Abstract
Trithorax (TRX) is a Drosophila SET domain protein that is required for the correct expression of homeotic genes. Here, we show that the TRX SET domain efficiently binds to core histones and nucleosomes. The primary target for the SET domain is histone H3 and binding requires the N-terminal histone tails. The previously described trxZ11 mutation changes a strictly conserved glycine in the SET domain to serine and causes homeotic transformations in the fly. We found that this mutation selectively interferes with histone binding, suggesting that histones represent a critical target during developmental gene regulation by TRX.
Keywords: Trithorax, SET domain, chromatin, histones, homeotic genes
The Polycomb group (PcG) of repressors and trithorax group (trxG) of activators target chromatin in order to “freeze” a mitotically stable pattern of gene expression and determined cell fate (Pirrotta 1998; Lyko and Paro 1999; Mahmoudi and Verrijzer 2001). The founding member of the trxG, the Drosophila trx gene, is required throughout development and controls the expression of several developmental regulators, including the homeotic genes (Ingham and Whittle 1980; Ingham 1985; Breen 1999). trx is related to the human Mixed Lineage Leukemia (MLL) gene, which is involved in translocations associated with the majority of cases of infant leukemias (Waring and Cleary 1997). TRX and MLL are part of a highly conserved regulatory network that is required for the correct expression of the homeotic selector genes and determination of segment identity in both mammals and Drosophila. They are very large proteins that contain structural motifs common to chromatin-associated factors such as PHD fingers and a C-terminal SET domain (Fig. 1A; Mazo et al. 1990; Stassen et al. 1995).
Figure 1.
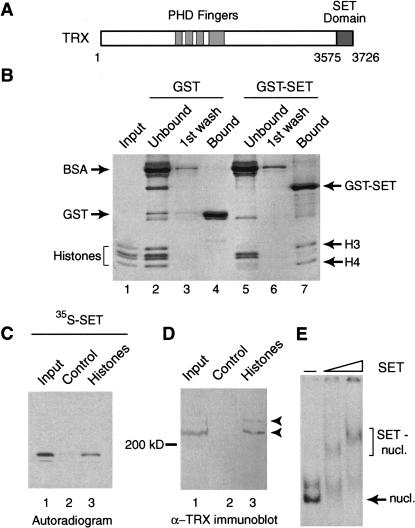
The TRX SET domain binds histones and nucleosomes. (A) Schematic representation of the domain structure of TRX. Ignoring other conserved regions, only the PHD fingers, and the C-terminal SET domain are indicated. (B) The TRX SET domain interacts preferentially with the H3–H4 tetramer. GST and GST–SET domain fusion proteins were immobilized on glutathione-Sepharose and incubated with Drosophila core histones. Unbound proteins and first wash fractions were TCA precipitated. Protein complexes were resolved by 15% SDS-PAGE and visualized by Coomassie staining. Input corresponds to 30% of the material used in the binding reactions. (C) The TRX SET domain binds to a histone-affinity matrix. BSA–Sepharose control beads (lane 2) and histone–Sepharose beads (lane 3) were incubated with radiolabeled SET domain. Protein complexes were resolved by 15% SDS-PAGE and bound proteins were detected by autoradiography. Lane 1 represents 5% of the input. (D) TRX protein interacts with immobilized histones. BSA–Sepharose control beads (lane 2) and histone–Sepharose beads (lane 3) were incubated with Drosophila nuclear extracts. Bound proteins were resolved by 7.5% SDS-PAGE, transferred to nitrocellulose, and probed with an α-TRX antibody. Lane 1 represents 5% of the input. (E) The TRX SET domain binds mononucleosomes. In vitro assembled mononucleosomes were incubated with either no protein (lane 1) or with increasing concentrations of SET domain (lanes 2 and 3) and resolved by 4% native PAGE.
