Abstract
The specificity of ubiquitin-mediated protein degradation with regards to the selection of substrates to be polyubiquitinated has only been determined rather recently. Substrate targeting by the N-end rule and HECT (homology to E6AP carboxyl terminus) domain ubiquitin ligases occurs through substrate-specific binding domains. In contrast, the SCF complex recruits substrates through a substrate adaptor protein, the F-box subunit. Despite evidence showing that Cdc20 and Cdh1 bind and activate the anaphase-promoting complex (APC) in a substrate-specific manner, there is no evidence that the activating protein and substrate interact directly; hence, no clear model exists for the mechanism of APC activation or recruitment of substrates. We show here that the activators Cdc20 and Cdh1 can associate with substrates via their N termini. In the absence of APC, Cdc20 and Cdh1 bind substrates reflecting Cdc20–APC and Cdh1–APC specificity. The N termini of Cdc20 and Cdh1 provide specificity functionally, as demonstrated by the generation of active chimeras that display the specificity corresponding to their N termini. Thus, Cdc20 and Cdh1 act as both substrate recognition and activating modules for APC.
Keywords: APC, Cdc20, Cdh1, substrate, binding
Ubiquitin-mediated protein degradation regulates important cellular processes from cell cycle progression to Wnt signaling (Peters 1998; for reviews, see Zachariae and Nasmyth 1999; Jackson et al. 2000). Attachment of a polyubiquitin chain to a protein targets that protein for degradation by the 26S proteasome (Coux et al. 1996; Baumeister et al. 1998). Ubiquitin becomes covalently attached to a substrate by interaction with a cascade of enzymes. The last in the series, a ubiquitin protein ligase (E3), transfers the ubiquitin to a lysine on the substrate protein (for review, see Hershko and Ciechanover 1998).
In the N-end rule pathway, extending from bacteria to humans, a destabilizing residue at the N terminus directs proteins for ubiquitination via the E3 N-recognin/Ubr1p (Saccharomyces cerevisiae) or E3α (in mammals). Direct binding of substrates to N-recognin/E3α is observed (Reiss et al. 1988; Gonda et al. 1989; Bartel et al. 1990; for review, see Varshavsky 1996). Similarly, HECT (homology to E6AP carboxyl terminus) domain E3s are highly conserved from yeast to vertebrates. They share homology in their C termini to E6AP and contain a conserved C-terminal cysteine residue that forms a thioester with ubiquitin; they bind substrates via their N termini (for review, see Jackson et al. 2000). E6AP targets the src kinases (Oda et al. 1999), HHR23A and HHR23B (Kumar et al. 1999). The HECT domain RSP5 in yeast targets Gap1 and Fur4 by interaction with its N-terminal WW domains (Hein et al. 1995; Rotin 1998; Wang et al. 1999). The fission yeast pub1 targets Cdc25 (Nefsky and Beach 1996). Xsmurf1 binds and targets Smad1 for degradation (Zhu et al. 1999).
Two ubiquitin protein ligases play a role in the cell cycle: the SCF (Skp1/Cullin/F-box) complex and the anaphase-promoting complex (APC) (Peters 1998; for reviews, see Zachariae and Nasmyth 1999; Jackson et al. 2000). Both complexes contain a protein with cullin homology (Cul1/Cdc53 in the SCF complex, APC2 in the APC) (Yu et al. 1998; Zachariae et al. 1998; Seol et al. 1999) and a RING-H2 finger protein, (Rbx1 in the SCF complex and APC11 in the APC) (Zachariae et al. 1998; Kamura et al. 1999; Skowyra et al. 1999; Gmachl et al. 2000; for review, see Tyers and Jorgensen 2000). Recruitment of substrates to the SCF complex occurs via the F-box subunit. F-box proteins interact with the Skp1 subunit of the SCF complex via the F-box motif (Bai et al. 1996). F-box proteins fall into three categories, those with WD repeats (Fbw), leucine-rich repeats (Fbl), or other domains (Fbx) (for review, see Jackson et al. 2000). Binding to substrates requires substrate phosphorylation and is thought to occur in the non-F-box domains (for review, see Jackson et al. 2000). Substrate binding has been shown to occur for Cdc4 via its WD domain (Skowyra et al. 1997), for Grr1 via its leucine rich repeats (Kishi and Yamao 1998; Hsiung et al. 2001), and for Skp2 via its other domain (Yeh et al. 2001). In some cases, however, the F-box protein may not be sufficient for substrate interaction; homo- or heterodimerization of the F-box or another factor may be required (for review, see Jackson et al. 2000).
The APC contains several additional subunits without a defined role; substrate interaction could occur through one of these proteins. APC activation requires its binding to the WD repeat containing proteins Cdc20 or Cdh1. The Cdc20- and Cdh1-bound forms of the APC demonstrate different substrate specificities (Fang et al. 1998a; Pfleger and Kirschner 2000). Cdc20–APC recognizes destruction box (D box) containing proteins (Glotzer et al. 1991; King et al. 1996; Fang et al. 1998a), and Cdh1–APC recognizes proteins containing either a D box or a KEN box (Pfleger and Kirschner 2000). No direct binding of activating protein to substrate has been demonstrated. In one case, the APC substrate Hsl1p, a Swe1p inhibitor, was identified through a two-hybrid interaction screen with Cdc20, but without addressing direct interaction (Burton and Solomon 2000). In another case, the putative substrate cyclin D was shown to bind to other APC subunits by αCdc27 immuno-precipitation analysis (Agami and Bernards 2000). More recently, the interaction between cyclin A–Cdk2 with the WD repeats of Cdc20 (Ohtoshi et al. 2000) or Cdh1 (Sorensen et al. 2001) has been reported for the recognition of Cdc20 and Cdh1 as substrates of cyclin A–Cdk2. Recruitment of substrate to the APC might occur through activating protein, through binding to one of the other subunits of APC, or through a combination of both.
In this report we present evidence that Cdc20 and Cdh1, the substrate-specific adaptors of the APC, interact with substrates in an APC-independent manner. We show that this interaction occurs through their N termini and does not require the WD repeats. We show that this binding does not require APC and can occur directly between purified proteins. Specificity is retained by the N terminus, as demonstrated by active chimeras between Cdc20 and Cdh1 when their N termini and WD repeats are swapped. N-terminal fragments of the activators are potent APC inhibitors both in vitro and in vivo, and we introduce a binding assay that could be used to identify additional APC targets.
