β-galactosidase | 127993 | 128101 | 14.19 | 14.47 | 3 [15] | 4 [15] | 1 [12] | 9466 | lac35A | 11.67 | 11.38 | 3 | 2 | 3 |
| GH51 α-N-arabinofuranosidase | 100251 | 127046 | 14.12 | 14.49 | 2 | 2 | 1 | 3651 | arb51A | 12.86 | 12.67 | 3 | 3 | 0 |
| FAD oxidoreductase | 122772 | 114192 | 14.08 | 14.22 | [2] | [1] | [0] | 138076 | 9.47 | 9.52 | 0 | 3 | 1 | |
| Highly expressed P. chrysosporium proteins and P. placenta homologs, if existent | ||||||||||||||
| GH61 | 121193 | cel61B | 15.61 | 13.02 | 1 | 2 | 0 | |||||||
| Transporter | 115604 | 14.24 | 13.98 | 136620 | 15.50 | 14.10 | 3 | 0 | 0 | |||||
| CDH | 11098 | cdh1 | 15.45 | 13.97 | 1 | 16 | 0 | |||||||
| GH6 CBHII exocellobiohydrolase | 133052 | cel6 | 15.41 | 13.62 | 2 | 7 | 0 | |||||||
| Elongation factor | 119872 | 47184 | 12.90 | 13.18 | 0 | 0 | 12 | 134660 | 15.19 | 13.86 | 2 | 4 | 13 | |
| CE1 feruloyl esterase | 126075 | axe1 | 15.05 | 12.80 | 4 | 6 | 0 | |||||||
| CE4 chitin acetylase | 116123 | 123580 | 13.06 | 13.58 | 124827 | 15.02 | 14.62 | 0 | 3 | 3 | ||||
| ATPase | 125396 | 117150 | 11.92 | 13.20 | 0 | 0 | [4] | 126811 | 15.00 | 14.26 | 3 | 0 | 8 | |
| GH12, endoglucanase | 121191 | 112669 | 14.04 | 14.41 | 7048 | cel12B | 14.99 | 13.73 | 3 | 2 | 0 | |||
| Oxalate decarboxylase | 46778 | 61132 | 12.33 | 11.90 | 121077* | 14.99 | 11.83 | 15 | 8 | 14 | ||||
| Methanol oxidase | 118723 | 129841 | 14.61 | 14.10 | 126879 | 14.93 | 14.37 | 10 | 0 | 0 | ||||
| GH5, endoglucanase | 117690 | 103675 | 13.81 | 13.31 | 6458 | cel5A | 14.91 | 12.37 | 0 | 5 | 0 | |||
| Catalase | 114720 | 123169 | 14.44 | 14.46 | [1] | [1] | [15] | 124398 | 14.82 | 14.14 | 2 | 2 | 7 | |
| Copper radical oxidase | 130302 | 104114 | 13.32 | 13.58 | 134241 | cro2 | 14.72 | 13.67 | 0 | 4 | 3 | |||
| GH28 polygalacturonase | 111730 | 43189 | 12.11 | 13.17 | [2] | [5] | [2] | 3805 | epg28A | 14.59 | 11.93 | 10 | 6 | 4 |
| Hypothetical | 127850 | 14.71 | 14.75 | 5659 | 14.56 | 13.33 | 3 | 3 | 2 | |||||
| Aldose 1-epimerase | 138479 | ale1 | 14.54 | 12.23 | 0 | 5 | 0 | |||||||
| Hypothetical | 138739 | 14.42 | 13.11 | 1 | 3 | 0 | ||||||||
Listing of genes with transcript signals >2 standard deviations (SD) above the genomewide average and with at least 2 unique peptides identified. For P. placenta, the BMA log2 average was 10.95, with an SD of 1.599, and the BMP value was 10.96, with an SD of 1.606. For P. chrysosporium, the BMA value was 11.55, with an SD of 1.417, and the BMP value was 11.549, with an SD of 1.391. Models with obvious inaccuracies and in need of manual editing are shown in bold. Asterisks indicate differentially regulated genes (>2-fold transcript accumulation; P < 0.05). All abbreviations are as in Tables 1 and 2.
