Abstract
Wild-type Arabidopsis plants maintain a relatively constant size over a wide range of temperatures. Here we show that this homeostasis requires the BONZAI1 (BON1) gene because bon1 null mutants make miniature fertile plants at 22°C but have wild-type appearance at 28°C. The expression of BON1 and a BON1-associated protein (BAP1) is modulated by temperature. Thus BON1 and BAP1 may have a direct role in regulating cell expansion and cell division at lower temperatures. BON1 contains a Ca2+-dependent phospholipid-binding domain and is associated with the plasma membrane. It belongs to the copine gene family, which is conserved from protozoa to humans. Our data suggest that this gene family may function in the pathway of membrane trafficking in response to external conditions.
Keywords: Homeostasis, Bonzai1, copine, BAP1, temperature, size
Multicellular organisms develop and maintain a relatively constant size and shape over a wide range of different environmental conditions. This homeostasis is accomplished both by extrinsic mechanisms (e.g., movements that can reestablish a constant environment) and by intrinsic mechanisms that alter cellular metabolism so that the organism can retain its morphology despite the altered environment. Animals can move to a new environment to maintain constant external conditions; whereas plants are anchored and can only bend in response to changes in environmental factors such as light, gravity, and mechanical force.
The existence of intrinsic homeostatic mechanisms suggests genetic control, which could be revealed by mutants that fail to maintain constant size and shape in response to an environmental perturbation. Recently, a genetic pathway has been discovered in Drosophila that regulates cell, organ, and body size. Mutations in genes apparently functioning in the insulin-signaling pathway result in miniature flies comprised of smaller and fewer cells (Oldham et al. 2000) under conditions where wild type maintains its standard size. These mutant flies phenocopy those wild type under starvation or overcrowding, which reveals a link between the control of morphology and nutritional conditions in the environment.
For plants these intrinsic genetic mechanisms must be essential in temperature homeostasis because these organisms are sessile and do not maintain constant body temperatures when the ambient temperature changes. Indeed, most plants maintain a relatively constant phenotype in varied temperatures. For example, Arabidopsis grows to similar size and morphology (less than a twofold change) over temperatures that range from 16°C to 30°C. This suggests the existence of genes whose function is to permit adaptation to temperature variation.
Studies on the tolerance of plants to freezing have revealed the mechanisms that contribute to survival at very low temperatures. Both physiological and genetic analyses point to the membrane systems as key to this homeostasis. Extreme cold results in the disruption of membrane function and death (Steponkus 1984). Plants can increase their tolerance to extreme cold by previous exposure to low nonfreezing temperatures, a phenomenon known as cold acclimation. A prominent feature of cold acclimation is an alteration of the membrane lipid composition and the accumulation of simple sugars and some hydrophilic peptides, both of which are thought to stabilize membranes to protect against freezing injury (Thomashow 1999). Genetic analysis has implicated the plasma membrane as key to thermotolerance. Mutants hypersensitive to freezing injury (sfr) are affected in the cryostability of the plasma membranes (Warren et al. 1996).
Our approach to identifying the genes involved in temperature homeostasis is to isolate Arabidopsis mutants unable to maintain size or shape over a broad temperature range. To this end we isolated a mutant, bonzai1 (bon1), that makes miniature plants at lower temperatures. The BON1 gene encodes one of the copines, a protein widely conserved in plants and animals. The BON1 protein is tightly associated with the plasma membrane and promotes aggregation of lipid vesicles in vitro. BON1 associates with another protein BAP1, which can suppress the bon1 phenotype when it is overexpressed. Our data suggest that the copine gene family may function in membrane trafficking and be transcriptionally regulated by the environmental conditions to which they are designed to respond.
Results
BON1 is a gene required for normal plant size at 22°C
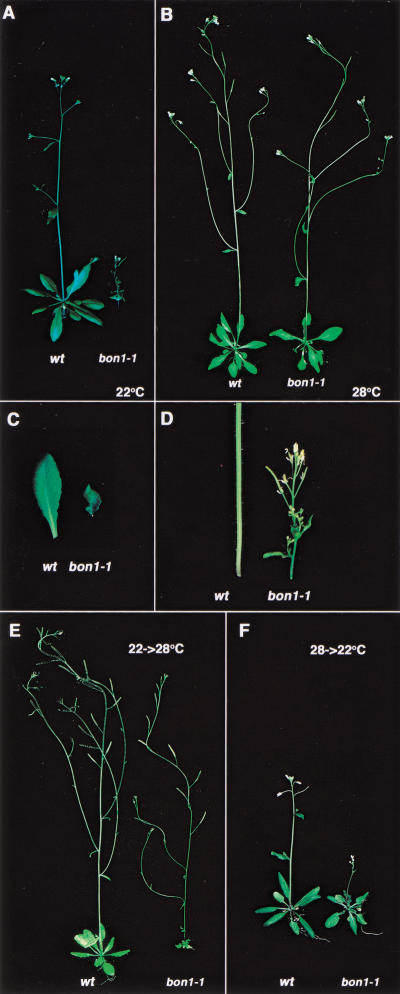
To obtain mutants that have a restricted temperature range for normal development, we screened for strains that were miniature at one temperature but not another. This screen uncovered a mutant, bon1, that exhibits a dwarf phenotype at 22°C (a normal growth temperature for wild type; Fig. 1A) but has a phenotype indistinguishable from that of the wild type at 28°C (Fig. 1B). This temperature dependent phenotype is not an allele-specific phenomenon because bon1-1 is a loss-of-function mutant (see below).
Figure 1.