Results
APC substrate-specific adaptors Cdc20 and Cdh1 specifically bind substrates
Cdc20–APC directs the ubiquitination of several D-box substrates in mitosis including the chromokinesin, Xkid (Funabiki and Murray 2000), the inhibitor of sister chromatid separation, securin (Holloway et al. 1993; Cohen-fix et al. 1996; Funabiki et al. 1996; Yamamoto et al. 1996; Zou et al. 1999), cyclin B (cyc B) (Glotzer et al. 1991), and an inhibitor of DNA replication, geminin (McGarry and Kirschner 1998). Cdh1–APC recognizes these four substrates at the end of mitosis and during G1, as well as the vertebrate NIMA related kinase Nek2, the p53-inducible KEN-box substrate mB99 (Utrera et al. 1998; Pfleger and Kirschner 2000), and at least four other KEN box-containing substrates (C. Pfleger and M.Kirschner, unpubl.). Information regarding the mechanism of interaction between APC and these substrates has been lacking.
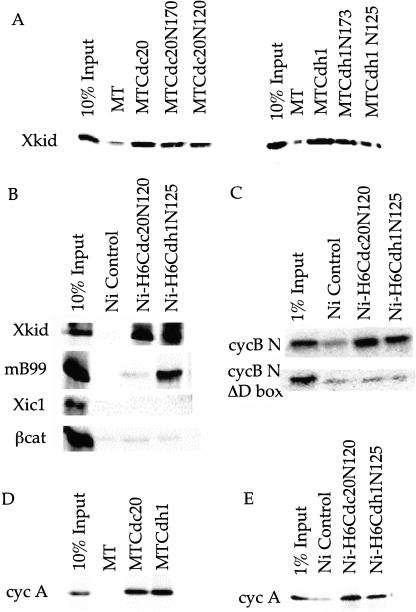
To investigate the interaction between APC and its substrates, we incubated cold in vitro translated myc-tagged hCdc20 or hCdh1 bound to α-myc coated beads with 35S-labeled in vitro translated Xkid, xsecurin, geminin, and mB99. Myc-tag alone (MT) does not bind any of these substrates. Myc-tagged Cdh1 (MTCdh1) binds Xkid, xsecurin, geminin, and mB99 (Fig. 1A), which contain D boxes or KEN boxes, to ∼10% of input. Myc-tagged hCdc20 (MTCdc20) binds the D box-containing substrates Xkid, xsecurin, and geminin (Fig. 1A), but not mB99 (Fig. 1A). Neither MTCdc20 nor MTCdh1 bind the SCF substrates Xic1 or β-cat (Fig. 1A) or over a dozen other non-APC substrates in this assay (data not shown). The failure of MTCdc20 or MTCdh1 to bind non-APC substrates, and the difference in binding of mB99 between MTCdc20 and MTCdh1 accurately reflects the differences in substrate specificity between Cdc20- and Cdh1-activated APC ubiquitin ligase activity.
Figure 1.
Myc-tagged full-length APC activators recover APC substrates with specificity in binding. (A) Cold in vitro translated MT, MTCdc20, and MTCdh1 bound to αmyc coated beads and incubated with 35S-labeled in vitro translated APC substrates Xkid, securin, geminin, and mB99, and the non-APC substrates β-cat and Xic1. The left-most lane of each panel indicates 10% of the in vitro translation material supplied in the binding assay. (B) Binding of MTCdc20 and MTCdh1 to αmyc beads, as above, incubated with 35S-labeled in vitro translated full-length Xenopus cyc B (top), xCdc2 (middle), or Xenopus cyc B cotranslated with xCdc2 (bottom). The left-most panel reflects 10% of the material supplied in each assay. (C) Binding of MTCdc20 and MTCdh1 to anti-myc beads, as above, incubated with 35S-labeled in vitro translated cyc B-N, cyc B-N ΔDbox, Cdc20N105, and Cdc20N90.
The facile success of recapitulating APC substrate specificity in a pull-down assay contrasts the lack of such reports, including our own previous difficulties. As discussed throughout this paper, this may be attributable to nonideal behavior of the cyc B substrate used in previous experiments. Extension to other D-box and KEN-box substrates resolves many of the problems. We repeated the pull-down assays in Figure 1A using full-length cyc B. In repeated experiments, we brought down little to no cyc B with MTCdc20, and varying levels of cyc B, much less than 10% (Fig. 1B, upper panel), with MTCdh1 where we saw very clear results with Xkid and B99. We therefore asked whether coexpression of full-length cyc B with the partner CDK would stimulate binding to Cdc20. As shown in Figure 1B (middle panel), Xenopus Cdc2 (xCdc2) by itself does not bind. However, cotranslation of cyc B with xCdc2 results in a dramatic increase in the binding of cyc B by both MTCdc20 and MTCdh1 (Fig. 1B, lower panel) despite the lack of binding of xCdc2. This may suggest an indirect effect of xCdc2 (through phosphorylation of cyc B or other induced conformational changes). Alternatively, Cdc20 and Cdh1 may displace cyc B from a cycB/Cdc2 complex.
Because we cannot rule out that the initial binding between cyc B and Cdc20 and Cdh1 occurs through an active cyc B/Cdc2 complex recognizing Cdc20 and Cdh1 as substrates, we repeated this assay using the N-terminal peptide of cyc B (cyc B-N). This fragment contains the D box, but lacks other functional domains of cyc B. The binding of this N-terminal peptide is much weaker than Xkid or mB99, binding to ∼1% of the input signal, but binding is observed to MTCdc20 and MTCdh1, but not to MT alone (Fig. 1C). In addition, Cdh1 binds the N-terminal 105 amino acids of Cdc20 (Cdc20N105) which contains the KEN box and is recognized by Cdh1 (Pfleger and Kirschner 2000) to ∼1%, whereas MTCdc20 and MT alone do not (Fig. 1C). The interaction between MTCdc20 and MTCdh1 and cyc B-N or Cdc20N105 relies on an intact D box or KEN box; a D box-deleted fragment of cyc B-N (cyc B-N ΔD box) and the N-terminal 90 amino acids of Cdc20 (Cdc20N90) lacking the KEN box do not bind in the same assay (Fig. 1C). Larger fragments of Cdc20 were not tested in this assay because of the possible problems of interpretation given the role of Cdc20 as an activator of APC. For example, we observed the ability of MTCdc20 and MTCdh1 to bind both full-length Cdc20 and Cdh1 in the pull-down assay (data not shown). Although both cyc B-N or Cdc20N105 and full-length cyc B or Cdc20 are all substrates for APC-mediated degradation, the N-terminal peptides show much shorter half-lives. We speculate that perhaps the weaker binding allows more rapid activator-substrate dissociation followed by more rapid degradation.