The SET domain is a highly conserved 130–150 amino acids motif initially recognized as a common element in chromatin regulators with opposing activities: the suppressor of position affect variegation Su(var)3-9, the PcG protein Enhancer of Zeste [E(z)], and TRX (Jenuwein et al. 1998). The SET domain has been implicated in a multitude of different protein–protein interactions and functions. The SET domains of MLL, yeast Set1p, and E(z) bind to myotubularin-related dual-specificity phosphatases and anti-phosphatases that modulate growth control (Cui et al. 1998). The TRX and MLL SET domains bind to the SNF5 component of the ATP-dependent remodeler SWI/SNF (Rozenblatt-Rosen et al. 1998) and mediate self-association (Rozovskaia et al. 2000). Furthermore, the SET domain of yeast Set1p binds the Mec3p checkpoint protein and has been implicated in DNA repair and telomere function (Corda et al. 1999). The Set1p SET domain alone suffices to mediate telomeric silencing, suggesting that it forms a functional unit (Nislow et al. 1997). Recently, it was shown that SUV39H1, the mammalian homolog of Su(var)3–9, selectively methylates lysine 9 of histone H3 (Rea et al. 2000; Jenuwein 2001). This modification creates a binding site for HP1 and thus can contribute to the propagation of a heterochromatin domain (Bannister et al. 2001; Lachner et al. 2001; Nakayama et al. 2001). Whereas the histone–methylase activity of SUV39H1 was critically dependent on the SET domain, additional protein domains were also required.
In contrast to SUV39H1, the SET domains of TRX and E(z) do not appear to mediate histone methylation (Rea et al. 2000; Jenuwein 2001). Thus, the TRX SET domain may form part of a methylase with a substrate other than histones or, alternatively, this SET domain may present a histone-recognizing module that is not a methylase. Here, we report that the TRX SET domain efficiently binds to core histones and to nucleosomes. We found that binding depends on the N-terminal histone tails and investigated the role of their covalent modifications. The effect of a homeotic mutation in the SET domain (trxZ11) on histone binding suggests that histone recognition constitutes an essential step during the in vivo control of gene expression by TRX.
Results and Discussion
The TRX SET domain binds core histones and nucleosomes
To assess whether the SET domain is a histone-recognizing module, we performed GST pull-down experiments using purified Drosophila core histones (Fig. 1B). Bound proteins were analyzed by SDS-PAGE followed by Coomassie staining. We found that the GST–SET domain fusion protein selectively retained histones H3 and H4, whereas binding to H2A and H2B was significantly weaker. Binding is efficient since almost all H3 and H4 is depleted from the unbound fraction (Fig. 1B, cf. lanes 5 and 7). The SET domain–histone interaction survived stringent washing conditions with a buffer containing 600 mM NaCl and 0.2% NP-40. Moreover, both the SET domain (isoelectric point of 8.6) and histones have a net positive charge at pH 7, arguing against a nonspecific electrostatic interaction. It should be noted that when higher amounts of histones were added, binding to all four core histones could be detected (see Fig. 2B, below). Next, we prepared an affinity-matrix by covalent coupling of either purified core histones or BSA to CNBr activated-Sepharose beads. We found that radiolabeled TRX SET domain efficiently associated with the histone–Sepharose but not with the BSA–Sepharose control matrix (Fig. 1C). The histone affinity matrix was used to investigate whether the endogenous TRX protein present in Drosophila embryo nuclear extracts could bind to histones. Bound protein fractions were resolved by SDS-PAGE and analyzed by immunoblotting using an antibody directed against TRX (Fig. 1D). As shown previously (Kuzin et al. 1994), TRX is present in nuclear extracts as processed polypeptides of a size greater than 200 kD (Fig. 1D, lane 1). Similar to the isolated SET domain, endogenous TRX is retained efficiently by the histone beads but not by the control matrix. Next, we examined binding of the SET domain to radiolabeled mononucleosomes in a mobility shift assay. Figure 1E reveals that the SET domain efficiently binds to mononucleosomes. The further retardation of SET–nucleosome complexes in the presence of higher amounts of the SET domain indicates binding of more than one SET molecule per nucleosome. Glycerol gradient sedimentation studies using nucleosomal arrays also revealed chromatin binding of the SET domain (data not shown). We conclude that the TRX SET domain is a histone-binding module that mediates association with chromatin.
Figure 2.