Phenotype of bon1-1. (A) Wild type (Col-0) and bon1-1 grown at 22°C. (B) Wild type and bon1-1 grown at 28°C. (C) Leaves of wild type and bon1-1 grown at 22°C. (D) Stems of wild type and bon1-1 grown at 22°C. (E) Plants were grown for 3 wk at 22°C and then transferred to 28°C for 10 d. Note that the small rosette in bon1-1 formed at 22°C. The stem that formed at 28°C is almost normal. (F) Plants were grown for 3 wk at 28°C and then transferred to 22°C for 10 d. Note that the rosette formed at 28°C is normal in bon1-1 but the stem and new leaves that formed at 22°C are miniature.
At 22°C, the bon1-1 leaves are greatly reduced in size and have a curved morphology (Fig. 1C). The inflorescence stem is thinner and shorter than the wild type (Fig. 1D). Despite the smaller size of the leaves and stems at 22°C, bon1-1 plants nevertheless develop relatively normal flowers and siliques and are completely fertile. bon1-1 does not have a dramatic effect on the timing of the developmental process. It bolts and sets seeds only slightly earlier than the wild type. The bon1 phenotype is genetically distinct from that of previously identified hormonal dwarf mutants such as axr1, det2, or ctr1. The bon1-1 defect cannot be rescued by the addition of hormones such as gibberellin or brassinosteroid or by mutations in hormonal signaling (data not shown). Moreover, the bon1-1 mutant is distinct from the cold-sensitive mutants deficient in the desaturation of the lipids in the membranes (Lightner et al. 1994) because bon1-1 does not have an altered lipid composition or altered composition of saturated fatty acids in the lipids (data not shown). These observations suggest that BON1 functions to maintain plant morphology through a novel mechanism.
BON1 is required to maintain cell size and number at 22°C
Using scanning electron microscope (SEM), we measured the width and length of the epidermal cells of the inflorescence stems grown at 22°C (Table 1). The length of the bon1-1 cells is approximately seven times shorter than that of the wild type, which accounts for most of the eightfold reduction in bon1 stem length at the nonpermissive temperature. A bon1-1 cell is about two-thirds as wide as the wild-type cell. There are also half as many cells per epidermal circumference in a bon1-1 mutant as there are in the wild type. Analysis of whole-stem cross sections revealed similar defects in the inner cells: There were fewer and smaller inner cells inside the stem. Reduction in the size of the pavement cells was observed by SEM on the leaf epidermis (data not shown). Taken together, these data suggest that the small size of bon1-1 at 22°C results from a reduction in both cell size and number. Because the number of cells on the outer surface is coordinated with the diameter of underlying cells, we cannot exclude the possibility that the bon1 effect on cell number is a consequence of its primary effect on cell size.
Table 1.
Cell size and number of stem epidermal cells in wild type and bon1-1
|
|
Stem length
|
Cell length
|
Cell number/stem file
|
Stem diameter
|
Cell width
|
Cell number/circumference
|
|---|---|---|---|---|---|---|
| Wild type | 18 cm | 288 ± 55 mm | ∼625 | 1.0 cm | 18 ± 2 mm | ∼174 |
| bon1-1 | 2.2 cm | 40 ± 9 mm | ∼550 | 0.38 cm | 12 ± 1 mm | ∼99 |
Temperature shift experiments were used to determine at what period of development BON1 function is required for growth. We found that when bon1-1 plants were shifted from 22°C to 28°C, the new stems and leaves that formed at 28°C had normal expansion and elongation when compared with the miniature stems and leaves formed when they were at 22°C. Conversely, after bon1-1 was shifted from 28°C to 22°C, the new leaves and stems that formed at 22°C had a miniature phenotype. Two examples are shown in Figure 1, E and F, in which the 3-week-old seedlings were shifted to another temperature for 10 d. These data show that BON1 is required continuously for normal growth of leaves and stems at 22°C.
BON1 is expressed in growing tissues and the expression is modulated by temperature
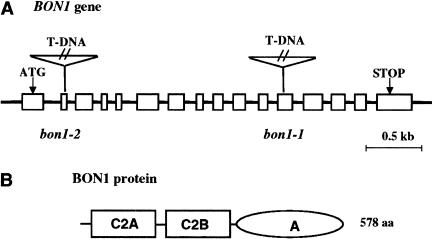
The BON1 gene was cloned on the basis of the T-DNA insertion that results in the bon1 phenotype. The mutation, which segregated as a single recessive trait in the F2 populations of a backcross to wild-type Columbia, was completely linked to the single T-DNA insertion in this mutant. We isolated a 6.5-kb wild-type genomic fragment flanking the T-DNA and a corresponding cDNA of this genomic fragment. Sequence alignment of the genomic fragment and the cDNA indicates that the BON1 gene is comprised of 16 exons and 15 introns (Fig. 2A). The T-DNA is inserted in the twelfth exon of the gene. The bon1-1 mutant defect was complemented either with the genomic fragment or a cDNA–GFP (green fluorescent protein) fusion, confirming the identity of the gene as BON1. Northern blot analysis showed that there was no wild-type RNA transcript of this gene in the mutant (data not shown), indicating that bon1-1 is a loss-of-function mutant. A second loss-of-function allele, bon1-2, (Fig. 2A) was subsequently isolated in the Wassilewskijas (WS) background. The WS bon1-2 allele appeared wild type at 22°C but had less elongated inflorescence stem at 16°C. When the WS bon1-2 was introgressed into Columbia background, it exhibited a phenotype similar to that of bon1-1. It is miniature at 22°C but wild-type at 28°C. Genetic analysis indicates that WS has a dominant suppressor of the bon1 mutation that is responsible for the modified phenotype of bon1-2 in this background (data not shown).
Figure 2.
Structure of the BON1 gene. (A) Genomic structure of the BON1 gene. The boxes represent the exons of the BON1 gene. The translation start and stop are indicated by ATG and STOP, respectively. The T-DNA insertion sites in bon1-1 and bon1-2 are in exons 2 and 12, respectively, as indicated. (B) Domains of BON1 protein. The BON1 protein has two C2 domains at the N terminus and one A domain at the C terminus.