Cellobiose dehydrogenase (CDH), an enzyme implicated in Fenton chemistry (reviewed in reference 2) and cellulose depolymerization (39), was highly expressed in BMP (log2 signal, 13.97; 16 unique peptides) (Table 3). In this connection, we also identified peptides corresponding to a putative aldose 1-epimerase (ALE1) in BMP but not in BMA (Table 2). Although peptides were absent (ALE1) or unconvincing (CDH) in BMA, transcript accumulation was significantly higher than that in BMP. The coordinate expression of ALE1 and CDH was previously noted in P. chrysosporium cultures containing microcrystalline cellulose as the sole carbon source (58). Possibly, these enzymes are physiologically connected through the generation of the β-anomer of cellobiose, the preferred substrate of CDH (22).
In contrast to the case for P. chrysosporium, significant (>2-fold) accumulation of glycoside hydrolase transcripts was not observed in BMA-grown P. placenta (Table 1), although the putative GH28 enzymes polygalacturonase (Ppl111730) and rhamnogalacturonase (Ppl58192) were upregulated in BMP (Table 1). However, high transcript levels and extracellular peptides were observed in many instances (Table 3; see Table S2 in the supplemental material). Among these, a potential GH5 endoglucanase (Ppl115648), albeit one without a cellulose binding domain, was highly expressed in P. placenta cultured in both BMA and BMP. This observation lends support to the possible involvement of an endo-acting 1,4-β-glucanase, but it should be mentioned that no other likely cellulases were expressed until day 14, when a second putative 1,4-β-glucanase (Ppl117690) appeared. The P. placenta genome contains two predicted GH61 genes, and both showed only modest transcript levels (log2 signal averages ranging from 8.86 to 9.21) and no peptides detected (see Table S2).
Although few P. placenta glycoside hydrolases exhibited significant (>2-fold) differential regulation on the lignocellulosic substrates, many oxidoreductase-encoding genes were substantially regulated. Transcripts corresponding to the copper radical oxidase gene cro5 were more abundant in BMA than in BMP, and peptides were detected in BMA and BMP (Table 1; Fig. 2). The predicted protein (Ppl56703) features 4 N-terminal repeats of a highly conserved domain (WSC; IPR013994) possibly involved in carbohydrate binding. Also accumulating in BMA relative to BMP were transcripts corresponding to polyphenol oxidase, a peroxidase, laccase, and a putative flavin-containing oxidoreductase.
Most surprising was the accumulation of oxidoreductase transcripts in BMP relative to those in BMA (Fig. 2 and 3). These included 15 genes encoding cytochrome P450s, a functionally diverse group of approximately 230 enzymes (∼2% of P. placenta gene models) (40). It has long been presumed that a subset of the 146 P450s of P. chrysosporium (41) are involved in the complete degradation of lignin metabolites, and progress has been made in understanding the regulation and reactions catalyzed by some of these (49, 63). Less is known regarding the P450s of P. placenta, although previous microarray analysis showed upregulation of transcripts corresponding to Ppl110015 and Ppl128850 in cultures containing microcrystalline cellulose relative to cultures in glucose medium (40). The P. chrysosporium ortholog of the former, Pchr130996, encodes a benzoate p-hydroxylase.
Little is known regarding the metabolism of softwood extractives, but consistent with the observed transcript profiles (Fig. 2), cytochrome P450s and dioxygenases have been proposed to be involved (35). In addition to extractives, hardwood and softwood species differ substantially in anatomical features and in lignin composition (1, 7), a factor long ago suggested as important in white rot decay (24). Significant alterations in availability of benzaldehydes and benzoic acid groups were recently shown to occur during P. placenta wood decay (64, 65), and these could be metabolized by P450-mediated hydroxylations (29). Differences between BMA and BMP lignin composition and/or accessibility might account for the observed P. placenta transcript profiles. Although the differences were not statistically significant, P. chrysosporium genes encoding putative P450s (Pchr129114, Pchr133311, and Pchr138612) and an aryl alcohol dehydrogenase (Pchr34489) accumulated >2-fold in BMP relative to BMA (see Table S2 in the supplemental material). Of course, P. chrysosporium and P. placenta differ sharply with respect to the completeness of lignin removal, but demethyloxylations (45) and benzaldehyde reductions (42, 64) are common transformations possibly reflected in the observed high expression of methanol oxidase and aryl alcohol dehydrogenase.