The N-terminal domains of Cdh1 and Cdc20 bind substrates
To identify the domains of Cdh1 and Cdc20 responsible for substrate binding, we repeated the substrate-binding assay using myc-tagged deletion mutants of hCdh1 and hCdc20. The N-terminal 170 amino acids (MTCdc20N170) and 120 amino acids of Cdc20 (MTCdc20N120) bind similar amounts of Xkid, as does full-length MTCdc20 (Fig. 2A); the N-terminal 173 amino acids (MTCdh1N173) and 125 amino acids of Cdh1 (MTCdh1N125) also bind similar amounts of Xkid (Fig. 2A) or mB99 (data not shown) as full-length MTCdh1. We also tested whether the C-terminal WD repeats of Cdc20 and Cdh1 interact with substrates. The isolated WD repeats of both Cdc20 (Cdc20Δ170) and Cdh1 (Cdh1Δ173) bind at least 10%–20% of substrate input in the binding assay, but also bind similar levels of other non-APC substrates (data not shown), indicating that this binding may be nonspecific. The self-association of Cdc20 and Cdh1 noted earlier requires the WD repeats of each protein (data not shown) and may reflect this nonspecific binding by the WD repeat region. Alternatively, self-association may play a role in activating APC. The stoichiometry of activator binding to APC has not been reported, and we have not determined if this homo- and hetero-dimerization is functionally required.
Figure 2.
N-terminal activator binds substrate with specificity. (A) Binding assays as above, using myc-tagged deletion mutants of Cdh1 and Cdc20. Myc-tagged fragments containing the N-terminal 120 amino acids of Cdc20 and the N-terminal 125 amino acids of Cdh1 were sufficient for binding of Xkid. (B) Bacterially expressed his-tagged N-terminal hCdc20 bound to nickel beads (Ni-H6Cdc20N120) binds Xkid, but not mB99. Bacterially expressed his-tagged N-terminal hCdh1 bound to nickel beads (Ni-H6Cdh1N125) binds both Xkid and mB99. Neither Ni-H6Cdc20N120 nor Ni-H6Cdh1N125 binds β-cat or Xic1. (C) Both N-terminal bacterially expressed hCdc20 and hCdh1 bind the N-terminal fragment of Xenopus cyclin B containing the D box, but fail to recover the D-box deleted N-terminal cyclin B. (D) In vitro-translated cyc A binds MTCdc20 and MTCdh1, but not to MT alone. Ten percent of input is shown in the left-most lane. (E) Bacterially expressed his-tagged N-terminal hCdc20 or N-terminal hCdh1 bind ∼1–5% of in vitro translated cyc A.
Bacterially expressed his-tagged N-terminal hCdc20 (H6Cdc20N120) and hCdh1 (H6Cdh1N125) bind substrates to a similar extent as the in vitro translated myc-tagged versions. Nickel-bound H6Cdc20N120 or H6Cdh1N125 binds significant in vitro translated Xkid as compared to a nickel bead control (Fig. 2B). Only nickel-bound H6hCdh1N125 binds significant amounts of the KEN-box substrate mB99. Neither H6Cdc20N120 nor H6Cdh1N125 bind the non-APC substrates Xic1 or β-cat (Fig. 2B) or several other non-APC substrates incubated in the nickel pull-down assay (data not shown). When incubated with the in vitro translation products of 10 pools of ∼30 clones each from a Xenopus dorsalized gastrula cDNA library, only one of the >300 proteins bound to H6hCdc20N120 and H6Cdh1N125. On sequencing, this was cyc B, indicating that these recombinant proteins recapitulate full-length specificity, rather than nonspecific interactions. As with in vitro translated MTCdc20 and MTCdh1, binding of cyc B-N is weak, but H6Cdc20N120 and H6Cdh1N125 bind detectable levels (typically between 1% and 5%) compared to a nickel bead control (Fig. 2C). This binding depends on an intact D box; cyc B-N ΔD box does not bind (Fig. 2C).
Recent reports showing pull-down assays indicate that cyclin A (cyc A) binds Cdc20 in its WD repeats (Ohtoshi et al. 2000) and Cdh1 via a cyclin-binding motif, R-X-L in the C terminus of Cdh1 (Sorensen et al. 2001). Alanine substitution of this cyclin-binding motif reduces the targeting of cyc A by Cdh1, but has no effect on the targeting of other substrates, consistent with the hypothesis that this interaction is not a general Cdh1-substrate interaction site, but specific for interaction with cyc A. In our binding assays, we see ∼30%–40% of cyc A input binding to full-length MTCdc20 and MTCdh1 (Fig. 2D). Binding to the N-terminal recombinant proteins is drastically reduced (to ∼1%) (Fig. 2E). This reduction could reflect the weaker binding seen for cyc B-N, or could indicate that cyc A binds Cdc20 and Cdh1 primarily through C-terminal regions attributable to recognizing the activators as substrates. Given the strong C-terminal binding, a strict requirement for D box recognition in the N terminus of cyc A may be unnecessary.
The N-terminal domains of Cdc20 and Cdh1 functionally inhibit APC activity in vitro and in vivo
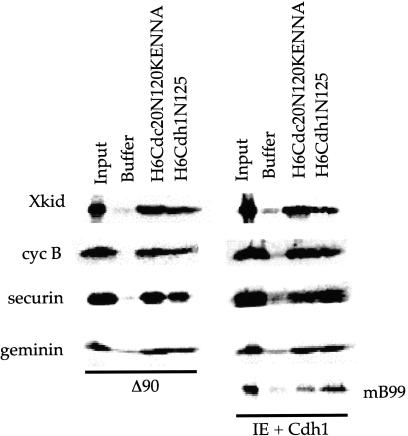
We would expect that if the N termini of Cdc20 and Cdh1 bind substrates, they should inhibit degradation. In other words, the Cdh1 N terminus should inhibit Cdh1–APC and Cdc20–APC activity towards D-box substrates and Cdh1–APC activity towards KEN-box substrates, whereas Cdc20–APC should inhibit APC activity towards only D-box substrates. Because of the internal KEN sequence in the Cdc20 N terminus, we used a KEN mutant peptide (H6Cdc20N120KENNA) that does not significantly compete for full-length Cdc20 degradation in Cdh1 extracts in the range of 5–10 μM (Pfleger and Kirschner 2000). Both the N termini of Cdc20 and Cdh1 block the degradation of D-box substrates Xkid, cyc B, securin, and geminin in mitotic extracts (Δ90) and Cdh1-supplemented extracts (Fig. 3). Only the N terminus of Cdh1 robustly blocks the degradation of the KEN-box substrate mB99, although a stabilizing effect is seen after the addition of N-terminal Cdc20 KEN mutant. Neither N-terminal fragment inhibits the dimerization of full-length activators in the binding assay (data not shown), and separate experiments in Figure 5 (see below) show that neither N-terminal fragment binds to APC; hence, they most likely cause inhibition by substrate sequestration. The stabilizing effect of Cdc20 and Cdh1 N-terminal peptides in extracts is seen for shorter fragments of 90 or 70 amino acid lengths, respectively (data not shown).
Figure 3.
N-terminal hCdc20 and hCdh1 inhibit APC in vitro. (A) In vitro degradation assay using Δ90 extracts or Cdh1 extracts of Xkid, cyc B, geminin, securin, and mB99 (Cdh1 extracts only) in the presence of buffer or 5 μM bacterially expressed H6Cdc20N120KENNA or H6Cdh1N125. Initial and 90 min time points are shown. Although we observe variability in half-lives between extracts, careful titration and kinetic analysis shows that half-lives are extended to ⋝60 min in the presence of 5 μM amounts of inhibitory N-terminal fragments.