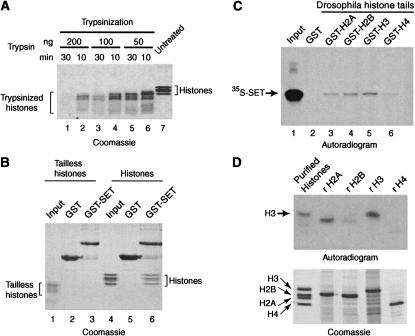
N-terminal histone tails are necessary for SET domain binding. (A) Purified Drosophila core histones were treated with trypsin as indicated and the extent of trypsinization was monitored by SDS-PAGE followed by Coomassie staining (lanes 1–6). Untreated core histones are shown in lane 7. The trypsinized histones shown in lane 4 (100 ng of trypsin for 10 min) were used in the experiment shown in B. (B) SET domain fails to bind tailless histones. GST alone (lanes 2 and 5) and GST–SET domain fusion protein (lanes 3 and 6) were immobilized on glutathione–Sepharose and incubated with either trypsinized histones (lanes 1–3) or untreated core histones (lanes 4–6). Lanes 1 and 4 contain 5% of the input material used in the binding reactions. Protein complexes were resolved by 15% SDS-PAGE and visualized by Coomassie staining. (C) The TRX SET domain binds the N-terminal histone tails. Drosophila histone tails expressed as GST-fusion proteins were immobilized on glutathione–Sepharose and were incubated in the presence of radiolabeled SET domain. Lane 1 represents 5% of the input material used in the binding reactions. Bound proteins were resolved by 15% SDS-PAGE and visualized by autoradiography. (D) Far-Western analysis of TRX SET domain histone binding. Drosophila purified core histones (lane 1) and each of the bacterially expressed recombinant Xenopus core histones (lanes 2– 5) were resolved by 15% SDS-PAGE and either stained with Coomassie (lower panel) or transferred to a nitrocellulose membrane and probed with radiolabeled SET domain (upper panel). Proteins bound to the filter were visualized by autoradiography.
The TRX SET domain recognizes the N-terminal histone tails
Histone tail domains are the protruding, flexible parts of the histones that mediate contacts between adjacent nucleosomes and interact with chromatin-associated proteins (Kingston and Narlikar 1999; Strahl and Allis 2000; Turner 2000; Jenuwein 2001). The unstructured histone tails are much more sensitive to digestion by trypsin than the globular domains and can be selectively removed by limited trypsinization (Fig. 2A). The fraction of trypsinized histones shown in lane 4 was used in a GST pull-down experiment to test the role of the tail domains in SET domain binding (Fig. 2B). In contrast to intact histones, the SET domain failed to recognize the tailless histones. Thus, the histone tails appear to be critical for binding by the TRX SET domain. To investigate whether they might also be sufficient, we assayed SET domain binding to Drosophila histone tail GST-fusion proteins (Georgel et al. 1997). Figure 2C shows modest binding to all the histone tails with a preference for the H3 tail domain, whereas binding to the H4 tail was very weak. Similar results were obtained using GST–yeast histone tail fusion proteins (data not shown).
As an additional approach to identify its target(s), we performed a far-Western analysis of SET domain binding to purified endogenous Drosophila histones and recombinant, bacterially expressed Xenopus histones (Luger et al. 1999). Histones were separated by SDS-PAGE and transferred to a nitrocellulose membrane that was probed with radiolabeled TRX SET domain. Autoradiography revealed strong binding to the endogenous histone H3 but only weak binding to the other endogenous histones (Fig. 2D). The recombinant histone H3 (rH3) is also recognized efficiently; but in addition, rH2A is also bound and weaker interactions with rH2B and rH4 were detected. Collectively, the results presented in Figures 1 and 2 strongly suggest that histone H3 is the main target for the TRX SET domain. Weaker association with the other histones was observed in some assays, suggesting they present secondary targets. Since SET domain binding to histone H4 is particularly weak, we assume that H4 is retained via its association with H3.
The histone tails are subjected to distinct posttranslational modifications that may constitute a “histone code” (Strahl and Allis 2000; Turner 2000) and can influence binding of specific chromatin-associated proteins. The bromodomain recognizes histone tails acetylated at specific lysine residues (Dhalluin et al. 1999; Jacobson et al. 2000), whereas binding of the chromodomain of heterochromatin protein 1 (HP1) depends on methylation of histone H3 on lysine 9 (Bannister et al. 2001; Jenuwein 2001; Lachner et al. 2001; Nakayama et al. 2001). To investigate the effect of histone modifications on SET domain binding, we performed pull-down experiments and probed the bound and unbound histone fractions with antibodies that recognize specific modifications. The input of the binding experiment was adjusted so that the bound and unbound fractions contained an approximately equal amount of histones as revealed by Western immunoblotting with a polyclonal serum raised against purified core histones (Fig. 3A). However, an antibody directed against acetylated lysines revealed a modest but reproducible enrichment for acetylated histones in the bound fraction. A similar enrichment was observed with antisera specific for histone H3 acetylated at lysine 9 or 14, respectively.