The BON1 gene is predicted to encode a protein of 578 amino acids. The N-terminal portion of the protein consists of two calcium-dependent phospholipid-binding C2 domains (C2A and C2B) and the C-terminal portion exhibits weak similarity to the A domain of integrin (Fig. 2B). The BON1 protein shows extensive homology to the copine gene family. Members of this family have been found in paramecium, worm, mouse, and human and are thought to be calcium-dependent phospholipid binding proteins (Creutz et al. 1998). In each organism there is more than one member of the gene family in the genome. Arabidopsis thaliana BON1 has two paralogs we have named BON2 and BON3. These paralogs are between 72% and 81% identical and 81% and 91% similar to each other at the amino acid level. These plant proteins are very similar to their human counterparts. Arabidopsis BON1 has 50% identity and 67% similarity to human copine I over the entire sequence.
Analysis of BON1 expression by Northern blots shows that BON1 is expressed at low levels, consistent with its low representation among the population of expression sequence tags (ESTs). Organ-specific Northern blots show that expression of BON1 in the leaves and stems is higher than that in roots or flowers (see below), which could explain why leaves and stems are the most severely affected tissues in the bon1-1 mutant.
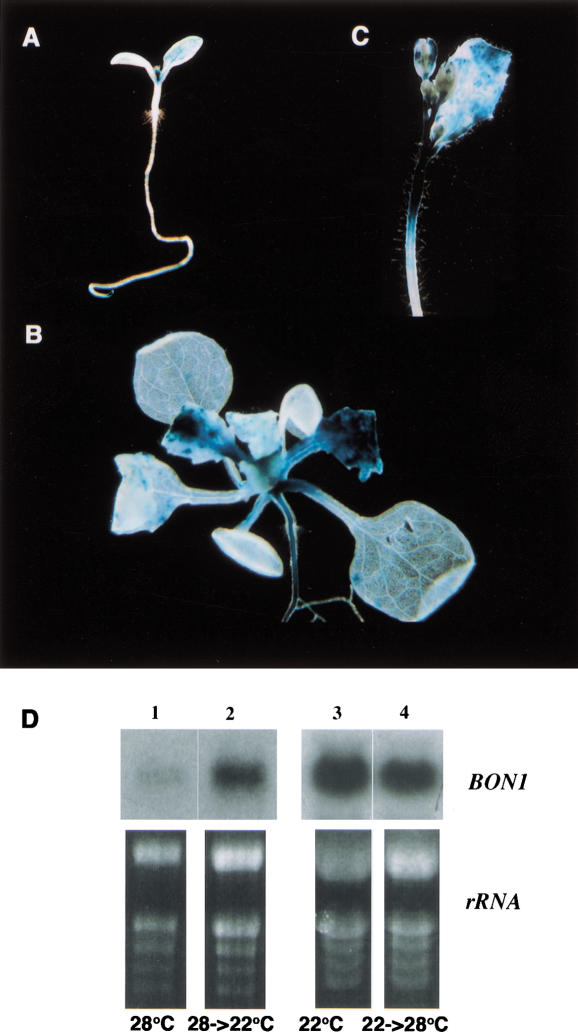
To further analyze the expression of BON1 at the tissue level, we made a translational fusion of BON1 with a beta-glucuronidase (GUS) reporter gene. GUS staining of the BON1–GUS transgenic plants shows that BON1 is more strongly expressed in growing tissues. It was expressed in younger leaves but not older ones (Fig. 3A,B). BON1 was also expressed in the inflorescence stems and was especially concentrated in the apical elongation portion of the stem (Fig. 3C). This expression pattern suggests a direct role of BON1 in regulating cell division and expansion.
Figure 3.
BON1 expression. (A–C) BON1-GUS expression pattern. Blue staining indicates the GUS expression. (A) Seedling right after germination. Expression is high in the first two leaves and the root tips. (B) Seedlings before bolting. Expression is seen in younger leaves not older ones. (C) Inflorescence stem. Expression is seen in the apical portion of the stem. (D) BON1 RNA expression modulated by temperature. RNA samples were isolated from 5-week-old plants, grown at 28°C (lane 1), and then shifted from 28°C to 22°C for 12 h (lane 2), grown at 22°C (lane 3) and then shifted from 22°C to 28°C for 12 h (lane 4).
The level of expression of BON1 RNA is regulated by temperature, especially during later development. We grew plants first at one temperature for one month and then moved half the population to another temperature for 12 h. The RNA level of BON1 increased approximately twofold when plants were shifted from 28°C to 22°C (Fig. 3D, lanes 1 and 2). A shift to lower temperature (16°C) gave about the same increase in the BON1 mRNA as the shift to 22°C. Conversely, there was a decrease in BON1 RNA level when plants were moved from 22°C to 28°C (Fig. 3D, lanes 3 and 4). In plants less than 2 wk old, BON1 had a relatively high expression level (as compared with older plants), and in these plants there was not much difference in the level of BON1 expression between those plants grown at 22°C and at 28°C.
BON1 is a phospholipid-binding protein associated with the plasma membrane
The availability of the cloned BON1 gene, coupled with the mutant phenotype, provides an avenue to determine the biological function of a copine gene family member. BON1 contains two C2 domains at the N terminus. C2 domains are Ca2+-dependent phospholipid-binding domains that confer calcium and/or phospholipid modulation on the activity of the associated domain (Kopka et al. 1998). In view of these considerations, we examined whether BON1 protein possesses phospholipid-binding activity in vitro.