Determining the precise role of these genes in lignocellulose degradation presents a challenge for future research, although perhaps a less daunting one than determining the role of the many interesting hypothetical proteins. Some of these are more precisely termed “proteins of unknown function,” including the 8 differentially regulated P. placenta gene products identified with LC-MS/MS as extracellular proteins and bearing no apparent homologs in P. chrysosporium (Table 2). Based on transcript levels and protein identification, these and other genes seem worthy targets for future investigation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (grant 2007-35504-18257 to the Forest Products Laboratory), by the Office of Science U.S. Department of Energy contract DE-AC02-05CH11231 to the Joint Genome Institute, and by the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DEFC02-07ER64494).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 6 May 2011.
REFERENCES
- 1. Adler E. 1977. Lignin chemistry. Past, present and future. Wood Sci. Technol. 11:169–218 [Google Scholar]
- 2. Baldrian P., Valaskova V. 2008. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 32:501–521 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee G., Scott-Craig J. S., Walton J. D. 2010. Improving enzymes for biomass conversion: a basic research perspective. Bioenerg. Res. 3:82–92 [Google Scholar]
- 4. Bao W., Lymar E., Renganathan V. 1994. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Biochem. Biotechnol. 42:642–646 [Google Scholar]
- 5. Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289–300 [Google Scholar]
- 6. Binder M., Hibbett D., Larsson K. H., Larsson E., Langer E. 2005. The phylogenetic distribution of resupinate forms in the homobasidiomycetes. Syst. Biodivers. 3:113–157 [Google Scholar]
- 7. Blanchette R. 1991. Delignification by wood-decay fungi. Annu. Rev. Phytopathol. 29:381–398 [Google Scholar]
- 8. Brazma A., et al. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29:365–371 [DOI] [PubMed] [Google Scholar]
- 9. Cohen R., Jensen K. A., Houtman C. J., Hammel K. E. 2002. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett. 531:483–488 [DOI] [PubMed] [Google Scholar]
- 10. Cowling E. B. 1961. Comparative biochemistry of the decay of sweetgum sapwood by white-rot and brown-rot fungi. Technical bulletin 1258. U.S. Department of Agriculture, Washington, DC.
- 11. Cowling E. B., Brown W. 1969. Structural features of cellulosic materials in relation to enzymatic hydrolysis, p. 152–187 In Hajny G. J., Reese E. T. (ed.), Cellulases and their applications. American Chemical Society advances in chemistry series 95. American Chemical Society, Washington, DC [Google Scholar]
- 12. Daniel G., et al. 2007. Characteristics of Gloeophyllum trabeum alcohol oxidase, an extracellular source of H2O2 in brown rot decay of wood. Appl. Environ. Microbiol. 73:6241–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Decelle B., Tsang A., Storm R. 2004. Cloning, functional expression and characterization of three Phanerochaete chrysosporium endo-1,4-β-xylanases. Curr. Genet. 46:166–175 [DOI] [PubMed] [Google Scholar]
- 14. Eriksson K.-E. L., Blanchette R. A., Ander P. 1990. Microbial and enzymatic degradation of wood and wood components. Springer-Verlag, Berlin, Germany [Google Scholar]
- 15. Hammel K. E., Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr. Opin. Plant Biol. 11:349–355 [DOI] [PubMed] [Google Scholar]
- 16. Harris P. V., et al. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49:3305–3316 [DOI] [PubMed] [Google Scholar]
- 17. Hart D. O., et al. 2000. Identification of Asp-130 as the catalytic nucleophile in the main alpha-galactosidase from Phanerochaete chrysosporium, a family 27 glycosyl hydrolase. Biochemistry 39:9826–9836 [DOI] [PubMed] [Google Scholar]
- 18. Henriksson G., et al. 1999. Endoglucanase 28 (Cel12A), a new Phanerochaete chrysosporium cellulase. Eur. J. Biochem. 259:88–95 [DOI] [PubMed] [Google Scholar]
- 19. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hibbett D. S., et al. 2007. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111:509–547 [DOI] [PubMed] [Google Scholar]