Figure 3.
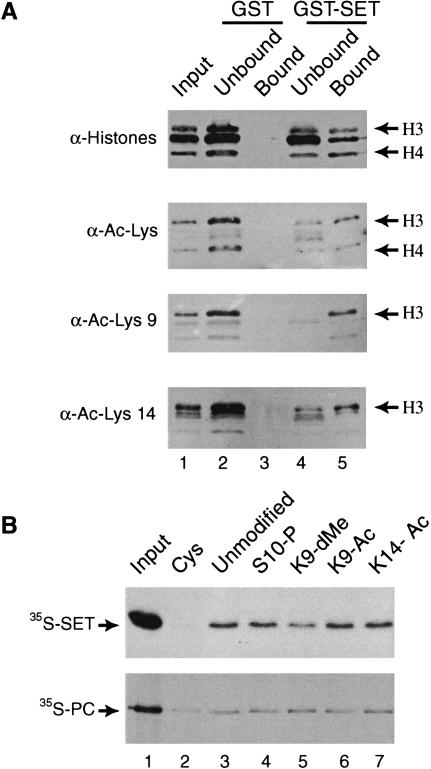
Histone tail modifications influence the SET domain binding affinity. (A) The GST–SET bound fraction is enriched in acetylated histone H3. GST–SET pull-down assays were performed in the presence of an excess of histones. Unbound material was TCA precipitated. Bound and unbound proteins were resolved by 15% SDS-PAGE, transferred to nitrocellulose membranes and probed with antibodies directed against core histones (top panel), acetylated lysine (α-Ac-Lys), acetylated histone H3 on lysine 9 (α-Ac-Lys9), and acetylated histone H3 on lysine 14 (α-Ac-Lys14). (B) Dimethylation of histone H3 on lysine 9 diminishes TRX SET binding. Control beads coupled with cysteine (Cys) or histone H3 tail peptides that were either unmodified, phosphorylated on serine 10 (S10-P), dimethylated on lysine 9 (K9-dMe), acetylated on lysines 9 (K9-Ac), or 14 (K14-Ac) were incubated with radiolabeled SET domain (upper panel) or Polycomb (lower panel). Bound proteins were resolved by SDS-PAGE and visualized by autoradiography. Input corresponds to 15% of the material used in the binding reactions.
To determine the effect of single modifications, we compared the binding of radiolabeled TRX SET domain to differentially modified histone H3 N-terminal peptides coupled to an affinity matrix. In agreement with our previous experiments, the H3 tail peptide was sufficient to mediate SET domain binding (Fig. 3B, lane 3). Acetylation at either position 9 or 14 resulted in only a minor increase in SET domain binding, as detected by PhosphorImager analysis. Thus, by themselves these modifications do not create a strong binding site for the TRX SET domain. However, methylation of lysine 9 results in an ∼twofold reduced affinity. In contrast, binding of the Drosophila Polycomb protein does not seem to be influenced by these modifications (See also Lachner et al. 2001). Collectively, these results suggest that the SET domain of the activator TRX preferentially interacts with hyperacetylated histones. However, a modification that is associated with silent heterochromatin, methylation of H3 lysine 9, diminishes TRX SET domain binding. Therefore, although the effects we observed were modest, they correlate well with the activating function of TRX and may assist in targeting TRX to euchromatin.