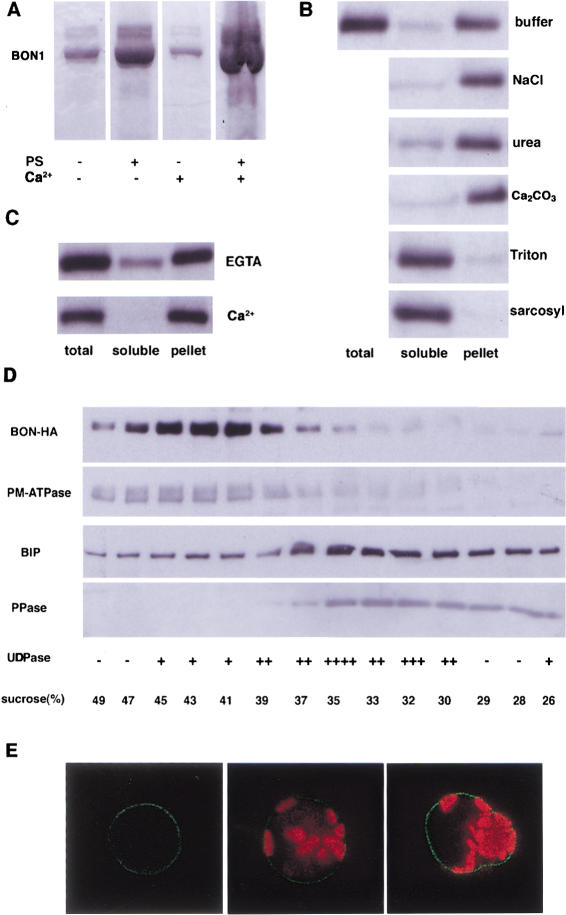
The binding assay utilized a tagged (6× His) BON1 protein that was expressed and purified from Escherichia coli. Purified recombinant BON1 protein was incubated with or without phospholipid and the mixture was pelleted by centrifugation. Phosphotidyl serine (PS) was used because C2 domains have been shown to bind to PS. The proteins associated with the lipids in the pellets were analyzed by electrophoresis on a gel. In the presence of lipid, three times more of BON1 protein was found in pellet (Fig. 4A), suggesting that BON1 was precipitated by its association with lipid. Addition of calcium to the protein lipid mixture yielded approximately six times more BON1 protein in the pellet (Fig. 6A, below). This could be due to an enhanced lipid-binding activity of BON1 by calcium or an increased lipid aggregation by calcium, which leads to more protein precipitation.
Figure 4.
Association of BON1 with membranes. (A) BON1 binds lipid. BON1 protein was mixed with (+) or without (−) phosphotidyl serine (PS) or calcium ion. The mixture was spun down and the protein in the pellet was run on SDS-PAGE and stained with Coomassie Blue. (B) BON1 is associated with membranes in the plants. Total proteins from BON1–HA plants were extracted and incubated with butter, buffer with 0.5 M NaCl, 2.5 M urea, 0.1 M Na2CO3, 1% Triton X-100, and 1% sacosyl, respectively, for 1 h. The soluble and pellet fractions were separated by ultracentrifugation. Total protein, soluble fractions, and pellet fractions were then separated by SDS-PAGE, blotted, and probed with anti-HA antibody. (C) Calcium stimulates the association of BON1 with membranes. Total proteins from BON1–HA plants were incubated with buffer containing EGTA (25 mM) or calcium ion (25 mM) before fractionation by centrifugation. (D) Subcellular fractionation by sucrose density centrifugation. The microsomal fraction from transgenic Arabidopsis containing BON1–HA was separated on a sucrose gradient of 25%–50%. Proteins from each fraction were separated on a 4%–20% SDS-PAGE gel and blotted to membranes. The blots were probed with anti-HA antibody and antibodies against marker proteins from different membranes: Plasma-membrane ATPase (PM-ATPase) for plasma membrane, BIP for endoplasmic reticulum, and pyrophosphatase (PPase) for vacoules. The activity of Golgi-specific UDPase activity was assayed (Schaller and DeWitt 1995) in each fraction and the strength of the activity was indicated from very strong (++++) to low (−). (E) BON1–GFP expression in Arabidopsis protoplasts. Leaf protoplasts were transformed with BON1–GFP fusion construct and the transient expression of GFP was observed under confocal microscopy. The green represents the GFP signal and the red represents chloroplast autofluorescence. The left image was with GFP signal alone and the right two images were overlays of both the GFP signal and the autofluorescence. GFP is mainly localized to the plasma membrane.
Figure 6.
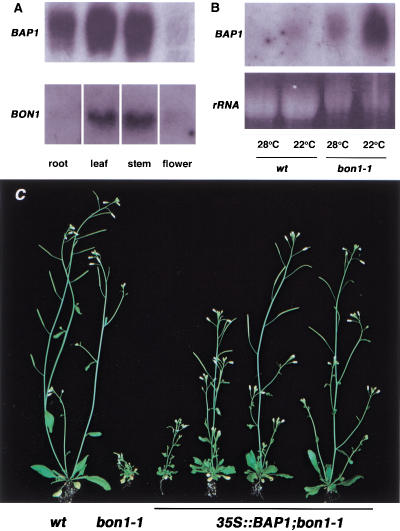
BAP1 expression and function. (A) Organ distribution of BAP1. Northern blot with total RNAs from different organs (root, leaf, stem, and flower). BON1 and BAP1 show higher expression in leaves and stems and BAP1 also has higher expression in roots. (B) BAP1 RNA expression in wild type and the bon1-1 mutant grown at 22°C and 28°C. (C) Suppression of the bon1-1 phenotype by BAP1 overexpression. Wild type and bon1-1 are on the left for comparison. Four independent transgenic lines of 35S::BAP1 in a bon1-1 background are on the right. The plants show varying degrees of suppression of bon1-1 phenotype by overexpression of BAP1.