Figure 5.
Cdc20 and Cdh1 N-terminal inhibitors do not bind APC. (A) Full-length hCdc20 and hCdh1 bind to iAPC immuno-purified from Xenopus extracts on αCdc27-coated beads, as do the first 170 amino acids of hCdc20 and 173 amino acids of hCdh1, however APC bound to αCdc27-coated beads fails to bind the N-terminal 120 amino acids of hCdc20 or 125 amino acids of hCdh1 above control beads. (B) Binding of full-length hCdc20 and hCdh1 to iAPC on beads in the presence of 5 or 10 μM N-terminal peptides of hCdc20 and hCdh1.
Xenopus embryos do not begin their own transcription until the mid-blastula transition (MBT); they rely on maternal messages for protein synthesis prior to MBT. Injection of RNA into embryos allows for expression of injected messages almost immediately. Injection of full-length MTCdc20 or MTCdh1 RNA into one cell of 2-cell Xenopus embryos had no effect on early divisions; however, injection of MTCdc20N120 or MTCdh1N125 RNA caused an arrest in the injected half of the embryo after one or two divisions. The cells remained large and ceased to divide whereas cells on the uninjected half continued to divide (Fig. 4). Propidium iodide staining of these embryos showed that cells on the injected side had extremely large nuclei that stained very intensely, reminiscent of the postMBT cells of embryos injected with protein of the Cdh1 inhibitor, MAD2L2 (Pfleger et al. 2001). Given the proportionality of propidium iodide staining and the size and intensity of the nuclear staining, these nuclei likely represent many rounds of DNA replication. This phenotype is consistent with results from other systems. Work in yeast has shown a requirement for APC activity in G1 to prevent uncontrolled entry into S phase (Irniger and Nasmyth 1997). Inhibition of the proteasome in sea urchin embryos results in overreplication and prevention of entry into mitosis (Kawahara et al. 2000). Inhibition of APC in cultured cells with a Cdh1 antibody leads to early entry into S phase and increased incorporation of BrdU, theoretically by accumulation of an unknown target protein involved in the initiation of DNA replication (Sorensen et al. 2000).
Figure 4.
Injection of RNAs encoding either N-terminal Cdc20 or Cdh1 block the early Xenopus embryonic divisions. Embryos at the 2-cell stage were injected in one cell with RNAs (500 pg) encoding either (A) MTCdc20, (B) MTCdc20N120, (C) MTCdh1, (D) MTCdh1N125, or (E) H20 and fixed at stage 6.5. Injection of MTCdh1N125 RNA results in cells that have enhanced nuclear staining. Embryos injected with MTCdhN125 RNAs (D) were fixed at stage 6.5 and stained with propidium iodide. Uninjected side (F) and injected side (G) of the same embryo shown at the same magnification. The cells on the injected side have much larger, more intensely staining nuclei (G) than the cells on the uninjected side (F).
Cdc20 and Cdh1 substrate binding does not require APC
Using an APC-binding assay (Fang et al. 1998a) in which 35S-labeled in vitro translated proteins are incubated with APC bound to αCdc27-coated beads, we tested the ability of hCdc20 and hCdh1 and various deletion mutants to interact with APC (Fig. 5A). APC beads bind substantial amounts of full-length hCdc20 or hCdh1 compared to a bead control. Similarly, the N-terminal regions extending to amino acid 170 of Cdc20 (Cdc20N170) or 173 of Cdh1 (Cdh1N173) bind to APC beads above control levels. Alanine substitution of possible Cdk1 phosphorylation sites in the N terminus of Cdh1 removes the ability of phosphorylation to cause dissociation of Cdh1 from the APC; furthermore, aspartate substitution of these residues blocks activation of APC by Cdh1 (Kramer et al. 2000), consistent with the N terminus contributing to the binding of Cdh1 to APC. In addition, recent work demonstrated the ability of two peptides consisting of amino acids 122–145 or from 166–187 of hCdc20 to inhibit the binding of full-length Cdc20 to APC (Zhang and Lees 2001), indicating a role for that region in binding, although their studies did not show binding of N-terminal fragments containing only this region to APC. Despite detectable binding of the N-terminal half of Cdc20 and Cdh1 to APC, Cdc20N120 and Cdh1N125 fail to bind APC under the same conditions (Fig. 5A). The isolated WD repeats of both hCdc20 and hCdh1 bind APC (data not shown), but we do not know if this binding represents a specific interaction, or whether it is nonspecific, as we saw for substrate binding.
Truncation of 50 amino acids of Cdc20 (from 170 to 120) or 48 amino acids from Cdh1 (from 173 to 125), retains substrate binding but loses the ability to bind APC. To test whether the truncated molecules merely have weaker binding, we asked whether the truncated regions would inhibit APC binding at higher concentrations by competition. Even at concentrations twice that at which we see complete stabilization of APC substrates in extracts, we see little to no effect on binding of full-length Cdc20 and Cdh1 (Fig. 5B). Furthermore, we see no interaction by gel filtration. In vitro translated MTCdc20N120 and MTCdh1N125 fractionate in a peak corresponding to protein of monomeric size both on a superdex 200 and on a superdex 75 column, far removed from fractions containing the APC (data not shown). In contrast, full-length in vitro translated Cdc20 and Cdh1 show two distinct peaks by gel filtration (superdex 200); one peak corresponds to a high molecular weight complex consistent with the migration of rabbit APC (from the reticulocyte lysate), and another peak corresponds to a size consistent with either monomer or dimer (data not shown). Lack of direct interaction between the substrate-binding domain of Cdc20 or Cdh1 and the APC suggests that the core APC complex from reticulocyte lysate is not required for the interaction of Cdc20 and Cdh1 with substrates, and that the in vitro and in vivo inhibition of APC by the N-terminal peptides does not result from interfering with productive binding of full-length activators and APC.
The interaction between substrate and activator is direct
We asked whether the activators bind substrate directly or whether they require another protein to mediate the interaction. It would be desirable to determine whether Cdc20 or Cdh1 directly interact with substrates in completely purified systems. Initially, we had difficulty expressing well-behaved versions of the substrates and the activators in sufficient quantities to allow for direct binding experiments. In earlier experiments with bacterially expressed cyc B-N (and its D-box mutant control) and Cdh1 and Cdc20 (data not shown), we detected no specific binding, and this thwarted our efforts to demonstrate the interactions. In our reinvestigation of this problem we found that the N-terminal cyc B peptide aggregated into complexes that migrated as a large complex (at least 80 kD or more) leaving a trailing peak to a monomeric size on gel filtration (data not shown). Aggregation was also observed when the N-terminal domain of cyc B was expressed in reticulocyte lysate and subjected to gel filtration (data not shown). Although it is an excellent and specific substrate for APC, the aggregation of cyc B-N makes direct binding experiments difficult to interpret.