The homeotic trxZ11 mutation interferes with histone binding
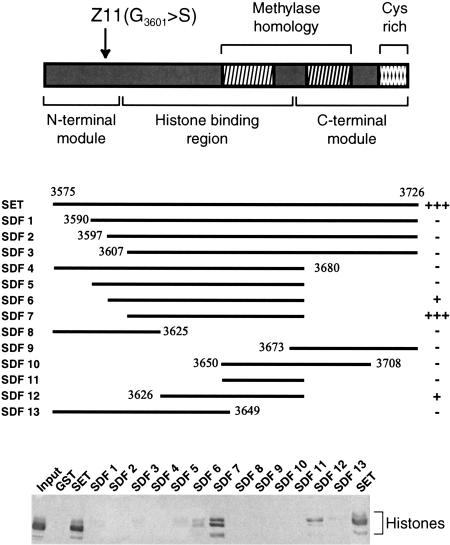
To define the regions of the TRX SET domain involved in histone recognition, we compared a series of deletion mutants in a histone binding experiment. Figure 4 shows that even small N-terminal deletions of the SET domain (SET deletion fragments SDF1, SDF2, and SDF3) abrogate histone binding. Likewise, a C-terminal deletion (SDF4) of the SET domain leads to loss of histone binding. Surprisingly, when this C-terminal deletion was combined with progressive N-terminal deletions (SDF5, SDF6, and SDF7) histone binding was restored. SDF7 comprising TRX residues 3607–3680 retained histones with an affinity comparable to that of the full-length TRX SET domain. Neither the N-terminal (SDF8) nor the C-terminal (SDF9) portions of the SET domain were able to bind histones by themselves. These regions might play a structural role in presenting the minimal histone binding region rather than contacting the histones directly. The smallest construct that was still able to bind significant amounts of histones ranged from amino acids 3626 to 3680 (SDF12). Interestingly, SDF7 and, in particular, SDF12 showed a stronger preference for binding to histone H3 compared to the other histones, than the full-length SET domain. There is no correlation between binding of the mutant versions of the SET domain and their net charge. For example, SDF11 and SDF12 both have an isoelectric point of 6.7, but SDF12 binds histones whereas SDF11 does not.
Figure 4.
Mapping of the TRX SET histone-binding domain. SET domain deletion fragments were tested for their ability to bind core histones in GST pull-down experiments. Protein complexes were resolved by 15% SDS-PAGE and analyzed by Western immunoblotting using anti-Drosophila histone antibodies The residues present in the SET deletion fragments (SDF1–13) are indicated. Two highly conserved blocks with homology to plant methylases, a C-terminal cysteine-rich region, and the position of the trxZ11 glycine to serine (G3601>S) mutation are schematically depicted. The identified minimal histone-binding domain and the N-terminal and C-terminal domains, which play a critical role in histone binding, are shown.
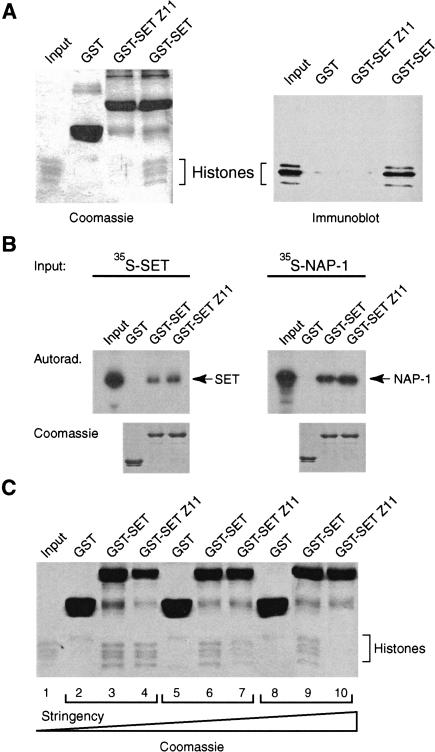
The involvement of the N-terminal region of the TRX SET domain in histone recognition is of great interest since this region harbors a developmental mutation. This mutation, trxZ11, causes the change of a highly conserved glycine (G 3601) to serine (Stassen et al. 1995). The trxZ11 mutation is a viable, strong hypomorph that gives rise to homeotic transformations in heterozygous flies (Breen 1999). Homozygotes display a much higher frequency of a wide variety of homeotic transformations, and in a hemizygous background trxZ11 causes lethality at the pupal stage. Therefore, the severity of the trxZ11 phenotype is subjected to dosage effects. To investigate whether the trxZ11 mutation affects the ability of the SET domain to bind histones, we compared the mutant (SETZ11) and wild-type SET domain in a histone binding experiment (Fig. 5A). In contrast to an approximately equal amount of wild-type SET domain, the SETZ11 failed to bind histones. The TRX SET domain self-associates (Rozovskaia et al. 2000), and we recently discovered that it binds the histone chaperone NAP-1 (K.R. Katsani and A.J. Kal, unpubl.). As shown in Figure 5B, neither self-association of the SET domain nor binding to NAP-1 is affected by the trxZ11 mutation, suggesting that the effect is selective and not due to a global misfolding of the SET domain. Interestingly, histone binding is strongly reduced due to the trxZ11 mutation but not completely abolished. Using milder assay conditions, we could still detect some residual histone binding by the SETZ11 domain (Fig. 5C). This finding agrees well with the observed gene dosage effects on the severity of the trxZ11 phenotype.