As the association of BON1 with lipids in vitro and its sequence homology suggest a membrane localization, we tested whether BON1 is associated with membranes in vivo by determining the localization of BON1 using transgenic plants expressing a HA-tagged BON1 gene. This BON1-HA gene is functional because it was able to complement the bon1-1 mutant (data not shown). Total proteins were separated into microsomal fraction and soluble fraction by ultracentrifugation. BON1-HA was found mainly in the microsomal fraction, indicating its associates with membranes in the cell (Fig. 4B). Several treatments were used to analyze this association. NaCl, urea, or carbonate (pH 11.5) did not alter the distribution of the protein significantly (Fig. 4B). These treatments are capable of stripping peripheral proteins from membranes. Reagents that dissolve membranes such as Triton X-100 and sarcosyl redistributed the protein to the soluble fraction (Fig. 4B). These observations suggest that BON1 is tightly bound to membranes even though the amino acid sequence of the protein does not predict any membrane-spanning domains. Although the association of BON1 with membranes was stable in the presence of EGTA (Fig. 4C), the addition of calcium ions partitioned all the BON1 protein to the membrane fraction (Fig. 4C). This indicates that calcium enhances the association of BON1 with membranes although it is not absolutely required.
To identify the membrane system with which BON1 is associated, we carried out cell fractionation to separate different endomembranes and the plasma membrane. BON1-HA was not detected in protein extracts from chloroplast, nuclei, or cell walls (data not shown) but was present in the microsomal fraction. The microsomal fraction of BON1-HA plants was separated on a 25% to 50% (w/v) sucrose gradient. The relative position of various membranes on the gradient was determined by assaying fractions for marker proteins specific to each membrane. The BON1-HA protein exhibited a distribution similar to that of the plasma membrane ATPase (Fig. 4D) but distinct from ER, vacuole, and Golgi.
The plasma membrane localization of BON1 indicated by cell fractionation was further supported by analysis in a transient expression system. A BON1–GFP fusion under a strong promoter was electroporated into Arabidopsis protoplasts and the expression of GFP was monitored 8 h later with confocal microscope. The GFP signal was mostly concentrated on the outer membrane of protoplasts (Fig. 4F), indicating a plasma-membrane localization.
BON1 enhances vesicle aggregation in vitro
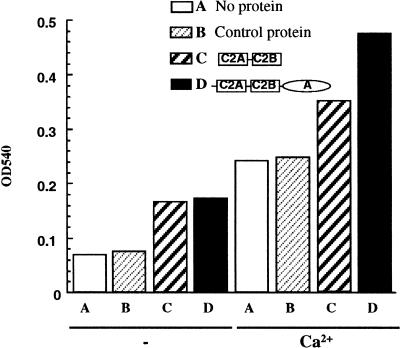
One mechanism by which BON1 could stimulate cell expansion and division would be to enhance membrane trafficking by facilitating the association and fusion of vesicles with the membrane. We tested the ability of BON1 to enhance vesicle aggregation in vitro using an assay that measures the turbidity of a lipid solution. Aggregation of the lipid vesicles is accompanied by an increase in turbidity (monitored by the absorbance at 540 nm). Constructs encoding the full-length BON1 protein, the N-terminal 2C2 domain, and the C-terminal A domain each with 6× His tags were expressed in E. coli and the resulting His-tagged proteins were purified. These recombinant proteins were incubated with PS or phosphotidyl inositol (PI). A difference in turbidity was observed with PS but not PI. Both the full-length BON1 protein and the 2C2 domain increased the absorbance of PS solution (Fig. 5), indicative of an activity that promotes vesicle aggregation. By contrast, the A domain of the BON1 protein had no effect on the turbidity (Fig. 5). As has been observed previously (Creutz et al. 1998), the addition of calcium alone stimulates the aggregation of vesicles. The addition of calcium in the presence of either the 2C2 domain or the full-length BON1 enhanced the aggregation considerably over the background stimulation observed in the presence of calcium (Fig. 5).
Figure 5.
Vesicle aggregation promoted by BON1 in vitro. Vesicle aggregation was monitored by the turbidity of the lipid. Phosphotidylserine was incubated with no protein (A), control protein (B), two C2 domains of BON1 (C), or full-length BON1 (D) in the presence (+) or absence (−) of calcium ion for half an hour and the turbidity of the lipid was measured by the absorbance at 540 nm.
BAP1 is a BON1 interacting protein
Our data indicates that BON1 may function to promote vesicle trafficking during cell expansion and division. It may do so via its interaction with other proteins; therefore, we looked for proteins that interact with BON1 using the yeast two-hybrid assay. We used the A domain (from Val 319 to Pro 578) as bait because it is the most likely segment of BON1 to be involved in protein–protein interactions (Creutz et al. 1998). A cDNA library made from the vegetative tissues of 3-week-old plants was screened for clones that signaled an interaction with this bait. Twenty-three positive clones rescued from the screen are the same gene, which we called the BON1 association protein 1 (BAP1).
BAP1 encodes a protein of 192 amino acids. The two-hybrid clone comprises the entire BAP1 except the first six amino acids. A BLAST search revealed that it has strong homology to another putative Arabidopsis protein in the database (we named this other protein BAL for BAP1-like). A SMART (Simple Modular Architecture Research Tool) search indicated that the N-terminal part of BAP1 (∼120 amino acids) has sequence homology and structural analogy to the C2 domain. The C2 domain family is very divergent (Rizo and Sudhof 1998), and the C2 domain of BAP1 is not significantly homologous to those of BON1 at the protein sequence level. The C-terminal 52 amino acids of BAP1 are 54% identical to those of BAL but do not show significant homology to any known motifs.
BAP1 has a similar function to BON1
BAP1 is expressed ubiquitously throughout the roots, leaves, stems, and flowers, with expression in leaves and stems relatively higher than in other parts of the plant (Fig. 6A). The expression of BAP1, like that of BON1, is modulated by temperature. There was an increase in BAP1 RNA when plants were shifted from a higher to lower temperature (Fig. 6B). Conversely, a higher temperature repressed BAP1 expression (Fig. 6B). Moreover, BAP1 expression is affected by BON1. In the bon1-1 mutant, there was more BAP1 transcript and the modulation of its expression by temperature was more pronounced than in wild type (Fig. 6B).