- 21. Hibbett D. S., Donoghue M. J. 2001. Analysis of character correlations among wood decay mechanisms, mating systems, and substrate ranges in homobasidiomycetes. Syst. Biol. 50:215–242 [PubMed] [Google Scholar]
- 22. Higham C. W., Gordon-Smith D., Dempsey C. E., Wood P. M. 1994. Direct 1H NMR evidence for conversion of β-d-cellobiose to cellobionolactone by cellobiose dehydrogenase from Phanerochaete chrysosporium. FEBS Lett. 351:128–132 [DOI] [PubMed] [Google Scholar]
- 23. Highley T. L. 1973. Influence of carbon source on cellulase activity of white rot and brown rot fungi. Wood Fiber 5:50–58 [Google Scholar]
- 24. Highley T. L. 1982. Influence of type and amount of lignin on decay by Coriolus versicolor. Can. J. For. Res. 12:435–438 [Google Scholar]
- 25. Holzbaur E., Tien M. 1988. Structure and regulation of a lignin peroxidase gene from Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 155:626–633 [DOI] [PubMed] [Google Scholar]
- 26. Irizarry R. A., et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 27. Ishida T., et al. 2009. Crystal structure of glycoside hydrolase family 55 β-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 284:10100–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishida T., Yaoi K., Hiyoshi A., Igarashi K., Samejima M. 2007. Substrate recognition by glycoside hydrolase family 74 xyloglucanase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 274:5727–5736 [DOI] [PubMed] [Google Scholar]
- 29. Kamada F., Abe S., Hiratsuka N., Wariishi H., Tanaka H. 2002. Mineralization of aromatic compounds by brown-rot basidiomycetes—mechanisms involved in initial attack on the aromatic ring. Microbiology 148:1939–1946 [DOI] [PubMed] [Google Scholar]
- 30. Kawai R., Igarashi K., Yoshida M., Kitaoka M., Samejima M. 2006. Hydrolysis of β-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-β-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 71:898–906 [DOI] [PubMed] [Google Scholar]
- 31. Kersten P., Cullen D. 2007. Extracellular oxidative systems of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Fungal Genet. Biol. 44:77–87 [DOI] [PubMed] [Google Scholar]
- 32. Kersten P. J. 1990. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc. Natl. Acad. Sci. U. S. A. 87:2936–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kersten P. J., Kirk T. K. 1987. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J. Bacteriol. 169:2195–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kirk T. K., Farrell R. L. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 35. Krings U., et al. 2009. Autooxidation versus biotransformation of alpha-pinene to flavors with Pleurotus sapidus: regioselective hydroperoxidation of alpha-pinene and stereoselective dehyrogenation of verbenol. J. Agric. Food Chem. 57:9944–9950 [DOI] [PubMed] [Google Scholar]
- 36. Li B., Nagalla S. R., Renganathan V. 1996. Cloning of a cDNA encoding cellobiose dehydrogenase, a hemoflavoenzyme from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macdonald J., et al. 2011. Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl. Environ. Microbiol. 77:3211–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahajan S., Master E. R. 2010. Proteomic characterization of lignocellulose-degrading enzymes secreted by Phanerochaete carnosa grown on spruce and microcrystalline cellulose. Appl. Microbiol. Biotechnol. 86:1903–1914 [DOI] [PubMed] [Google Scholar]
- 39. Mansfield S. D., deJong E., Saddler J. N. 1997. Cellobiose dehydrogenase, an active agent in cellulose depolymerization. Appl. Environ. Microbiol. 63:3804–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martinez D., et al. 2009. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. U. S. A. 106:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez D., et al. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695–700 [DOI] [PubMed] [Google Scholar]
- 42. Matsuzaki F., Shimizu M., Wariishi H. 2008. Proteomic and metabolomic analyses of the white-rot fungus Phanerochaete chrysosporium exposed to exogenous benzoic acid. J. Proteome Res. 7:2342–2350 [DOI] [PubMed] [Google Scholar]
- 43. Munoz I. G., et al. 2001. Family 7 cellobiohydrolases from Phanerochaete chrysosporium: crystal structure of the catalytic module of Cel7D (CBH58) at 1.32 A resolution and homology models of the isozymes. J. Mol. Biol. 314:1097–1111 [DOI] [PubMed] [Google Scholar]