Figure 5.
The trxZ11 mutation impedes histone binding. (A) The histone-binding ability of equal amounts of GST–SET domain fusion protein containing the trxZ11 mutation GST–SET Z11 and wild-type GST–SET domain was compared in a pull-down experiment (see Fig. 1 legend ). Bound proteins were resolved by 15% SDS-PAGE and visualized by Coomassie staining or by Western immunoblotting using anti-histone antibodies. (B) The effect of the trxZ11 mutation on other protein–protein interactions was tested in a GST pull-down experiment. GST–SET domain or GST–SET Z11 fusion proteins were immobilized on glutathione–Sepharose and incubated with radiolabeled TRX SET domain or radiolabeled NAP-1 protein, respectively. Bound proteins were resolved by 15% SDS-PAGE and visualized by autoradiography (top panel). The bottom panels show the Coomassie stain of the same gels. (C) The histone binding experiment shown in A was repeated using variable binding conditions. (Lanes 2–4) 75 mM KCl, 0.02% NP-40; (lanes 5–7) 200 mM KCl, 0.02% NP-40; (lanes 8–10) 200 mM KCl, 0.08% NP-40. The increasing stringency of the binding conditions is indicated by a triangle (bottom). Final washes were with a buffer containing 600 mM NaCl and 0.2% NP-40.
Role of the SET domain in gene regulation
Unlike the heterochromatic silencing factor SUV39H1, the activator TRX appears to be unable to methylate histone tails (Rea et al. 2000; C.P. Verrijzer, unpubl.). This failure, however, is not due to the inability to bind histones. Thus, rather than an active enzymatic core, the TRX SET domain might act as a histone binding module that anchors TRX to the chromatin template. Similar to anti-phosphatases, the TRX SET domain may bind the substrate (i.e., the histone tails) and block modification by related active enzymes such as SUV39H1, thus preventing HP1 recruitment. Such a mechanism may contribute to the antagonistic activities of the silencer SUV39H1 and the activator TRX. Our results indicate that modification of the histone tails can influence binding of the TRX SET domain and may contribute to directing TRX to active chromatin. TRX is a developmental regulator that is essential for the normal expression of multiple homeotic genes. This function is impaired by the trxZ11 mutation in the SET domain resulting in homeotic transformations (Stassen et al. 1995; Breen 1999). An analogous mutation in the SET domain of yeast Set1p causes a defect in telomeric silencing, supporting the notion that this mutation interferes with a highly conserved function (Nislow et al. 1997). Although other SET domain functions may also be affected, our results show that the trxZ11 mutation incapacitates its ability to bind histones. This finding provides a molecular explanation for the aberrant development of trxZ11 flies and implies that histone recognition by the TRX SET domain is essential for the in vivo functioning of TRX.
Materials and methods
DNA constructs
Details of cloning procedures are available upon request. Briefly, PCR fragments, encoding either the TRX SET domain (amino acids 3575– 3726; Stassen et al. 1995) or various deletion fragments, were cloned in-frame in a pGEX–2TK (Pharmacia) derived plasmid (pGEX–2TKN). The trxZ11 mutation was generated by PCR-based site-directed mutagenesis. TRX residues that were encoded by the various SDF deletion constructs are indicated in Figure 4. Templates for in vitro transcription/translation of the SET domain, NAP-1, and PC were generated by cloning of the corresponding full-length coding sequences into the pTβSTOP vector. The GST–Drosophila histone tail fusion and recombinant histones constructs were kindly provided by C. Wu (Georgel et al. 1997) and T. Richmond (Luger et al. 1999), respectively.