To identify whether BAP1 has a similar function to that of BON1, we expressed the BAP1 gene under the CaMV 35S promoter and introduced the transgene into the bon1-1 mutant. We reasoned that if the two proteins were involved in the same event, then overexpression of BAP1 might suppress the mutant defects of the bon1 mutant. Of 18 transgenic lines analyzed, 9 showed suppression to varying degrees, ranging from good (1) to moderate (3) to weak (5) (Fig. 6C). Those plants that were partially suppressed had more elongated stems and more expanded leaves than the bon1 mutant. Thus overexpression of BAP1 can suppress the defect in bon1, but the extent of suppression may be modified by the site of transgene integration or transgene silencing.
Discussion
BON1 and BAP1 maintain homeostasis in plant growth
Our studies have uncovered two genes whose function is required to maintain the size of Arabidopsis at relatively low temperature. Loss of BON1 function leads to miniature plants at ambient temperature due to a reduction in both cell expansion and division but has no obvious phenotype at high temperature. A second gene is a BON1 interacting protein, BAP1, whose overexpression partially suppresses the bon1 phenotype, establishing a functional connection between these two genes.
The bon1 phenotype differs from that of a typical cold-sensitive mutation in a vital gene. Null mutations in a vital gene would be lethal at all temperatures; whereas BON1 function and BON1 protein are required to maintain normal plant size at low temperature. Mutants with a conditional cold-sensitive phenotype similar to that of bon1 have been identified previously (Tsukaya et al. 1993; Akamatsu et al. 1999). These mutants (asculis1, asculis3, and asculis4) grow normally at 28°C and have reduced stem elongation and leaf expansion at the lower temperature. They could be involved in the same genetic pathway as BON1 or in a parallel pathway that maintains size at low temperature. However, such conclusions are premature because neither the identity of the genes nor the nature of the mutations responsible for these phenotypes are known.
Further support for the role of BON1 and BAP1 in homeostasis at low temperature comes from analysis of the transcriptional pattern of the two genes. First, BON1 is expressed more strongly in growing tissues than in mature tissues. Second, both BON1 and BAP1 expression are under temperature control: Both have elevated expression when plants were shifted from high to low temperature. Moreover, the enhancement of BAP1 expression in response to low temperature is more drastic in the bon1 background than in the wild type. These data suggest that BON1 and BAP1 are required for normal growth at lower temperature and that they are up-regulated under the conditions in which their function is required. There appears to be an additional compensatory stimulation of BAP1 expression when there is insufficient BON1.
The function of Arabidopsis BON1 and the copine family
Our data on BON1 provide important insights into the function of the copine gene family of which BON1 is a member. The copine family includes members from protozoa to humans, but, despite its evolutionary conservation, little is known about the function of its members. Previous work in paramecium and mice showed that copines have lipid-binding activity and promote the aggregation of vesicles (Creutz et al. 1998). It was suggested that the paramecium copine might be involved in vesicle trafficking because of its association with secretory vesicles (Creutz et al. 1998). In mice the N-copine expressed in the hippocampus is induced by kainate stimulation. The protein is detected in neurons, both in the cell bodies and dendrites (Nakayama et al. 1999). Kainate has been reported to stimulate long-term potentiation and it was therefore surmised that N-copine has a role in synaptic plasticity (Nakayama et al. 1998).
Our data indicate that the BON1 protein may regulate membrane biogenesis and cell wall remodeling through exocytosis, which ultimately controls cell size. All copines have a similar domain structure: The C2 N-terminal domain and a C-terminal A domain. Like the paramecium and mice copines, the full-length Arabidopsis BON1 protein and the truncated C2 domain bind phospholipids and the binding is stimulated by calcium. Furthermore, BON1 promotes aggregation of lipid vesicles in vitro. Therefore, all copines may have similar biochemical activities in addition to structural similarities. Previous work provided only a few hints as to the subcellular localization of copines. N-copine was localized to postsynaptic membranes where synaptic plasticity occurs (Nakayama et al. 1999) and one of the chromaffin granule-binding proteins (chromobindin 17) is likely to be a copine (Creutz et al. 1998). Our data argue that BON1 protein is mainly associated with the plasma membrane.
One model that takes account of all our data is that the copine BON1 and its interacting protein BAP1 function directly in fusion of vesicles with plasma membrane at low temperature—a process that would contribute both to membrane growth and wall expansion. Exocytosis is required for cell growth in many organisms and in plant cells a decrease in exocytosis has been shown to decrease cell size (Carroll et al. 1998). In other systems, the core machinery for vesicle docking and fusion has been identified (Jahn and Sudhof 1999). However, it is not well understood how the basic fusion machinery is regulated. As fusion is a temperature-dependent process (Weber et al. 1998), the copine/BAP1 proteins may be required to accelerate a process that occurs less rapidly at low temperature. Indeed, some of the mutations defective in membrane fusion are cold-sensitive in yeast (Lehman et al. 1999; Otte et al. 2001).
BON1 could function in temperature homeostasis either by acting catalytically (increasing the fusion of vesicles with the membrane) or structurally (by associating with the plasma membrane to maintain membrane function at low temperature). Recently, a protein kinase activity was attributed to the A domain of human copine III (Caudell et al. 2000), suggesting that these proteins have enzymatic function. Copines in other organisms may function similarly to accelerate vesicle trafficking in response to specific environmental signals. For example, the neuronal copines could accelerate membrane trafficking at the synapse upon chemical stimulation, thereby enhancing synaptic transmission.