The lack of success with bacterially expressed and purified protein in contrast with in vitro translated protein could be explained by the fact that: (1) interaction between activator and substrate requires another factor; (2) the bacterial proteins lack some crucial modification or are folded improperly; or (3) the already weak cyc B-N interaction is too weak to detect conclusively with partially misfolded bacterial protein. The major techniques to address direct interaction, which include equilibrium dialysis, analytical ultracentrifugation, differential spectroscopy, or isothermal titrating calorimetry, require large amounts of monodisperse protein species.
Even though the N-terminal domains of Cdc20 and Cdh1 do not detectably interact with APC, we wished to rule out any role of APC (including a transient role) in the process of substrate binding. We depleted APC from reticulocyte lysate using αCdc27-coated beads before in vitro translating myc-tagged cold activator and 35S-labeled substrate. Substrate binding was completely unaffected (data not shown).
To eliminate other large molecules that may play a role in substrate binding, we further purified and characterized three substrates made by in vitro translation. We fractionated 35S-labeled in vitro translated Xkid, geminin, and xsecurin on a superdex 200 gel filtration column. Unlike cyc B-N, these substrates showed no aggregation; each fractionated as a distinct peak corresponding to a monomer (or at most a dimer) molecular mass (data not shown). For example, xsecurin migrated at an apparent molecular mass of ∼20–30 kD. When we incubated the peak fractions of Xkid, geminin, or securin with nickel-bound H6Cdc20N120 or H6Cdh1N125, all three bound to beads containing either H6Cdc20N120 or H6Cdh1N125, but did not bind to a nickel bead control (Fig. 6A). These experiments rule out the role of even catalytic amounts of APC and make it unlikely that any large molecular components carried over from the reticulocyte extracts participated in the binding interaction.
Figure 6.
N-terminal hCdc20 and hCdh1 bind substrates directly. (A) 35S-labeled in vitro translated Xkid, geminin, and securin, each isolated by gel filtration over a Superdex 200 column incubated in the presence of Nickel beads, or bacterially expressed H6Cdc20N120 or H6Cdh1N125 bound to nickel beads. (B) Pull-down assays using bacterially expressed H6Cdc20N120 or H6Cdh1N125 cross-linked to Pierce UltraLink beads incubated with bacterially expressed and purified human H6-securin, baculovirus-expressed and -purified mB99, or bacterially expressed and purified Xenopus H6-Ubcx. Visualization of his-tagged proteins was achieved following Western transfer and blotting with a QIAGEN α-pentahis antibody. Five percent input is shown for H6securin, and 10% input is shown for H6-mB99 and H6-Ubcx.
Having determined for several substrates that binding of the Cdc20 and Cdh1 N termini to substrates occurs in an APC-independent manner, we again attempted to show direct binding using purified proteins other than cyc B. We cross-linked bacterially expressed and purified H6Cdc20N120 and H6Cdh1N125 to Pierce UltraLink beads and incubated them with bacterially expressed and purified human H6-securin or baculovirus-expressed and purified H6-mB99. H6-securin bound to both H6Cdc20N120 beads and H6Cdh1N125 beads, but not to control beads (Fig. 6B). Typically, the amount bound reflected 5% of the input or more (5% input shown). In parallel experiments, H6Cdh1N125 beads bound 5%–10% of H6-mB99 protein (10% input shown), whereas little or no binding was seen to control beads or HcCdc20N120 beads (Fig. 6B). Interestingly, under less stringent washing conditions, H6Cdc20N120 beads bound detectable levels of mB99 above control beads although these levels were significantly lower than H6Cdh1N125 beads (data not shown). This binding could reflect low-level recognition of one of several R-X-X-L motifs in mB99 (one of which, at position 364, is conserved in the human sequence). Alternatively, Cdc20 may recognize the mB99 KEN sequence at lower affinity than the D box which may be out-competed by other proteins present in the reticulocyte lysate but absent in the direct binding assay; this might also explain the slight stabilizing effect of H6Cdc20N120KENNA on mB99 in the degradation assay. Neither H6Cdc20N120 beads nor H6Cdh1N125 beads bind bacterially expressed and purified H6-Ubcx (Fig. 6B) or purified BSA above control beads (data not shown), indicating that this interaction is specific.
The N termini of Cdc20 and Cdh1 confer substrate specificity in functional assays
The best evidence that the N termini functionally direct specificity would be to show that switching domains in functional protein switches substrate specificity. Ubiquitination of APC substrates can be reconstituted in vitro using a purified system (Fang et al. 1998a) containing immunopurified APC on beads activated by full-length Cdc20 or Cdh1. We created chimeric hCdc20 and hCdh1 proteins by exchanging the WD repeats (shown schematically in Fig. 7A). Both chimeric proteins ubiquitinate cyc B-N (Fig. 7B), indicating that they retained activity. Each chimera showed a slight reduction in the average polyubiquitin chain length compared to wild-type Cdc20 and Cdh1 (higher molecular weight polyubiquitined conjugates formed, but lower molecular weight conjugates were also enriched), with comparable activity to each other. Neither the N-terminal regions (Cdc20N170 and Cdh1N173) (Fig. 7B) nor the C-terminal WD repeats (Cdc20Δ170 and Cdh1Δ173) (data not shown) are competent to activate APC for polyubiquitination of cyc B-N in this purified system; only monoubiquitinated species (which form in the presence of iAPC alone) form. Only the chimera containing the N terminus of Cdh1, NCdh1–CCdc20 (bold letters indicate the N and C termini), ubiquitinates the Cdh1 KEN-box substrate hCdc20N170 (Fig. 7C). As with cyc B-N, the average polyubiquitin chain length is slightly lower than for wild-type Cdh1, however significant polyubiquitination occurs. Because both chimeras showed activity for the D box, but only NCdh1–CCdc20 showed activity for the KEN box, either (1) the WD repeats bind the D box and the N terminus of Cdh1 binds the KEN box, or (2) the specificity of activator chimera is mediated by its N terminus and not its C terminus. Given the behavior of the Cdc20 and Cdh1 N termini in the binding assays, we find it more likely that the N terminus of each molecule directs specific binding to substrates.
Figure 7.
Functional specificity is encoded by the N-terminal domain of hCdc20 and hCdh1. (A) Schematic showing the design of chimeric proteins exchanging the amino-termini and WD repeats of hCdc20 and hCdh1. (B) Ubiquitination assay using in vitro translated Cdh1, Cdc20, NCdh1–CCdc20, Cdc20N170, NCdc20–CCdh1, and Cdh1N173 to activate APC for ubiquitination of 35S-labeled cyc B-N. (C) Ubiquitination assay using Cdh1, Cdc20, NCdh1–CCdc20, and NCdc20–CCdh1 to activate APC for ubiquitination of 35S-labeled KEN-box containing Cdc20N170. An asterisk marks a background band present in the in vitro translation products in B and C. (D) Alignment of Cdch1 and Cdc20 family members from human (h), Xenopus (x), and Drosophila (Dm). Conserved residues of the DR(F/Y)IPXR are in bold type and are boxed.