- 44. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 45. Niemenmaa O., Uusi-Rauva A., Hatakka A. 2007. Demethoxylation of [O(14)CH (3)]-labelled lignin model compounds by the brown-rot fungi Gloeophyllum trabeum and Poria (Postia) placenta. Biodegradation 19:555–565 [DOI] [PubMed] [Google Scholar]
- 46. Ravalason H., et al. 2008. Secretome analysis of Phanerochaete chrysosporium strain CIRM-BRFM41 grown on softwood. Appl. Microbiol. Biotechnol. 80:719–733 [DOI] [PubMed] [Google Scholar]
- 47. Sato S., Liu F., Koc H., Tien M. 2007. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 153:3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 49. Subramanian V., Yadav J. 2008. Regulation and heterologous expression of P450 enzyme system components of the white rot fungus Phanerochaete chrysosporium. Enzyme Microb. Technol. 43:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tanaka H., et al. 2007. Characterization of a hydroxyl-radical-producing glycoprotein and its presumptive genes from the white-rot basidiomycete Phanerochaete chrysosporium. J. Biotechnol. 128:500–511 [DOI] [PubMed] [Google Scholar]
- 51. Tempelaars C., Birch P., Sims P., Broda P. 1994. Isolation, characterization, and analysis of the cbhII gene of Phanerochaete chrysosporium. Appl. Environ. Microbiol. 60:4387–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tien M., Kirk T. K. 1984. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc. Natl. Acad. Sci. U. S. A. 81:2280–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uzcategui E., Johansson G., Ek B., Pettersson G. 1991. The 1,4-d-glucan glucanohydrolases from Phanerochaete chrysosporium. Re-assessment of their significance in cellulose degradation mechanisms. J. Biotechnol. 21:143–160 [DOI] [PubMed] [Google Scholar]
- 54. Uzcategui E., et al. 1991. Pilot-scale production and purification of the cellulolytic enzyme system from the white-rot fungus Phanerochaete chrysosporium. Biotechnol. Appl. Biochem. 13:323–334 [Google Scholar]
- 55. Uzcategui E., Ruiz A., Montesino R., Johansson G., Pettersson G. 1991. The 1,4-β-d-glucan cellobiohydrolase from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J. Biotechnol. 19:271–286 [DOI] [PubMed] [Google Scholar]
- 56. Vanden Wymelenberg A., et al. 2009. Transcriptome and secretome analysis of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl. Environ. Microbiol. 75:4058–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vanden Wymelenberg A., et al. 2010. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 76:3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vanden Wymelenberg A., et al. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17–34 [DOI] [PubMed] [Google Scholar]
- 59. Vanden Wymelenberg A., et al. 2006. Structure, organization, and transcriptional regulation of a family of copper radical oxidase genes in the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 72:4871–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wariishi H., Valli K., Gold M. H. 1992. Mn oxidation by manganese peroxidase from Phanerochaete chrysosporium: kinetic mechanisms and role of chelators. J. Biol. Chem. 267:23688–23695 [PubMed] [Google Scholar]
- 61. Worrall J. J., Anagnost S. E., Zabel R. A. 1997. Comparison of wood decay among diverse lignicolous fungi. Mycologia 89:199–219 [Google Scholar]
- 62. Xu G., Goodell B. 2001. Mechanisms of wood degradation by brown-rot fungi: chelator-mediated cellulose degradation and binding of iron by cellulose. J. Biotechnol. 87:43–57 [DOI] [PubMed] [Google Scholar]
- 63. Yadav J. S., Doddapaneni H., Subramanian V. 2006. P450ome of the white rot fungus Phanerochaete chrysosporium: structure, evolution and regulation of expression of genomic P450 clusters. Biochem. Soc. Trans. 34:1165–1169 [DOI] [PubMed] [Google Scholar]
- 64. Yelle D., Wei D., Ralph J., Hammel K. E. 2011. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot fungus Postia placenta. Environ. Microbiol. 13:1091–1100 [DOI] [PubMed] [Google Scholar]
- 65. Yelle D. J., Ralph J., Lu F., Hammel K. E. 2008. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 10:1844–1849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.