Protein procedures
Recombinant GST-fusion proteins were expressed in Escherichia coli BL 21 and purified using standard procedures. 35S-radiolabeled proteins were expressed using TNT rabbit reticulocyte lysates (Promega). Drosophila core histones (Bulger and Kadonaga 1994) and Drosophila nuclear extracts (Kal et al. 2000) were prepared as described. The histone-affinity matrix was prepared by covalent coupling of ∼1 mg core histones or BSA to 1 mL of CNBr activated-Sepharose (Pharmacia). Peptides were coupled to SulfoLink Coupling Gel (Pierce). The GST pull-down experiments were performed according to Jiménez et al. (1999) in binding buffer (20 mM HEPES-KOH at pH 7.6, 2.5 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 mM DTT) containing 200 mM KCl and 0.08% NP-40 followed by a series of washes with RIPA buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 0.2% NP-40) containing 600 mM NaCl. Other protein–protein interaction assays were performed using similar procedures. For peptide pull-downs, binding was allowed for 1 h at room temperature in a buffer containing 150 mM NaCl and washes performed in the presence of 1% NP-40. Far-Western analysis was performed as described (Kal et al. 2000). The anti-histone polyclonal antiserum was generated by immunizing rabbits with purified Drosophila core histones. The various other anti-histone antibodies used were purchased from Upstate Biotechnologies. The anti-TRX antiserum was raised against the first 270 N-terminal amino acids. Histone H3 tail peptides were a generous gift from T. Jenuwein (Lachner et al. 2001). Mononucleosomes were assembled by salt dialysis on a 196-bp 5S DNA fragment (Owen-Hughes and Workman 1996). For the mobility shift assay, mononucleosomes and purified SET domain incubated for 45 min at room temperature in 20 mM HEPES-KOH (pH 7.6), 50 mM NaCl, 5% glycerol, 100 μg/mL BSA, 2 mM DTT, and resolved by PAGE on a 4% native gel.
Acknowledgments
We are very grateful to T. Jenuwein and M. Lachner for advice and the generous gift of peptides; and C. Wu, M. Grunstein, T. Richmond, T. Owen-Hughes, K. Ura, and A. Mazo for the gift of plasmids. We also thank J. Muñoz, G. Chalkley, and J. van der Knaap for reagents; and J. Svejstrup, L. Comai, L. Fradkin, J. Dorsman, E. Kalkhoven, N. Little, and lab members for critical reading of the manuscript. This work was supported in part by an EC grant (HPRN CT2000 00078) to C.P.V.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL verrijzer@lumc.nl; FAX 31-71-527-6284.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.201901.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Breen TR. Mutant alleles of the Drosophila trithorax gene produce common and unusual homeotic and other developmental phenotypes. Genetics. 1999;152:319–344. doi: 10.1093/genetics/152.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Kadonaga JT. Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol Genet. 1994;5:241–262. [Google Scholar]
- Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, Gilson E, Geli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW. A clonal analysis of the requirement for the trithorax gene in the diversification of segments in Drosophila. J Embryol Exp Morphol. 1985;89:349–365. [PubMed] [Google Scholar]
- Ingham PW, Whittle R. Trithorax: A new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol Gen Genet. 1980;179:607–614. [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–273. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP. The Drosophila Brahma complex is an essential coactivator for the trithorax group protein zeste. Genes & Dev. 2000;14:1058–1071. [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes & Dev. 1994;8:2478–2490. doi: 10.1101/gad.8.20.2478. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Lyko F, Paro R. Chromosomal elements conferring epigenetic inheritance. Bioessays. 1999;21:824–832. doi: 10.1002/(SICI)1521-1878(199910)21:10<824::AID-BIES4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Verrijzer CP. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben Simchon L, Croce CM, Mazo A, et al. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- Stassen MJ, Bailey D, Nelson S, Chinwalla V, Harte PJ. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech Dev. 1995;52:209–223. doi: 10.1016/0925-4773(95)00402-m. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Waring PM, Cleary ML. Disruption of a homolog of trithorax by 11q23 translocations: Leukemogenic and transcriptional implications. Curr Top Microbiol Immunol. 1997;220:1–23. doi: 10.1007/978-3-642-60479-9_1. [DOI] [PubMed] [Google Scholar]