The localization of substrate binding to the N-terminal region may seem surprising given the overlapping specificity (in terms of D box recognition) of Cdc20 and Cdh1 and the lack of sequence homology in this region. Despite extensive N-terminal sequence diversity, one motif is highly conserved between the Cdc20 and Cdh1 sequences throughout different species, DR(F/Y)IPXRX∼45–75(K/R)XL (Fig. 7C). In Cdc20 sequences, the RYIPX4(K/R)XL portion of the motif repeats (Fig. 7C).
Discussion
The APC displays different substrate specificities depending on its association with the activating proteins Cdc20 and Cdh1. Cdc20 binds and activates APC during mitosis, whereas Cdh1 binds and activates APC late in mitosis and during G1 (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998a). The selective association with activator confers APC specificity; the Cdc20- and Cdh1-bound forms have both shared and specific substrates, although unlike the F-box proteins of the SCF complex, there has been no evidence for direct interaction between these proteins and substrates.
Two obvious models could explain how Cdc20–APC and Cdh1–APC target both shared and distinct substrates. Rather than directly binding and recruiting substrate to APC, as occurs in the case of the recruitment of substrates by the F-box protein in the SCF, Cdc20 and Cdh1 might induce a conformational change in the APC core complex, exposing a different set of substrate-interacting subunits. Such a model would explain the difference in specificity conferred by Cdc20 and Cdh1, as well as explain the need for so many subunits. One APC subunit might specifically bind KEN box-containing proteins whereas another specifically binds D box-containing proteins. Alternatively, Cdc20 and Cdh1 might interact directly with substrate proteins, recruiting them to the APC. This model requires that Cdc20 bind substrates containing a well-defined D box, whereas Cdh1 bind both well- and less well-defined D-box motifs as well as the KEN box. Binding several motifs might require different domains of Cdh1, or might reflect an underlying structural similarity between the motifs accommodated in Cdh1 but not in Cdc20, for example by a “bump and hole” theory proposed earlier (Pfleger and Kirschner 2000).
Most investigations of APC–substrate interactions have used cyc B. The initial analysis of cyc B targeting and degradation revealed the D box (Glotzer et al. 1991; King et al. 1996), and the N terminus of Xenopus cyc B has been used widely in analysis of APC activity. This fragment makes a much better substrate than full-length cyc B in terms of conjugation to ubiquitin in a purified system and the kinetics of degradation in extracts. The lack of success in binding assays by our own lab and others might reflect weak binding of this substrate; in particular, weak binding could promote product dissociation as in many enzymatic reactions. Rapid release of ubiquitinated substrates would lead to more rapid presentation to the proteasome and therefore more rapid degradation. Even very weak binding can be detected using several biochemical techniques. These techniques generally require monodisperse populations of proteins. However, gel filtration of both in vitro translated and bacterially expressed cyc B-N showed that this peptide aggregated, making binding experiments difficult to perform or interpret.
By expanding binding experiments to include several other APC substrates, we now present evidence that Cdc20 and Cdh1 each bind their substrates. Binding reflects Cdc20–APC and Cdh1–APC specificity. Cdc20 binds the APC substrate Xkid, but not B99, whereas Cdh1 binds both Xkid and B99. Neither Cdc20 nor Cdh1 binds the SCF substrates β-cat or Xic1, or over a dozen non-APC substrates incubated in the assay, including the D box-deleted form of N-terminal cyc B. Consistent with our binding observations, recent binding experiments by the Solomon group have shown that binding of Hsl1p to Cdc20 (Burton and Solomon 2000, 2001) require the D box, and binding of Hsl1p to Cdh1 requires the KEN box (Burton and Solomon 2001). Furthermore, the binding of Hsl1p to Cdh1 is direct (Burton and Solomon 2001).
We have localized a region sufficient for specific binding of substrates to the N termini of both Cdc20 and Cdh1, although we cannot rule out a role for the WD repeats in the full-length context. Furthermore, binding is not mediated by APC. Binding is retained in a specific manner in APC-depleted extracts and by bacterially expressed N-terminal fragments of Cdc20 and Cdh1 incubated with the substrates Xkid, geminin, and securin isolated from APC by gel filtration. The securin protein used in these assays migrated at its expected small molecular weight by gel filtration. Any contaminating activity would have to elute both in fractions containing Xkid (molecular mass of 70 kD) as well as in the later fractions containing the smaller protein, securin, suggesting that binding of the activator to substrate is direct. To confirm this, we show that these bacterial N termini interact with bacterially expressed securin, and the bacterially expressed Cdh1 N terminus interacts with baculovirus-expressed and purified mB99, whereas neither N-terminal Cdc20 nor Cdh1 bind the nonsubstrates H6-Ubcx or BSA above control levels. Furthermore, the yeast substrate Hsl1p directly interacts with purified baculovirus-expressed full-length Cdh1 and purified Hsl1p expressed in bacteria (Burton and Solomon 2001).
We have shown that the N termini of Cdc20 and Cdh1 direct the binding to substrate functionally by the behavior of active chimeric proteins exchanging the N termini and WD repeats between Cdc20 and Cdh1. The specificity of the chimera reflects the specificity of its N terminus, not its C-terminal WD repeats, contrary to the Fbw subset of F-box proteins of the SCF complex which require the WD repeats for substrate interaction (Skowyra et al. 1997; for review, see Jackson et al. 2000).
Although the extreme N termini of Cdc20 and Cdh1 that bind substrates fail to bind APC, N-terminal fragments containing 50 or 48 amino acids more of Cdc20 and Cdh1, respectively, do retain the ability to bind APC. We have not determined if these fragments or full-length Cdc20 and Cdh1 can bind substrate and APC concurrently. If they cannot, activator might transfer substrate to another APC subunit on binding APC. The cryo-electron microscopy-determined structure of APC (Gieffers et al. 2001) indicates a large inner cavity that might serve as a reaction chamber to hold the substrate during the ubiquitination reaction. Cdc20 and Cdh1 might deposit substrates into this cavity for catalysis. However, N-terminal fragments capable of both substrate-binding and APC-binding fail to activate APC. We have observed the ability of Cdc20 and Cdh1 to dimerize, requiring the WD repeats, but we have not established if this dimerization is functionally required, although dimerization of certain F-box proteins is required for substrate binding (for review, see Jackson et al. 2000). One molecule of activating protein could bind substrate (either directly or indirectly) while another interacts with the APC core complex. Substrate-bound activator could be bound by the WD repeats of APC-bound activator, bringing substrate to APC for ubiquitination.