A second possibility is that BON1 has a role in altering the phospholipid content of the membrane. Indeed, there is an Arabidopsis cold-sensitive mutant whose defect appears to result from a greater proportion of saturated fatty acids in the membrane (Lightner et al. 1994). However, the lipid composition of the bon1 mutant shows no significant difference from that of the wild type. As these experiments were performed on whole plants, we cannot rule out the possibility that the loss of BON1 function results in the subtle alteration of lipid composition in a specific membrane. An exact functional assignment for BON1 in Arabidopsis is complicated by the presence of homologs of the BON1 gene. Once mutations in these other BON genes are obtained, the phenotype of strains lacking the function of the entire copine family can be determined.
Materials and methods
Isolation and characterization of the bon1 mutant
Several mutant pools from the Arabidopsis Biological Resource Center were screened for leaf expansion mutants. Approximately 300 plants were grown per flat (26 cm × 52 cm) for phenotypic screening. bon1-1 was isolated from the activation tagging lines in Col background (Weigel et al. 2000).
For scanning microscopy, plant material was fixed overnight at 4°C in 3% glutaraldehyde in 25 mM phosphated buffer (pH 7.0), dehydrated through ethanol serials, critical-point dried, gold sputter-coated, and examined with a scanning electron microscope. Images were taken and the cell dimensions were analyzed using NIH imaging program.
Cloning of the BON1 gene
A 0.5-kb genomic fragment outside the left border of the T-DNA was rescued by using Universal GenomeWalker kit (Clontech). Sequence of this 0.5-kb fragment revealed its location on TAC clone K22G18. A 6.5-kb BamHI fragment containing the gene was subcloned from this TAC clone. Using the same 0.5-kb fragment, we isolated a cDNA from an Arabidopsis cDNA library CD4-14 obtained from the Arabidopsis Biological Resource Center.
Constructs
For complementation, a 6.5-kb BamHI genomic fragment of BON1 was cloned into pCGN1548 (McBride and Summerfelt 1990) and pCGN-NOS with 3′-NOS added in pCGN1548. For BON1–GFP fusion, full-length cDNA of this gene was cloned into a GFP expression vector (Chiu et al. 1996). For BON1–GUS fusion, a 5-kb BamHI/BglII fragment was cloned into PZP212 vector (Diener et al. 2000). For BON1–HA fusion, a BamHI site was introduced by PCR method to the genomic BON1 gene to replace the stop codon. The 3 × HA epitope was amplified and ligated into the BamHI site at the last codon of BON1 so that it is in frame with BON1. The BON1–HA fusion was then inserted into pCGN–NOS.
RNA and DNA analysis
Standard molecular techniques were used (Sambrook et al. 1989). Total RNA was prepared using Tri Reagent (Molecular Research Center).
Plant transformation
Plasmids were introduced into agrobacterium strain ASE (Fraley et al. 1985) or GV3101 (Koncz and Schell 1986) by electroporation, and transformed into wild-type Col-0, bon1-1/+ or bon1-1 by the floral dip method (Clough and Bent 1998).
GUS staining
Plant material was prefixed on ice in 90% acetone for 15 min, rinsed in staining buffer without X-Gluc (cyclohexylammonium salt), infiltrated with staining solution [50 mM NaPO4 at pH 7.2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 2 mM X-Gluc)] under vacuum for 15 min, and incubated at 37°C overnight. Materials were then dehydrated in an ethanol series.
Protein analysis
For total protein extraction, rosettes were homogenized in extraction buffer (12% sucrose, 100 mM Tris-HCl at pH 7.5, 1 mM EDTA, 2 mM DTT, and proteinase inhibitor cocktail from Calbiochem) with chilled mortar and pestle. The mixture was spun at 15,000 rpm in a microcentrifuge for 5 min to remove insoluble debris. To assay the effects of different reagents on the association of BON1 with membrane, supernatant was incubated for 1 h on ice with equal volume of buffer to make the final concentration of 0.5 M NaCl, 0.1 M Na2CO3, 2.5 M urea, 1% Triton X-100, and 1% sarcosyl, respectively. The soluble and pellet fractions were separated by centrifuging the supernatant at 100,000g for 1 h.
Western blot
Proteins were run on 4%–20% Ready-gel (Bio Rad) and transferred to PVDF membrane (NEN Life Science Products). Membranes were blocked with 5% nonfat dry milk in PBT (phosphate-buffered saline with 0.1% Tween 20), incubated with primary and then secondary antibodies in 2% milk, and detected with ECL (Amershan Pharmacia Biotech). HA antibody was purchased from BAbCO.
Membrane fractionation and analysis
The procedure was adapted from a method described previously (Schaller and DeWitt 1995). Briefly, 3-week-old leaves grown in liquid medium of MS salt, 2% sucrose, and B5 vitamins were homogenized in extraction buffer (50 mM Tris at pH 8, 2 mM EDTA, 20% glycerol, 1 mM DTT, and the proteinase inhibitor cocktail). Homogenate was filtered through Miracloth (Calbiochem) and centrifuged at 5000g for 5 min to remove organelles and debris. Supernatants were centrifuged at 100,000g for 1 h to pellet microsomal membranes. Membrane pellets were resuspended in buffer (25 mM Tris at pH 7.5, 10% sucrose, 2 mM EDTA, 1 mM DTT, and proteinase inhibitors) at 0.625 mL/10 g starting material. Microsomes (0.5 mL) were then layered onto a 15 mL sucrose gradient of 25%–50% and centrifuged at 100,000g for 16 h. Then 1-mL fractions were collected and analyzed. Fractions containing ER, vacuole, and PM were identified by Western blot using marker enzymes. Fractions containing Golgi were identified by enyzmatic analysis of Triton-stimulated UDPase.