If one molecule of activating protein interacts with both APC and substrate concurrently, the WD repeats may provide a means to stabilize specific interactions. We observed binding of the WD repeats to both APC and to substrates, but we failed to show specificity in those interactions; we cannot rule out that such interactions occur. The lack of selectivity of the WD repeats for substrate and APC binding may reflect nonspecific stabilization of either substrate-interaction or APC-interaction once specific binding has occurred in the N-terminal region, or, alternatively, the WD repeats may initiate nonspecific interaction with other proteins randomly, but they are only retained if specific interaction with the N terminus follows.
In addition to binding substrates, the N-terminal fragments of hCdc20 and hCdh1 inhibit Cdc20 and Cdh1 activity both in vitro and in vivo. This inhibition does not occur by inhibiting dimerization of activator, or by inhibiting binding of full-length activator to APC. We hypothesize that the N-terminal fragments sequester substrate from functionally active APC. Substrate binding to the N-terminal region which are more divergent than the carboxyl termini, may indicate a role for the motif DR(F/Y)IPXRX∼45−75(K/R)XL which is highly conserved in the N-terminal region of Cdc20 and Cdh1 in species from yeast to humans. Alternatively, despite lower sequence homology in the N termini, structural aspects may be conserved in this domain.
The MAD2 related protein, MAD2L2 (also referred to as MAD2B) inhibits Cdh1–APC similar to the MAD2 inhibition of Cdc20 (Pfleger et al. 2001). Neither MAD2 nor MAD2L2 inhibit binding of Cdc20 to APC, or Cdh1 to APC, respectively (Fang et al. 1998b; Pfleger et al. 2001), or the binding of Cdc20 and Cdh1 to substrates (Pfleger et al. 2001), but actually inhibit the release of substrate bound to Cdc20 and Cdh1, respectively (Pfleger et al. 2001). This inhibition of substrate release does not require the WD repeats of Cdc20 and Cdh1, but is seen for Cdc20N170 and Cdh1N173 (data not shown), both of which contain the DR(F/Y)IPXRX∼45−75(K/R)XL motif. This sequence is required for activation of APC by Cdc20 and Cdh1; alanine substitution of this first part of this motif, DR(F/Y)IPXR to AAAAAXA, in both Cdc20 and Cdh1 results in proteins unable to activate APC (C. Pfleger, unpubl.). The initial (K/R)XL region of this motif has been identified recently as a conserved portion of the MAD2-binding region of Cdc20 (Zhang and Lees 2001; H. Yu, pers. comm.). Future studies should address the functional role of the DR(F/Y)IPXR motif.
Our study of the interaction of APC activators and their substrates has helped us understand the mechanism of APC-mediated ubiquitination. In addition, dominant negative versions of Cdc20 and Cdh1 have been used to study the importance of these activators in extracts. Biochemical and structural studies of direct interaction between activator and substrate require monodisperse populations of substrate not possible with cyc B-N. Once such populations of D-box and KEN-box regions from other substrates are generated, further atomic level structural analysis will be possible. In particular, Burton and Solomon (2001) have shown direct binding between full-length Cdh1 and a fragment of Hsl1p. Further understanding of the biological function of the APC will proceed not only through mechanistic studies, but also through the identification of additional substrates. In addition to previous methods of identifying substrates by global degradation screens using Xenopus extracts (McGarry and Kirschner 1998; Zou et al. 1999) which do not support global degradation of all substrates (C. Pfleger, unpubl.), a binding-based screen may reveal additional substrates by bypassing the requirement for global degradation, potentially identifying substrates that are degraded locally or compartmentally. Using this substrate-binding assay, we have identified two other proteins as potential APC substrates (C. Pfleger, unpubl.).
Recent work has revealed a family of Cdh1 homologs with distinct tissue distributions and substrate specificities (Y. Wan and M. Kirschner, in prep.). These additional Cdc20 and Cdh1-like molecules could expand the number of substrates targeted by APC as well as allow for tissue-specific or developmental-specific patterns of substrate specificity. Therefore APC, like the SCF complexes, possesses several substrate-specific adaptors that function in various cellular and developmental processes.
Materials and methods
Pull-down assays using in vitro translated Myc-tagged proteins
Cold in vitro translated myc-tagged proteins were bound to 9E10 anti-myc coupled beads purchased from Santa Cruz. After this prebinding, [35S]methionine-labeled substrate was added, in addition to cycloheximide to 100 μg/mL and BSA to 1 mg/mL in binding buffer (50 mM Hepes at pH 7.7, 50 mM NaCl, 1mM MgCl2, 1 mM EDTA, and 0.2% Tween-20). Following a 2 h incubation, samples were loaded onto minicolumns on a vacuum assembly and washed four times with 1 mL of binding buffer and eluted with heated sample buffer.
Depletion of APC from reticulocyte lysate
αCdc27 antibody coated Affi-prep Protein A beads were added to one-fourth the volume to reticulocyte lysate and incubated at 4°C for 2 h. Depleted reticulocyte lysate was used to translate both cold myc-tagged proteins and [35S]methionine-labeled substrates.
Expression and purification of recombinant proteins
N-terminal Cdc20 and Cdh1 peptides were cloned into a pET 28 bacterial expression vector tagging them with an N-terminal His tag. Protein was expressed in bacteria, purified on nickel beads, and eluted with high imidazole. Gel filtration isolated monodisperse forms of the proteins from aggregated material. Purified His-tagged securin was a gift from Hui Zou (Harvard Medical School, Boston, MA). mB99 was cloned into a pFastbac vector containing a His tag; baculovirus-expressed protein was purified from Sf9 cells on nickel beads and eluted with high imidazole.
Pull-down assays using recombinant His-tagged proteins bound to Ni-beads
Nickel-bound His-tagged N-terminal peptides of bacterially expressed recombinant Cdc20 or Cdh1 were incubated with 35S-labeled in vitro translated substrate proteins in 50 mM Tris (pH 7.5), 100 mM KCl. The beads were washed using the minicolumns and vacuum manifold described above in 50 mM Tris (pH 7.5), 300 mM KCl, 20 mM imidazole, and a final wash with the concentration of imidazole increased to 30 mM. His-tagged peptides were eluted from the nickel beads by increasing the imidazole to a concentration of 500 mM.
Pull-down assays using immunopurified APC
Immunopurified interphase APC (iAPC) on αCdc27 beads was incubated with 35S-labeled in vitro translated Cdc20 or Cdh1 wild-type or mutant proteins at room temperature for 1 h on a TOMY shaker. Beads were washed five times with 1 mL binding buffer in minicolumns on a vacuum manifold and eluted as described for the MT binding assay. As a control, beads coated with preimmune serum were subjected to the same assay.