Arabidopsis protoplast transient expression and confocal microscopy
Leaf mesophyll protoplasts were isolated from 3-week-old Arabidopsis and were transfected by a modified polyethylene glycol method as described previously (Abel and Theologis 1994). Ten thousand protoplasts were transfected with 10 μg of DNA and then incubated for 9 h at 23°C before subcelluar localization was determined with a confocal microscope. Confocal laser scanning micrographs of transfected protoplasts were recorded using a Leica microscope (Leica Microsystems) equipped with a laser scanning unit (TCS NT).
Protein expression in E. coli
Full-length 2C2 domain (M1 to G308) and A domain (V319 to P578) were amplified from BON1 cDNA and cloned into a vector pKS11 (under T7 promoter, with 6 × His tags at the N terminus). The constructs were transformed into BL21plyS (Novagen). Protein was induced with 1 mM IPTG for 10 h at 25°C. Proteins were purified with Ni-NTA agarose beads (QIAGEN) according to instructions.
Two-hybrid screen
DNA was prepared from the Arabidopsis thaliana MATCHMAKER cDNA library in E. coli (Clontech). The library complexity was 3 × 106. DNA was transformed into yeast strain PJ69-4α (James et al. 1996) and 2.5 × 107 yeast transformants were pooled as the library. The bait (A domain of BON1) was cloned between RI and BamHI sites of pGBD-C2 and transformed into PJ49-4a. The library α cells were mated with the bait a cells. A screening protocol (Robertson et al. 2000) was followed. Diploid cells (∼108) were plated onto SC Ade− and His− medium supplemented with 40 mM of 3-AT. About 80 clones retested positively were chosen for plasmid rescue.
Acknowledgments
We are grateful for the generosity and help of C. Benning and C. Xu in lipid analysis, G.E. Schaller and M. Randlett in cell fractionation experiment, and J. Sheen in GFP transient expression. We thank G.E. Schaller and R. Gaxiola for comments on the manuscript, N. Watson and Keck foundation for electron microscopy support, S. Johnson for assistance in protein domain searching, P. Rea for antibody to PPase, M. Sussman for antibody to PM-ATPase, and M. Chrispeels for antibody to BIP. We also thank Arabidopsis Biological Resources for the DNA and seed stocks. This work is supported by National Science Foundation Grant No. MCB-9974451 to G.R.F. and Jane Coffin Childs Memorial Fund to J.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
Sequence data described in this paper have been submitted to GenBank under accession nos. AY045764 and AY045765.
Footnotes
E-MAIL gfink@wi.mit.edu; FAX (617) 258-9872.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.918101.
References
- Abel S, Theologis A. Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 1994;5:421–427. doi: 10.1111/j.1365-313x.1994.00421.x. [DOI] [PubMed] [Google Scholar]
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda Y. Expression of endoxyloglucan transferase genes inacaulis mutants of Arabidopsis. Plant Physiol. 1999;121:715–722. doi: 10.1104/pp.121.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AD, Moyen C, Van Kesteren P, Tooke F, Battey NH, Brownlee C. Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. Plant Cell. 1998;108:1267–1276. doi: 10.1105/tpc.10.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudell EG, Caudell JJ, Tang CH, Yu TK, Frederick MJ, Grimm EA. Characterization of human copine III as a phosphoprotein with associated kinase activity. Biochemistry. 2000;39:13034–13043. doi: 10.1021/bi001250v. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, Beisson J, Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell. 2000;12:853–870. doi: 10.1105/tpc.12.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick CL, Hoffman NL, Sanders PR. The sev system—A new disarmed TI-plasmid vector system for plant transformation. Biotechnology. 1985;3:629–635. [Google Scholar]
- Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Kopka J, Pical C, Hetherington AM, Muller-Rober B. Ca2+/phospholipid-binding (C2) domain in multiple plant proteins: Novel components of the calcium-sensing apparatus. Plant Mol Biol. 1998;36:627–637. doi: 10.1023/a:1005915020760. [DOI] [PubMed] [Google Scholar]
- Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner J, James DW, Jr, Dooner HK, Browes J. Altered body morphology is caused by increased stearate levels in a mutant of Arabidopsis. Plant J. 1994;6:401–412. [Google Scholar]
- McBride KE, Summerfelt KR. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Yaoi T, Yasui M, Kuwajima G. N-copine: A novel two C2-domain-containing protein with neuronal activity-regulated expression. FEBS Lett. 1998;428:80–84. doi: 10.1016/s0014-5793(98)00497-9. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Yaoi T, Kuwajima G. Localization and subcellular distribution of N-copine in mouse brain. J Neurochem. 1999;72:373–379. doi: 10.1046/j.1471-4159.1999.0720373.x. [DOI] [PubMed] [Google Scholar]
- Oldham S, Bohni R, Stocker H, Brogiolo W, Hafen E. Genetic control of size in Drosophila. Philos Trans R Soc Lond B Biol Sci. 2000;355:945–952. doi: 10.1098/rstb.2000.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S, Belden WJ, Heidtman M, Liu J, Jensen ON, Barlowe C. Erv41p and Erv46p. New components of COPII vesicles involved in transport between the ER and Golgi complex. J Cell Biol. 2001;152:503–518. doi: 10.1083/jcb.152.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Robertson L S, Causton HC, Young RA, Fink GR. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc Natl Acad Sci. 2000;97:5984–5988. doi: 10.1073/pnas.100113397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE, DeWitt ND. Analysis of the H(+)-ATPase and other proteins of the Arabidopsis plasma membrane. Meth Cell Biol. 1995;50:129–148. [PubMed] [Google Scholar]
- Steponkus PL. Role of plasma membrane in freeezing injury and cold acclimation. Annu Rev Plant Physiol. 1984;35:543–584. [Google Scholar]
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Naito S, Redei GP, Komeda Y. A new class of mutants in Arabidopsis thaliana, acaulis1, affecting the development of both inflorescences and leaves. Development. 1993;118:751–764. [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R. Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;111:1011–1019. doi: 10.1104/pp.111.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1014. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]