Pull-down assays using recombinant proteins cross-linked to beads
Bacterially expressed and purified H6Cdc20N120 or H6Cdh1N125 were cross-linked to Pierce UltraLink beads at the same molar concentrations. Control beads were treated to identical conditions. Cross-linked beads were incubated with purified proteins for 2 h at room temperature on an inverting rocker. Samples were loaded onto minicolumns attached to a vacuum manifold and subjected to 2–3 washes with XB containing 300 mM NaCl, 0.1% Tween, 2–3 washes with XB containing 500 mM NaCl, 0.1% Tween, two washes of XB, and then eluted with heated sample buffer. Samples were run on a gel and visualized by either Coomassie staining or Western transfer and detection by QIAGEN αpenta-His antibody.
Preparation of low speed supernatant interphase extracts
Extracts were prepared as described previously (Murray 1991; Pfleger and Kirschner 2000).
In vitro ubiquitination assays
Ubiquitination assays were as described previously (Fang et al. 1998a; Pfleger and Kirschner 2000). APC on αCdc27 beads was bound to Cdc20 or Cdh1 purified from baculovirus-infected Sf9 cells (Fang et al. 1998a; Pfleger and Kirschner 2000) or in vitro translated Cdc20, Cdh1, and chimeras. 50 μg/mL Ubcx, 1.25 mg/mL ubiquitin, 200 μg/mL recombinant E1, 0.1 mg/mL cycloheximide, 1–5 μM ubiquitin aldehyde was added in the presence of 35S-labeled substrate.
Construction of chimeric Cdc20 and Cdh1
A SalI site was inserted by changing the nucleotide sequence but preserving the amino acid sequence VD at position 190 in Cdc20 and 193 in Cdh1, a region of high homology just after the start of the first WD repeat. PCR fragments corresponding to the region between the N terminus and the SalI site of each protein were ligated to the region between the SalI site and the C terminus of the other protein and into a pCS2+-based vector in a position for in vitro translation using the Sp6 primer.
RNA and DNA injection into Xenopus embryos
The indicated amounts of RNA or DNA were injected into one cell of two cell Xenopus embryos.
Capped RNA was synthesized from linearized plasmid DNA templates. As the cap analog (GpppG) is a potent inhibitor of translation, care was taken to remove all unincorporated cap. Messenger RNAs were resuspended in water to 100–500 ng/μL. Embryos were fixed in MEMFA (0.1 M MOPS at pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde), washed three times with PBT (PBS plus 0.2% Tween-20), and incubated at 37°C for 2 h with Propidium Iodide (10 ug/mL) in the presence of RNAse A (50 μg/mL). Propidium Iodide-stained embryos were washed three times with PBT, dehydrated for 5 min in methanol, and mounted in 2:1 benzyl benzoate/benzyl alcohol.
Acknowledgments
We thank Janet Burton and Mark Solomon for communicating their unpublished data. We also thank Hui Zou and Adrian Salic for important discussions and suggestions throughout the course of this work; purified bacterially expressed securin protein was generously provided by Hui Zou. We also thank Teresita Bernal, Louise Evans, and Leslie Mayer for technical assistance. E. Lee is a Merck fellow supported by the Helen Hay Whitney Foundation. We thank the National Institute of General Medical Science (GM26875 and GM39023) for their support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL marc@hms.harvard.edu; FAX (617) 432-2250.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.918201.
References
- Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Mathias N, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes & Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998a;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- ————— The checkpoint protein MAD2 and the mitotic regulator Cdc20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998b;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus Chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Dube P, Harris JR, Stark H, Peters JM. Three-dimensional structure of the Anaphase-Promoting Complex. Mol Cell. 2001;7:907–913. doi: 10.1016/s1097-2765(01)00234-9. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–137. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci. 2000;97:8973–8978. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda DK, Bachmair A, Wunning I, Tobias JW, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- Hein C, Speingael JY, Volland C, Haguenauer-Tsapis R, Andre B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;74:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol Cell Biol. 2001;21:2506–2520. doi: 10.1128/MCB.21.7.2506-2520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kalein WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kawahara H, Philipova R, Yokosawa H, Patel R, Tanaka K, Whitaker M. Inhibiting proteasome activity causes overreplication of DNA and blocks entry into mitosis in sea urchin embryos. J Cell Sci. 2000;113:2659–2670. doi: 10.1242/jcs.113.15.2659. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Yamao F. An essential function of Grr1 for the degradation of Cln2 is to act as a binding core that links Cln2 to Skp1. J Cell Sci. 1998;111:3655–3661. doi: 10.1242/jcs.111.24.3655. [DOI] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell-cycle extracts. Meth Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nefsky B, Beach D. Pub1 acts as an E6-AP-like protein ubiquitin ligase in the degradation of cdc25. EMBO J. 1996;15:1301–1312. [PMC free article] [PubMed] [Google Scholar]
- Oda H, Kumar S, Howley PM. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtoshi A, Maeda T, Higashi H, Ashizawa S, Hatakeyama M. Human p55Cdc20 associates with Cyclin A and is phosphorylated by the Cyclin A–Cdk2 complex. Biochem Biophys Res Comm. 2000;268:530–534. doi: 10.1006/bbrc.2000.2167. [DOI] [PubMed] [Google Scholar]
- Peters JM. SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes & Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pfleger CM, Salic A, Lee E, Kirschner MW. Inhibition of Cdh1–APC by the MAD2-related protein MAD2L2: A novel mechanism for regulating Cdh1. Genes & Dev. 2001;15:1759–1764. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y, Kaim D, Hershko A. Specificity of binding of N-terminal residues of proteins to ubiquitin-protein ligase. Use of amino acid derivatives to characterize specific binding sites. J Biol Chem. 1988;263:2693–2798. [PubMed] [Google Scholar]
- Rotin D. WW (WWP) domains: From structure to function. Curr Top Microbiol Immunol. 1998;228:115–133. doi: 10.1007/978-3-642-80481-6_5. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer J, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig K, Tyers M, Elledge SJ, Harper JW. F-box proteins are components of E3 complexes and act as receptors to recruit phosphorylated substrates for ubiquitination. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. Non-periodic activity of the human Anaphase-Promoting Complex-Cdh1 ubiquitin ligase results in continuous DNA synthesis uncoupled from mitosis. Mol Cell Biol. 2000;20:7613–7623. doi: 10.1128/mcb.20.20.7613-7623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A Conserved cyclin-binding domain determines functional interplay between Anaphase-Promoting Complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol Cell Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P. Proteolysis and the cell cycle: With this RING I do thee destroy. Curr Opin Gen Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Utrera R, Collavin L, Lazarevic D, Delia D, Schneider C. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 1998;17:5015–5025. doi: 10.1093/emboj/17.17.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: Functions, mysteries, uses. Proc Natl Acad Sci. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. Cdc20 and Cdh1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KH, Kondo T, Zheng J, Tsvetkov M, Blair LM, Zhang H. The f-box protein skp2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem Biophys Res Comm. 2001;281:884–890. doi: 10.1006/bbrc.2001.4442. [DOI] [PubMed] [Google Scholar]
- Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: Identification of a subunit to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and Anaphase-Promoting Complex: Model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]